Abstract
Two porcine Escherichia coli isolates harbored the cfr gene on conjugative plasmids of 38,405 bp (pGXEC6) and 41,646 bp (pGXEC3). In these two plasmids, the cfr gene was located within a 4,612-bp region containing a tnpA-IS26-cfr-IS26-Δhyp element. Plasmid pGXEC3 was almost identical to pGXEC6 except for a 3,235-bp ISEcp1-blaCTX-M-14b insertion. The colocation of the multiresistance cfr gene with an extended-spectrum-β-lactamase gene on a conjugative plasmid may support the dissemination of these genes by coselection.
TEXT
During the past decade, the transferable multiresistance gene cfr, which confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A (1), has been identified mainly in Gram-positive bacteria, including Staphylococcus spp., Bacillus spp., Enterococcus spp., Macrococcus caseolyticus, Jeotgalicoccus pinnipedialis, and Streptococcus suis (2, 3). However, a small number of reports have described the presence of the gene cfr in Gram-negative bacteria, such as Proteus vulgaris (4) and Escherichia coli (5–7). The global spread of this gene in bacteria of human and animal origin has gained particular public health attention, since cfr confers resistance to linezolid, the last-resort antibiotic against vancomycin-resistant enterococci, methicillin-resistant Staphylococcus aureus, and penicillin-resistant pneumococci (2, 8). In Enterobacteriaceae from human and animal sources, extended-spectrum-β-lactamase (ESBL) genes have emerged during recent years globally (9). Since the year 2000, ESBL genes of the type blaCTX-M have become the predominant ESBL genes (10, 11). Similar to the cfr gene (2), ESBL genes are often located on plasmids, including plasmids that carry additional resistance genes which may facilitate the dissemination and persistence of the ESBL genes (8, 11–13).
So far, a single conjugative IncA/C broad-host-range plasmid of E. coli, designated pSCEC2 (6), has been described, in which the cfr gene was colocated with the resistance genes tet(A) (tetracycline resistance), strA-strB (streptomycin resistance), and sul2 (sulfonamide resistance). In the present study, two novel cfr-carrying plasmids, pGXEC6 and pGXEC3, were identified, one of which also harbored a blaCTX-M14b gene. These two plasmids were completely sequenced to gain insight into their genetic relatedness and understand how they may have developed.
Thirty-one pericardial fluid swab samples collected from diseased swine from June 2013 to January 2014 in four different commercial pig farms in the Guangxi Province, China, were inoculated on brain heart infusion agar plates supplemented with 10 mg/liter florfenicol. Two cfr-positive E. coli isolates, designated GXEC6 and GXEC3, were obtained from samples from two different pig farms, farm A (a breeding pig farm) and farm B (a fattening pig farm), respectively. The antimicrobial usage records for these two pig farms indicated that aside from ceftriaxone, which was used for prophylactic purposes at farm A, other antimicrobial agents, such as ampicillin, florfenicol, sulfamethoxazole, amikacin, enrofloxacin, lincomycin, and tiamulin, were extensively used to prevent or treat bacterial infections at the two pig farms. The two E. coli isolates were typed by multilocus sequence typing (MLST) (see http://mlst.warwick.ac.uk) and subjected to pulsed-field gel electrophoresis (PFGE) (14). Conjugation experiments by filter mating and S1-PFGE combined with Southern blotting (15) were performed as described previously (15). PCR-based replicon typing (PBRT) followed previously published recommendations (16). The two cfr-positive original E. coli isolates and their transconjugants were tested for their susceptibility to amikacin, ampicillin, ceftazidime, chloramphenicol, enrofloxacin, florfenicol, gentamicin, sulfamethoxazole, and tetracycline by broth microdilution according to CLSI recommendations (17, 18). ESBL confirmatory tests also followed CLSI recommendations (18). The cfr-carrying plasmids of the transconjugants were sequenced using the 454 Life Sciences GS FLX system (Roche). The gaps between the contigs were closed by PCR, and the whole-plasmid sequences were draft assembled using the SeqMan software from DNAStar (Lasergene, Madison, WI, USA). The AatII, BstEII, ScaI, and SnaBI restriction profiles of cfr-carrying plasmids were determined and compared with the plasmid sequences to confirm the correct assembly of the contigs. The plasmid sequences were annotated using the Vector NTI program, and the putative coding sequences were identified using the ORF Finder program (http://www.ncbi.nlm.nih.gov/projects/gorf/). Sequence comparisons were performed using BLASTN and BLASTP (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Although isolates GXEC6 and GXEC3 originated from different farms, PFGE analysis revealed two related XbaI patterns. This observation may be explained by the fact that fattening pig farm B, from which isolate GXEC3 originated, imports weaned piglets from pig-breeding farm A, from which isolate GXEC6 was obtained. MLST identified each isolate as sequence type (ST) 746, and according to the MLST database, this ST was found in E. coli isolates of human and horse origins from Egypt and Germany, respectively. Moreover, a recent study reported an ST746 chicken E. coli isolate from China coharboring the blaCTX-M-55 and blaCTX-M-123 genes (19). The cfr gene was transferable by conjugation from these two original E. coli isolates to the recipient E. coli strain J53. Transconjugants were obtained with a high frequency (∼2 × 10−3 to 3 × 10−3 per donor cell). S1-PFGE of the transconjugants revealed approximate sizes of the cfr-carrying plasmids of 38 kb (pGXEC6) and 41 kb (pGXEC3). None of the two cfr-carrying plasmids produced amplicons by PBRT using the 18 primer pairs described previously (16). While the pGXEC6-carrying transconjugant conferred resistance to only chloramphenicol and florfenicol, the pGXEC3-transconjugant was also resistant to ampicillin and ceftazidime and exhibited an ESBL phenotype (Table 1).
TABLE 1.
MICs for E. coli GXEC3 and GXEC6 and the corresponding cfr-positive E. coli J53 transconjugants
| Strain | MIC (mg/liter) ofa: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| AMP | TAZ | GEN | AMK | CHL | FFC | ENO | TET | SMZ | |
| GXEC3 | ≥256 | 64 | 64 | 16 | 256 | 128 | ≥32 | 128 | 32 |
| E. coli J53 (pGXEC3)b | 256 | 32 | 2 | 2 | 128 | 128 | 0.06 | 2 | 16 |
| GXEC6 | 8 | 1 | 64 | 8 | 256 | 128 | ≥32 | 128 | ≥512 |
| E. coli J53 (pGXEC6)c | 4 | 0.06 | 2 | 2 | 128 | 128 | 0.06 | 2 | 16 |
| E. coli J53 | 4 | 0.06 | 1 | 2 | 2 | 1 | 0.03 | 2 | 8 |
AMP, ampicillin; TAZ, ceftazidime; GEN, gentamicin; AMK, amikacin; CHL, chloramphenicol; FFC, florfenicol; ENO, enrofloxacin; TET, tetracycline; SMZ, sulfamethoxazole.
E. coli J53 transconjugants of GXEC3.
E. coli J53 transconjugants of GXEC6.
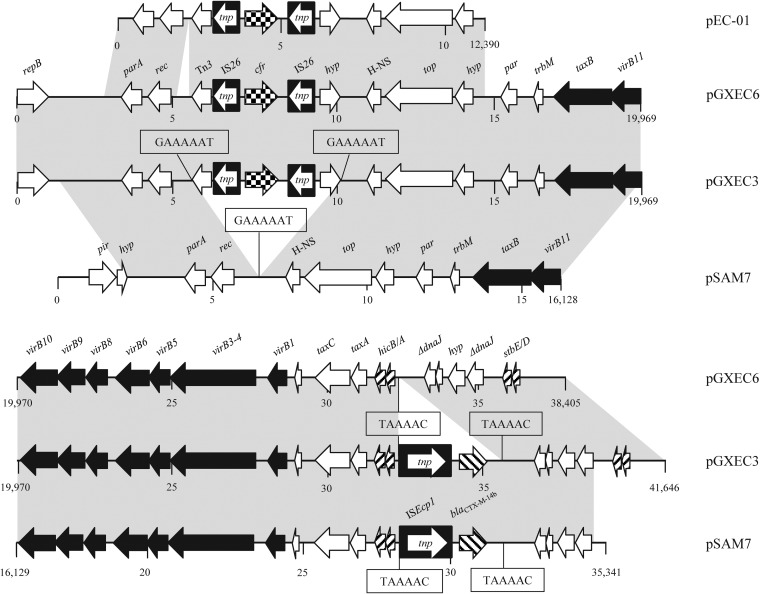
Plasmid pGXEC3 had a size of 41,646 bp, while that of pGXEC6 was 38,405 bp. Comparative sequence analysis of the two plasmids revealed that a 3,241-bp fragment was inserted upstream of the hicA gene in pGXEC3 (Fig. 1), whereas the remaining parts of the two plasmids were virtually identical except for several nucleotide substitutions. The insertion included an intact 3,235-bp ISEcp1-blaCTX-M-14b transposition unit and one 6-bp direct repeat (Fig. 1). The detection of a functionally intact blaCTX-M-14b gene explained the observed ESBL phenotype. In these two plasmids, the cfr gene was located in a 4,612-bp segment, which included a 1,545-bp cfr-carrying central region that was identical to the corresponding sequence of plasmid pEC-01 from porcine E. coli (5) (Fig. 1). Moreover, comparative analysis of the 13,122-bp cfr-carrying fragment of pGXEC6/pGXEC3 and the 12,390-bp cfr-carrying fragment of pEC-01 revealed >99% nucleotide sequence identity, except for a 732-bp insertion in pGXEC6/pGXEC3 at positions 5041 to 5772. The cfr gene was flanked by two identical copies of the insertion sequence IS26 located in the same orientation. Further database searches revealed that pGXEC3 and pGXEC6 were related to the recently described IncX4 blaCTX-M-14b-carrying plasmid pSAM7 from bovine E. coli, isolated in 2008 in the United Kingdom (20), except for the cfr-carrying segment, the repB plasmid replication gene region, and a segment containing the stbDE toxin/antitoxin genes (Fig. 1). It is tempting to assume that the 4,612-bp region consisting of tnpA-IS26-cfr-IS26-Δhyp may represent a transposition unit which has integrated into a pSAM7-like backbone to give rise to pGXEC3. Since ISEcp1 is able to mobilize itself and adjacent resistance genes, such as blaCTX-M-type genes (21–23), it is possible that plasmids pGXEC3 and pGXEC6 have developed from one another by either a gain or loss of the ISEcp1-blaCTX-M-14b segment.
FIG 1.
Linear comparison of plasmids pGXEC6 and pGXEC3 with pEC-01 from E. coli (GenBank accession no. JN982327) and pSAM7 from E. coli (GenBank accession no. JX981514). The arrows indicate the positions and directions of the transcription of the genes. Gray shading indicates >99% nucleotide sequence identity. Direct repeats are shown in boxes. Δ indicates a truncated gene. A distance scale in kb is displayed below the plasmid sequences.
To the best of our knowledge, this is the first report of the cfr gene and an ESBL gene (blaCTX-M-14b) being colocated on a conjugative plasmid in E. coli. Given the widespread use of penicillins in veterinary medicine (24), the occurrence and dissemination of plasmids carrying cfr and ESBL genes among Gram-negative bacteria need further surveillance.
Nucleotide sequence accession numbers.
The sequences of plasmids pGXEC3 and pGXEC6 have been deposited in GenBank under accession no. KM580532 and KM580533, respectively.
ACKNOWLEDGMENTS
This work was funded by the national 973 program (grant 2012CB518801), the National Natural Science Foundation of China (grant 31201862), and the Central Public-Interest Scientific Institution Basal Research Fund (grant 0302014016). The contribution of S.S. was funded by the German Federal Ministry of Education and Research (BMBF) through the German Aerospace Center (DLR), grant 01KI1313D (RESET II) and grant 01KI1301D (MedVet-Staph II).
REFERENCES
- 1.Long KS, Poehlsgaard J, Kehrenberg C, Schwarz S, Vester B. 2006. The Cfr rRNA methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics. Antimicrob Agents Chemother 50:2500–2505. doi: 10.1128/AAC.00131-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shen J, Wang Y, Schwarz S. 2013. Presence and dissemination of the multiresistance gene cfr in Gram-positive and Gram-negative bacteria. J Antimicrob Chemother 68:1697–1706. doi: 10.1093/jac/dkt092. [DOI] [PubMed] [Google Scholar]
- 3.Wang Y, Li D, Song L, Liu Y, He T, Liu H, Wu C, Schwarz S, Shen J. 2013. First report of the multiresistance gene cfr in Streptococcus suis. Antimicrob Agents Chemother 57:4061–4063. doi: 10.1128/AAC.00713-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y, Wang Y, Wu CM, Schwarz S, Shen Z, Zhang W, Zhang Q, Shen JZ. 2011. Detection of the staphylococcal multiresistance gene cfr in Proteus vulgaris of food animal origin. J Antimicrob Chemother 66:2521–2526. doi: 10.1093/jac/dkr322. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y, He T, Schwarz S, Zhou D, Shen Z, Wu C, Wang Y, Ma L, Zhang Q, Shen J. 2012. Detection of the staphylococcal multiresistance gene cfr in Escherichia coli of domestic animal origin. J Antimicrob Chemother 67:1094–1098. doi: 10.1093/jac/dks020. [DOI] [PubMed] [Google Scholar]
- 6.Zhang WJ, Xu XR, Schwarz S, Wang XM, Dai L, Zheng HJ, Liu S. 2014. Characterization of the IncA/C plasmid pSCEC2 from Escherichia coli of swine origin that harbours the multiresistance gene cfr. J Antimicrob Chemother 69:385–389. doi: 10.1093/jac/dkt355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng H, Sun J, Ma J, Li L, Fang LX, Zhang Q, Liu YH, Liao XP. 2014. Identification of the multi-resistance gene cfr in Escherichia coli isolates of animal origin. PLoS One 9:e102378. doi: 10.1371/journal.pone.0102378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Witte W, Cuny C. 2011. Emergence and spread of cfr-mediated multiresistance in staphylococci: an interdisciplinary challenge. Future Microbiol 6:925–931. doi: 10.2217/fmb.11.69. [DOI] [PubMed] [Google Scholar]
- 9.Smet A, Martel A, Persoons D, Dewulf J, Heyndrickx M, Herman L, Haesebrouck F, Butaye P. 2010. Broad-spectrum beta-lactamases among Enterobacteriaceae of animal origin: molecular aspects, mobility and impact on public health. FEMS Microbiol Rev 34:295–316. doi: 10.1111/j.1574-6976.2009.00198.x. [DOI] [PubMed] [Google Scholar]
- 10.Bush K, Jacoby GA. 2010. Updated functional classification of beta-lactamases. Antimicrob Agents Chemother 54:969–976. doi: 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naseer U, Sundsfjord A. 2011. The CTX-M conundrum: dissemination of plasmids and Escherichia coli clones. Microb Drug Resist 17:83–97. doi: 10.1089/mdr.2010.0132. [DOI] [PubMed] [Google Scholar]
- 12.Wang J, Stephan R, Power K, Yan Q, Hächler H, Fanning S. 2014. Nucleotide sequences of 16 transmissible plasmids identified in nine multidrug-resistant Escherichia coli isolates expressing an ESBL phenotype isolated from food-producing animals and healthy humans. J Antimicrob Chemother 69:2658–2668. doi: 10.1093/jac/dku206. [DOI] [PubMed] [Google Scholar]
- 13.Wu G, Day MJ, Mafura MT, Nunez-Garcia J, Fenner JJ, Sharma M, van Essen-Zandbergen A, Rodríguez I, Dierikx C, Kadlec K, Schink AK, Wain J, Helmuth R, Guerra B, Schwarz S, Threlfall J, Woodward MJ, Woodford N, Coldham N, Mevius D. 2013. Comparative analysis of ESBL-positive Escherichia coli isolates from animals and humans from the UK, The Netherlands and Germany. PLoS One 8:e75392. doi: 10.1371/journal.pone.0075392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hansen KH, Bortolaia V, Damborg P, Guardabassi L. 2014. Strain diversity of CTX-M-producing Enterobacteriaceae in individual pigs: insights into the dynamics of shedding during the production cycle. Appl Environ Microbiol 80:6620–6626. doi: 10.1128/AEM.01730-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang WJ, Lu Z, Schwarz S, Zhang RM, Wang XM, Si W, Yu S, Chen L, Liu S. 2013. Complete sequence of the blaNDM-1-carrying plasmid pNDM-AB from Acinetobacter baumannii of food animal origin. J Antimicrob Chemother 68:1681–1682. doi: 10.1093/jac/dkt066. [DOI] [PubMed] [Google Scholar]
- 16.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 17.Clinical and Laboratory Standards Institute. 2013. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals—4th ed. Approved standard VET01-A4 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 18.Clinical and Laboratory Standards Institute. 2013. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals, 2nd informational supplement. CLSI VET01-S2 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 19.He D, Partridge SR, Shen J, Zeng Z, Liu L, Rao L, Lv L, Liu JH. 2013. CTX-M-123, a novel hybrid of the CTX-M-1 and CTX-M-9 group beta-lactamases recovered from Escherichia coli isolates in China. Antimicrob Agents Chemother 57:4068–4071. doi: 10.1128/AAC.00541-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stokes MO, Abuoun M, Umur S, Wu G, Partridge SR, Mevius DJ, Coldham NG, Fielder MD. 2013. Complete sequence of pSAM7, an IncX4 plasmid carrying a novel blaCTX-M-14b transposition unit isolated from Escherichia coli and Enterobacter cloacae from cattle. Antimicrob Agents Chemother 57:4590–4594. doi: 10.1128/AAC.01157-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toleman MA, Walsh TR. 2010. ISCR elements are key players in IncA/C plasmid evolution. Antimicrob Agents Chemother 54:3534. doi: 10.1128/AAC.00383-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freire Martin I, AbuOun M, Reichel R, La Ragione RM, Woodward MJ. 2014. Sequence analysis of a CTX-M-1 IncI1 plasmid found in Salmonella 4,5,12:i:-, Escherichia coli and Klebsiella pneumoniae on a UK pig farm. J Antimicrob Chemother 69:2098–2101. doi: 10.1093/jac/dku098. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Song C, Duan G, Zhu J, Yang H, Xi Y, Fan Q. 2013. Transposition of ISEcp1 modulates blaCTX-M-55-mediated Shigella flexneri resistance to cefalothin. Int J Antimicrob Agents 42:507–512. doi: 10.1016/j.ijantimicag.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 24.Silley P, Simjee S, Schwarz S. 2012. Surveillance and monitoring of antimicrobial resistance and antibiotic consumption in humans and animals. Rev Sci Tech 31:105–120. [DOI] [PubMed] [Google Scholar]