Abstract
Respiratory syncytial virus (RSV) is the leading cause of acute lower respiratory tract infections in young children and other high-risk populations. RSV nucleoprotein (N) is essential for virus assembly and replication as part of the viral ribonucleoprotein (RNP) complex. RSV604 was a putative N inhibitor in phase 2 clinical trials whose molecular mechanism of action (MoA) was not well understood. This study investigated the cell line-dependent potency of RSV604 and demonstrated its direct binding to the N protein in vitro, providing the first evidence of direct target engagement for this class of inhibitors reported to date. The affinity of RSV604 N binding was not affected by RSV604 resistance mutations in the N protein. RSV604 engaged in two different MoAs in HeLa cells, inhibiting both RSV RNA synthesis and the infectivity of released virus. The lack of inhibition of viral RNA synthesis in some cell lines explained the cell-type-dependent potency of the inhibitor. RSV604 did not inhibit viral RNA synthesis in the RSV subgenomic replicon cells or in the cell-free RNP assay, suggesting that it might act prior to viral replication complex formation. RSV604 did not alter N protein localization in the infected cells. Taken together, these results provide new insights leading to an understanding of the MoAs of RSV604 and other similar N inhibitors.
INTRODUCTION
Respiratory syncytial virus (RSV) is an enveloped, nonsegmented, negative-sense RNA virus in the family Paramyxoviridae. It is the leading cause of acute lower respiratory tract infections (ALRI) in young children and other high-risk populations worldwide (1). RSV infection has accounted for an estimated 34 million cases of ALRI and over 66,000 deaths among children younger than 5 years in developing countries each year (2). The association of RSV infection with morbidity in elderly patient populations with underlying chronic illnesses (e.g., chronic obstructive pulmonary disease, asthma, and immunocompromised patients) has also been increasingly recognized in recent years (3, 4). Despite decades of research and drug development endeavors, no effective RSV treatment or vaccine is available (1). Ribavirin, a panantiviral and the only approved small-molecule therapy against RSV, has an unclear molecular mechanism of action (MoA) and restricted clinical utility due to its toxicity, limited efficacy, and complexity of use (5). Immunoprophylaxis with RSV-neutralizing antibodies is effective only as a preventive measure, but not as a therapeutic treatment (5).
In the past 2 decades, RSV inhibitors in clinical development for RSV therapeutic treatment have primarily targeted viral fusion to prevent infection of new cells. Other early clinical candidates included the small interfering RNA (ALN-RSV01) (6, 7), nucleoside inhibitor (ALS-8176) (8), and nucleoprotein (N) inhibitor (RSV604) (9). These inhibitors have yet to show reported therapeutic efficacy in the clinic against natural RSV infections (10). In two recent phase 2a RSV challenge studies with adult volunteers, the fusion inhibitor GS-5806 (Gilead) and the nucleotide inhibitor ALS-8176 (Alios) demonstrated a reduction in disease symptoms and viral load when dosed at a relatively low viral load and before the onset of symptoms (11). In a phase 2a trial in stem cell transplant patients with RSV infection, RSV604 reduced the viral load and symptoms in the subset of patients with RSV604 plasma exposure reaching 1× the 90% effective concentration (EC90), though not in those with less plasma exposure (12).
Of the 11 RSV-encoded proteins, N is one of the most conserved structural proteins and is essential for virus encapsidation by coating the entire viral RNA genome to form the ribonucleoprotein (RNP). Although poorly understood, this multifunctional protein was also found to play an important role in viral RNA replication, mRNA transcription, and virus assembly (3). RSV604, discovered through chemical optimization of an RSV high-throughput screen hit, was reported to be an N inhibitor based on the identification of resistance mutations in the N gene in the escape viruses (9, 13). However, the MoA and mechanism of resistance for the compound were unclear. The MoA of another class of negative-strand RNA virus nucleoprotein inhibitors, aryl piperazine amides, such as nucleozin, is through inducing nucleoprotein aggregation and altering its localization in influenza virus-infected cells (14–16). The function of nucleoprotein is well conserved among negative-strand RNA viruses.
In this study, we dissected the steps in the RSV replication cycle that are impacted by RSV604, demonstrated direct target-inhibitor binding, investigated its cell-type-dependent activity, and analyzed the effects of resistance mutations on target engagement in vitro. These results provide new insights into the RSV N inhibitor MoA, which may facilitate future drug discovery and application of RSV604 as a tool to investigate the role of the N protein in RSV replication.
MATERIALS AND METHODS
Cells and virus.
HeLa, HEp-2, and BHK-21 cells were cultured according to ATCC instructions. HeLa cell-derived RSV subgenomic replicon cells (Apath) were cultured as previously described (17, 18). RSV A2 (ATCC) was propagated, and viral titers were determined by a 50% tissue culture infective dose (TCID50) assay in HEp-2 cells, as previously reported (19).
Compounds.
RSV604 and the RSV L inhibitor AZ-27 were synthesized in house as previously described (9, 13, 18), solubilized in dimethyl sulfoxide (DMSO) to a 10 mM concentration, and serially diluted to the desired compound concentrations, with a final DMSO concentration of 0.1% (vol/vol) in assay medium for testing. AZ-27, which inhibits viral RNA synthesis, served as a control in this study.
Compound uptake analysis.
The HeLa and BHK-21 cells were seeded at 106 cells/well in 6-well plates and incubated with DMSO, RSV604, or AZ-27 at 37°C. The cells were then washed 3 times with Dulbecco's PBS (DPBS) at 2, 4, and 24 h posttreatment and lysed with lysis buffer (150 mM NaCl, 1.0% NP-40, 50 mM Tris-Cl [pH 8.0]) for 10 min at 4°C. Compound in the cell lysates was analyzed using an Agilent 6490 Triple Quadrupole Mass Spectrometer (MS) equipped with an Agilent 1290 liquid chromatograph (LC) (Agilent Technologies) coupled with the Waters Acquity HSS T3 column (2.1 by 50 mm; 1.8 μm). Mobile phases of water (0.1% formic acid) and acetonitrile (0.1% formic acid) were applied in the gradient elution mode. The following multiple-reaction-monitoring transitions were monitored for the analytes of interest in positive-ion mode: RSV604 (+) 389.0→235.0, AZ-27 (+) 634.3→201.1. Calibration curves were acquired by plotting the observed compound concentrations against the known concentrations for each condition. The R2 value for each standard curve was over 0.99.
RSV ELISA.
The RSV enzyme-linked immunosorbent assay (ELISA) was performed as previously described to measure compound potency (18). Briefly, HeLa, BHK-21, or HEp-2 cells seeded in 96-well plates were infected with RSV A2 at a multiplicity of infection (MOI) of 0.1 in the presence of compound for 3 days, followed by quantitation of expressed RSV F protein by ELISA to determine the compound's EC50 and EC90.
Cytotoxicity assay.
The effects of compounds on cell viability were measured by the standard format previously described (18). Briefly, cells were seeded and incubated with compound under the RSV ELISA conditions in the absence of virus infection, followed by CellTiter-Glo substrate (Promega) addition and luminescence detection to determine the compound's 50% cytotoxic concentration (CC50).
RSV replicon assay.
The RSV replicon assay was performed as previously described (20). Briefly, RSV replicon cells were cultured in the presence of compound or DMSO (control) for 2 days, followed by addition of EnduRen substrate (Promega) to measure the replicon luciferase reporter signal.
RSV qRT-PCR assay.
HeLa, BHK-21, or RSV replicon cells were seeded at 5 × 104 cells/well in 24-well plates overnight, followed by incubation with medium containing DMSO (control), RSV604, or AZ-27 at 1× the EC90 at 37°C. EC90 values calculated from RSV ELISA and replicon assays of the same cell lines were used in this and subsequent immunofluorescence studies, except for RSV604, where only the EC90 from HeLa cell ELISA was measurable, and therefore, its value was used for all cell lines (RSV604, 14 μM [HeLa]; AZ-27, 0.1 μM [HeLa], 3.5 μM [BHK-21], and 4 μM [RSV replicon]). Two hours after the compound incubation, HeLa and BHK-21 cells were infected with RSV A2 at an MOI of 0.1 for 2 h, followed by 3 DPBS washes and incubation with compound-containing medium. At the indicated time points postinfection/compound incubation, total RNAs were isolated from the cells, followed by quantitative reverse transcription-PCR (qRT-PCR) quantitation according to a previously described protocol (21). Briefly, the first-strand cDNA synthesis and quantitative PCR (qPCR) used primers recognizing the N region of the negative-strand RSV RNA genome, with negative-sense viral RNA transcripts serving as standards and processed along with the samples. Intracellular RSV RNA copy numbers were expressed as the normalized RNA copy number per nanogram of total RNA. The amount of released virus in the culture supernatants was measured by the same qRT-PCR protocol described above and normalized as the released viral RNA copy number per milliliter.
RNP assay.
The cell extracts enriched in the RSV RNP complex were prepared as previously described (22). Briefly, HEp-2 cells were infected with RSV A2 at an MOI of 10 for 2 h, followed by removal of the virus-containing medium and culture in DMEM supplemented with 2% fetal bovine serum for 21 h. The cells were then incubated for 1 h in the presence of d-actinomycin added to the medium (2 μg/ml) to inhibit host polymerases activities prior to preparation of the viral RNP-containing cell extracts as previously described (22). Viral RNA synthesis in the RNP complex was measured in a 50-μl reaction mixture containing 5 μl cell extract in the reaction buffer (5 μCi [32P]CTP; 0.5 mM [each] ATP, GTP, and UTP; 5 μM CTP; 4 mM MgCl2; 3 mM dithiothreitol [DTT]; 0.5 mM EGTA; 120 mM potassium acetate [KAc]; 10 mM Tris-HCl [pH 7.5]; 5% glycerol; 50 μg/ml bovine serum albumin [BSA]) in the presence of 2% DMSO, 0.02 μg/μl anti-N MAb (RSV1C3; Abcam), or 10 μM RSV604. After 2 h of incubation at 30°C, the reactions were stopped by adding 250 μl ice-cold stopping buffer (10 mM Tris, pH 7.5, 500 mM NaNH3, 2 mM EDTA, 0.1 μg/ml tRNA), followed by viral RNA extraction with 300 μl of phenol-chloroform-isoamyl alcohol (50:49:1), precipitation with an equal volume of isopropanol, and centrifugation for 30 min at 15,000 rpm. The pellets were washed once with ice-cold 70% ethanol, dissolved in 1× formaldehyde loading buffer (AM8546G; Life Technologies), heated for 5 min at 70°C, and run on a 6% phosphonoacetic acid (PAA)-urea gel, followed by overnight exposure to Amersham Hyperfilm MP.
Immunofluorescence.
Subconfluent (80%) HeLa or BHK-21 cell monolayers grown on glass coverslips were treated with DMSO (control), RSV604 (1× EC90), or AZ-27 (1× EC90) for 24 h. The cells were washed with ice-cold PBS and fixed with 4% paraformaldehyde for at least 10 min at room temperature, followed by the permeabilization of cell membranes with 0.2% Triton X-100 for 5 min. The fixed cells were washed thoroughly with PBS and then incubated for 30 min in blocking buffer (Blockaid; Life Technologies), followed by immunostaining with anti-F (MAB8262X; Millipore), N (MAB858-3B-5; Millipore), or M2-1 (ab94805; Abcam) antibody. Bound antibodies were detected with species-specific fluorochrome-conjugated secondary antibodies (AS-21381 and A-21240; Life Technologies). All the antibodies were diluted 1:100 in 1% BSA in PBS. Coverslips were mounted in fluorescent mounting medium with DAPI (4′,6-diamidino-2-phenylindole) (Life Technologies) and analyzed by confocal laser scanning microscopy, as described previously (23, 24). Images of more than 20 cells were analyzed for each sample.
RSV N protein production.
The construction of an RSV His6-tagged N(13-391) protein expression plasmid and expression and purification of the N protein were described previously (25). Briefly, a pET-28b-based plasmid (pJT1128) encoding N-terminal His6-tagged RSV N protein (amino acids 13 to 391; with a 12-amino-acid truncation at the N terminus) and a pET-30a-based plasmid encoding glutathione S-transferase (GST)-tagged RSV full-length P protein (amino acids 1 to 241), both codon optimized for expression in Escherichia coli, were constructed and cotransformed into E. coli to facilitate N protein overproduction. E. coli cells carrying the plasmids were induced by IPTG (isopropyl-β-d-thiogalactopyranoside) and harvested for N protein purification using HiTrap Ni2+ column (GE Healthcare Life Sciences) affinity chromatography, followed by size exclusion chromatography. The concentrated N protein was eluted with elution buffer (50 mM Tris-HCl [pH 8.0], 0.5 M NaCl, 1 mM EDTA, 1 mM DTT, and 5% glycerol) and stored at −80°C. Similar RSV N expression plasmids encoding single (L139I) or double (L139I plus I129L) amino acid substitutions were engineered through site-directed mutagenesis, and the recombinant mutant N proteins were produced as described above. The purified proteins were characterized by SDS-PAGE and LC-MS, and the concentrations were determined by the Bradford method. The final yield of purified N protein was ∼30 mg from 4 liters of cell culture. RSV P was not recovered.
SPR.
Surface plasmon resonance (SPR) experiments were performed on a Biacore T200 using Series S NTA sensor chips (GE Healthcare) and His6-RSV N protein. The immobilization of RSV N protein was performed using standard nickel capture chemistry in a binding buffer [50 mM HEPES (pH 7.5), 150 mM NaCl, 50 μM EDTA, 1 mM tris(2-carboxyethyl)phosphine (TCEP), 0.005% T-20 (pH 7.5)]. Varying concentrations of RSV604 binding to the immobilized RSV N protein were then monitored using the same buffer. The data were analyzed using Biacore T200 Evaluation Software V 2.0.
RESULTS
Cell line-dependent RSV inhibition.
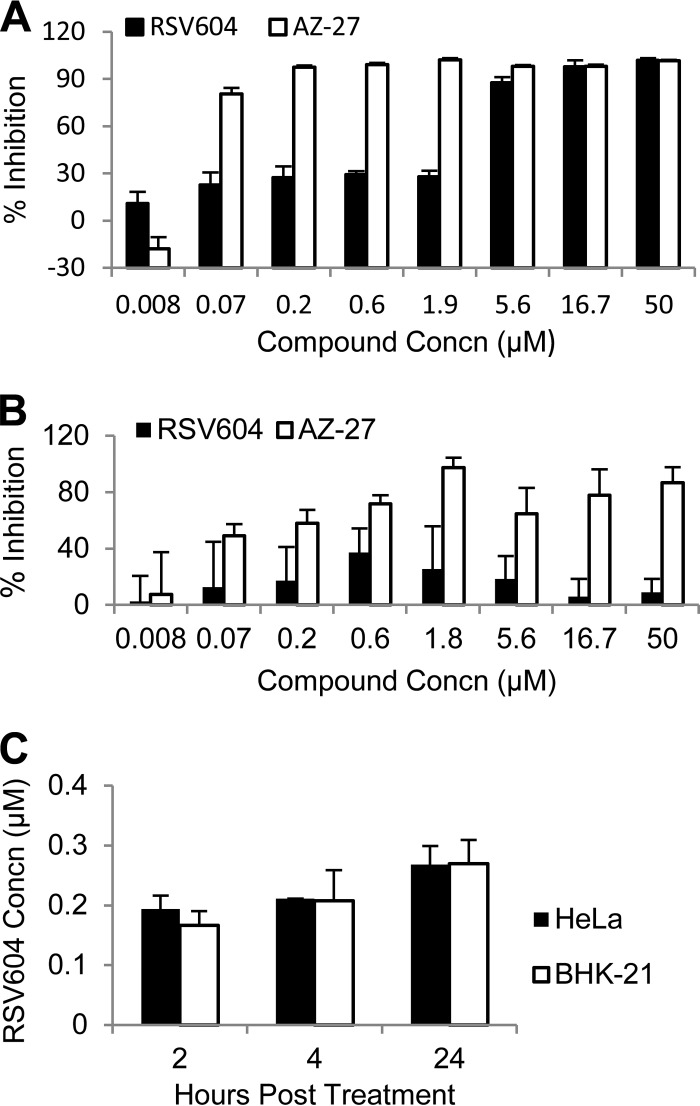
The anti-RSV activity of RSV604 was examined in a 3-day RSV A2 infection assay using several cell lines (Fig. 1A and B). The compound demonstrated potency against RSV infection in multiple cell types, including HeLa and HEp-2 cells, with an EC50 (∼2 μM) similar to the reported values (Table 1 and Fig. 1A) (9, 18). Interestingly, RSV604 showed minimal activity against RSV infection in BHK-21 cells, in contrast to the control compound, AZ-27, an L inhibitor, which maintained its potency in all cell types tested (Table 1 and Fig. 1B) (18). To investigate whether this cell line-dependent activity was due to differences in compound penetration and stability in the different cell lines, the compound concentrations within the HeLa and BHK-21 cells were determined by LC-MS analysis after 2- to 24-h culture in the assay medium containing 10 μM RSV604 (Fig. 1C). The cells showed similar compound absorptions and accumulations at all the time points examined, suggesting that the cell line-dependent potency is not due to lack of RSV604 penetration into BHK-21 cells.
FIG 1.
(A and B) Potency of RSV604 against RSV A2 infection in HeLa (A) and BHK-21 (B) cells. The data shown are percent inhibition of the RSV signal (means and standard deviations [SD]; n = 3) following a 3-day infection at an MOI of 0.1 as measured by RSV ELISA. The RSV L inhibitor AZ-27 served as a positive control. (C) RSV604 absorption into HeLa and BHK-21 cells. The cells were harvested at the indicated times after compound treatment at 10 μM. The compound concentration (Concn) in each cell lysate was determined by LC-MS analysis. The data shown are means and SD (n = 2).
TABLE 1.
Activities of RSV604 and AZ-27 against RSV in different cell lines
| Cell line | RSV604a |
AZ-27a |
||
|---|---|---|---|---|
| EC50 (μM) | CC50 (μM) | EC50 (μM) | CC50 (μM) | |
| HeLa | 2 ± 0.5 | >50 | 0.04 ± 0.02 | >50 |
| HEp-2 | 1.8 ± 0.15 | >50 | 0.01 ± 0.005 | >50 |
| BHK-21 | >50 | >50 | 0.05 ± 0.03 | >50 |
The EC50 was measured by RSV ELISA following 3-day RSV A2 infection in the presence of compound in HeLa, HEp-2, and BHK-21 cells. The CC50 was measured by cytotoxicity assay in parallel with the EC50 assays. The values shown are means ± SD (n ≥ 3).
RSV N binding in vitro.
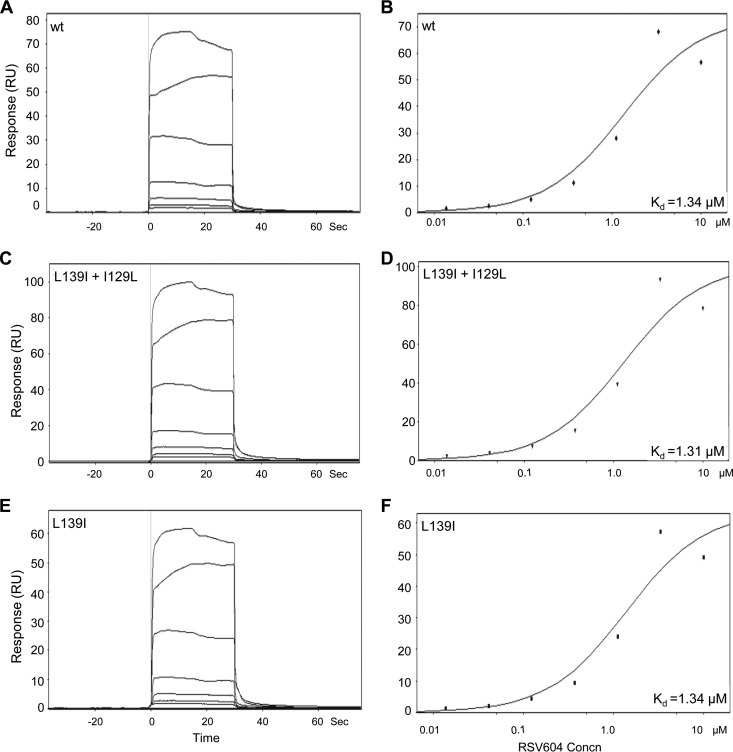
The cell line-dependent activity of RSV604 raised the question of whether the RSV N protein is the direct target of the inhibitor, as the existing evidence supporting N as the target was indirect and based only on resistant mutations mapping to the gene encoding the N protein (9). To test for a possible direct compound-target interaction, a recombinant RSV N protein with a 12-amino-acid truncation at the N terminus was expressed in E. coli to enable the production of soluble N protein (25, 26). Compound binding to the purified N protein monomer was then measured by SPR (Fig. 2). RSV604 directly bound to RSV N in SPR analysis with a binding affinity comparable to its cellular potency against RSV (Kd = 1.6 ± 0.4 μM; n = 3), supporting the concept that N is a direct target of the inhibitor. The previously reported RSV604-resistant N amino acid substitutions (L139I and I129L) were engineered into the recombinant N protein to study their effects on compound-target engagement (9). Interestingly, introduction of these single or double amino acid substitutions into the N protein did not alter the binding affinity for RSV604 in the SPR analysis (Kd = 1.34 ± 0.04 μM; n = 2), suggesting that resistance may rely on a mechanism other than loss of compound binding to the target (Fig. 2).
FIG 2.
Direct binding between RSV604 and RSV N protein as measured by SPR analysis. (A, C, and E) Sensorgrams representing direct-binding kinetics for RSV604 against wild-type (wt) and mutant RSV N shown in response units (RU) as a function of time with increasing concentrations of RSV604. (B, D, and F) Plots of signals at equilibrium versus the concentrations of RSV604 to determine affinity. The data shown are from a representative SPR experiment using the indicated wt or mutant (L139I plus I129L or L139I) recombinant RSV N protein (amino acids 13 to 391).
Inhibition of N function.
RSV N is a multifunctional protein involved in viral RNA synthesis and encapsidation and also interacting with viral and host proteins (27). To begin to understand the effect of RSV604 on these functions, viral RNA synthesis and the infectivity of released virus were examined in the presence of the compound.
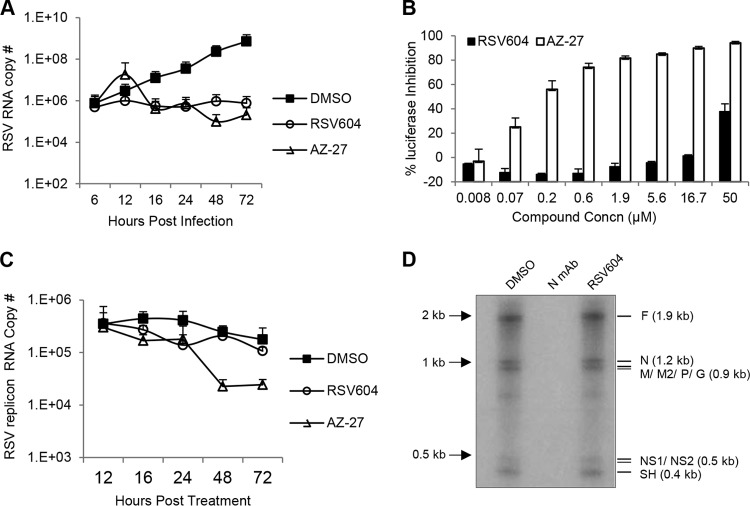
Intracellular RNA synthesis of RSV A2 was analyzed by qRT-PCR at different time points postinfection at an MOI of 0.1 in the presence of RSV604 (1× EC90, from RSV ELISA in HeLa cells) or AZ-27 (1× EC90, from RSV ELISA in the corresponding cell lines) (Fig. 3). In HeLa cells, similar to the L polymerase inhibitor AZ-27, RSV604 significantly reduced intracellular viral RNA copy numbers at multiple time points compared to the DMSO control (Fig. 3A), suggesting that RSV604 blocks viral RNA synthesis during RSV infection. In a time-of-addition study, RSV604 also maintained its potency when first added 6 h postinfection, supporting a postentry MoA (data not shown). Surprisingly, in the RSV replicon cells with established subgenomic viral RNA transcription and replication (17), RSV604 did not affect viral replicon RNA synthesis (Fig. 3B and C). To further investigate its role in viral RNA synthesis, RSV604 was tested in a cell-free RNP assay using crude RSV replication complex isolated from RSV A2-infected cells. RSV604 did not block viral RNA synthesis catalyzed by the preformed replication complex, unlike the anti-N antibody, which bound to the N protein in the RNP and completely inhibited viral RNA synthesis (Fig. 3D). Together, these data suggest that RSV604 may impact de novo viral RNA synthesis but not the already established replication complex within the cell.
FIG 3.
(A) Effect of RSV604 on intracellular viral replication in HeLa cells. HeLa cells were infected with RSV A2 at an MOI of 0.1 in the presence of DMSO, RSV604 (1× EC90), or AZ-27 (1× EC90). The quantity of RSV RNA in the infected cells was determined by RT-qPCR at the indicated times postinfection. (B and C) Effect of RSV604 on viral RNA synthesis in RSV replicon cells. Following compound treatment at the same concentrations as for panel A, percent inhibition of the replicon luciferase reporter activity (B) and intracellular viral replicon RNA copy numbers (C) was determined by luciferase assay at 48 h and by qRT-PCR assay at the indicated time points, respectively. The data shown are means and SD (n ≥ 2). (D) Effect of RSV604 on viral RNA synthesis in the RNP assay. Viral RNA synthesis in the RSV RNP replication complex was visualized using [32P]UTP substrate in a 2-h reaction in the presence of DMSO (control), anti-N monoclonal antibody (MAb), or RSV604 (10 μM), followed by 6% PAA-urea gel electrophoresis. The RSV transcripts are labeled based on their expected sizes (lengths in nucleotides). The crude RSV RNP fraction used in this reaction was isolated from RSV A2-infected HEp-2 cells.
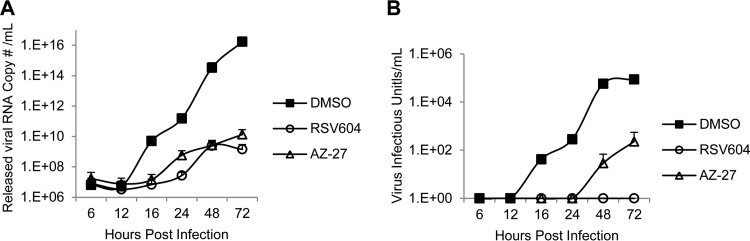
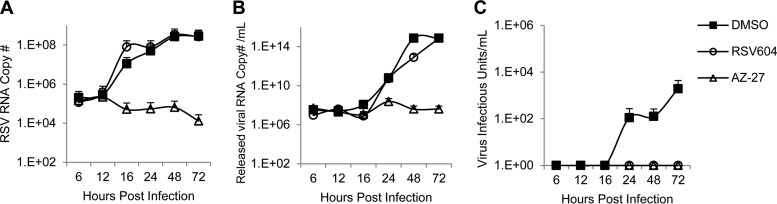
To investigate the effects of RSV604 on infectious virus assembly and release, the amount of released virus and its infectivity during a 72-h infection were analyzed by qRT-PCR and TCID50 assay, respectively. Similar to AZ-27, RSV604 delayed progeny virus release and also reduced the amount of virus released from HeLa cells, as expected for an inhibitor of viral RNA synthesis (Fig. 4A). Interestingly, the virus released in the presence of RSV604 completely lost its infectivity, whereas AZ-27 had minimal impact (Fig. 4B). These results demonstrated that RSV604 could block both viral RNA synthesis and the infectivity of the released virus. Whether failing to act on either of these mechanisms contributes to the cell-type-dependent potency of the compound (Fig. 1) was then investigated. In a similar experiment following RSV A2 infection of BHK-21 cells, RSV604 showed no effect on viral RNA synthesis or the amount of virus released, in contrast to the HeLa cell infection data (Fig. 5A and B). However, it abolished the infectivity of the released virus, similar to its effect in HeLa cells (Fig. 5C). These data suggested that the ability of RSV604 to inhibit viral RNA synthesis was cell line dependent, which largely determined its potency under the assay conditions, whereas its ability to reduce virus infectivity appeared to be independent of the cell type.
FIG 4.
Effect of RSV604 on virus release and infectivity in HeLa cells. HeLa cells were infected with RSV A2 at an MOI of 0.1 in the presence of DMSO (control), RSV604 (1× EC90), or AZ-27 (1× EC90). (A) The culture supernatants containing the released virus were collected at the indicated times, and the amounts of released virus were measured by qRT-PCR for RSV RNA. (B) Released virus infectivity was determined by TCID50 assay. The data shown are means and SD (n = 2).
FIG 5.
Effect of RSV604 on viral RNA synthesis (A), virus release (B), and infectivity (C) in BHK-21 cells. BHK-21 cells were infected with RSV A2 at an MOI of 0.1 in the presence of DMSO (control), RSV604 (1× EC90, as in RSV ELISA in HeLa cells) or AZ-27 (1× EC90, as in RSV ELISA in BHK-21 cells). (A) The quantity of RSV RNA in the infected cells was determined by qRT-PCR at the indicated time points. (B) The amount of released virus was determined by qRT-PCR quantitation of the RSV RNA in the culture medium supernatants. (C) The infectivity of the virus released into the culture supernatants was determined by TCID50 assay. The data shown are means and SD (n = 2).
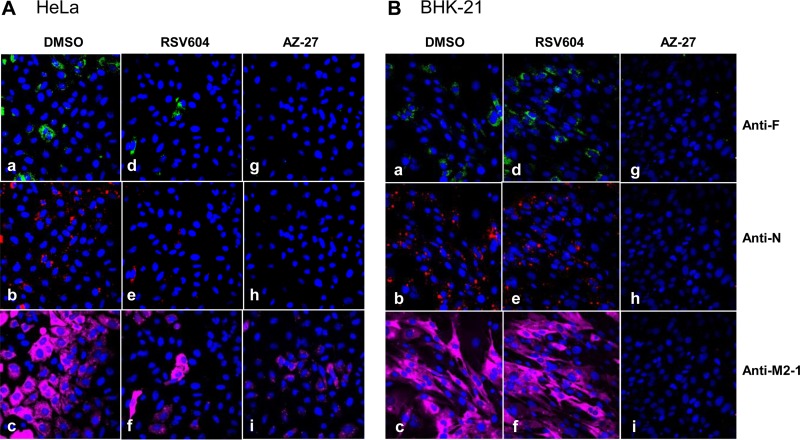
Effect on viral protein localization.
The effect of RSV604 on viral protein intracellular localization and accumulation was examined by confocal laser scanning microscopy. HeLa and BHK-21 cells infected by RSV A2 in the presence of DMSO, RSV604, or AZ-27 were analyzed by indirect immunofluorescence staining with anti-F, anti-N, and anti-M2-1 antibodies. At 24 h postinfection, N protein was detected in the cytoplasmic inclusions, F protein was near the cell surface, and M2-1 was diffusely spread inside the infected cells, as previously described (Fig. 6) (23). RSV604 treatment significantly reduced viral protein expression in HeLa cells (Fig. 6A). In contrast, RSV604 did not inhibit viral protein expression in BHK-21 cells (Fig. 6B), consistent with its lack of inhibition of viral RNA synthesis in the cell line (Fig. 1 and 5), whereas AZ-27 reduced F, N, and M2-1 expression in both cell types, as expected (Fig. 6). Interestingly, although RSV604 impacted the assembly and release of infectious virus particles (Fig. 4 and 5), no effect on viral protein distribution/localization was detected in the infected cells (Fig. 6). Similar results were obtained at other time points postinfection in HeLa and BHK-21 cells and the RSV replicon cells treated with RSV604 (data not shown).
FIG 6.
Effects of RSV604 and AZ-27 on RSV F, N, and M2-1 protein expression and localization. HeLa and BHK-21 cells were infected with RSV A2 at an MOI of 10 in the presence of DMSO (a, b, and c), RSV604 (1× EC90, as in RSV ELISA in HeLa cells) (d, e, and f), or AZ-27 (1× EC90, as in RSV ELISA in the corresponding cell lines) (g, h, and i). The cells were fixed 24 h postinfection, followed by immunostaining with anti-F, anti-N, or anti-M2-1 antibody, and visualized under confocal microscopy. The data shown are from a representative experiment.
DISCUSSION
This study characterized the mechanism of action of RSV604 and demonstrated for the first time that RSV604 bound directly to RSV N protein in vitro. This is important for validating N as the target of the inhibitor, as RSV604 showed cell line-dependent activity that was found to be independent of compound penetration into the cells. Investigating the impact of RSV604 on N function revealed that it could block both RSV RNA synthesis and the infectivity of released virus in a susceptible cell line. These two MoAs appeared to be separable, as RSV604 did not inhibit viral RNA synthesis in BHK-21 cells while still maintaining the ability to reduce the infectivity of virus released from the cells.
These results provide new insights into the RSV604 MoA and at the same time raise new questions that merit further investigation. Reduction of viral infectivity did not appear to be sufficient to retain RSV604 potency in the BHK-21 infection assay. Whether this suggests that the infectivity of virus spread through cell-to-cell fusion was less impacted remains to be determined. The mechanism for the cell line-dependent inhibition of viral RNA synthesis by RSV604 is still not known. A possible explanation is that a host factor(s) is involved, and variations in the host factor expression level, sequence, and/or structure among cell lines from different origins may contribute to this cell line-dependent inhibition. The RSV604 binding site on the N protein is also not yet defined. The known resistance mutations in the N gene were mapped to a region encoding a surface pocket exposed to solvent and with unclear function, away from the N oligomerization and RNA binding regions (28). Whether this is the RSV604 and host factor-binding pocket remains to be determined. Interestingly, the resistance mutations did not affect RSV604 and N protein binding in vitro, suggesting that the mechanism of resistance may not rely on reducing inhibitor-target binding, and thus, these mutations may or may not be located at the inhibitor binding site. Additional investigations, such as defining the inhibitor-N complex structure, will be needed to address these questions.
Side-by-side comparison of RSV604 with an RSV L polymerase inhibitor (AZ-27) in an infection time course study identified similar patterns of inhibition of viral RNA synthesis in HeLa cells by the two compounds. It is well established that the RSV L, N, M2-1, and P proteins are essential for viral RNA synthesis in cells (23, 29, 30). Interestingly, RSV604 did not significantly affect viral RNA synthesis in the RSV replicon cells nor in the viral replication complex isolated from infected cells. Since both of these systems measure inhibition of an established viral replication complex, one hypothesis could be that RSV604 may prevent the de novo assembly of the functional viral replication complex but have no impact on preformed replication complexes, which in turn affects viral replication and potentially also the assembly of infectious virus particles. Alternatively, RSV604 may indirectly impact viral RNA synthesis in infected cells through an unknown mechanism not captured by the replicon and RNP systems. These possibilities merit further study, which may also help to better understand the role of RSV N protein at the different stages of viral RNA transcription and replication.
The novel finding of a second MoA for RSV604, inhibition of released virus infectivity, highlights the potential for this class of inhibitors in delivering efficacy and suppressing resistance by having dual mechanisms of inhibition within one molecule. The details of this mechanism remain to be clarified. They may include, but are not limited to, the possibility that RSV604 induces aberrant assembly of virus RNP and particles or that the bound compound may be encapsidated into progeny virus and interfere with N function in the subsequent round of infection. It would be interesting to examine whether the RNP assembled in the presence of RSV604 is functional and whether RSV604 is incorporated into virions, which we have not determined. RSV604 did not appear to affect N protein distribution and localization in the infected cells, suggesting that its MoA differs from that of the influenza virus nucleoprotein inhibitor nucleozin, which induces nucleoprotein aggregation and alters its intracellular localization (14–16). A new RSV N inhibitor with improved potency and physical properties would be desirable to reach the full potential for this class of RSV inhibitors.
ACKNOWLEDGMENTS
We thank Melinda Foulk for compound synthesis, Wendy Salmon and the Imaging Facility at Whitehead Institute for Biomedical Research for assistance with confocal microscopy study, Paul Miller for scientific discussions and manuscript editing, and Ewan Dunn and Ed Buurman for constructive comments on the manuscript.
All authors were employees of AstraZeneca when conducting this work.
REFERENCES
- 1.Borchers AT, Chang C, Gershwin ME, Gershwin LJ. 2013. Respiratory syncytial virus—a comprehensive review. Clin Rev Allergy Immunol 45:331–379. doi: 10.1007/s12016-013-8368-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nair H, Simoes EA, Rudan I, Gessner BD, Azziz-Baumgartner E, Zhang JS, Feikin DR, Mackenzie GA, Moisi JC, Roca A, Baggett HC, Zaman SM, Singleton RJ, Lucero MG, Chandran A, Gentile A, Cohen C, Krishnan A, Bhutta ZA, Arguedas A, Clara AW, Andrade AL, Ope M, Ruvinsky RO, Hortal M, McCracken JP, Madhi SA, Bruce N, Qazi SA, Morris SS, El Arifeen S, Weber MW, Scott JA, Brooks WA, Breiman RF, Campbell H, Severe Acute Lower Respiratory Infections Working Group . 2013. Global and regional burden of hospital admissions for severe acute lower respiratory infections in young children in 2010: a systematic analysis. Lancet 381:1380–1390. doi: 10.1016/S0140-6736(12)61901-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collins PL, Fearns R, Graham BS. 2013. Respiratory syncytial virus: virology, reverse genetics, and pathogenesis of disease. Curr Top Microbiol Immunol 372:3–38. doi: 10.1007/978-3-642-38919-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falsey AR, Hennessey PA, Formica MA, Cox C, Walsh EE. 2005. Respiratory syncytial virus infection in elderly and high-risk adults. N Engl J Med 352:1749–1759. doi: 10.1056/NEJMoa043951. [DOI] [PubMed] [Google Scholar]
- 5.Roymans D, Koul A. 2011. Treatment of respiratory syncytial virus infection: past, present and future. In Resch B. (ed), Human Respiratory Syncytial Virus Infection. InTech. [Google Scholar]
- 6.DeVincenzo J, Lambkin-Williams R, Wilkinson T, Cehelsky J, Nochur S, Walsh E, Meyers R, Gollob J, Vaishnaw A. 2010. A randomized, double-blind, placebo-controlled study of an RNAi-based therapy directed against respiratory syncytial virus. Proc Natl Acad Sci U S A 107:8800–8805. doi: 10.1073/pnas.0912186107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zamora MR, Budev M, Rolfe M, Gottlieb J, Humar A, Devincenzo J, Vaishnaw A, Cehelsky J, Albert G, Nochur S, Gollob JA, Glanville AR. 2011. RNA interference therapy in lung transplant patients infected with respiratory syncytial virus. Am J Respir Crit Care Med 183:531–538. doi: 10.1164/rccm.201003-0422OC. [DOI] [PubMed] [Google Scholar]
- 8.Debing Y, Jochmans D, Neyts J. 2013. Intervention strategies for emerging viruses: use of antivirals. Curr Opin Virol 3:217–224. doi: 10.1016/j.coviro.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chapman J, Abbott E, Alber DG, Baxter RC, Bithell SK, Henderson EA, Carter MC, Chambers P, Chubb A, Cockerill GS, Collins PL, Dowdell VC, Keegan SJ, Kelsey RD, Lockyer MJ, Luongo C, Najarro P, Pickles RJ, Simmonds M, Taylor D, Tyms S, Wilson LJ, Powell KL. 2007. RSV604, a novel inhibitor of respiratory syncytial virus replication. Antimicrob Agents Chemother 51:3346–3353. doi: 10.1128/AAC.00211-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roymans D, Koul A. 2010. Respiratory syncytial virus: a prioritized or neglected target? Future Med Chem 2:1523–1527. doi: 10.4155/fmc.10.235. [DOI] [PubMed] [Google Scholar]
- 11.DeVincenzo JP, Whitley RJ, Mackman RL, Scaglioni-Weinlich C, Harrison L, Farrell E, McBride S, Lambkin-Williams R, Jordan R, Xin Y, Ramanathan S, O'Riordan T, Lewis SA, Li X, Toback SL, Lin SL, Chien JW. 2014. Oral GS-5806 activity in a respiratory syncytial virus challenge study. New Engl J Med 371:711–722. doi: 10.1056/NEJMoa1401184. [DOI] [PubMed] [Google Scholar]
- 12.Marty FM, Chemaly RF, Liakopoulou E, Dent JC, PK. 2007. A double-blind, randomised, placebo-controlled study to evaluate the safety and efficacy of RSV604 in adults with respiratory syncytial virus (RSV) infection following stem cell transplantation. IX International Symposium on Respiratory Viral Infections, Hong Kong, China. [Google Scholar]
- 13.Henderson EA, Alber DG, Baxter RC, Bithell SK, Budworth J, Carter MC, Chubb A, Cockerill GS, Dowdell VC, Fraser IJ, Harris RA, Keegan SJ, Kelsey RD, Lumley JA, Stables JN, Weerasekera N, Wilson LJ, Powell KL. 2007. 1,4-Benzodiazepines as inhibitors of respiratory syncytial virus. The identification of a clinical candidate. J Med Chem 50:1685–1692. doi: 10.1021/jm060747l. [DOI] [PubMed] [Google Scholar]
- 14.Kao RY, Yang D, Lau LS, Tsui WH, Hu L, Dai J, Chan MP, Chan CM, Wang P, Zheng BJ, Sun J, Huang JD, Madar J, Chen G, Chen H, Guan Y, Yuen KY. 2010. Identification of influenza A nucleoprotein as an antiviral target. Nat Biotechnol 28:600–605. doi: 10.1038/nbt.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amorim MJ, Kao RY, Digard P. 2013. Nucleozin targets cytoplasmic trafficking of viral ribonucleoprotein-Rab11 complexes in influenza A virus infection. J Virol 87:4694–4703. doi: 10.1128/JVI.03123-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cianci C, Gerritz SW, Deminie C, Krystal M. 2013. Influenza nucleoprotein: promising target for antiviral chemotherapy. Antivir Chem Chemother 23:77–91. doi: 10.3851/IMP2235. [DOI] [PubMed] [Google Scholar]
- 17.Malykhina O, Yednak MA, Collins PL, Olivo PD, Peeples ME. 2011. A respiratory syncytial virus replicon that is noncytotoxic and capable of long-term foreign gene expression. J Virol 85:4792–4801. doi: 10.1128/JVI.02399-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tiong-Yip CL, Aschenbrenner L, Johnson KD, McLaughlin RE, Fan J, Challa S, Xiong H, Yu Q. 2014. Characterization of a respiratory syncytial virus L protein inhibitor. Antimicrob Agents Chemother 58:3867–3873. doi: 10.1128/AAC.02540-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graham BS, Perkins MD, Wright PF, Karzon DT. 1988. Primary respiratory syncytial virus infection in mice. J Med Virol 26:153–162. doi: 10.1002/jmv.1890260207. [DOI] [PubMed] [Google Scholar]
- 20.Tiong-Yip CL, Plant H, Sharpe P, Fan J, Rich K, Gorseth E, Yu Q. 2014. Development of a high-throughput replicon assay for the identification of respiratory syncytial virus inhibitors. Antiviral Res 101:75–81. doi: 10.1016/j.antiviral.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 21.Bannister R, Rodrigues D, Murray EJ, Laxton C, Westby M, Bright H. 2010. Use of a highly sensitive strand-specific quantitative PCR to identify abortive replication in the mouse model of respiratory syncytial virus disease. Virol J 7:250. doi: 10.1186/1743-422X-7-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mason SW, Lawetz C, Gaudette Y, Do F, Scouten E, Lagace L, Simoneau B, Liuzzi M. 2004. Polyadenylation-dependent screening assay for respiratory syncytial virus RNA transcriptase activity and identification of an inhibitor. Nucleic Acids Res 32:4758–4767. doi: 10.1093/nar/gkh809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li D, Jans DA, Bardin PG, Meanger J, Mills J, Ghildyal R. 2008. Association of respiratory syncytial virus M protein with viral nucleocapsids is mediated by the M2-1 protein. J Virol 82:8863–8870. doi: 10.1128/JVI.00343-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Magliano D, Marshall JA, Bowden DS, Vardaxis N, Meanger J, Lee JY. 1998. Rubella virus replication complexes are virus-modified lysosomes. Virology 240:57–63. doi: 10.1006/viro.1997.8906. [DOI] [PubMed] [Google Scholar]
- 25.Shapiro AB, Gao N, O'Connell N, Hu J, Thresher J, Gu RF, Overman R, Hardern IM, Sproat GG. 2014. Quantitative investigation of the affinity of human respiratory syncytial virus phosphoprotein C-terminus binding to nucleocapsid protein. Virol J 11:191. doi: 10.1186/s12985-014-0191-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.El Omari K, Scott K, Dhaliwal B, Ren J, Abrescia NG, Budworth J, Lockyer M, Powell KL, Hawkins AR, Stammers DK. 2008. Crystallization and preliminary X-ray analysis of the human respiratory syncytial virus nucleocapsid protein. Acta Crystallogr Sect F Struct Biol Cryst Commun 64:1019–1023. doi: 10.1107/S1744309108031059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hacking D, Hull J. 2002. Respiratory syncytial virus—viral biology and the host response. J Infect 45:18–24. doi: 10.1053/jinf.2002.1015. [DOI] [PubMed] [Google Scholar]
- 28.Tawar RG, Duquerroy S, Vonrhein C, Varela PF, Damier-Piolle L, Castagne N, MacLellan K, Bedouelle H, Bricogne G, Bhella D, Eleouet JF, Rey FA. 2009. Crystal structure of a nucleocapsid-like nucleoprotein-RNA complex of respiratory syncytial virus. Science 326:1279–1283. doi: 10.1126/science.1177634. [DOI] [PubMed] [Google Scholar]
- 29.Carromeu C, Simabuco FM, Tamura RE, Farinha Arcieri LE, Ventura AM. 2007. Intracellular localization of human respiratory syncytial virus L protein. Arch Virol 152:2259–2263. doi: 10.1007/s00705-007-1048-4. [DOI] [PubMed] [Google Scholar]
- 30.Garcia J, Garcia-Barreno B, Vivo A, Melero JA. 1993. Cytoplasmic inclusions of respiratory syncytial virus-infected cells: formation of inclusion bodies in transfected cells that coexpress the nucleoprotein, the phosphoprotein, and the 22K protein. Virology 195:243–247. doi: 10.1006/viro.1993.1366. [DOI] [PubMed] [Google Scholar]