Abstract
The objective of this study was to assess the phenotypic susceptibility of HIV-1 subtype C isolates, with nonnucleoside reverse transcriptase inhibitor (NNRTI) resistance-associated amino acid changes, to newer NNRTIs. A panel of 52 site-directed mutants and 38 clinically derived HIV-1 subtype C clones was created, and the isolates were assessed for phenotypic susceptibility to etravirine (ETR), rilpivirine (RPV), efavirenz (EFV), and nevirapine (NVP) in an in vitro single-cycle phenotypic assay. The amino acid substitutions E138Q/R, Y181I/V, and M230L conferred high-level resistance to ETR, while K101P and Y181I/V conferred high-level resistance to RPV. Y181C, a major NNRTI resistance-associated amino acid substitution, caused decreased susceptibility to ETR and, to a lesser extent, RPV when combined with other mutations. These included N348I and T369I, amino acid changes in the connection domain that are not generally assessed during resistance testing. However, the prevalence of these genotypes among subtype C sequences was, in most cases, <1%. The more common EFV/NVP resistance-associated substitutions, such as K103N, V106M, and G190A, had no major impact on ETR or RPV susceptibility. The low-level resistance to RPV and ETR conferred by E138K was not significantly enhanced in the presence of M184V/I, unlike for EFV and NVP. Among patient samples, 97% were resistant to EFV and/or NVP, while only 24% and 16% were resistant to ETR and RPV, respectively. Overall, only a few, relatively rare NNRTI resistance-associated amino acid substitutions caused resistance to ETR and/or RPV in an HIV-1 subtype C background, suggesting that these newer NNRTIs would be effective in NVP/EFV-experienced HIV-1 subtype C-infected patients.
INTRODUCTION
Highly active antiretroviral therapy (HAART), which comprises the concomitant use of multiple potent antiretroviral drugs, has contributed to a significant decrease in the morbidity and mortality of people infected with HIV-1 (1). The failure of HAART through the acquisition of HIV drug resistance-associated substitutions that cause a decrease in viral susceptibility is usually due to poor adherence and insufficient drug concentrations. Although more than two-thirds of the global HIV-1 infections occur in sub-Saharan African countries, where HIV-1 infections are dominated by non-B subtypes, HAART regimens have largely been developed and tested against HIV-1 subtype B isolates (2). Subtype C accounts for almost half of all global infections and dominates the epidemic in southern Africa (3). While HAART agents are effective against all subtypes (4), greatly aiding the global response to HIV infection, specific resistance mutations and disparities in drug susceptibilities can differ by subtype (5). Examples include the K65R (6) and V106M (7) resistance-associated amino acid substitutions, which develop more frequently under drug pressure in HIV-1 subtype C than subtype B.
Nonnucleoside reverse transcriptase (RT) inhibitors (NNRTIs) are a component of most first-line HAART regimens. For efavirenz (EFV) and nevirapine (NVP), the genetic barrier to the development of resistance-associated mutations and a loss of potency is low. The accumulation of EFV and NVP resistance-associated mutations is rapid, often occurring within 3 months of virologic failure (8). Recent studies in South Africa have shown that up to 80% of patients who fail EFV or NVP therapy develop NNRTI resistance-associated mutations (9–12). Furthermore, the substantial cross-resistance between these two drugs makes their sequential use after virologic failure inappropriate.
Etravirine (ETR; TMC125) (13–15) and rilpivirine (RPV; TMC278) (16, 17) are diarylpyrimidine (DAPY) NNRTIs with resistance profiles that only partially overlap those of EFV and NVP (18, 19). The efficacy of ETR was assessed in the DUET-1 and DUET-2 trials with treatment-experienced patients, while RPV was assessed in treatment-naive patients in the ECHO and THRIVE trials (13–17, 20). Both ETR and RPV suppress viral replication irrespective of HIV-1 subtype and have shown activity against clinically relevant mutants (19, 21–23). However, data on the sequential use of these agents and cross-resistance have mostly been derived from subtype B-infected cohorts and often as part of clinical trials, with little information on how HIV-1 subtype C isolates from first-line NNRTI treatment failures might respond (22, 24). In this study, using clinical samples and site-directed mutants, we investigated the phenotypic impact of NNRTI resistance-associated amino acid substitutions on ETR and RPV susceptibility in an HIV-1 subtype C background since these drugs are likely to be increasingly used in South Africa.
MATERIALS AND METHODS
HIV-1 subtype C sequences.
A total of 1,433 HIV-1 subtype C sequences from patients exposed to EFV or NVP were used to determine the prevalence of different NNRTI resistance-associated mutations. Sequences were obtained from the South African Treatment and Resistance Network (SATuRN) database (http://www.bioafrica.net/regadb/) (n = 766) (9) and the Stanford HIV Drug Resistance Database (HIVdb) (http://hivdb.stanford.edu/) (n = 667). Only sequences with NNRTI resistance-associated mutations were included. A covariation analysis was performed to identify significantly associated pairs of NNRTI resistance-associated mutations as previously described (25). Briefly, a Jaccard similarity coefficient was employed to identify pairwise correlations among NNRTI resistance-associated mutations. Holm's correction was used to control for the familywise error rate of multiple pairwise comparisons.
Clinical samples for phenotyping.
Patient samples (n = 38) from the South African Virological Evaluation (SAVE) study (9) and a workplace HIV program within the mining industry (26) and samples submitted for routine genotyping were selected for resistance phenotyping (27). Samples were selected on the basis of availability and the presence of one or more NNRTI resistance-associated mutations. Ethical approval for drug resistance testing on these samples was provided by the Committee for Research on Human Subjects (University of the Witwatersrand, Johannesburg, South Africa) and the Regional Medical Ethics Board (Karolinska Institutet, Stockholm, Sweden).
HIV-1 expression vectors.
A retroviral vector-based, single-round infection system was used for in vitro phenotyping (28–32). For this, a 3,500-bp fragment spanning gag and part of the integrase gene (bp 737 to 4,403 of strain HXB2) of the HIV-1 subtype B p8.9NSX vector (30) was replaced with the subtype C MJ4 proviral genome (GenBank accession number AF321523.1) (31, 33). NNRTI resistance-associated mutations were introduced into the p8.MJ4 plasmid using a QuikChange II XL site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA) in conjunction with mutagenesis primers synthesized by Integrated DNA Technologies (Coralville, IA, USA). Plasmids were transformed into Escherichia coli XL10-Gold ultracompetent cells (Stratagene, La Jolla, CA, USA) by a standard heat shock method. Individual clones were sequenced by population-based sequencing using a BigDye Terminator (v3.1) cycle sequencing kit (Applied Biosystems, Foster City, CA, USA) and an ABI Prism 3130xl genetic analyzer (Applied Biosystems, Foster City, CA, USA). The Stanford HIV drug resistance algorithm was used for quality assessment of sequences and mutation recognition (34). Mutants with double NNRTI resistance-associated mutations were created by successive rounds of site-directed mutagenesis in p8.MJ4.
For the phenotypic assessment of patient samples, the p8.MJ4 expression vector was modified by the introduction of an HpaI restriction site toward the end of the protease (PR) gene (bp 2496 of HXB2) by site-directed mutagenesis. In combination with an intrinsic HpaI restriction site in the reverse transcriptase gene of MJ4 (bp 3650 in HXB2), this allowed the excision of an ∼1,154-bp fragment from p8.MJ4 with HpaI endonuclease (Roche, Basel, Switzerland) and the religation of the blunt vector ends with a Rapid DNA Dephos & Ligation kit (Roche, Basel, Switzerland) to produce p8.MJ4ΔRT.
Construction of patient-derived plasmids.
The viral RNA from patient plasma samples was reverse transcribed with a ThermoScript RT-PCR system (Invitrogen, Life Technologies Corporation, Carlsbad, CA, USA) and an ∼1,154-kb reverse transcriptase fragment amplified by nested PCR using an Expand High-FidelityPlus PCR system (Roche, Basel, Switzerland). The second-round PCR primers (primers HpaI-F [5′-CCTACACCTGTTAACATAATTGGAAGRAATATGTTGAC-3′] and HpaI-R [5′-CAGCCTCTGTTAACTGTTTTACATCATTAGTGTG-3′) introduced an HpaI restriction site (the underlined GTTAAC sequence) at the 5′ and 3′ ends of the PCR amplicons. The p8.MJ4ΔRT vector and amplicons were restricted with the HpaI endonuclease (Roche, Basel, Switzerland) and ligated using the Rapid DNA Dephos & Ligation kit (Roche, Basel, Switzerland) to introduce patient-derived amplicons into p8.MJ4ΔRT. Ligation products were transformed into E. coli XL10-Gold ultracompetent cells (Stratagene, La Jolla, CA, USA) by a standard heat shock procedure. The plasmid inserts were sequenced, and the sequences were compared to the sequence of the original amplicon.
Single-cycle nonreplicating phenotypic assay.
Three plasmids were utilized for the production of virus-like particles: pMD.G (29) to facilitate viral entry, pCSFLW (35) for the detection of viral integration by bioluminescence, and p8.MJ4 (or p8.MJ4ΔRT) for the expression of HIV-1 subtype C PR-RT. Plasmids were cotransfected into HEK293T cells, in which cotransfection was facilitated with either the Fugene6 transfection reagent (Roche, Basel, Switzerland) (30) or polyethylenimine (PEI; Polysciences, Inc., Warrington, PA, USA). Virus-containing supernatants were harvested at 48 and 72 h posttransfection. Viral titers were determined by infecting HEK293T cells with serial dilutions of virus-containing supernatants. After 48 h, the expression of firefly luciferase was determined with the Bright-Glo luciferase assay substrate (Promega, Fitchburg, WI, USA) on a Victor3 multilabel reader (PerkinElmer, Waltham, MA, USA). Virus inocula were standardized to produce a luminescence of 1 × 104 to 1 × 105 relative light units (RLU) after 48 h of incubation with HEK293T cells in drug-free medium.
For drug susceptibility assays, duplicate serial dilutions of all four NNRTIs (ETR, RPV, EFV, and NVP) were prepared in 96-well cell culture plates to which standardized virus inocula and HEK293T cells were added. The MJ4 virus was included in each assay run as a wild-type control. After incubation for 48 h, the expression of firefly luciferase was determined as described above. Each virus was screened in at least two independent assays for each drug, and the 50% effective concentration (EC50) values were calculated. Fold change (FC) values were determined by reference to the EC50s for the MJ4 wild-type control virus for each drug. The lower FC cutoff value for each drug was determined using the 99th percentile of the average EC50 for MJ4 wild-type virus, assessed in multiple repeat screens of each drug. Samples were classified as susceptible if the FC value fell below the FC cutoff values for ETR (FC cutoff value, 3.6), RPV (FC cutoff value, 2.6), EFV (FC cutoff value, 3.8), or NVP (FC cutoff value, 2.8). For resistance, an FC of ≥10 was used as the upper cutoff for all 4 drugs, although in many cases these levels exceeded >40. Samples with FC values between the lower and upper assay cutoff values were considered to show reduced susceptibility. It is important to note that the cutoff values used in this analysis are not linked to clinical correlates or outcomes. Furthermore, the classifications of susceptible, reduced susceptibility, and resistant are used merely to rank the responses for the NNRTIs used in our assay.
RESULTS
Prevalence of NNRTI resistance-associated mutations in the subtype C data set.
Sequence data from 1,433 HIV-1 subtype C-infected patients who failed first-line NNRTI treatment and developed NNRTI resistance-associated mutations were used in this study. The majority of these patients were treated with EFV (63.3%) or NVP (30.7%), with 3% being exposed to both drugs (Table 1). Less than 1% of patients were treated with other NNRTIs (delavirdine and MKC422), and for 2.7% the NNRTI treatment was not known. Most patients from the SATuRN database were treated with EFV, while those from the Stanford database were more biased toward NVP use.
TABLE 1.
HIV-1 subtype C sequences with NNRTI resistance-associated mutations used in this study
| NNRTI treatment(s) | No. (%) of patients |
||
|---|---|---|---|
| SATuRN database | Stanford database | Total | |
| EFV | 630 (82.2) | 278 (41.6) | 908 (63.3) |
| NVP | 124 (16.1) | 316 (47.4) | 440 (30.7) |
| NVP, EFV | 12 (1.6) | 31 (4.6) | 43 (3.0) |
| Unknown | 39 (5.8) | 39 (2.7) | |
| Othera | 3 (0.4) | 3 (0.2) | |
| Total | 766 | 667 | 1,433 |
The patients received delavirdine (DLV) or MKC422.
A total of 30 published NNRTI resistance-associated mutations (36) were identified in this data set. The EFV/NVP-selected amino acid substitutions K103N, V106M, Y181C, and G190A had the highest overall prevalence: 52%, 25%, 15%, and 15% of samples, respectively (Fig. 1). V90I, A98G, L100I, K101E/H/P, K103S, V108I, E138A/K, V179D, Y188C/L, G190S, H221Y, P225H, and M230L were identified in 1% to 10% of sequences. The remaining 9 substitutions, V106A/I, E138G/Q/R, V179T, Y181I/V, and Y188H, had prevalences of <1%. Overall, the frequencies of resistance-associated amino acid substitutions were similar between the Stanford and SATuRN data sets, despite the differences in treatment histories.
FIG 1.
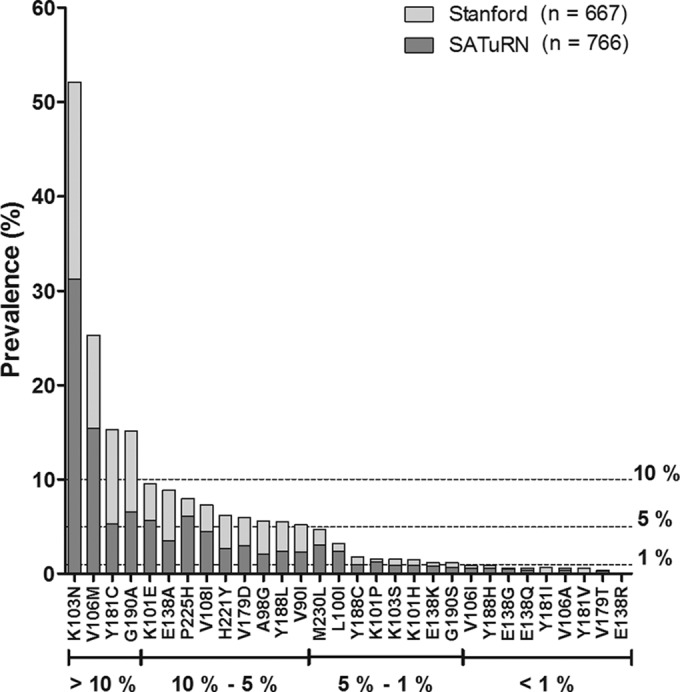
Prevalence of NNRTI resistance-associated amino acid substitutions among first-line treatment failures. Sequences from 1,433 HIV-1 subtype C-infected patients failing first-line therapy with NNRTI resistance-associated mutations were obtained from the SATuRN (n = 766; light gray bars) and Stanford (n = 667; dark gray bars) databases. Amino acid substitutions are grouped according to prevalence: >10%, 5% to 10%, 1% to 5%, and <1%.
Single site-directed mutations and NNRTI phenotypic susceptibility.
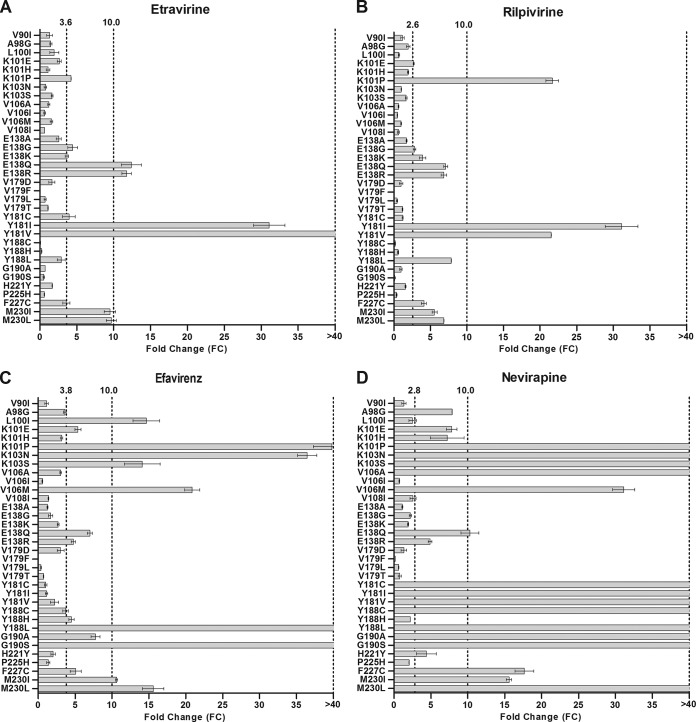
Thirty-four NNRTI resistance-associated mutations were introduced by site-directed mutagenesis into an HIV-1 subtype C expression vector, and the activities of the NNRTIs against the mutants were tested in a single-cycle nonreplicating phenotypic assay. The mutations included the 30 amino acid substitutions identified in our data set plus an additional 4 changes (V179F, V179L, F227C, and M230I) shown by others to be associated with ETR/RPV resistance (36).
Of these 34 single amino acid substitutions, only Y181I and Y181V caused high levels of resistance (FC > 40) to both ETR (Fig. 2A) and RPV (Fig. 2B). E138Q/R and M230I/L caused ∼10-fold resistance to ETR but had a lesser effect on susceptibility to RPV, whereas K101P and Y188L caused resistance to RPV but not to ETR. None of the remaining substitutions, including those frequently associated with EFV and/or NVP resistance, caused a significant reduction in susceptibility to ETR or RPV.
FIG 2.
Phenotypic resistance of viruses with single amino acid substitutions associated with resistance to the NNRTIs ETR, RPV, EFV, and NVP. Single NNRTI resistance-associated mutations (n = 34) were introduced into the p8.MJ4 subtype C HIV-1 expression plasmid by site-directed mutagenesis. Phenotypic susceptibilities to ETR (A), RPV (B), EFV (C), and NVP (D) were determined using a single-cycle nonreplicative phenotypic assay. The lower cutoff values determined for this assay are indicated for each drug. Columns indicate mean values, while error bars indicate the standard errors of the means (SEMs). Each mutant was tested in duplicate in at least two independent experiments.
For EFV and NVP, the K101P, K103N/S, V106M, Y188L, G190S, and M230I/L amino acid substitutions caused resistance to both drugs (Fig. 2C and D). In addition, V106A, E138Q, Y181C/I/V, Y188C, G190A, and F227C caused resistance to NVP. Susceptibility to EFV was not affected by the V106A, Y181C/I/V, or Y188C substitution, but E138Q, G190A, and F227C caused reduced susceptibility. L100I caused resistance to EFV but did not affect the susceptibility to NVP. For the remainder, only A98G, K101E/H, E138R, and H221Y reduced the susceptibility to NVP, while only K101E and E138R reduced the susceptibility to EFV.
Effect of NNRTI resistance-associated combinations on NNRTI susceptibility.
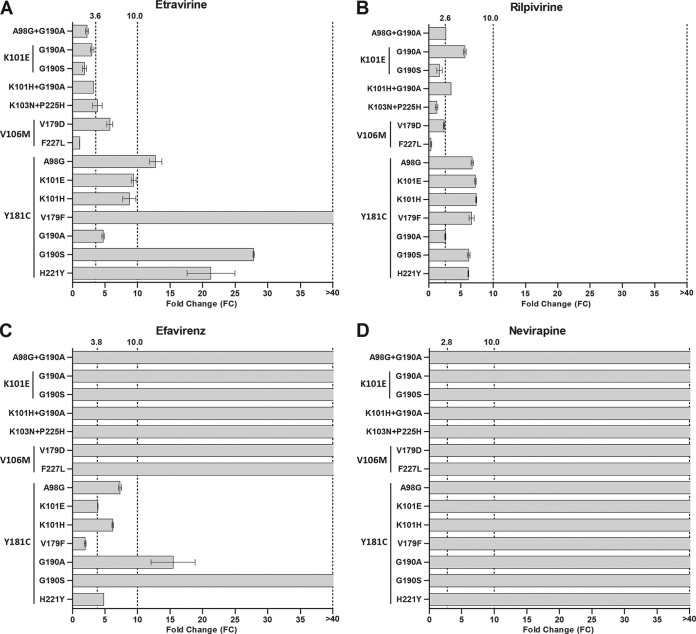
Five pairs of significantly (P < 0.0001) associated amino acid substitutions were identified in the subtype C data set: K101E-G190A, K103N-P225H, V106M-V179D, V106M-F227L, and Y181C-H221Y (Table 2). These 5 combinations occurred in 4 to 8% of the subtype C sequences from patients treated with EFV and/or NVP. In addition, 10 positively correlated pairs of ETR resistance-associated substitutions from a similar analysis of subtype B were selected (25) (Table 2). The K101E-G190A combination, identified in our subtype C analysis, was also identified in the subtype B analysis, with a similar prevalence being detected in both subtypes (5.6% and 3.4%, respectively). None of the remaining 9 combinations identified in the subtype B data set, all of which had a low prevalence (0.2% to 6.5%), were observed in our subtype C data set.
TABLE 2.
Pairs of positively correlated NNRTI resistance-associated amino acid substitutions
| Combination | P value for association | Prevalence (%) |
|---|---|---|
| Subtype C data seta | ||
| K101E-G190A | <0.0001 | 5.6 |
| K103N-P225H | <0.0001 | 7.7 |
| V106M-V179D | <0.0001 | 4.4 |
| V106M-F227L | <0.0001 | 4.0 |
| Y181C-H221Y | <0.0001 | 3.5 |
| Subtype B data setb | ||
| Y181C-G190A | <0.0001 | 6.5 |
| K101E-G190A | <0.0001 | 3.4 |
| K101E-Y181C | <0.0001 | 2.7 |
| K101E-G190S | <0.0001 | 1.0 |
| H101H-G190A | <0.0001 | 0.9 |
| A98G-Y181C | <0.0001 | 2.4 |
| K101H-Y181C | <0.0001 | 0.7 |
| A98G-G190A | <0.0001 | 1.6 |
| Y181C-G190S | <0.0001 | 0.9 |
| V179F-Y181C | <0.0001 | 0.2 |
From 1,433 subtype C sequences with NNRTI resistance-associated mutations from the SATuRN and Stanford databases.
From 13,039 published subtype B sequences (25).
All 14 significantly associated amino acid substitution pairs (5 subtype C and 9 subtype B amino acid substitution pairs) were generated, and the isolates were tested for phenotypic NNRTI resistance (Fig. 3). The seven NNRTI resistance-associated combinations that lacked Y181C had no major impact on ETR or RPV susceptibility (Fig. 3A and B). Combinations containing Y181C had a greater effect on ETR susceptibility than RPV susceptibility, particularly those containing A98G, V179F, G190S, and H221Y. Mutants containing Y181C had a minor effect on EFV susceptibility, except for combinations with G190A/S (Fig. 3C). Other combinations with G190A/S or V106M, as well as the K103N-P225H combination, caused high-level resistance to EFV. All double NNRTI resistance-associated mutations (with or without Y181C) caused high-level resistance to NVP (Fig. 3D). These data indicate that for the most frequently selected double NNRTI resistance-associated substitutions, there was considerably less resistance to ETR than to EVP and NVP and no high-level resistance to RPV.
FIG 3.
Phenotypic resistance to ETR, RPV, EFV, and NVP of viruses with double resistance-associated amino acid substitutions. Positively associated double mutations (n = 14) were introduced into the p8.MJ4 subtype C HIV-1 expression plasmid. Phenotypic susceptibilities to ETR (A), RPV (B), EFV (C), and NVP (D) were determined with a single-cycle nonreplicative phenotypic assay. The lower cutoff values determined for this assay are indicated for each drug. Columns indicate mean values, while error bars indicate the standard errors of the means (SEMs). Each mutant was tested in duplicate in at least two independent experiments.
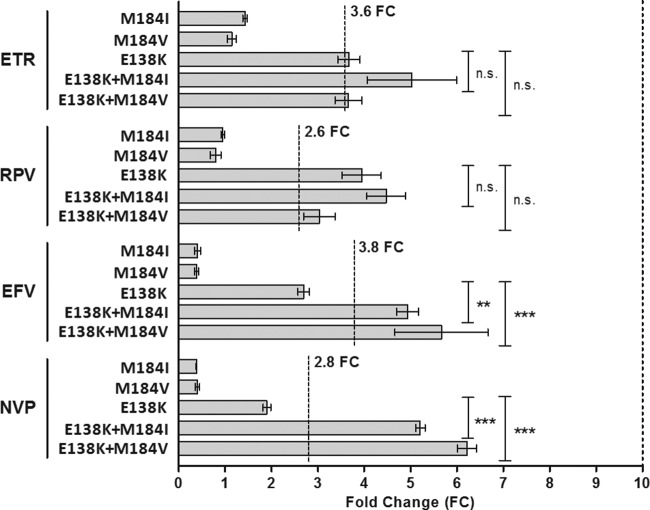
Impact of the M184I/V NRTI amino acid substitution on NNRTI susceptibility.
The frequent selection of the nucleoside reverse transcriptase inhibitor (NRTI) resistance-associated amino acid substitution M184I/V with E138K in the ECHO and THRIVE trials implicated this combination in reduced susceptibility to RPV (22, 37). To investigate this in a subtype C background, E138K with M184I or M184V was created in p8.MJ4 and the clones were tested for phenotypic susceptibility. The single M184I and M184V substitutions did not reduce susceptibility to any of the NNRTIs, while the single E138K substitution caused reduced susceptibility to RPV, as expected (Fig. 4). A low level of reduction in ETR susceptibility was also observed with this amino acid substitution. However, the E138K-M184I/V combination reduced susceptibility to all four NNRTIs. Thus, despite E138K-M184V/I being associated with RPV resistance, the largest decreases in susceptibility were observed for EFV and NVP, and we also noted a minor effect on ETR susceptibility.
FIG 4.
Impact of E138K and M184IV on phenotypic susceptibility to ETR, RPV, EFV, and NVP. The phenotypic susceptibility to ETR, RPV, EFV, and NVP of site-directed mutants containing amino acid substitutions M184I, M184V, E138K, and their combinations were determined. The lower cutoff values determined for this assay are indicated for each drug. Columns indicate mean values, while error bars indicate the standard errors of the mean (SEMs). Each mutant was tested in duplicate in at least two independent experiments. n.s., not significant; **, P < 0.01 (2-way analysis of variance); ***, P < 0.001 (2-way analysis of variance).
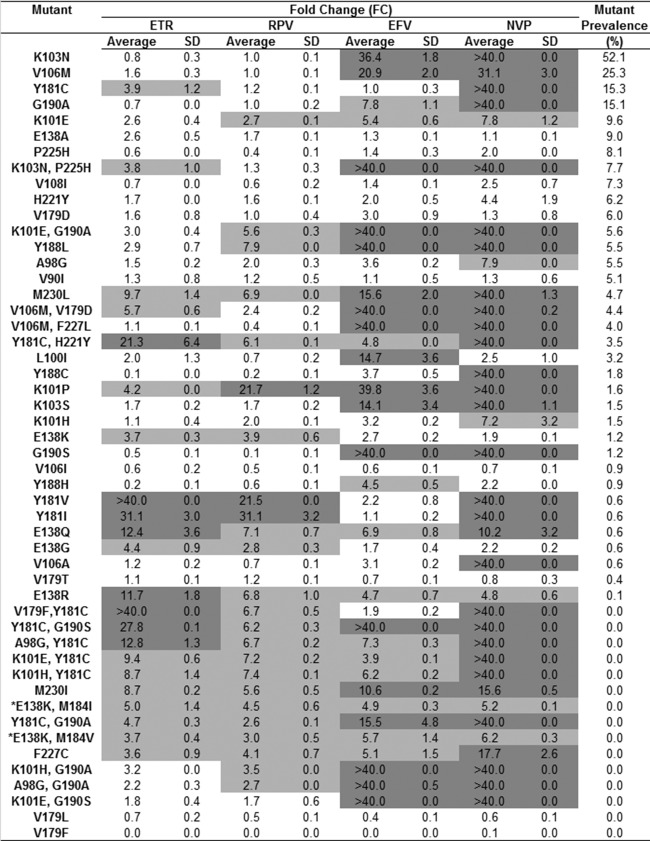
Sensitivity of mutants with prevalent NNRTI resistance-associated mutations to ETR and RPV.
To gauge the likely impact of NNRTI resistance-associated mutations on ETR and RPV efficacy, the phenotypic data for the susceptibility of all the single and double mutants to each drug were ranked according to mutation prevalence in the subtype C data set (Fig. 5). The most prevalent mutations (>10%) caused no major reduction in ETR or RPV susceptibility and included the signature EFV- and NVP-associated substitutions K103N, V106M, Y181C, and G190A. All mutations that caused high-level resistance to ETR or RPV were present at a ≤3.5% prevalence. For ETR this included Y181C-H221Y, Y181V/I, and E138Q/R, while for RPV this was restricted to K101P and Y181V/I. While a significant number of mutants showed reduced susceptibility to ETR and RPV, these were not detected in our HIV-1 subtype C data set. Overall, of the 50 mutants tested, far fewer showed high-level resistance to RPV (n = 3, 6%) and ETR (n = 8, 16%) than to EFV (n = 18, 36%) and NVP (n = 30, 60%).
FIG 5.
Phenotypic susceptibility of mutants with single and double NNRTI resistance-associated mutations. The in vitro phenotypic responses of mutants with site-directed single and double NNRTI resistance-associated mutations, ranked according to the prevalence of the mutation among the 1,433 sequences with NNRTI resistance-associated mutations used in this analysis, are summarized. Some mutations (*) were not observed among the sequences but were included, as they are implicated in ETR and RPV resistance. Light gray shading, FCs of >3.6 (ETR), >2.6 (RPV), >3.8 (EFV), and >2.8 (NVP); dark gray shading, FC of >10.
Susceptibility of clinical samples to ETR and RPV.
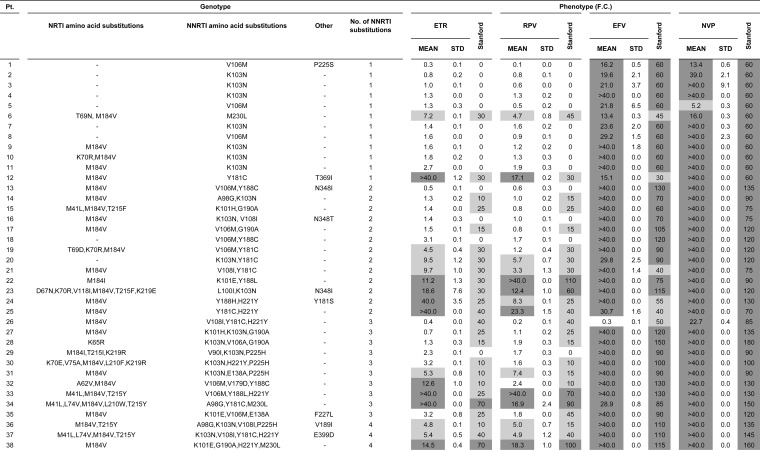
In order to assess whether samples with resistance mutations from patients who failed EFV or NVP treatment would be susceptible to ETR and RPV, we tested 38 clinical patient samples using our in-house phenotypic assay. For this, the reverse transcriptase gene was amplified from the plasma RNA of each patient and inserted into the p8.MJ4 vector. Clones with one (n = 12), two (n = 13), three (n = 10), or four (n = 3) NNRTI resistance-associated mutations were selected. This panel included sequences with the major EFV/NVP-related NNRTI resistance-associated substitutions K103N (47.3%), V106M (26.3%), Y181C (18.4%), and G190A (13.2%).
As expected, all samples showed high-level resistance to EFV and NVP, except for one sample that was susceptible to EFV, while another sample showed a reduced susceptibility to NVP (Fig. 6). In contrast, more than half the samples were fully susceptible to ETR (n = 22/38, 58%) and RPV (n = 24/38, 63%), with less than a quarter showing high-level resistance to these newer NNRTIs. Resistance to ETR and RPV was generally associated with the number of mutations. Thus, of the 12 patients with single NNRTI resistance-associated substitutions, including K103N and V106M, 10 (83%) remained susceptible to ETR and RPV. The patient with M230L as the single NNRTI resistance-associated substitution (patient 6) showed reduced susceptibility, as expected. One patient (patient 12) showed high-level resistance to all NNRTIs but had only a single Y181C amino acid substitution, which confers resistance to NVP exclusively. Overall, the presence of more than one NNRTI resistance-associated mutation was associated with a higher risk of resistance, although some samples with 3 or 4 NNRTI resistance-associated mutations remained susceptible or showed only reduced susceptibility to ETR and RPV.
FIG 6.
Genotypic and phenotypic analysis of samples from EFV/NVP-experienced patients. Patient samples with NNRTI resistance-associated NNRTI resistance-associated amino acid substitutions were screened for NNRTI susceptibility in an in vitro phenotypic assay. The Stanford HIV Drug Resistance Database genotypic resistance scores are included for reference purposes. Light gray shading, FCs of >3.6 (ETR), >2.6 (RPV), >3.8 (EFV), and >2.8 (NVP); dark gray shading, FC of >10. Pt. patient number; STD, standard deviation.
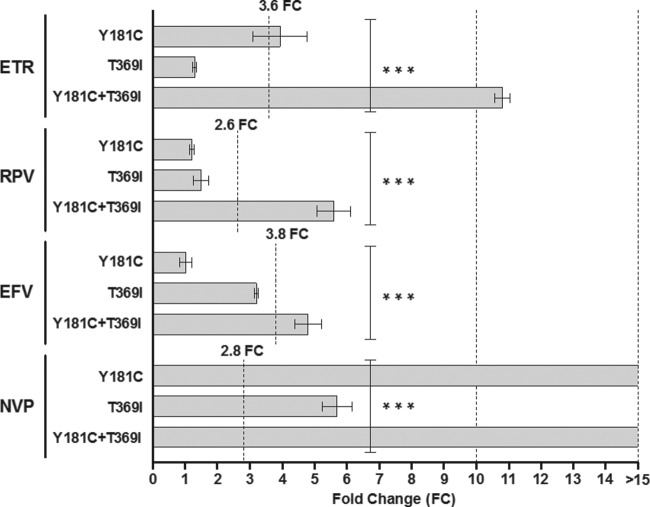
Closer inspection of the sample from patient 12 revealed the presence of the connection domain (CN) substitution T369I, in addition to Y181C. In order to determine if this mutation was responsible for the high level of resistance to all NNRTIs, two site-directed mutations (T369I and Y181C-T369I) were created in p8.MJ4. Phenotypic testing showed that T369I alone had no impact on susceptibility to ETR, RPV, or EFV but conferred decreased susceptibility to NVP (Fig. 7). However, when T369I was combined with Y181C, a significant decrease in susceptibility to ETR, RPV, and EFV was observed. Since Y181C confers high-level resistance to NVP, it was not possible to see any additional effect of T369I on susceptibility to NVP. These data suggest that for ETR, RPV, and EFV, the T369I connection domain amino acid substitution has a major impact on NNRTI susceptibility when present in combination with Y181C.
FIG 7.
Contribution of T369I to NNRTI resistance. Site-directed mutants containing the T369I amino acid substitution, an amino acid substitution found in the connection domain of patient 12 (Fig. 6), in combination with Y181C were tested for susceptibility to ETR, RPV, EFV, and NVP. The lower cutoff values are indicated for each drug. Columns indicate mean values, while error bars indicate the standard errors of the means (SEMs). Each mutant was tested in duplicate in at least two independent experiments. ***, P < 0.001 (2-way analysis of variance).
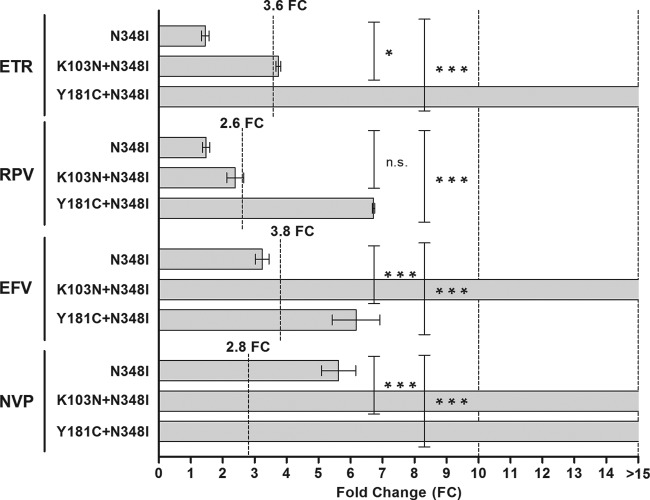
A second connection domain amino acid substitution (N348I) was observed in 2 patient samples. The N348I site-directed mutant was susceptible to all NNRTIs except NVP (FC, 5.6) (Fig. 8). However, the mutant with K103N, which singly has no effect on susceptibility to ETR (Fig. 5), showed reduced susceptibility when N348I was combined with K103N. Similarly, combinations with Y181C resulted in significant effects, particularly on susceptibility to ETR. This, once again, confirmed the negative influence of combinations with Y181C on ETR and RPV susceptibility.
FIG 8.
Contribution of N348I to NNRTI resistance. Site-directed mutants containing the N348I amino acid substitution, an amino acid substitution in the connection domain of some patients (Fig. 6), in combination with K103N or Y181C were tested for susceptibility to ETR, RPV, EFV, and NVP. The lower cutoff values are indicated for each drug. Columns indicate mean values, while error bars indicate the standard errors of the mean (SEMs). Each mutant was tested in duplicate in at least two independent experiments. n.s., not significant; *, P < 0.05 (2-way analysis of variance); ***, P < 0.001 (2-way analysis of variance).
DISCUSSION
In this study, we investigated the potential cross-resistance between NNRTIs in the context of HIV-1 subtype C. We showed that the prevalent single NNRTI resistance-associated amino acid substitutions, such as K103N, V106M, Y181C, or G190A that cause high-level resistance to EFV and/or NVP had no effect on susceptibility to ETR and RPV. Combinations of such amino acid substitutions, particularly those containing Y181C, were more likely to cause resistance to ETR and RPV, but such genotypes occurred rarely in subtype C data sets. Among samples from patients failing EFV/NVP-based therapies, more than half retained full susceptibility to ETR and/or RPV. This suggests that ETR and RPV would be suitable alternatives for subtype C patients failing EFV- or NVP-containing regimens.
Our data largely correspond with previously published data on site-directed mutations in HIV-1 subtype B (19, 21, 22, 38). Similar to others, we demonstrated the contribution of K101P, E138K/Q/R, Y181I/V, and M230I/L to ETR and/or RPV resistance (19, 22–24, 38, 39). However, some of our subtype C mutants showed a level of resistance to ETR and RPV slightly higher than that reported for subtype B mutants. These included E138Q/R and M230I/L mutations for ETR and E138K/Q/R, Y188L, and M230I/L mutations for RPV. Only E138Q/R changed the classification for ETR from susceptible in subtype B to resistant in subtype C. For RPV, E138K/Q/R, Y188L, and M230I/L also changed the classification from susceptible to reduced susceptibility. Differences in susceptibility between subtypes B and C with substitutions at position 138, including E138A, which did not confer resistance in our study, were also noted in another study (40).
In a comprehensive analysis of genotype-phenotype correlations performed predominantly on clinical subtype B sequences, amino acid substitutions K101P, Y181I/V, Y188L, and M230L have also been shown to contribute toward decreased susceptibility or resistance to ETR/RPV (41). The K101P amino acid substitution conferred a level of reduction in ETR susceptibility higher than that found in the current study. Some additional amino acid substitutions were also shown to contribute to ETR/RPV resistance in the genotype-phenotype subtype B correlation: A98G, L100I, and V179T affected both ETR and RPV susceptibility, while E138A and Y188H reduced susceptibility to RPV only. The F227C amino acid substitution conferred resistance to ETR and RPV in the genotype-phenotype correlation, while only a small reduction in susceptibility was observed in our study. However, unlike E138K/Q in the current study, no change in the susceptibility to ETR/RPV was observed in isolates with these mutations in the subtype B genotype-phenotype correlation. The effect of E138R on ETR/RPV susceptibility was not reported in that study.
There are a few possible explanations for the discrepancies observed between our data for subtype C and the published data for subtype B. First, the variation between different assay types (replicating versus nonreplicating, method of quantitation, etc.) could make some assays more sensitive at detecting phenotypic resistance than others (42). Second, polymorphic amino acid residues that inherently differ between virus isolates could lead to a range of phenotypic responses to the same drug (43). In the current study, only a single subtype C reference isolate was used, and the phenotypic responses observed could be isolate specific. Third, the variation in responses could be subtype specific, and several studies have shown differences between subtypes regarding both replication capacity and phenotypic susceptibility (44–46). The screening of additional subtype C isolates could validate the phenotypic differences that were observed between our subtype C mutants and the published subtype B mutants.
The NNRTI binding pocket is flexible, which allows similar but structurally unique NNRTIs to bind to the same compartment on HIV-1 RT (47). EFV and NVP are structurally related, and substantial cross-resistance exists between these two drugs. For this reason, the recycling of EFV/NVP after first-line treatment failure is not advised (48). Since the data sets and samples used in this study were derived from patients with NVP and EFV treatment failures, the high levels of resistance seen confirm the limited efficacy of these older NNRTIs against circulating strains. ETR and RPV are structurally distinct from EFV and NVP and are effective against most EFV- and/or NVP-resistant strains (49). This was confirmed in our study, where we showed, using site-directed mutants, that the most prevalent EFV- and NVP-related NNRTI resistance-associated substitutions caused no decrease in ETR or RPV susceptibility. Overall, the number of NNRTI resistance-associated substitutions that conferred resistance to ETR/RPV substantially was lower than the number that conferred resistance to EFV/NVP.
The Y181C amino acid substitution, which is one of the most prevalent NNRTI resistance-associated substitutions (50), had no phenotypic impact on ETR or RPV susceptibility. However, combinations of Y181C with other NNRTI resistance-associated substitutions conferred variable degrees of resistance to ETR and, to a lesser extent, RPV. This was observed in both the double mutants and clinical patient samples and has also been reported for HIV-1 subtype B (21, 22, 51, 52). This suggests that RPV may show higher levels of efficacy against viruses with Y181C and that ETR may not be a suitable alternative in patients that harbor Y181C. However, these data are based on an arbitrary in vitro cutoff, and so it is unclear whether this would hold true in a clinical setting.
The E138K amino acid change causes low-level resistance to RPV that is moderately increased in the presence of the NRTI resistance-associated substitution M184I but not M184V in subtype B (22, 53). Some studies have suggested a mutual compensatory role of these amino acid changes regarding enzyme processivity (54–56), while others have shown that they confer a decrease in viral fitness (53). While we showed that E138K also conferred low-level resistance to RPV and ETR, we did not observe a significantly enhanced increase when it was combined with M184I/V. However, we did notice a significant increase when this combination was tested against EFV and NVP. Resistance to EFV was also observed in the ECHO and THRIVE trials at week 48 in patients with E138K-M184I/V, although resistance to NVP occurred less often (22). Thus, NRTI resistance-associated substitutions, such as M184I/V, can impact susceptibility to NNRTIs, and as such, the E138K substitution needs to be taken into account before switching to RPV.
Although the resistance-associated mutations have been clearly defined for ETR and RPV, some additional mutations that fall outside the scope of traditional genotypic resistance testing have been identified. Mutations in the connection domain (CN) of HIV-1 reverse transcriptase have been shown to influence mutational pathways and contribute to resistance to both NRTIs and NNRTIs (57). The frequently selected N348I amino acid substitution, also observed in some of our patient samples, has been shown to decrease NVP susceptibility (58). When it occurs in combination with other NNRTI resistance-associated amino acid substitutions, such as K103N and Y181C, N348I has been shown to contribute to EFV, ETR, and RPV resistance (59, 60). In addition, RPV resistance is enhanced by this substitution in the background of E138K (61). We have confirmed in HIV-1 subtype C that N348I alone does not affect ETR/RPV susceptibility but can significantly reduce ETR and/or RPV susceptibility when it occurs in combination with other NNRTI resistance-associated substitutions, particularly Y181C. A less common CN amino acid change, T369I, was observed in conjunction with Y181C in one of our phenotypically resistant patient samples. Previous publications have shown that T369I can influence EFV, NVP, and ETR susceptibility in subtype B (62, 63). Reconstruction of the mutations in p8.MJ4 demonstrated that T369I significantly decreased susceptibility to all NNRTIs in subtype C, including RPV, when it occurred in combination with Y181C. However, by itself, T369I reduced susceptibility to NVP only. The Stanford HIV Drug Resistance Database reports a prevalence of 15.5% and 2.1% for N348I and T369I, respectively, among HIV-1 subtype C NNRTI treatment failures. While T369I is unlikely to be a major factor contributing to ETR or RPV resistance due to its low prevalence, N348I may play a more significant role. It may therefore be important in future to genotype regions outside those genotyped in standard resistance tests.
Mutations shown to cause resistance to ETR and/or RPV had a low prevalence (<6%) in our data set, and the prevalence was even lower (<2%) for those causing high-level resistance. According to the Stanford HIV Drug Resistance Database, the prevalences of these amino acid substitutions are similar in subtypes B and C, except for M230L, which is more prevalent in subtype C (4.7%) than subtype B (0.9%). The E138A amino acid substitution, which also occurs more frequently in subtype C (9% versus 3% in subtype B), was not shown in our assay to decrease susceptibility to any of the NNRTIs, as has been reported by others (40). All double mutations that caused resistance to ETR and/or RPV in subtype C had a prevalence of <8%, and only 5 of the 14 tested combinations were actually observed in our subtype C data sets.
In conclusion, we have shown that ETR and RPV are active against HIV-1 subtype C isolates with the prevalent EFV/NVP resistance-related mutations and would therefore be expected to be efficacious in the treatment of HIV-1 subtype C in patients that fail an EFV- or NVP-based regimen. However, the presence of Y181C, especially after failure on an NVP-based treatment, could hamper the effectiveness of ETR and, to a lesser extent, RPV. Overall, the development of newer NNRTIs with the ability to retain activity against clinically relevant mutant strains has kept this important class of drugs in the front line of antiviral therapies.
ACKNOWLEDGMENTS
This work was supported by a Research Trust Development Grant from the National Health Laboratory Service (NHLS), research grant 11/03 from the Poliomyelitis Research Foundations (PRF), and grant AI06858 from the National Institute of Allergy and Infectious Diseases (NIAID).
We thank Gillian Hunt and Jessica Radzio for helpful comments on the manuscript. Plasmids p8.9 and pMDG were supplied by Didier Trono (École Polytechnique Fédérale de Lausanne, Lausanne, Switzerland), and plasmid pCSFLW was supplied by Nigel Temperton (University College London, London, United Kingdom). Efavirenz and nevirapine were supplied by the National Institutes of Health (NIH) AIDS Research and Reference Reagent Program. Etravirine and rilpivirine were supplied by Janssen Pharmaceutical Companies.
REFERENCES
- 1.Hankins C. 2013. Overview of the current state of the epidemic. Curr HIV/AIDS Rep 10:113–123. doi: 10.1007/s11904-013-0156-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Santos AF, Soares MA. 2010. HIV genetic diversity and drug resistance. Viruses 2:503–531. doi: 10.3390/v2020503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hemelaar J, Gouws E, Ghys PD, Osmanov S. 2011. Global trends in molecular epidemiology of HIV-1 during 2000-2007. AIDS 25:679–689. doi: 10.1097/QAD.0b013e328342ff93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geretti AM, Harrison L, Green H, Sabin C, Hill T, Fearnhill E, Pillay D, Dunn D, UK Collaborative Group on HIV Drug Resistance. 2009. Effect of HIV-1 subtype on virologic and immunologic response to starting highly active antiretroviral therapy. Clin Infect Dis 48:1296–1305. doi: 10.1086/598502. [DOI] [PubMed] [Google Scholar]
- 5.Lai MT, Lu M, Felock PJ, Hrin RC, Wang YJ, Yan Y, Munshi S, McGaughey GB, Tynebor RM, Tucker TJ, Williams TM, Grobler JA, Hazuda DJ, McKenna PM, Miller MD. 2010. Distinct mutation pathways of non-subtype B HIV-1 during in vitro resistance selection with nonnucleoside reverse transcriptase inhibitors. Antimicrob Agents Chemother 54:4812–4824. doi: 10.1128/AAC.00829-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Invernizzi CF, Coutsinos D, Oliveira M, Moisi D, Brenner BG, Wainberg MA. 2009. Signature nucleotide polymorphisms at positions 64 and 65 in reverse transcriptase favor the selection of the K65R resistance mutation in HIV-1 subtype C. J Infect Dis 200:1202–1206. doi: 10.1086/605894. [DOI] [PubMed] [Google Scholar]
- 7.Brenner B, Turner D, Oliveira M, Moisi D, Detorio M, Carobene M, Marlink RG, Schapiro J, Roger M, Wainberg MA. 2003. A V106M mutation in HIV-1 clade C viruses exposed to efavirenz confers cross-resistance to non-nucleoside reverse transcriptase inhibitors. AIDS 17:F1–F5. doi: 10.1097/00002030-200301030-00001. [DOI] [PubMed] [Google Scholar]
- 8.Cozzi-Lepri A, Paredes Phillips AN, Clotet B, Kjaer J, Von Wyl V, Kronborg G, Castagna A, Bogner JR, Lundgren JD, EuroSIDA in EuroCoord. 2012. The rate of accumulation of nonnucleoside reverse transcriptase inhibitor (NNRTI) resistance in patients kept on a virologically failing regimen containing an NNRTI. HIV Med 13:62–72. doi: 10.1111/j.1468-1293.2011.00943.x. [DOI] [PubMed] [Google Scholar]
- 9.El-Khatib Z, Ekstrom AM, Ledwaba J, Mohapi L, Laher F, Karstaedt A, Charalambous S, Petzold M, Katzenstein D, Morris L. 2010. Viremia and drug resistance among HIV-1 patients on antiretroviral treatment: a cross-sectional study in Soweto, South Africa. AIDS 24:1679–1687. doi: 10.1097/QAD.0b013e32833a097b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Orrell C, Walensky RP, Losina E, Pitt J, Freedberg KA, Wood R. 2009. HIV type-1 clade C resistance genotypes in treatment-naive patients and after first virological failure in a large community antiretroviral therapy programme. Antivir Ther 14:523–531. [PMC free article] [PubMed] [Google Scholar]
- 11.van Zyl GU, van der Merwe L, Claassen M, Zeier M, Preiser W. 2011. Antiretroviral resistance patterns and factors associated with resistance in adult patients failing NNRTI-based regimens in the Western Cape, South Africa. J Med Virol 83:1764–1769. doi: 10.1002/jmv.22189. [DOI] [PubMed] [Google Scholar]
- 12.Marconi VC, Sunpath H, Lu Z, Gordon M, Koranteng-Apeagyei K, Hampton J, Carpenter S, Giddy J, Ross D, Holst H, Losina E, Walker BD, Kuritzkes DR. 2008. Prevalence of HIV-1 drug resistance after failure of a first highly active antiretroviral therapy regimen in KwaZulu Natal, South Africa. Clin Infect Dis 46:1589–1597. doi: 10.1086/587109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lazzarin A, Campbell T, Clotet B, Johnson M, Katlama C, Moll A, Towner W, Trottier B, Peeters M, Vingerhoets J, de Smedt G, Baeten B, Beets G, Sinha R, Woodfall B. 2007. Efficacy and safety of TMC125 (etravirine) in treatment-experienced HIV-1-infected patients in DUET-2: 24-week results from a randomised, double-blind, placebo-controlled trial. Lancet 370:39–48. doi: 10.1016/S0140-6736(07)61048-4. [DOI] [PubMed] [Google Scholar]
- 14.Katlama C, Haubrich R, Lalezari J, Lazzarin A, Madruga JV, Molina JM, Schechter M, Peeters M, Picchio G, Vingerhoets J, Woodfall B, De Smedt G. 2009. Efficacy and safety of etravirine in treatment-experienced, HIV-1 patients: pooled 48 week analysis of two randomized, controlled trials. AIDS 23:2289–2300. doi: 10.1097/QAD.0b013e3283316a5e. [DOI] [PubMed] [Google Scholar]
- 15.Madruga JV, Cahn P, Grinsztejn B, Haubrich R, Lalezari J, Mills A, Pialoux G, Wilkin T, Peeters M, Vingerhoets J, de Smedt G, Leopold L, Trefiglio R, Woodfall B. 2007. Efficacy and safety of TMC125 (etravirine) in treatment-experienced HIV-1-infected patients in DUET-1: 24-week results from a randomised, double-blind, placebo-controlled trial. Lancet 370:29–38. doi: 10.1016/S0140-6736(07)61047-2. [DOI] [PubMed] [Google Scholar]
- 16.Molina JM, Cahn P, Grinsztejn B, Lazzarin A, Mills A, Saag M, Supparatpinyo K, Walmsley S, Crauwels H, Rimsky LT, Vanveggel S, Boven K, ECHO Study Group. 2011. Rilpivirine versus efavirenz with tenofovir and emtricitabine in treatment-naive adults infected with HIV-1 (ECHO): a phase 3 randomised double-blind active-controlled trial. Lancet 378:238–246. doi: 10.1016/S0140-6736(11)60936-7. [DOI] [PubMed] [Google Scholar]
- 17.Cohen CJ, Andrade-Villanueva J, Clotet B, Fourie J, Johnson MA, Ruxrungtham K, Wu H, Zorrilla C, Crauwels H, Rimsky LT, Vanveggel S, Boven K, THRIVE Study Group . 2011. Rilpivirine versus efavirenz with two background nucleoside or nucleotide reverse transcriptase inhibitors in treatment-naive adults infected with HIV-1 (THRIVE): a phase 3, randomised, non-inferiority trial. Lancet 378:229–237. doi: 10.1016/S0140-6736(11)60983-5. [DOI] [PubMed] [Google Scholar]
- 18.Vingerhoets J, Azijn H, Fransen E, De Baere I, Smeulders L, Jochmans D, Andries K, Pauwels R, de Bethune MP. 2005. TMC125 displays a high genetic barrier to the development of resistance: evidence from in vitro selection experiments. J Virol 79:12773–12782. doi: 10.1128/JVI.79.20.12773-12782.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Azijn H, Tirry I, Vingerhoets J, de Bethune MP, Kraus G, Boven K, Jochmans D, Van Craenenbroeck E, Picchio G, Rimsky LT. 2010. TMC278, a next-generation nonnucleoside reverse transcriptase inhibitor (NNRTI), active against wild-type and NNRTI-resistant HIV-1. Antimicrob Agents Chemother 54:718–727. doi: 10.1128/AAC.00986-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katlama C, Clotet B, Mills A, Trottier B, Molina JM, Grinsztejn B, Towner W, Haubrich R, Nijs S, Vingerhoets J, Woodfall B, Witek J. 2010. Efficacy and safety of etravirine at week 96 in treatment-experienced HIV type-1-infected patients in the DUET-1 and DUET-2 trials. Antivir Ther 15:1045–1052. doi: 10.3851/IMP1662. [DOI] [PubMed] [Google Scholar]
- 21.Vingerhoets J, Azijn H, Tambuyzer L, Dierynck I, De Meyer S, Rimsky L, Nijs S, De Smedt G, de Bethune MP, Picchio G. 2010. Short communication: activity of etravirine on different HIV type 1 subtypes: in vitro susceptibility in treatment-naive patients and week 48 pooled DUET study data. AIDS Res Hum Retroviruses 26:621–624. doi: 10.1089/aid.2009.0239. [DOI] [PubMed] [Google Scholar]
- 22.Rimsky L, Vingerhoets J, Van Eygen V, Eron J, Clotet B, Hoogstoel A, Boven K, Picchio G. 2012. Genotypic and phenotypic characterization of HIV-1 isolates obtained from patients on rilpivirine therapy experiencing virologic failure in the phase 3 ECHO and THRIVE studies: 48-week analysis. J Acquir Immune Defic Syndr 59:39–46. doi: 10.1097/QAI.0b013e31823df4da. [DOI] [PubMed] [Google Scholar]
- 23.Tambuyzer L, Azijn H, Rimsky LT, Vingerhoets J, Lecocq P, Kraus G, Picchio G, de Bethune MP. 2009. Compilation and prevalence of mutations associated with resistance to non-nucleoside reverse transcriptase inhibitors. Antivir Ther 14:103–109. [PubMed] [Google Scholar]
- 24.Vingerhoets J, Tambuyzer L, Azijn H, Hoogstoel A, Nijs S, Peeters M, de Bethune MP, De Smedt G, Woodfall B, Picchio G. 2010. Resistance profile of etravirine: combined analysis of baseline genotypic and phenotypic data from the randomized, controlled phase III clinical studies. AIDS 24:503–514. doi: 10.1097/QAD.0b013e32833677ac. [DOI] [PubMed] [Google Scholar]
- 25.Reuman EC, Rhee SY, Holmes SP, Shafer RW. 2010. Constrained patterns of covariation and clustering of HIV-1 non-nucleoside reverse transcriptase inhibitor resistance mutations. J Antimicrob Chemother 65:1477–1485. doi: 10.1093/jac/dkq140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoffmann CJ, Charalambous S, Sim J, Ledwaba J, Schwikkard G, Chaisson RE, Fielding KL, Churchyard GJ, Morris L, Grant AD. 2009. Viremia, resuppression, and time to resistance in human immunodeficiency virus (HIV) subtype C during first-line antiretroviral therapy in South Africa. Clin Infect Dis 49:1928–1935. doi: 10.1086/648444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pillay V, Pillay C, Kantor R, Venter F, Levin L, Morris L. 2008. HIV type 1 subtype C drug resistance among pediatric and adult South African patients failing antiretroviral therapy. AIDS Res Hum Retroviruses 24:1449–1454. doi: 10.1089/aid.2008.0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ikeda Y, Ylinen LM, Kahar-Bador M, Towers GJ. 2004. Influence of gag on human immunodeficiency virus type 1 species-specific tropism. J Virol 78:11816–11822. doi: 10.1128/JVI.78.21.11816-11822.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage FH, Verma IM, Trono D. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 30.Parry CM, Kohli A, Boinett CJ, Towers GJ, McCormick AL, Pillay D. 2009. Gag determinants of fitness and drug susceptibility in protease inhibitor-resistant human immunodeficiency virus type 1. J Virol 83:9094–9101. doi: 10.1128/JVI.02356-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gupta RK, Kohli A, McCormick AL, Towers GJ, Pillay D, Parry CM. 2010. Full-length HIV-1 Gag determines protease inhibitor susceptibility within in vitro assays. AIDS 24:1651–1655. doi: 10.1097/QAD.0b013e3283398216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mbisa JL, Gupta RK, Kabamba D, Mulenga V, Kalumbi M, Chintu C, Parry CM, Gibb DM, Walker SA, Cane PA, Pillay D. 2011. The evolution of HIV-1 reverse transcriptase in route to acquisition of Q151M multi-drug resistance is complex and involves mutations in multiple domains. Retrovirology 8:31. doi: 10.1186/1742-4690-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ndung'u T, Renjifo B, Essex M. 2001. Construction and analysis of an infectious human immunodeficiency virus type 1 subtype C molecular clone. J Virol 75:4964–4972. doi: 10.1128/JVI.75.11.4964-4972.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu TF, Shafer RW. 2006. Web resources for HIV type 1 genotypic-resistance test interpretation. Clin Infect Dis 42:1608–1618. doi: 10.1086/503914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wright E, Temperton NJ, Marston DA, McElhinney LM, Fooks AR, Weiss RA. 2008. Investigating antibody neutralization of lyssaviruses using lentiviral pseudotypes: a cross-species comparison. J Gen Virol 89:2204–2213. doi: 10.1099/vir.0.2008/000349-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson VA, Calvez V, Günthard HF, Paredes R, Pillay D, Shafer RW, Wensing AM, Richman DD. 2013. Update of the drug resistance mutations in HIV-1: March 2013. Top Antiviral Med 2:4–12. [PMC free article] [PubMed] [Google Scholar]
- 37.Rimsky L, Van Eygen V, Hoogstoel A, Stevens M, Boven K, Picchio G, Vingerhoets J. 2013. 96-week resistance analyses of rilpivirine in treatment-naive, HIV-1-infected adults from the ECHO and THRIVE phase III trials. Antivir Ther 18:967–977. doi: 10.3851/IMP2636. [DOI] [PubMed] [Google Scholar]
- 38.Tambuyzer L, Vingerhoets J, Azijn H, Daems B, Nijs S, de Bethune MP, Picchio G. 2010. Characterization of genotypic and phenotypic changes in HIV-1-infected patients with virologic failure on an etravirine-containing regimen in the DUET-1 and DUET-2 clinical studies. AIDS Res Hum Retroviruses 26:1197–1205. doi: 10.1089/aid.2009.0302. [DOI] [PubMed] [Google Scholar]
- 39.Xu HT, Colby-Germinario SP, Asahchop EL, Oliveira M, McCallum M, Schader SM, Han Y, Quan Y, Sarafianos SG, Wainberg MA. 2013. Effect of mutations at position E138 in HIV-1 reverse transcriptase and their interactions with the M184I mutation on defining patterns of resistance to nonnucleoside reverse transcriptase inhibitors rilpivirine and etravirine. Antimicrob Agents Chemother 57:3100–3109. doi: 10.1128/AAC.00348-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sluis-Cremer N, Jordan MR, Huber K, Wallis CL, Bertagnolio S, Mellors JW, Parkin NT, Harrigan PR. 2014. E138A in HIV-1 reverse transcriptase is more common in subtype C than B: implications for rilpivirine use in resource-limited settings. Antiviral Res 107:31–34. doi: 10.1016/j.antiviral.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Melikian GL, Rhee SY, Varghese V, Porter D, White K, Taylor J, Towner W, Troia P, Burack J, Dejesus E, Robbins GK, Razzeca K, Kagan R, Liu TF, Fessel WJ, Israelski D, Shafer RW. 2014. Non-nucleoside reverse transcriptase inhibitor (NNRTI) cross-resistance: implications for preclinical evaluation of novel NNRTIs and clinical genotypic resistance testing. J Antimicrob Chemother 69:12–20. doi: 10.1093/jac/dkt316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang J, Rhee SY, Taylor J, Shafer RW. 2005. Comparison of the precision and sensitivity of the Antivirogram and PhenoSense HIV drug susceptibility assays. J Acquir Immune Defic Syndr 38:439–444. doi: 10.1097/01.qai.0000147526.64863.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parkin NT, Hellmann NS, Whitcomb JM, Kiss L, Chappey C, Petropoulos CJ. 2004. Natural variation of drug susceptibility in wild-type human immunodeficiency virus type 1. Antimicrob Agents Chemother 48:437–443. doi: 10.1128/AAC.48.2.437-443.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Armstrong KL, Lee TH, Essex M. 2011. Replicative fitness costs of nonnucleoside reverse transcriptase inhibitor drug resistance mutations on HIV subtype C. Antimicrob Agents Chemother 55:2146–2153. doi: 10.1128/AAC.01505-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Buckheit RW., Jr 2004. Understanding HIV resistance, fitness, replication capacity and compensation: targeting viral fitness as a therapeutic strategy. Expert Opin Investig Drugs 13:933–958. doi: 10.1517/13543784.13.8.933. [DOI] [PubMed] [Google Scholar]
- 46.Wainberg MA, Brenner BG. 2012. The impact of HIV genetic polymorphisms and subtype differences on the occurrence of resistance to antiretroviral drugs. Mol Biol Int 2012:256982. doi: 10.1155/2012/256982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Das K, Clark AD Jr, Lewi PJ, Heeres J, De Jonge MR, Koymans LM, Vinkers HM, Daeyaert F, Ludovici DW, Kukla MJ, De Corte B, Kavash RW, Ho CY, Ye H, Lichtenstein MA, Andries K, Pauwels R, De Bethune MP, Boyer PL, Clark P, Hughes SH, Janssen PA, Arnold E. 2004. Roles of conformational and positional adaptability in structure-based design of TMC125-R165335 (etravirine) and related non-nucleoside reverse transcriptase inhibitors that are highly potent and effective against wild-type and drug-resistant HIV-1 variants. J Med Chem 47:2550–2560. doi: 10.1021/jm030558s. [DOI] [PubMed] [Google Scholar]
- 48.Antinori A, Zaccarelli M, Cingolani A, Forbici F, Rizzo MG, Trotta MP, Di Giambenedetto S, Narciso P, Ammassari A, Girardi E, De Luca A, Perno CF. 2002. Cross-resistance among nonnucleoside reverse transcriptase inhibitors limits recycling efavirenz after nevirapine failure. AIDS Res Hum Retroviruses 18:835–838. doi: 10.1089/08892220260190308. [DOI] [PubMed] [Google Scholar]
- 49.Das K, Lewi PJ, Hughes SH, Arnold E. 2005. Crystallography and the design of anti-AIDS drugs: conformational flexibility and positional adaptability are important in the design of non-nucleoside HIV-1 reverse transcriptase inhibitors. Prog Biophys Mol Biol 88:209–231. doi: 10.1016/j.pbiomolbio.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 50.Wallis CL, Mellors JW, Venter WD, Sanne I, Stevens W. 2010. Varied patterns of HIV-1 drug resistance on failing first-line antiretroviral therapy in South Africa. J Acquir Immune Defic Syndr 53:480–484. doi: 10.1097/QAI.0b013e3181bc478b. [DOI] [PubMed] [Google Scholar]
- 51.Ruxrungtham K, Pedro RJ, Latiff GH, Conradie F, Domingo P, Lupo S, Pumpradit W, Vingerhoets JH, Peeters M, Peeters I, Kakuda TN, De Smedt G, Woodfall B. 2008. Impact of reverse transcriptase resistance on the efficacy of TMC125 (etravirine) with two nucleoside reverse transcriptase inhibitors in protease inhibitor-naive, nonnucleoside reverse transcriptase inhibitor-experienced patients: study TMC125-C227. HIV Med 9:883–896. doi: 10.1111/j.1468-1293.2008.00644.x. [DOI] [PubMed] [Google Scholar]
- 52.Xu H, Quan Y, Brenner BG, Bar-Magen T, Oliveira M, Schader SM, Wainberg MA. 2009. Human immunodeficiency virus type 1 recombinant reverse transcriptase enzymes containing the G190A and Y181C resistance mutations remain sensitive to etravirine. Antimicrob Agents Chemother 53:4667–4672. doi: 10.1128/AAC.00800-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kulkarni R, Babaoglu K, Lansdon EB, Rimsky L, Van Eygen V, Picchio G, Svarovskaia E, Miller MD, White KL. 2012. The HIV-1 reverse transcriptase M184I mutation enhances the E138K-associated resistance to rilpivirine and decreases viral fitness. J Acquir Immune Defic Syndr 59:47–54. doi: 10.1097/QAI.0b013e31823aca74. [DOI] [PubMed] [Google Scholar]
- 54.Hu Z, Kuritzkes DR. 2011. Interaction of reverse transcriptase (RT) mutations conferring resistance to lamivudine and etravirine: effects on fitness and RT activity of human immunodeficiency virus type 1. J Virol 85:11309–11314. doi: 10.1128/JVI.05578-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu HT, Asahchop EL, Oliveira M, Quashie PK, Quan Y, Brenner BG, Wainberg MA. 2011. Compensation by the E138K mutation in HIV-1 reverse transcriptase for deficits in viral replication capacity and enzyme processivity associated with the M184I/V mutations. J Virol 85:11300–11308. doi: 10.1128/JVI.05584-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu HT, Oliveira M, Quashie PK, McCallum M, Han Y, Quan Y, Brenner BG, Wainberg MA. 2012. Subunit-selective mutational analysis and tissue culture evaluations of the interactions of the E138K and M184I mutations in HIV-1 reverse transcriptase. J Virol 86:8422–8431. doi: 10.1128/JVI.00271-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Delviks-Frankenberry KA, Lengruber RB, Santos AF, Silveira JM, Soares MA, Kearney MF, Maldarelli F, Pathak VK. 2013. Connection subdomain mutations in HIV-1 subtype-C treatment-experienced patients enhance NRTI and NNRTI drug resistance. Virology 435:433–441. doi: 10.1016/j.virol.2012.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yap SH, Sheen CW, Fahey J, Zanin M, Tyssen D, Lima VD, Wynhoven B, Kuiper M, Sluis-Cremer N, Harrigan PR, Tachedjian G. 2007. N348I in the connection domain of HIV-1 reverse transcriptase confers zidovudine and nevirapine resistance. PLoS Med 4:e335. doi: 10.1371/journal.pmed.0040335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Menendez-Arias L, Betancor G, Matamoros T. 2011. HIV-1 reverse transcriptase connection subdomain mutations involved in resistance to approved non-nucleoside inhibitors. Antiviral Res 92:139–149. doi: 10.1016/j.antiviral.2011.08.020. [DOI] [PubMed] [Google Scholar]
- 60.Brehm JH, Koontz DL, Wallis CL, Shutt KA, Sanne I, Wood R, McIntyre JA, Stevens WS, Sluis-Cremer N, Mellors JW, CIPRA-SA Project 1 Study Team. 2012. Frequent emergence of N348I in HIV-1 subtype C reverse transcriptase with failure of initial therapy reduces susceptibility to reverse-transcriptase inhibitors. Clin Infect Dis 55:737–745. doi: 10.1093/cid/cis501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu HT, Colby-Germinario SP, Oliveira M, Han Y, Quan Y, Zanichelli V, Wainberg MA. 2014. The connection domain mutation N348I in HIV-1 reverse transcriptase enhances resistance to etravirine and rilpivirine but restricts the emergence of the E138K resistance mutation by diminishing viral replication capacity. J Virol 88:1536–1547. doi: 10.1128/JVI.02904-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gupta S, Fransen S, Paxinos EE, Stawiski E, Huang W, Petropoulos CJ. 2010. Combinations of mutations in the connection domain of human immunodeficiency virus type 1 reverse transcriptase: assessing the impact on nucleoside and nonnucleoside reverse transcriptase inhibitor resistance. Antimicrob Agents Chemother 54:1973–1980. doi: 10.1128/AAC.00870-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gupta S, Vingerhoets J, Fransen S, Tambuyzer L, Azijn H, Frantzell A, Paredes R, Coakley E, Nijs S, Clotet B, Petropoulos CJ, Schapiro J, Huang W, Picchio G. 2011. Connection domain mutations in HIV-1 reverse transcriptase do not impact etravirine susceptibility and virologic responses to etravirine-containing regimens. Antimicrob Agents Chemother 55:2872–2879. doi: 10.1128/AAC.01695-10. [DOI] [PMC free article] [PubMed] [Google Scholar]