Abstract
GSK1322322 is a novel antibacterial agent under development, and it has known antibacterial activities against multidrug-resistant respiratory and skin pathogens through its inhibition of the bacterial peptide deformylase. Here, we used next-generation sequencing (NGS) of the bacterial 16S rRNA genes from stool samples collected from 61 healthy volunteers at the predosing and end-of-study time points to determine the effects of GSK1322322 on the gastrointestinal (GI) microbiota in a phase I, randomized, double-blind, and placebo-controlled study. GSK1322322 was administered either intravenously (i.v.) only or in an oral-i.v. combination in single- and repeat-dose-escalation infusions. Analysis of the 16S rRNA sequence data found no significant changes in the relative abundances of GI operational taxonomic units (OTUs) between the prestudy and end-of-study samples for either the placebo- or i.v.-only-treated subjects. However, oral-i.v. treatment resulted in significant decreases in some bacterial taxa, the Firmicutes and Bacteroidales, and increases in others, the Betaproteobacteria, Gammaproteobacteria, and Bifidobacteriaceae. Microbiome diversity plots clearly differentiated the end-of-study oral-i.v.-dosed samples from all others collected. The changes in genome function as inferred from species composition suggest an increase in bacterial transporter and xenobiotic metabolism pathways in these samples. A phylogenetic analysis of the peptide deformylase protein sequences collected from the published genomes of clinical isolates previously tested for GSK1322322 in vitro susceptibility and GI bacterial reference genomes suggests that antibiotic target homology is one of several factors that influences the response of GI microbiota to this antibiotic. Our study shows that dosing regimen and target class are important factors when considering the impact of antibiotic usage on GI microbiota. (This clinical trial was registered at the GlaxoSmithKline Clinical Study Register under study identifier PDF 113376.)
INTRODUCTION
The roles of microbial organisms in human health and well-being are complex and multifaceted. Maintaining proper symbiosis between the human host and our endogenous gastrointestinal (GI) microbial ecosystem, or microbiome, is essential for a well-functioning immune system and may delay the onset of many chronic diseases (1, 2). Recently, there has been increased interest in and concern about the potential damaging effects of prolonged antibiotic use on the microbiome (3, 4, 5), since oral dosing can significantly alter gut microbial communities (6, 7). Human epidemiological studies suggest that long-term intake of antibiotics is potentially linked with later-life onset of inflammatory bowel diseases (8, 9), asthma (10), and Clostridium difficile infections (11). Animal studies involving the dosing of different antibiotics show a potential disruption in intestinal homeostasis based on changes in the gut microbiota composition and bacterial metabolites (12, 13). With the increasing global threat of drug-resistant bacteria, new antibiotics are urgently needed. However, recent studies suggest that more systematic analyses of the potential effects of these agents on the human microbiome should be carried out during drug development.
Community-acquired bacterial pneumonia (CABP) is the leading cause of infectious death in developed countries, with mortality rates of 16 to 47% in subjects hospitalized with CABP who progress to intense care unit (ICU) care (14). GSK1322322 is a new antibiotic with a novel mechanism of action that is effective against predominant CABP pathogens, including Streptococcus pneumoniae, Haemophilus influenzae, Moraxella catarrhalis, Mycoplasma pneumoniae, Chlamydia pneumoniae, Legionella spp., methicillin-resistant Staphylococcus aureus (MRSA), and methicillin-susceptible S. aureus (MSSA) (15). As a new synthetic molecule of the hydrazide class, GSK1322322 is a potent inhibitor of bacterial peptide deformylase (PDF), a highly conserved and essential bacterial metalloprotease that matures newly synthesized peptides by N-formyl group cleavage (16, 17). GSK1322322 (500 to 1,500 mg) was shown to be well tolerated in a randomized, double-blinded, placebo-controlled, six-cohort phase I clinical trial with 62 healthy volunteers with repeat oral-intravenous (i.v.) combination or i.v.-only dosing for 5 to 6 days (18, 19).
Targeted DNA sequencing of the bacterial 16S rRNA gene regions using next-generation sequencing (NGS) platforms, such as Illumina MiSeq, has been shown to be highly useful for cataloguing microbiome diversity (20, 21). In this study, we report the results of a microbiome analysis of stool samples taken as an exploratory research arm of the GSK1322322 dose-ranging phase I clinical trial in healthy volunteers (18). Fecal samples were collected with consent from all volunteers at the predosing and postdosing end-of-study time points. Subsequently, DNA was extracted, and Illumina MiSeq DNA sequencing analysis of the 16S rRNA variable region 4 (V4) was performed. The changes in the microbiome community before and after dosing, as well as across dosing regimens, were determined.
GSK1322322 has a specific mode of action against a single conserved target, the PDF protein. There are two distinct yet evolutionarily related classes of PDF protein: class I, which is found in all Gram-negative bacteria, some Gram-positive bacteria, and eukaryotic mitochondria and chloroplast organelles, and class II, which is found exclusively in Gram-positive bacteria (16, 17). Given the growing number of bacterial genome sequences, including those of human gut microbes (22), and the availability of extensive GSK1322322 MIC profiling data against bacterial pathogens (15), we attempted to determine the relationship between antibacterial target conservation and the observed trends in microbiome shifts from clinical subjects using molecular evolutionary analyses.
MATERIALS AND METHODS
Clinical study design.
The study design and subject population were described previously by Naderer et al. (18, 23). Briefly, adults age 18 to 65 years with a body mass index (BMI) of 18.5 to 29.9 kg/m2 and in generally good health were eligible for enrollment. Female volunteers of non-child-bearing potential were also eligible. Single-dose oral tablet (500 mg each) doses of GSK1322322 (for a total of 1,000 or 1,500 mg) were administered in two cohorts (B and C) to determine absolute bioavailability, mean absorption time, and systemic exposure of oral tablet administration (Table 1). Single-dose escalation of i.v. GSK1322322 from 500 to 3,000 mg was administered in six cohorts (A1, A2, B, C, D, and E) to determine tolerability, dose proportionality, urinary excretion, and systemic exposure. The highest dosages, 2,000 mg and 3,000 mg, were administered in i.v.-only formulations. Repeat-dose escalation of i.v. GSK1322322 from 500 to 1,500 mg was administered in six cohorts (A1, A2, B, C, F1, and F2) to evaluate tolerability, the accumulation ratio, time invariance, and systemic exposure. In addition, two formulations for i.v. administration of GSK1322322 were evaluated for safety and tolerability: a free-base formulation (in cohorts A1, A2, B, and C) and a more stable mesylate salt preparation (in cohorts D, E, F1, and F2).
TABLE 1.
Overview of dosing regimen and clinical study designa
| Period | Cohort | No. active/no. of placebo | Dose (mg) protocolb | Infusion duration/rate (min) |
|---|---|---|---|---|
| 1 | A1 | 4/2 | Single dose, 500 i.v. GSK1322322/placebo followed by BID for 4 days | 60 |
| A2 | 4/2 | Single dose, 500 i.v. GSK1322322/placebo followed by BID for 4 days | 30 | |
| 2 | B | 6/2 | Initial single dose 1,000 oral GSK1322322/placebo; initial single dose 1,000 i.v. GSK1322322/placebo, followed by BID for 4 days | 60 |
| 3 | C | 18/3 | Initial single dose 1,500 oral GSK1322322/placebo; initial single dose 1,500 i.v. GSK1322322/placebo, followed by BID for 4 days | 60 |
| 4 | D | 3/1 | Single dose 2,000 i.v. GSK1322322/placebo | 60 |
| 5 | E | 3/1 | Single dose 3,000 i.v. GSK1322322/placebo | 60 |
| 6 | F1 | 4/2 | 1,000 i.v. GSK1322322/placebo BID for 4 days | 60 |
| F2 | 4/2 | 1,000 i.v. GSK1322322/placebo BID for 4 days | 60 |
For further details on volunteer demographics, characteristics, and dosing regimen, see Table 1 in Naderer et al. (18).
i.v., intravenous; BID, two times a day.
Volunteers were admitted to the study unit the day before drug administration and discharged after the study procedures were completed on day 3, 5, or 7, depending on the cohort. All oral doses were administered following an overnight fast of at least 10 h. Standardized meals were served daily while volunteers were housed within the unit. Each volunteer provided written informed consent. The study was approved by an institutional review board (Western International Review Board, Olympia, WA) and was conducted in accordance with good clinical practice and the Declaration of Helsinki. Four volunteers withdrew from the study due to mild adverse events (AEs) (two patients with irritation, one patient with urticaria at the i.v. infusion site, and one patient with moderate pruritic rash) (23).
Fecal sample collection and DNA extraction.
With the consent of the subjects, stool samples were collected during this study for measuring the microbiome. The samples were collected predose and at the end of the study treatment. The stool samples were collected with a sterile spoon, transferred into a prelabeled stool collection tube containing 8 ml of stool DNA stabilizer, mixed by shaking, and then immediately stored frozen at −20°C prior to shipment. Care was taken to minimize any sample contamination and reduce prolonged exposure to air.
For DNA extraction, the frozen stool samples were completely thawed, and DNA was isolated from approximately 1.4 ml of each homogenized sample using the PSP Spin Stool DNA Plus kit (Invitek, Berlin, Germany). DNA was isolated, as per the manufacturer's instructions. Each DNA sample was quantified by spectrophotometry (NanoDrop, ND-1000; Thermo Scientific, DE, USA) prior to PCR amplification.
PCR amplification and DNA sequencing of bacterial 16S rRNA genes.
Multiplex bar-coded primers were used for paired-end sequencing of the 16S rRNA variable 4 (V4) region on the Illumina MiSeq platform (Illumina, San Diego, CA). We used the 16S rRNA V4 region primers 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′), as recommended by Caporaso et al. (24), for maximal coverage of bacterial phylogeny. The 16S rRNA gene primers were combined with the appropriate barcode and linker oligonucleotides. Illumina amplicon library generation was performed as described previously (24), except for the additional steps of purification of the PCR products after amplification by AMPure (Beckman Coulter, Brea, CA) and quantification by spectrophotometry (NanoDrop, ND-1000; Thermo Scientific, DE), followed by normalization using SequalPrep (Life Technologies, Carlsbad, CA). The amplified bar-coded DNAs from 119 samples were then pooled. The samples were controlled for quality and quantity using an Agilent Bioanalyzer (Santa Clara, CA) chip for the absence of primer-dimers and accurate sizing of product, as well as quantified using quantitative PCR (qPCR) (Kapa Biosystems, Wilmington, MA). The samples were diluted to a final dilution of 7 pM, combined at a 95:05 ratio with 7 pM of PhiX control, and run on a MiSeq 2 × 150 cycle run. The Illumina sequencer instrument, reagents (MiSeq reagent kit version 2 300 cycle), and pooled samples were prepared according to Illumina MiSeq protocols. Data collection and base calling were performed on the MiSeq instrument using CASAVA 1.8 (Illumina). After the removal of sequences that failed the Illumina quality filtering, the reads were converted to the FASTQ format. Sequence quality using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) was determined before and after demultiplexing of the samples.
Microbiome data analysis.
Additional quality filtering and analyses of multiplexed DNA sequencing reads from the forward primer were performed using the software QIIME 1.7 (25). The reads were truncated at the base preceding the first low-quality stretch, and only reads of ≥75 bases long were retained. The reads were discarded if the sequence contained one or more ambiguous base calls or if the barcode sequence contained any mismatch errors. PCR chimera filtering was accomplished using usearch version 6.1 (26). The closed-reference QIIME protocol was used with the UCLUST method (26) to select operational taxonomic units (OTUs). The sequences with ≥97% identity were clustered together. A representative sequence from each cluster was used to identify the bacterial taxa from the May 2013 edition of the Greengenes 16S rRNA database (27, 28). OTUs containing fewer than two sequences were discarded, and sequences with <60% similarity to those in the Greengenes database were also discarded to remove potential contaminants from the data set.
The OTU table was rarefied to a depth of 33,582 sequences, and the resultant table was used for diversity analyses, as per the recommended guidelines. β-Diversity was estimated using the UniFrac metric to calculate the distances between the samples and visualized by principal coordinate analysis (PCoA) (29). Analyses of variance (ANOVAs) of the treatment effects and each OTU category (L2 [phylum] to L7 [OTU]) were calculated using QIIME 1.7 (25) and custom R scripts.
Potential changes in the microbiome at the functional level were determined using the software PICRUSt (30), with default settings, and the Kyoto Encyclopedia of Genes and Genomes (KEGG) database release 70.0 (31, 32), and they were visualized using STAMP (33). The human-specific pathways were removed from the results to focus on true bacterial pathways. Bonferroni-corrected P values of <0.05 were used to determine the statistical significances for all analyses.
Phylogenetic analyses.
Phylogenetic reconstruction of the peptide deformylase sequences from pathogenic and human intestinal bacterial species began with downloading all gastrointestinal bacterial reference genomes from the NIH Human Microbiome Project (http://www.hmpdacc.org/) and creating local BLAST databases (34) of bacterial species found by our 16S rRNA gene sequencing in any subject fecal sample. PDF homologs in gut microbial genomes were found by text searching the gene annotations or homology (BLASTp E value ≤ 1.0e−10) to known PDF query sequences from Escherichia coli strain K-12 and S. aureus strain Mu50. Further pathogenic and nonpathogenic bacterial PDF sequences were obtained from NCBI based on the availability of published GSK1322322 MIC data (15), and a nonredundant sequence FASTA file was created for multiple-sequence alignment using Clustal W version 1.83 (35). The multiple-sequence alignment was edited manually for a final alignment of 108 amino acids and 366 OTUs (sequences). Phylogenetic trees were reconstructed by the distance neighbor-joining (NJ) method using the program MEGA6 with the JTT model and 1,000 bootstrap replicates (36). All trees were visualized using the program FigTree version 1.4.0 (http://tree.bio.ed.ac.uk/software/figtree).
RESULTS
16S rRNA amplicon DNA sequencing depth and community diversity.
A total of 119 stool samples from 62 subjects were processed for DNA purification, 16S rRNA gene amplification, and amplicon DNA sequencing. A single oral-i.v. combination-treated subject (with both prestudy and end-of-study samples collected) was removed because of failed PCR DNA amplification, leaving 61 subjects and 117 samples. No sample failed DNA sequencing, with the fewest reads assigned to any sample being 33,586 and the median being 64,935 reads. After preprocessing, the entire data set contained 20,744,830 sequences, of which 20,447,605 were assigned to OTUs. A total of 4,662 distinct OTUs were detected among the 117 samples.
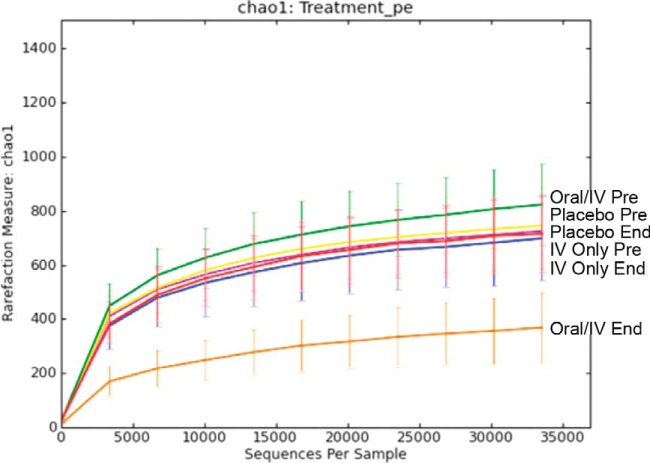
A comparison of α-diversity rarefaction curves (chao1 OTU richness estimation) showed overall high levels of biodiversity for the samples categorized based on dosing regimen (placebo, oral-i.v. GSK1322322 combination, and GSK1322322 by i.v. only) and time of collection (prestudy and end of study) (Fig. 1). The curves show a plateau trend, suggesting that the depth of coverage was sufficient to capture most of the OTU proportional abundance. However, the curves for the samples categorized as oral-i.v. combination GSK1322322 dosing at the end of the study showed markedly lower chao1 values, reflecting lower OTU richness in this sample subset.
FIG 1.
α-Diversity rarefaction plots using chao1 measure of within-group diversity. Plots are shown for prestudy (Pre) and end-of-study (End) samples collected from each of the three treatment groups, placebo, i.v. only, and oral-i.v. dosing. The analysis and figure were generated using the software QIIME version 1.7 (25).
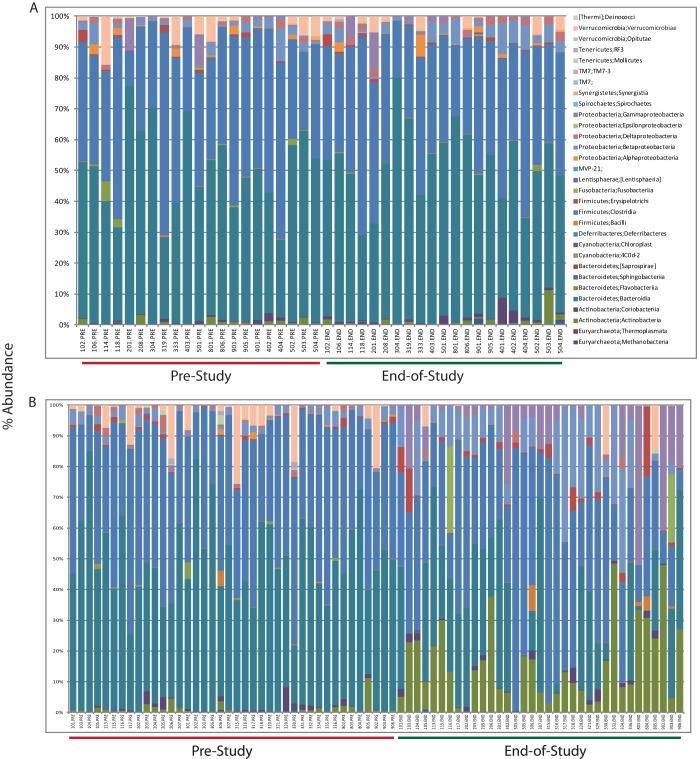
Using ANOVAs with multiple test corrections, we evaluated the potential occurrences of relative abundances with the available subject metadata variables, which included subject identifier, gender, age, body mass index (BMI), dosing levels (in mg), treatment (placebo, i.v. only, or oral-i.v. combination), and minor diarrhea adverse events (GSK1322322 was well tolerated, and no subjects withdrew from the study due to severe adverse events [18, 23]). No significant differences in the bacterial relative abundances at any OTU level (L2 through L7) were found for the subject variables of age, gender, BMI, adverse events, or subject identifier. Comparisons of prestudy versus end-of-study distributions of bacterial relative abundances at all OTU levels showed no significant differences among the subjects receiving the placebo or i.v.-only dosing regimens (Fig. 2A). However, in similar comparisons between prestudy versus the end of the study, sample plots of relative bacterial taxon abundance show that several OTUs were markedly changed for subjects receiving oral-i.v. combination regimens (Fig. 2B). ANOVAs with Bonferroni's correction for multiple tests revealed significant (P value ≤ 0.05) increases and decreases in specific OTUs (Table 2). The bacterial families showing significant decreases in relative abundances include various members of the phyla Firmicutes, such as Ruminococcaceae, Lachnospiraceae, and other Clostridiales families. In addition, various Bacteroidales families were reduced. Some specific species identified as having decreases in relative abundances were Faecalibacterium prausnitzii, Parabacteroides distasonis, Bacteroides uniformis, and Blautia obeum. Other OTUs increased under oral-i.v. GSK1322322 treatment, which included members of the Betaproteobacteria (Sutterella spp.), Gammaproteobacteria (Enterobacter spp.), and Bifidobacteriaceae. Of the Bifidobacteriaceae, some specific species identified were Bifidobacterium pseudolongum and Bifidobacterium adolescentis.
FIG 2.
Histogram of proportional changes in bacterial OTU abundance at the class level. The samples are arranged in matched order for prestudy and end-of-study samples from the same subject for placebo and i.v.-only dosing (A) and oral-i.v. dosing regimens (B). The analyses and figures were generated using the software QIIME version 1.7 (25).
TABLE 2.
Significantly changed gut bacterial operational taxonomic units in subjects receiving oral-i.v.-administered GSK1322322 in prestudy versus end-of-study comparisons
| Operational taxonomic unitsa | Mean proportional abundance |
Percent change | P valueb | |
|---|---|---|---|---|
| Prestudy | End of study | |||
| Firmicutes; Clostridia; Clostridiales; Ruminococcaceae; Faecalibacterium prausnitzii | 0.050749 | 0.002567 | −4.82 | 1.2582E−09 |
| Firmicutes; Clostridia; Clostridiales; Ruminococcaceae | 0.113500 | 0.035066 | −7.85 | 7.5614E−07 |
| Bacteroidetes; Bacteroidia; Bacteroidales[Odoribacteraceae]; Odoribacter | 0.003950 | 0.000007 | −0.4 | 8.4503E−07 |
| Firmicutes; Clostridia; Clostridiales; Ruminococcaceae; Faecalibacterium | 0.000044 | 0.000002 | −0.01 | 2.2920E−05 |
| Bacteroidetes; Bacteroidia; Bacteroidales; Porphyromonadaceae; Parabacteroides distasonis | 0.008073 | 0.000515 | −0.76 | 3.5451E−05 |
| Firmicutes; Clostridia; Clostridiales; Lachnospiraceae; Blautia | 0.004165 | 0.001283 | −0.29 | 6.6706E−05 |
| Proteobacteria; Betaproteobacteria; Burkholderiales; Alcaligenaceae; Sutterella | 0.022233 | 0.116435 | +9.43 | 2.4570E−04 |
| Actinobacteria; Actinobacteria; Bifidobacteriales; Bifidobacteriaceae; Bifidobacterium pseudolongum | 0.000253 | 0.001713 | +0.15 | 3.9237E−04 |
| Actinobacteria; Actinobacteria; Bifidobacteriales; Bifidobacteriaceae; Bifidobacterium adolescentis | 0.008518 | 0.084296 | +7.58 | 1.1377E−03 |
| Bacteroidetes; Bacteroidia; Bacteroidales[Barnesiellaceae] | 0.013507 | 0.000128 | −1.34 | 1.3239E−03 |
| Firmicutes; Clostridia; Clostridiales[Mogibacteriaceae] | 0.000516 | 0.000086 | −0.05 | 1.6670E−03 |
| Firmicutes; Clostridia; Clostridiales; Lachnospiraceae | 0.053939 | 0.130409 | +7.65 | 3.0110E−03 |
| Actinobacteria; Actinobacteria; Bifidobacteriales; Bifidobacteriaceae; Bifidobacterium | 0.001409 | 0.065791 | +6.44 | 6.4763E−03 |
| Bacteroidetes; Bacteroidia; Bacteroidales[Odoribacteraceae]; Butyricimonas | 0.002951 | 0.000330 | −0.27 | 2.0858E−02 |
| Proteobacteria; Gammaproteobacteria; Enterobacteriales; Enterobacteriaceae | 0.000889 | 0.074874 | +7.4 | 2.5116E−02 |
| Bacteroidetes; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides uniformis | 0.049349 | 0.008082 | −4.13 | 2.5985E−02 |
| Firmicutes; Clostridia; Clostridiales; Lachnospiraceae; Anaerostipes | 0.000668 | 0.000074 | −0.06 | 4.1837E−02 |
| Firmicutes; Clostridia; Clostridiales; Lachnospiraceae; Blautia obeum | 0.000066 | 0.000010 | −0.01 | 4.6599E−02 |
OTU determination at the L7 level in QIIME, as described in Materials and Methods.
Bonferroni-corrected P values for multiple tests.
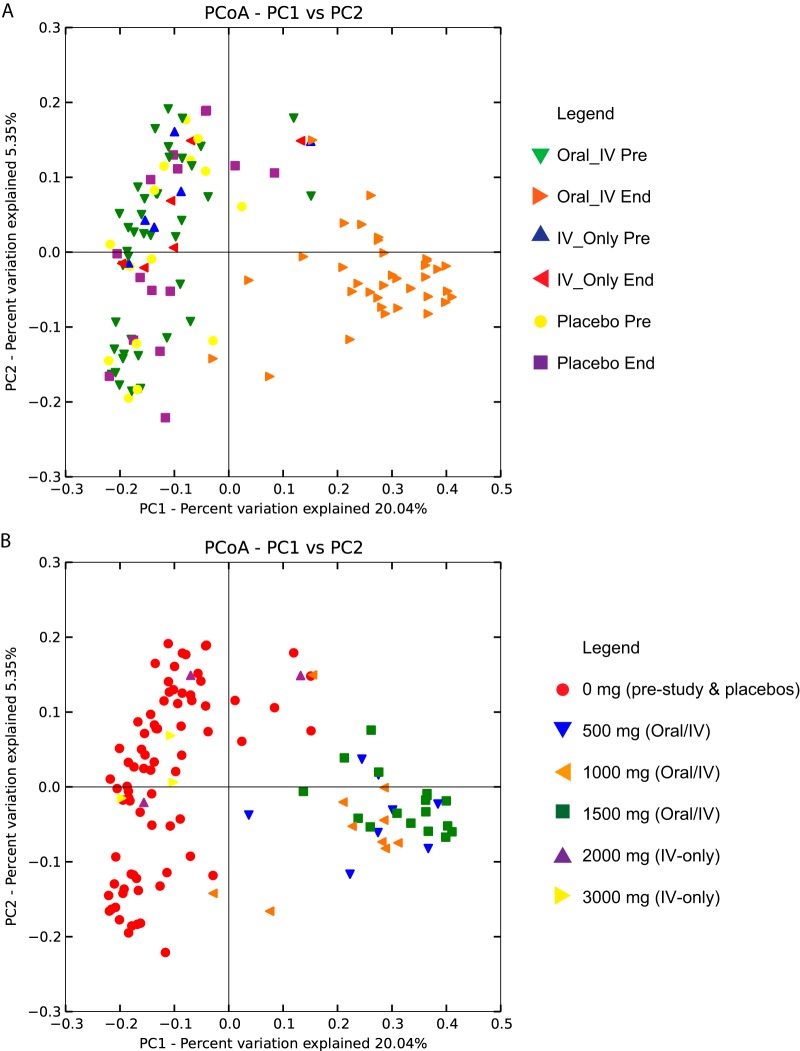
We assessed the overall differences between the bacterial communities of the samples, as partitioned by all available metadata variables (i.e., age, gender, BMI, subject identifier, adverse events, dose level, and treatment) using β-diversity indices calculated for unweighted UniFrac measures of OTU phylogenetic distance (37). Principal coordinate analysis (PCoA) plots of β-diversity indices revealed that the clearest separation of bacterial communities occurred for either dosing regimen (mg) or treatment (i.e., oral-i.v., i.v. only, and placebo). Here, the end-of-study samples from the subjects receiving the oral-i.v. dosing regimen formed a well-separated cluster from all the prestudy samples, regardless of treatment, as well as end-of-study samples from either the placebo or i.v.-only-treated subjects (Fig. 3A and B).
FIG 3.
Principal coordinate analysis (PCoA) plots of β-diversity measures of between-group microbiota community diversity based on treatment (placebo, i.v. only, and oral-i.v.) and sample collection time (prestudy or end of study) (A) and dosage (B). Note that highest two doses were administered by i.v. only. The analysis and figure were generated using the software QIIME version 1.7 (25).
Functional and phylogenomic analyses.
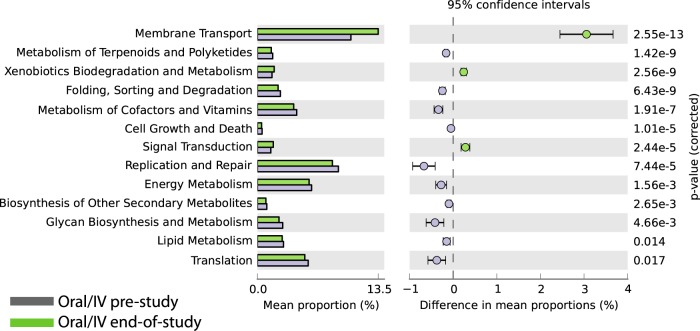
We looked for potential functional changes in oral-i.v.-dosed bacterial communities by comparing prestudy to end-of-study matched samples using the software PICRUSt (30), which uses 16S rRNA sequence profiles to estimate metagenomic content based on reference bacterial genomes and the KEGG pathway database (32). The most significant increased pathway representations in the end-of-study samples were membrane transport, which includes multidrug transporters, xenobiotic metabolism and degradation, and signal transduction (Fig. 4). The pathways with decreased presence in the samples were metabolism of terpenoids and polyketides, protein folding, sorting and degradation, and metabolism of cofactors and vitamins.
FIG 4.
Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) predictions of the functional composition of oral-i.v. prestudy (blue bars) and end-of-study (green bars) metagenome using 16S rRNA gene data and a database of reference genomes (30). The KEGG database (31) functional categories are shown with the displayed histograms and P value determinations, as calculated by the STAMP software (33).
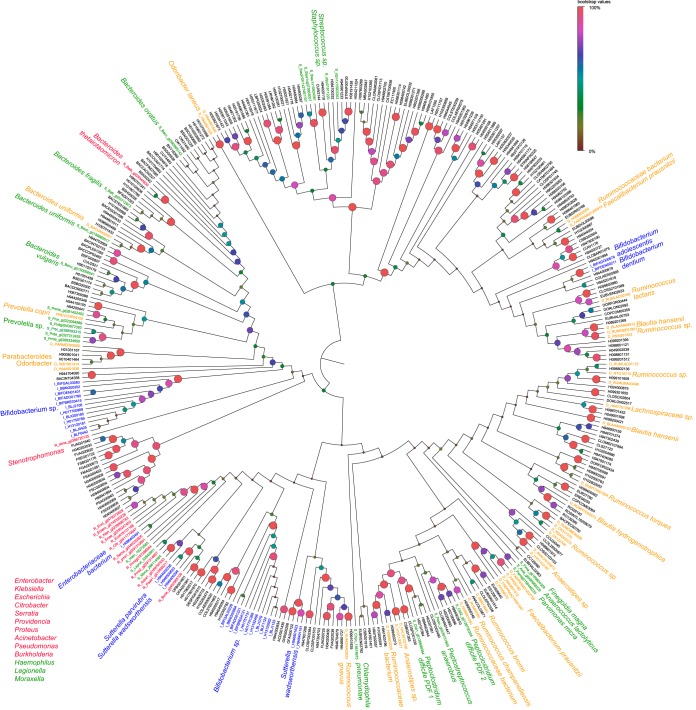
We also reconstructed a composite phylogenetic tree of publically available PDF amino acid sequences from the gut microbiota detected in our study, along with PDF proteins from bacterial pathogens for which we had available MIC (micrograms per milliliter) in vitro data (15) (Fig. 5). Many bacterial genomes contain two or more PDF-encoding genes (16). However, in most bacterial pathogens, only the pdfA gene is essential for growth, and other gene copies are nonredundant and cannot compensate for a loss of pdfA function (17). Whenever possible, we edited our final multiple-sequence alignment to known pdfA homologs, although this was not comprehensive, since multicopy pdf genes from most gut bacteria have not been fully annotated; thus, some species appear more than once in the tree. While we used the closed-reference method in QIIME for OTU identification against 16S rRNA sequences with known species associations, only a subset of those OTUs have corresponding complete reference genome sequences for obtaining their PDF proteins. Our final edited alignment of PDF proteins was 108 amino acids in length and contained a total of 366 sequences, of which 41 were from bacterial pathogens associated with known MIC data for GSK1322322, and the remainder were from gut bacterial reference genomes.
FIG 5.
Phylogenetic tree of peptide deformylase amino acid sequences for species of gastrointestinal (GI) microbiota detected in this study and bacterial pathogens with available MIC data for GSK1322322 (15). Bacterial pathogens were classified as low susceptible (red) or high susceptible (green) according to an MIC cutoff of ≥8.0 µg/ml (see Materials and Methods). GI bacteria were classified by changes in oral-i.v. end-of-study compared to prestudy samples as having a statistically significant increase (blue) or decrease (orange) in the relative abundances, or as no change (black). The phylogenetic tree was reconstructed using the neighbor-joining method with the JTT option in the software MEGA6 (36). Support for nodes in 1,000 bootstrap replicates is indicated by increased sizes as well as weighting to the red color spectrum of the circles at the nodes.
In the phylogenetic tree, there were several clades in which pathogenic bacterial susceptibility (MIC ≥ 8.0 µg/ml) to GSK1322322 and gut bacterial species abundances showed similar trends. For example, several Proteobacteria species, such as those belonging to Enterobacter, had low in vitro susceptibilities to GSK1322322 and also showed increases in their relative abundances in the end-of-study samples from oral-i.v.-dosed subjects (Fig. 5). Conversely, in vitro MIC testing revealed Prevotella species to be susceptible to GSK1322322 (MIC ≤ 8.0), and this group also had similar decreases in oral-i.v.-dosed subjects. Many diverse groups of bacteria for which there were no closely related PDF homologs from the tested bacterial pathogens showed significant increases or decreases in relative abundances. Moreover, most of the bacterial OTUs represented in the phylogenetic analyses appeared to be unaffected by GSK1322322 administration, despite the universal presence of the PDF protein-coding genes in their genomes.
DISCUSSION
In this study, we evaluated the effects of a novel antibacterial drug, GSK1322322, on the human gut microbiota in a randomized dose-escalating phase I clinical trial with matched placebo controls in healthy volunteers. Our study adds to the growing body of literature on the potential short- and long-term effects of antibiotics on the gut microbiome in several important ways. First, our longitudinal interventional study design with matching prestudy and end-of-study samples from the same subjects on oral-i.v., i.v.-only, and placebo dosing regimens allowed us to evaluate the potential effects of GSK1322322 on the microbiome in terms of changes within individuals as well as by the drug delivery method. Previous studies on the effects of antibiotics on human microbiota have compared population cohorts delineated by treatment or nontreatment, which, although informative, can be confounded by interindividual variation in personal microbiome compositions (38). Other reports of the longitudinal monitoring of microbiome changes over time under antibiotic treatment have been limited to much smaller numbers of human subjects than those used in our study (39).
Second, GSK1322322 is a new generation of antibiotics with a very specific mechanism of action and a known bacterial target, peptide deformylase (PDF), which is essential for bacterial survival and proliferation (40). Additional insights are gained from the fact that PDF is an unexploited bacterial target, and there has been little clinical exposure to PDF inhibitors (41). Therefore, we were able to evaluate the effects of GSK1322322 on the microbiota in terms of the potential variation in the target sequence and pathways across bacterial taxa without the confounding effects of selection for resistance due to the historical clinical usage of this class of drugs.
We found the most significant effect on microbiota was the method of drug administration. No significant differences in relative bacterial abundances were observed for either the placebo- or i.v.-only-treated subjects. However, significant changes in relative bacterial abundances and distinct β-diversity clustering were found for the oral-i.v. combination treatment. Absolute bacterial abundance cannot be determined by 16S rRNA amplicon sequencing, given the efforts to normalize the sample DNA amounts required for NGS. Assays of select bacterial species using gene-specific quantitative PCR (qPCR) reactions would be required to determine absolute bacterial abundance (which was not performed as part of this study). However, it is known that the use of antibiotics with bioavailability in the intestine will result in lower bacterial cell counts, depending on the type of drug, its dosage, and level of exposure (42). For GSK1322322, the absolute bioavailabilities of the current tablet formulation were 69% and 56% at 1,000 mg and 1,500 mg, respectively, under fasted conditions (18, 23). Comparisons of the α-rarefaction curves (Fig. 2) show a much lower bacterial diversity in the end-of-study samples from oral-i.v.-dosed subjects than those dosed with i.v. only, which suggests that GSK1322322 taken orally does have certain antibacterial activity in the gut. However, systemic levels of i.v.-only-administered GSK1322322 have no significant impact on the gut microbiome, similar to the results seen in placebo-treated subjects.
In the growing discourse over the potentially negative effects of long-term antibiotic exposure on beneficial microbes in our GI tract, it is important to consider the route of drug administration into the human body (3). To our knowledge, there have not been similar comparative studies in humans on the effects on the gut microbiome of an antibiotic when administered by different modes. Zhang et al. (43) inoculated mice with antibiotic-resistant tet(M)-carrying Enterobacter spp. or blaCMY-2-carrying E. coli and then administered different doses of either tetracycline hydrochloride or ampicillin by oral and i.v. routes. They found that mice receiving orally administered antibiotics had significantly higher levels of antibiotic-resistant bacterial strains, while i.v.-treated animals had negligible changes. These differences might be due to renal versus gastrointestinal tract secretion of i.v. and orally administered antibiotics, respectively. The greatest current medical need for new antibiotics is to treat emerging drug-resistant bacterial strains in hospitalized critical care patients. In this patient population, i.v. administration is the most likely delivery route for these future drugs, which will potentially minimize their impact on the GI microbiome and mitigate any selection pressure favoring the propagation of antibiotic resistance genes.
Oral-i.v.-administered GSK1322322 did produce some significant changes in the gut microbial ecosystems of those subjects, as evidenced from the overall distinct clustering of the β-diversity measures in the PCoA plots (Fig. 3), as well as the ANOVAs of the relative abundances of OTUs (Table 2). The bacterial families showing significant decreases in relative abundances include various members of the phylum Firmicutes, such as Ruminococcaceae, Lachnospiraceae, and other Clostridiales families, which correlates with the antibacterial activity of GSK1322322 against Gram-positive (Firmicutes) bacterial pathogens, such as S. aureus and S. pneumoniae. Bacteroidales were also reduced in our analysis. Some specific species identified as showing decreases in relative abundances were F. prausnitzii, P. distasonis, B. uniformis, and B. obeum. Other OTUs that increased under oral-i.v. GSK1322322 dosing included members of the Betaproteobacteria (Sutterella spp.), Gammaproteobacteria (Enterobacter spp.), and Bifidobacteriaceae (the species B. pseudolongum, B. adolescentis, and others).
Whether or not GSK1322322 has a negative, neutral, or positive effect on gut microbiota communities is difficult to assess, mainly because our knowledge of the interplay between specific GI tract bacterial species and human health is still evolving. For example, F. prausnitzii has been perceived to have a protective role in the gut and in minimizing the effects of Crohn's disease (44); however, recent clinical studies have questioned the causative association of this species with disease improvement (45). Bacteroides spp., such as B. fragilis, have been shown to produce polysaccharides that have beneficial immunomodulatory effects (46), while others, such as P. distasonis, can be carriers of multidrug resistance loci (47). Proteobacterial species, including Enterobacter spp., have been associated with instigating proinflammatory cascades leading to various disease pathologies (as reviewed in reference 48). Conversely, Bifidobacterium spp. have potential immunomodulatory properties, and several strains are actively being developed as probiotics (49). Regardless, GSK1322322 was well tolerated in this phase I clinical trial subject group, and gastrointestinal AEs, as monitored, were uncommon and did not prevent volunteers from completing the dosing regimen (18, 23).
GSK1322322 engages a highly specific target found in all bacterial species, namely, PDF, although the compound is not universal in its bactericidal effects (15). Our phylogenomic analysis shows that the spectrum of susceptibility to GSK1322322 among gastrointestinal tract microbiota can be partially explained or predicted from in vitro MIC data from bacterial pathogens. For example, Enterobacter spp. increase in the oral-i.v. end-of-study samples correlated with the lower susceptibility of Enterobacter spp. and other Gram negatives with closely related PDF proteins, as measured in vitro. However, many species were unaffected in terms of relative abundance, while others increased or decreased. In part, the distribution might be based on the divergence of the targeted PDF amino acid sequences, as well as species-specific differences in cell wall permeability, unknown intrinsic resistance mechanisms, or microenvironment exposure to the compound.
The functional analyses summarized in Fig. 4 suggest that the greatest changes occurred in pathways potentially involving resistance mechanisms, such as efflux pumps, xenobiotic (antibiotic) metabolism, and adaptive resistance via lowering of the growth rate (i.e., slowing of metabolic pathways and DNA repair) (reviewed in references 50, 51, and 52). However, our functional analyses are highly provisional, insofar as the computational method employed by PICRUSt infers metagenomic content indirectly based on computed linkages between 16S rRNA gene signatures and reference bacterial genomes (30). Direct metagenomic DNA sequencing will be required to substantiate the inferences of the genomic potential of the microbiome made here in this study.
The concurrent increase in some bacterial groups with the decrease of others for the end-of-study oral-i.v. dosing regimen of GSK1322322 potentially reflects dynamic changes in the gastrointestinal tract microbial ecosystem caused by antibiotic exposure. Dethlefsen and Relman (39) characterized the microbiome from three volunteers taking two courses of the antibiotic ciprofloxacin for 10 months and found profound shifts in the overall gut microbial ecosystem, reflecting potential niche replacement of one bacterial species or group by another. Similar to terrestrial and aquatic ecosystems, certain bacterial species might have more prominent roles in shaping the overall composition of the microbiome and be potential “keystone” species; alterations of their abundances might lead to new niches being available for exploitation by lower-abundance species that might be from the same or different taxonomic groups (53, 54). We can only speculate that some of the dynamic changes in bacterial species abundances induced by oral-i.v. GSK1322322 dosing regimens might reflect a disruption in interspecies interactions. While beyond the scope of the present study, it would also be interesting to monitor how the observed changes in bacterial species are reversible over time, especially the resilience of the microbiome to return to its predosing state.
Our study shows specific changes in the microbiome associated with the novel antibacterial compound GSK1322322. Importantly, the effects are mitigated by the drug delivery method, and no significant changes in the relative abundances of any bacterial taxa were observed for subjects treated with the i.v.-only dosing regimen. Dosing with an oral component did result in significant changes in some bacterial taxa and shifted overall bacterial community diversity measures away from those of samples taken from subjects either prestudy for placebo, i.v.-only, and oral-i.v. dosing or at the end of the study for placebo and i.v.-only dosing. However, while the changes in the microbiota were subtle, the differences were significant and specific to particular bacterial groups. Future microbiome analyses involving different antibiotics in clinical trials will be valuable for understanding potential interactions between antibiotics and human gastrointestinal microbial communities.
ACKNOWLEDGMENTS
The original clinical trial and this microbiome study were entirely funded by GlaxoSmithKline.
We all worked for GlaxoSmithKline at the time this work was performed.
REFERENCES
- 1.Blaser M, Bork P, Fraser C, Knight R, Wang J. 2013. The microbiome explored: recent insights and future challenges. Nat Rev Microbiol 11:213–217. doi: 10.1038/nrmicro2973. [DOI] [PubMed] [Google Scholar]
- 2.Holmes E, Li JV, Marchesi JR, Nicholson JK. 2012. Gut microbiota composition and activity in relation to host metabolic phenotype and disease risk. Cell Metab 16:559–564. doi: 10.1016/j.cmet.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 3.Zeissig S, Blumberg RS. 2014. Life at the beginning: perturbation of the microbiota by antibiotics in early life and its role in health and disease. Nat Immunol 15:307–310. doi: 10.1038/ni.2847. [DOI] [PubMed] [Google Scholar]
- 4.Blaser MJ, Falkow S. 2009. What are the consequences of the disappearing human microbiota? Nat Rev Microbiol 7:887–894. doi: 10.1038/nrmicro2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blaser M. 2011. Antibiotic overuse: stop the killing of beneficial bacteria. Nature 476:393–394. doi: 10.1038/476393a. [DOI] [PubMed] [Google Scholar]
- 6.Pérez-Cobas AE, Artacho A, Knecht H, Ferrús ML, Friedrichs A, Ott SJ, Moya A, Latorre A, Gosalbes MJ. 2013. Differential effects of antibiotic therapy on the structure and function of human gut microbiota. PLoS One 8:e80201. doi: 10.1371/journal.pone.0080201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pérez-Cobas AE, Gosalbes MJ, Friedrichs A, Knecht H, Artacho A, Eismann K, Otto W, Rojo D, Bargiela R, von Bergen M, Neulinger SC, Däumer C, Heinsen FA, Latorre A, Barbas C, Seifert J, dos Santos VM, Ott SJ, Ferrer M, Moya A. 2013. Gut microbiota disturbance during antibiotic therapy: a multi-omic approach. Gut 62:1591–1601. doi: 10.1136/gutjnl-2012-303184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shaw SY, Blanchard JF, Bernstein CN. 2011. Association between the use of antibiotics and new diagnoses of Crohn's disease and ulcerative colitis. Am J Gastroenterol 106:2133–2142. doi: 10.1038/ajg.2011.304. [DOI] [PubMed] [Google Scholar]
- 9.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, Essers J, Mitrovic M, Ning K, Cleynen I, Theatre E, Spain SL, Raychaudhuri S, Goyette P, Wei Z, Abraham C, Achkar JP, Ahmad T, Amininejad L, Ananthakrishnan AN, Andersen V, Andrews JM, Baidoo L, Balschun T, Bampton PA, Bitton A, Boucher G, Brand S, Büning C, Cohain A, Cichon S, D'Amato M, De Jong D, Devaney KL, Dubinsky M, Edwards C, Ellinghaus D, Ferguson LR, Franchimont D, Fransen K, Gearry R, Georges M, Gieger C, Glas J, Haritunians T, Hart A, et al. 2012. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Semic-Jusufagic A, Belgrave D, Pickles A, Telcian AG, Bakhsoliani E, Sykes A, Simpson A, Johnston SL, Custovic A. 2014. Assessing the association of early life antibiotic prescription with asthma exacerbations, impaired antiviral immunity, and genetic variants in 17q21: a population-based birth cohort study. Lancet Respir Med 2:621–630. doi: 10.1016/S2213-2600(14)70096-7. [DOI] [PubMed] [Google Scholar]
- 11.Peterfreund GL, Vandivier LE, Sinha R, Marozsan AJ, Olson WC, Zhu J, Bushman FD. 2012. Succession in the gut microbiome following antibiotic and antibody therapies for Clostridium difficile. PLoS One 7:e46966. doi: 10.1371/journal.pone.0046966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho I, Yamanishi S, Cox L, Methé BA, Zavadil J, Li K, Gao Z, Mahana D, Raju K, Teitler I, Li H, Alekseyenko AV, Blaser MJ. 2012. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature 488:621–626. doi: 10.1038/nature11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Membrez M, Blancher F, Jaquet M, Bibiloni R, Cani PD, Burcelin RG, Corthesy I, Macé K, Chou CJ. 2008. Gut microbiota modulation with norfloxacin and ampicillin enhances glucose tolerance in mice. FASEB J 22:2416–2426. doi: 10.1096/fj.07-102723. [DOI] [PubMed] [Google Scholar]
- 14.Jacobs MR, Felmingham D, Appelbaum PC, Grüneberg RN, Alexander Project Group . 2003. The Alexander Project 1998–2000: susceptibility of pathogens isolated from community-acquired respiratory tract infection to commonly used antimicrobial agents. J Antimicrob Chemother 52:229–246. doi: 10.1093/jac/dkg321. [DOI] [PubMed] [Google Scholar]
- 15.O'Dwyer K, Hackel M, Hightower S, Hoban D, Bouchillon S, Qin D, Aubart K, Zalacain M, Butler D. 2013. Comparative analysis of the antibacterial activity of a novel peptide deformylase inhibitor, GSK1322322. Antimicrob Agents Chemother 57:2333–2342. doi: 10.1128/AAC.02566-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guilloteau JP, Mathieu M, Giglione C, Blanc V, Dupuy A, Chevrier M, Gil P, Famechon A, Meinnel T, Mikol V. 2002. The crystal structures of four peptide deformylases bound to the antibiotic actinonin reveal two distinct types: a platform for the structure-based design of antibacterial agents. J Mol Biol 320:951–962. doi: 10.1016/S0022-2836(02)00549-1. [DOI] [PubMed] [Google Scholar]
- 17.Huang J, Van Aller GS, Taylor AN, Kerrigan JJ, Liu WS, Trulli JM, Lai Z, Holmes D, Aubart KM, Brown JR, Zalacain M. 2006. Phylogenomic and biochemical characterization of three Legionella pneumophila polypeptide deformylases. J Bacteriol 188:5249–5257. doi: 10.1128/JB.00866-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naderer OJ, Jones LS, Zhu J, Kurtinecz M, Dumont E. 2013. Safety, tolerability, and pharmacokinetics of oral and intravenous administration of GSK1322322, a peptide deformylase inhibitor. J Clin Pharmacol 53:1168–1176. doi: 10.1002/jcph.150. [DOI] [PubMed] [Google Scholar]
- 19.Naderer OJ, Dumont E, Zhu J, Kurtinecz M, Jones LS. 2013. Single-dose safety, tolerability, and pharmacokinetics of the antibiotic GSK1322322, a novel peptide deformylase inhibitor. Antimicrob Agents Chemother 57:2005–2009. doi: 10.1128/AAC.01779-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.NIH HMP Working Group, Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, Bonazzi V, McEwen JE, Wetterstrand KA, Deal C, Baker CC, Di Francesco V, Howcroft TK, Karp RW, Lunsford RD, Wellington CR, Belachew T, Wright M, Giblin C, David H, Mills M, Salomon R, Mullins C, Akolkar B, Begg L, Davis C, Grandison L, Humble M, Khalsa J, Little AR, Peavy H, Pontzer C, Portnoy M, Sayre MH, Starke-Reed P, Zakhari S, Read J, Watson B, Guyer M. 2009. The NIH Human Microbiome Project. Genome Res 19:2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Proctor LM. 2011. The Human Microbiome Project in 2011 and beyond. Cell Host Microbe 10:287–291. doi: 10.1016/j.chom.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 22.Human Microbiome Project Consortium. 2012. A framework for human microbiome research. Nature 486:215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naderer OJ, Dumont E, Zhu J, Kurtinecz M, Jones LS. 2013. Safety, tolerability and pharmacokinetics of repeat dosing of the antibiotic GSK1322322, a peptide deformylase inhibitor: a randomized placebo-controlled study. J Antimicrob Chemother 68:1901–1909. doi: 10.1093/jac/dkt097. [DOI] [PubMed] [Google Scholar]
- 24.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Navas-Molina JA, Peralta-Sánchez JM, González A, McMurdie PJ, Vázquez-Baeza Y, Xu Z, Ursell LK, Lauber C, Zhou H, Song SJ, Huntley J, Ackermann GL, Berg-Lyons D, Holmes S, Caporaso JG, Knight R. 2013. Advancing our understanding of the human microbiome using QIIME. Methods Enzymol 531:371–444. doi: 10.1016/B978-0-12-407863-5.00019-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 27.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. 2012. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R. 2011. UniFrac: an effective distance metric for microbial community comparison. ISME J 5:169–172. doi: 10.1038/ismej.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C. 2013. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. 2014. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res 42:D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanehisa M, Goto S. 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parks DH, Beiko RG. 2010. Identifying biologically relevant differences between metagenomic communities. Bioinformatics 26:715–721. doi: 10.1093/bioinformatics/btq041. [DOI] [PubMed] [Google Scholar]
- 34.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson JD, Gibson TJ, Higgins DG. 2002. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics Chapter 2:Unit 2.3. doi: 10.1002/0471250953.bi0203s00. [DOI] [PubMed] [Google Scholar]
- 36.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lozupone CA, Hamady M, Kelley ST, Knight R. 2007. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol 73:1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fouhy F, Guinane CM, Hussey S, Wall R, Ryan CA, Dempsey EM, Murphy B, Ross RP, Fitzgerald GF, Stanton C, Cotter PD. 2012. High-throughput sequencing reveals the incomplete, short-term recovery of infant gut microbiota following parenteral antibiotic treatment with ampicillin and gentamicin. Antimicrob Agents Chemother 56:5811–5820. doi: 10.1128/AAC.00789-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dethlefsen L, Relman DA. 2011. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A 108(Suppl 1):S4554–S4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Butler D, Chen D, O'Dwyer K, Lewandowski T, Aubart K, Zalacain M. 2014. Potent sub-MIC effect of GSK1322322 and other peptide deformylase inhibitors on in vitro growth of Staphylococcus aureus. Antimicrob Agents Chemother 58:290–296. doi: 10.1128/AAC.01292-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sutcliffe JA. 2011. Antibiotics in development targeting protein synthesis. Ann N Y Acad Sci 1241:122–152. doi: 10.1111/j.1749-6632.2011.06323.x. [DOI] [PubMed] [Google Scholar]
- 42.Panda S, El Khader I, Casellas F, López Vivancos J, García Cors M, Santiago A, Cuenca S, Guarner F, Manichanh C. 2014. Short-term effect of antibiotics on human gut microbiota. PLoS One 9:e95476. doi: 10.1371/journal.pone.0095476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang L, Huang Y, Zhou Y, Buckley T, Wang HH. 2013. Antibiotic administration routes significantly influence the levels of antibiotic resistance in gut microbiota. Antimicrob Agents Chemother 57:3659–3666. doi: 10.1128/AAC.00670-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jia W, Whitehead RN, Griffiths L, Dawson C, Waring RH, Ramsden DB, Hunter JO, Cole JA. 2010. Is the abundance of Faecalibacterium prausnitzii relevant to Crohn's disease? FEMS Microbiol Lett 310:138–144. doi: 10.1111/j.1574-6968.2010.02057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gerasimidis K, Bertz M, Hanske L, Junick J, Biskou O, Aguilera M, Garrick V, Russell RK, Blaut M, McGrogan P, Edwards CA. 2014. Decline in presumptively protective gut bacterial species and metabolites are paradoxically associated with disease improvement in pediatric Crohn's disease during enteral nutrition. Inflamm Bowel Dis 20:861–871. doi: 10.1097/MIB.0000000000000023. [DOI] [PubMed] [Google Scholar]
- 46.Mazmanian SK, Round JL, Kasper DL. 2008. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 47.Molina J, Barrantes G, Quesada-Gómez C, Rodríguez C, Rodríguez -Cavallini E. 2014. Phenotypic and genotypic characterization of multi-drug resistant Bacteroides, Parabacteroidesspp., and Pseudoflavonifractor from a Costa Rican hospital. Microb Drug Resist 20:478–484. doi: 10.1089/mdr.2013.0180. [DOI] [PubMed] [Google Scholar]
- 48.Winter SE, Lopez CA, Bäumler AJ. 2013. The dynamics of gut-associated microbial communities during inflammation. EMBO Rep 14:319–327. doi: 10.1038/embor.2013.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Groeger D, O'Mahony L, Murphy EF, Bourke JF, Dinan TG, Kiely B, Shanahan F, Quigley EM. 2013. Bifidobacterium infantis 35624 modulates host inflammatory processes beyond the gut. Gut Microbes 4:325–339. doi: 10.4161/gmic.25487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Andersson DI, Hughes D. 2014. Microbiological effects of sublethal levels of antibiotics. Nat Rev Microbiol 12:465–478. doi: 10.1038/nrmicro3270. [DOI] [PubMed] [Google Scholar]
- 51.Delmar JA, Su CC, Yu EW. 2014. Bacterial multidrug efflux transporters. Annu Rev Biophys 43:93–117. doi: 10.1146/annurev-biophys-051013-022855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cox G, Wright GD. 2013. Intrinsic antibiotic resistance: mechanisms, origins, challenges and solutions. Int J Med Microbiol 303:287–292. doi: 10.1016/j.ijmm.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 53.Hajishengallis G, Darveau RP, Curtis MA. 2012. The keystone-pathogen hypothesis. Nat Rev Microbiol 10:717–725. doi: 10.1038/nrmicro2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ze X, Le Mougen F, Duncan SH, Louis P, Flint HJ. 2013. Some are more equal than others: the role of “keystone” species in the degradation of recalcitrant substrates. Gut Microbes 4:236–240. doi: 10.4161/gmic.23998. [DOI] [PMC free article] [PubMed] [Google Scholar]