LETTER
The incidence of infections by Enterobacteriaceae carrying the carbapenemase gene showing a paradoxical phenotype of resistance to virtually all ß-lactams, including meropenem but not including imipenem, is increasing (1, 2, 3). Here we report the first isolation of Klebsiella pneumoniae carrying blaOXA-181 in Japan showing a similar stealth-type resistance phenotype.
In April 2010, a man was admitted to a hospital in Mumbai, India, with unidentified multiple organ failures. In June 2010, he was transferred to an intensive care unit (ICU) in Hiroshima, Japan. K. pneumoniae MS5166 was isolated from the urine sample of the patient. This isolate was resistant to almost all ß-lactams, including meropenem, but was susceptible to imipenem following CLSI criteria (4): the MIC was 2 μg/ml for imipenem and 8 μg/ml for meropenem using the broth microdilution method. Further, MS5166 was resistant to aminoglycosides and fluoroquinolones. The MIC values (μg/ml) were as follows: for amikacin, >32; for gentamicin, >8; for tobramycin, >8; for ciprofloxacin, >2; and for levofloxacin, >4. The result of the metallo-β-lactamase phenotype test (5) was negative. PCR for carbapenemase genes performed using 11 universal primer sets (6) gave a positive result for the gene encoding OXA-48-like protein. Direct sequencing of the amplicon indicated it is 100% identical to blaOXA-181, differing from blaOXA-48 by four amino acid substitutions.
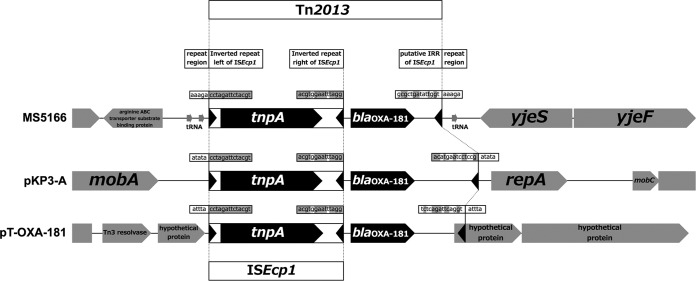
The draft genome sequence of strain MS5166 generated using Illumina MiSeq (Nextera paired-end library; 3,971,486 bp; 62.33-fold coverage), assembled using CLC Genomics Workbench (CLC bio, Cambridge, MA), and generated using OSLay (7) indicated that blaOXA-181 is on the chromosome (Fig. 1).
FIG 1.
Structural comparison of strains MS5166, pKP3-A, and pT-OXA-181 in the ISEcp1-blaOXA-181 region (GenBank accession no. AB972272, JN205800, and JQ996150, respectively). Open reading frames (ORFs) are represented by pentagons. Nucleotide letters in squares represent direct or inverted repeats (shaded base pairs are identical, and white base pairs are different), and target site duplications (repeat regions) are represented in squares. Tn2013 in MS5166 was found to be located between bp 438346 and bp 438347 on the chromosome of the K. pneumoniae NTUH-K2044 genome (GenBank accession no. NC012731).
ISEcp1 is located upstream of blaOXA-181 as reported previously (8, 9). blaOXA-181 is located on a deleted version of Tn2013 (2,961 bp), flanked by a 5-bp duplication of the target site (AAAGA) in the MS5166 chromosome between the arginine ABC transporter substrate-binding protein gene and the yjeS gene (Fig. 1). A putative inverted repeat right (IRR) of ISEcp1 revealed weak identity with its original IRR sequence (8). Potron et al. previously showed that Tn2013 integrated into the Escherichia coli chromosome upon conjugation in vitro (8). This is the first clinical case of chromosomally integrated Tn2013 shown to be ISEcp1 associated with the blaOXA-181 gene in K. pneumoniae.
A ResFinder search (10) of the draft genome sequence of MS5166 identified blaCTX-M-15, blaTEM-1A, blaSHV-11, armA, aadA2, aac(6′)-Ib, aadA1, and aac(6′)Ib-cr genes in addition to the blaOXA-181 gene. Multilocus sequence typing showed that MS5166 belongs to ST43, a “minor” cluster producing OXA-181 and CTX-M-15 in India (11).
The catalytic activities (kcat/Km) of OXA-181 for imipenem were reportedly 20 times higher than those for meropenem (12). We therefore sought a mechanism other than OXA-181 to explain the imipenem-susceptible, meropenem-resistant phenotype of MS5166. We identified a 14-bp deletion in ompK35 leaving a 22-amino-acid sequence with a truncation of the large C-terminal region and a frameshift mutation in ompK36 causing addition of two amino acids in loop 3 generating the variant OmpK36V (13). A clinical K. pneumoniae isolate producing extended-spectrum β-lactamase (ESBL) and OmpK36V but lacking OmpK35 showed the imipenem-susceptible, meropenem-resistant phenotype (13). Therefore, the absence of OmpK35 and the frameshift mutation in OmpK36 may be the mechanisms for this peculiar carbapenem resistance phenotype in ESBL- and OXA-181-producing K. pneumoniae MS5166.
Nucleotide sequence accession number.
The partial sequence of MS5166 containing blaOXA-181 was deposited in GenBank (accession no. AB972272).
ACKNOWLEDGMENT
We thank Jim Nelson for editorial assistance.
REFERENCES
- 1.Shigemoto N, Kuwahara R, Kayama S, Shimizu W, Onodera M, Yokozaki M, Hisatsune J, Kato F, Ohge H, Sugai M. 2012. Emergence in Japan of an imipenem-susceptible, meropenem-resistant Klebsiella pneumoniae carrying blaIMP-6. Diagn Microbiol Infect Dis 72:109–112. doi: 10.1016/j.diagmicrobio.2011.09.019. [DOI] [PubMed] [Google Scholar]
- 2.Shigemoto N, Kayama S, Kuwahara R, Hisatsune J, Kato F, Nishio H, Yamasaki K, Wada Y, Sueda T, Ohge H, Sugai M. 2013. A novel metallo-β-lactamase, IMP-34, in Klebsiella isolates with decreased resistance to imipenem. Diagn Microbiol Infect Dis 76:119–121. doi: 10.1016/j.diagmicrobio.2013.02.030. [DOI] [PubMed] [Google Scholar]
- 3.Castanheira M, Deshpande LM, Mathai D, Bell JM, Jones RN, Mendes RE. 2011. Early dissemination of NDM-1- and OXA-181-producing Enterobacteriaceae in Indian hospitals: report from the SENTRY Antimicrobial Surveillance Program, 2006–2007. Antimicrob Agents Chemother 55:1274–1278. doi: 10.1128/AAC.01497-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute (CLSI). 2013. Performance standards for antimicrobial susceptibility testing; 23th informational supplement (M100-S23). CLSI, Wayne, PA. [Google Scholar]
- 5.Kouda S, Ohara M, Onodera M, Fujiue Y, Sasaki M, Kohara T, Kashiyama S, Hayashida S, Harino T, Tsuji T, Itaha H, Gotoh N, Matsubara A, Usui T, Sugai M. 2009. Increased prevalence and clonal dissemination of multidrug-resistant Pseudomonas aeruginosa with the blaIMP-1 gene cassette in Hiroshima. J Antimicrob Chemother 64:46–51. doi: 10.1093/jac/dkp142. [DOI] [PubMed] [Google Scholar]
- 6.Poirel L, Walsh TR, Cuvillier V, Nordmann P. 2011. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis 70:119–123. doi: 10.1016/j.diagmicrobio.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 7.Richter DC, Schuster SC, Huson DH. 2007. OSLay: optimal syntenic layout of unfinished assemblies. Bioinformatics 23:1573–1579. doi: 10.1093/bioinformatics/btm153. [DOI] [PubMed] [Google Scholar]
- 8.Potron A, Nordmann P, Lafeuille E, Al Maskari Z, Al Rashdi F, Poirel L. 2011. Characterization of OXA-181, a carbapenem-hydrolyzing class D β-lactamase from Klebsiella pneumoniae. Antimicrob Agents Chemother 55:4896–4899. doi: 10.1128/AAC.00481-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Villa L, Carattoli A, Nordmann P, Carta C, Poirel L. 2013. Complete sequence of the IncT-type plasmid pT-OXA-181 carrying the blaOXA-181 carbapenemase gene from Citrobacter freundii. Antimicrob Agents Chemother 57:1965–1967. doi: 10.1128/AAC.01297-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lascols C, Peirano G, Hackel M, Laupland KB, Pitout JDD. 2013. Surveillance and molecular epidemiology of Klebsiella pneumoniae isolates that produce carbapenemases: first report of OXA-48-like enzymes in North America. Antimicrob Agents Chemother 57:130–136. doi: 10.1128/AAC.01686-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Potron A, Rondinaud E, Poirel L, Belmonte O, Boyer S, Camiade S, Nordmann P. 2013. Genetic and biochemical characterization of OXA-232, a carbapenem-hydrolysing class D β-lactamase from Enterobacteriaceae. Int J Antimicrob Agents 41:325–329. doi: 10.1016/j.ijantimicag.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 13.García-Fernández A, Miriagou V, Papagiannitsis CC, Giordano A, Venditti M, Mancini C, Carattoli A. 2010. An ertapenem-resistant extended-spectrum-β-lactamase-producing Klebsiella pneumoniae clone carries a novel OmpK36 porin variant. Antimicrob Agents Chemother 54:4178–4184. doi: 10.1128/AAC.01301-09. [DOI] [PMC free article] [PubMed] [Google Scholar]