Abstract
The development of direct-acting antiviral agents is a promising therapeutic advance in the treatment of hepatitis C virus (HCV) infection. However, rapid emergence of drug resistance can limit efficacy and lead to cross-resistance among members of the same drug class. ABT-450 is an efficacious inhibitor of HCV NS3/4A protease, with 50% effective concentration values of 1.0, 0.21, 5.3, 19, 0.09, and 0.69 nM against stable HCV replicons with NS3 protease from genotypes 1a, 1b, 2a, 3a, 4a, and 6a, respectively. In vitro, the most common amino acid variants selected by ABT-450 in genotype 1 were located in NS3 at positions 155, 156, and 168, with the D168Y variant conferring the highest level of resistance to ABT-450 in both genotype 1a and 1b replicons (219- and 337-fold, respectively). In a 3-day monotherapy study with HCV genotype 1-infected patients, ABT-450 was coadministered with ritonavir, a cytochrome P450 3A4 inhibitor shown previously to markedly increase peak, trough, and overall drug exposures of ABT-450. A mean maximum HCV RNA decline of 4.02 log10 was observed at the end of the 3-day dosing period across all doses. The most common variants selected in these patients were R155K and D168V in genotype 1a and D168V in genotype 1b. However, selection of resistant variants was significantly reduced at the highest ABT-450 dose compared to lower doses. These findings were informative for the subsequent evaluation of ABT-450 in combination with additional drug classes in clinical trials in HCV-infected patients. (Study M11-602 is registered at ClinicalTrials.gov under registration no. NCT01074008.)
INTRODUCTION
Hepatitis C virus (HCV) infection is a global health problem, with 160 to 180 million individuals infected worldwide (1, 2). Chronic HCV infection can lead to serious liver disease, including cirrhosis, liver failure, and hepatocellular carcinoma. There are 7 major HCV genotypes, which differ in their geographic distribution, disease progression, and response to therapy (3). In the United States, Europe, and Japan, genotype 1 is the most prevalent genotype, and globally it accounts for approximately 60% of HCV infections (4).
Therapy for those infected with HCV genotype 1 improved with the approval of the NS3/4A protease inhibitors (PIs) telaprevir, boceprevir, and, more recently, simeprevir (5–10). Although the addition of a PI to pegylated interferon (pegIFN) and ribavirin (RBV) therapy significantly improved sustained virologic response (SVR) rates compared to those with pegIFN/RBV therapy alone, IFN-based therapies are associated with treatment-limiting toxicities (11). In addition, there are many patients who are ineligible for IFN-based treatment due to comorbidities such as depression (12). Early clinical trials with these PIs also demonstrated that drug resistance developed within days after initiation of treatment (13–15). The rapid selection of resistant variants is facilitated by a high rate of virus production and the infidelity of the HCV RNA polymerase (16). Thus, there is a need for effective treatments for HCV genotype 1 infection that eliminate the need for IFN while increasing SVR rates and reducing the development of resistance.
HCV is a positive-sense single-stranded RNA virus with a genome that consists of a single large open reading frame flanked by 5′ and 3′ untranslated regions. The large open reading frame is translated into a polyprotein, which is subsequently proteolytically processed into the proteins that are necessary for viral replication (17). The viral NS3/4A protease is essential for this process and is a validated drug target, as evidenced by the approval of the linear peptidomimetic covalent inhibitors telaprevir and boceprevir and the macrocyclic noncovalent peptidomimetic inhibitor simeprevir. ABT-450, identified by AbbVie and Enanta as a lead compound, is a macrocyclic noncovalent peptidomimetic inhibitor of HCV NS3/4A protease that is in clinical development at AbbVie for use in combination with the NS5A inhibitor ombitasvir (formerly known as ABT-267) (described in an accompanying article [18]) and the nonnucleoside NS5B polymerase inhibitor dasabuvir (formerly known as ABT-333) (W. Kati, G. Koev, M. Irvin, J. Beyer, Y. Liu, P. Krishnan, T. Reisch, R. Mondal, R. Wagner, A. Molla, C. Maring, and C. Collins, submitted for publication), with or without RBV for the treatment of chronic HCV infection (19–23).
ABT-450 is metabolized primarily by cytochrome P450 (CYP) 3A4. In a prior study of healthy volunteers, coadministration of ABT-450 with a low dose of the CYP3A4 inhibitor ritonavir (combination denoted ABT-450/r) dramatically increased peak, trough, and overall ABT-450 plasma concentrations as well as half-life, resulting in sustained high plasma levels with once-daily dosing (24, 25). In this paper, we report the mean viral load declines and resistance-associated variants (RAVs) observed after 3 days of therapy in HCV genotype 1-infected patients treated with 1 of 3 doses of ABT-450 (50 mg, 100 mg, or 200 mg) combined with ritonavir at 100 mg (50/100 mg, 100/100 mg, or 200/100 mg). We also report the in vitro inhibitory activity of ABT-450 against wild-type genotypes 1a, 1b, 2a, 3a, 4a, and 6a (genotype 5 has not been evaluated, as attempts to generate a functional chimeric replicon containing NS3 protease from genotype 5 have been unsuccessful), as well as its in vitro resistance profile in genotype 1.
MATERIALS AND METHODS
Compound.
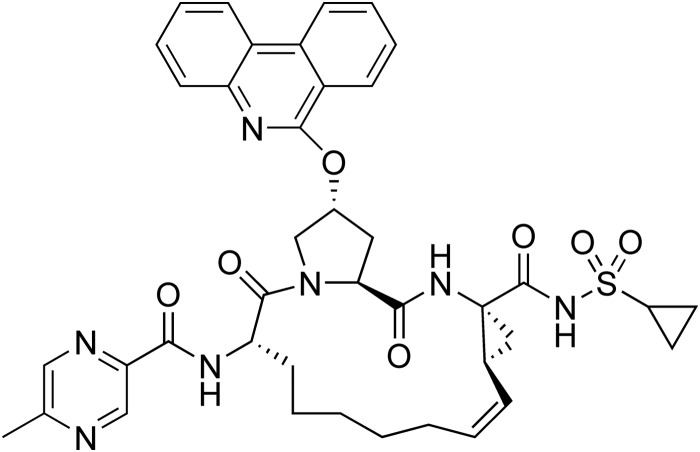
ABT-450, (2R,6S,12Z,13aS,14aR,16aS)-N-(cyclopropylsulfonyl)-6-{[(5-methylpyrazin-2-yl)carbonyl]amino}-5,16-dioxo-2-(phenanthridin-6-yloxy)-1,2,3,6,7,8,9,10,11,13a,14,15,16,16a-tetradecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecine-14a(5H)-carboxamidehydrate (Fig. 1), was synthesized at AbbVie (North Chicago, IL) (26).
FIG 1.
Chemical structure of ABT-450.
Replicon cell lines.
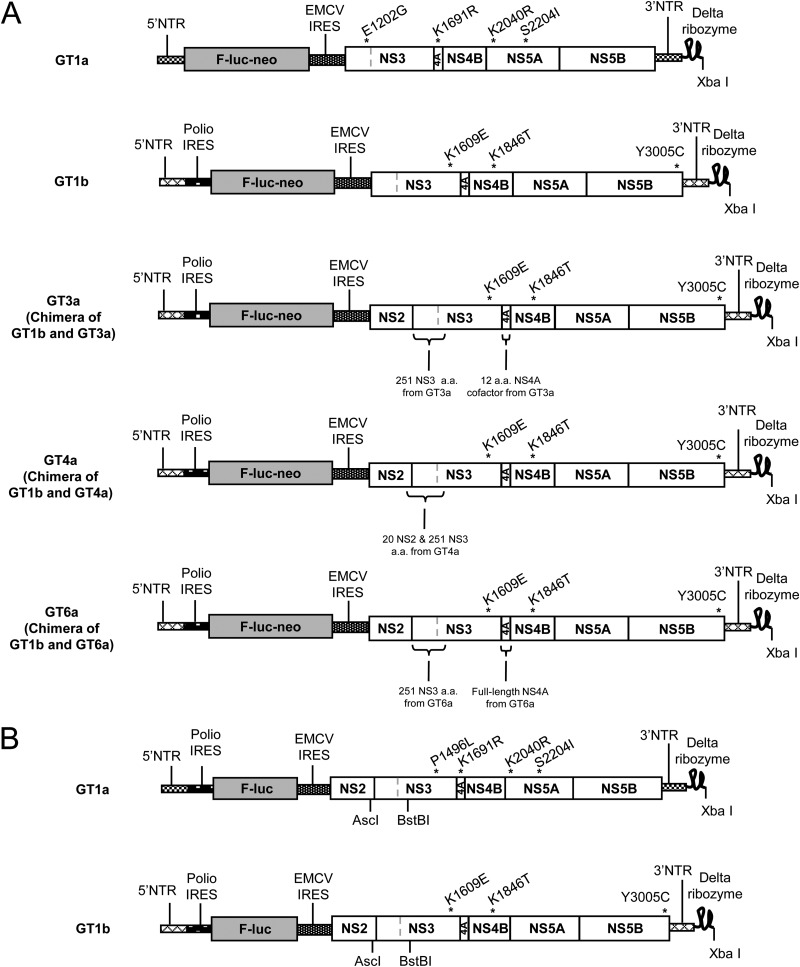
The genotype 1a and 1b stable subgenomic replicon cell lines used for compound characterization in cell culture are derived from HCV strains 1a-H77 and 1b-Con1 (GenBank accession numbers NC_004102 and AJ238799, respectively) (Fig. 2A). Both constructs are bicistronic subgenomic replicons similar in structure to those described by Lohmann et al. (27). The genotype 1a replicon contains the 5′ nontranslated region (NTR) from 1a-H77 followed by a firefly luciferase reporter gene and the neomycin phosphotransferase (Neo) gene, which together comprise the first cistron of the bicistronic replicon construct. This is followed by the encephalomyocarditis virus (EMCV) internal ribosome entry site (IRES) and the second cistron containing the 1a-H77 NS3-NS5B-coding region with adaptive mutations encoding E1202G, K1691R, K2040R, and S2204I and finally by the 1a-H77 3′ NTR. The 1b-Con1 replicon construct is similar in structure to the 1a-H77 replicon; however, the 5′ NTR, NS3-NS5B-coding region, and 3′ NTR are derived from 1b-Con1. The adaptive mutations in the 1b-Con1 replicon are those encoding K1609E, K1846T, and Y3005C, and this replicon construct contains a poliovirus IRES between the HCV 5′ NTR and the firefly luciferase gene (28).
FIG 2.
Schematic diagrams of HCV replicons used in this study. (A) Replicons for stable cell lines. (B) Replicons for transient assays. C, cysteine; E, glutamic acid; delta ribozyme, ribozyme from hepatitis delta virus; E, glutamic acid; EMCV, encephalomyocarditis virus; F-luc, firefly luciferase; G, glycine; I, isoleucine; IRES, internal ribosome entry site; K, lysine; L, leucine; Neo, neomycin phosphotransferase gene; NTR, nontranslated region; P, proline; polio, poliovirus; R, arginine; S, serine; T, threonine, Y, tyrosine.
In addition to the genotype 1a and 1b replicons, chimeric replicons in the 1b-Con1 background were generated by insertion of the region encoding the first 251 amino acids of NS3 from genotype 3a, 4a, or 6a in place of the corresponding region in a 1b-Con1 NS2-NS5B replicon that contained adaptive mutations encoding the same amino acid changes as found in the 1b-Con1 NS3-NS5B replicon described above (Fig. 2A). In the genotype 3a replicon, the region encoding NS4A amino acids 21 to 32 (numbered relative to NS4A open reading frame) of the 1b-Con1 backbone was also replaced with the corresponding residues from genotype 3a. In the genotype 4a replicon, the region encoding the 20 amino acids at the C terminus of NS2 from the 1b-Con1 backbone was replaced with the corresponding region from genotype 4a. In the genotype 6a replicon, the region encoding the entire 54 amino acids of NS4A was also replaced with the corresponding region from genotype 6a. The genotype 3a and 4a sequences were each derived from a population sequence from a single treatment-naive HCV-infected patient. The NS3 and NS4A genes of the genotype 6a replicon were synthetically constructed based on generation of a consensus sequence derived from the alignment of 15 sequences in GenBank. Stably replicating cell lines were generated by transfecting RNA transcribed in vitro from each of the replicon constructs into a Huh7-derived cell line and selecting for individual colonies using 400 μg/ml G418 (29). Activity of ABT-450 against genotype 2a JFH-1 (GenBank accession number AB047639) was determined at Southern Research Institute (Birmingham, AL) with a quantitative reverse transcriptase PCR (qRT-PCR) assay using a subgenomic replicon that did not contain a luciferase reporter (30, 31).
Antiviral activity in cell culture.
Replicon cell lines were maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 100 IU/ml penicillin, 100 μg/ml streptomycin, and 200 μg/ml G418, all of which were from Invitrogen (Carlsbad, CA), as well as 10% (vol/vol) fetal bovine serum (FBS) (Atlanta Biologicals, Flowery Branch, GA). The inhibitory effect of ABT-450 was evaluated by incubating replicon-containing cells in the presence of a series of ABT-450 dilutions for 3 days in the same medium containing 5% FBS, followed by measurement of firefly luciferase activity using the luciferase assay system (Promega, Madison, WI). In assays measuring inhibitory activity in the presence of human plasma, the medium contained 40% human plasma (Bioreclamation, Westbury, NY) and 5% FBS. The percent inhibition of HCV RNA replication was calculated for each compound concentration, and the 50% effective concentration (EC50) was calculated using nonlinear regression sigmoidal dose-response variable slope curve fitting to the 4-parameter logistic equation (32) and GraphPad Prism 4 software. The cytotoxicity of ABT-450 was determined by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (Sigma-Aldrich, St. Louis, MO) colorimetric assay (33). The 50% cytotoxicity concentration (CC50) was calculated using nonlinear regression sigmoidal dose-response variable slope curve fitting as described above.
In vitro resistance selection.
The 1a-H77 and 1b-Con1 replicon cell lines (105 cells) were plated in 150-mm cell culture plates and grown in the presence of G418 (400 μg/ml) and ABT-450 at a concentration that was 10-, 100-, or 500-fold above the EC50 for the respective cell line. After approximately 3 weeks of treatment, most cells were cleared of replicon RNA and therefore were unable to survive in the G418-containing medium. The cells containing resistant replicon variants survived and formed colonies that were isolated and further expanded. In order to characterize resistant replicon variants, total RNA was extracted from the expanded colonies, the NS3 protease-coding region was amplified by RT-PCR using gene-specific primers, and the nucleotide sequence of the amplified sample was determined.
Antiviral activity against a panel of resistant mutants.
The 1a-H77 and 1b-Con1 subgenomic replicon shuttle vector constructs used for introduction of mutations of interest in the NS3 gene were similar to the replicon cell line constructs described above, but in both cases the Neo gene was not present, and the HCV NS2 gene was inserted between the EMCV IRES and the NS3 gene (Fig. 2B). In addition, the 1a-H77 replicon construct had the adaptive mutation in NS3 protease encoding E1202G replaced with one encoding P1496L in NS3 helicase. An AscI restriction site was introduced into the NS2 gene 62 nucleotides upstream of the 5′ end of the NS3 gene, and a BstBI restriction site was introduced within the helicase domain of NS3 after the NS3 amino acid 251 codon. The introduction of these restriction sites did not result in an amino acid insertion or change in either the genotype 1a or 1b replicon. Mutations encoding resistance-associated variants were introduced by site-directed mutagenesis and confirmed by sequence analysis. Subgenomic replicon RNA was generated by linearization of plasmid DNA followed by in vitro transcription. Replicon RNA was transfected into Huh7-derived cells, and inhibition of replication of the HCV replicon by ABT-450 was measured using the luciferase assay as described above, except that cells were incubated for 4 days rather than 3 days prior to lysis. Replication efficiency was calculated as a percentage of wild-type replication using the following equation: 100 ×[(mutant 4-day luciferase activity/wild-type 4-day luciferase activity)/(mutant 4-h luciferase activity/wild-type 4-h luciferase activity)].
Antiviral activity against a panel of genotype 1a and 1b isolates.
The HCV 1a-H77 and 1b-Con1 replicon shuttle vectors described above were used to generate replicons containing NS3 genes from a panel of genotype 1a and 1b patient isolates for assessing the activity of ABT-450. These replicon shuttle vectors allowed insertion of the region encoding the complete NS3 protease domain without adaptive mutations. The region encoding the C-terminal 20 amino acids of NS2 and amino acids 1 to 251 of NS3 was amplified by RT-PCR of viral RNA from genotype 1a and 1b isolates using primers incorporating AscI and BstBI restriction sites. The amplified products were inserted into the appropriate shuttle vector, and the EC50s of ABT-450 were evaluated in transient assays as described above.
Clinical study design.
Study M11-602 (ClinicalTrials.gov registration no. NCT01074008) was a randomized, multiple-dose, placebo-controlled, blinded (active versus placebo), dose-ranging phase 2a clinical trial to explore the safety, tolerability, pharmacokinetics (PK), and antiviral activity of three direct-acting antiviral agents (DAAs), i.e., ABT-450/r and 2 nonnucleoside inhibitors of HCV NS5B polymerase, dasabuvir and ABT-072, in genotype 1 HCV-infected treatment-naive patients in the United States, including Puerto Rico (34). All of the patients provided written informed consent. The study was performed in accordance with Good Clinical Practice guidelines and the principles of the Declaration of Helsinki, and the study protocol was approved by the relevant institutional review boards and regulatory agencies. In this study, 75 patients meeting all eligibility criteria and none of the exclusion criteria were randomized to receive various doses of ABT-450/r, dasabuvir, or ABT-072. Only data from the 24 patients treated with ABT-450/r are discussed in this report. Eligibility criteria for study M11-602 included the following: age of 18 to 65 years, body mass index (BMI) of ≥18 and <35 kg/m2, chronic HCV genotype 1 infection for at least 6 months prior to study enrollment, plasma HCV RNA level of ≥100,000 IU/ml at screening, liver biopsy within the past 3 years with histology consistent with HCV-induced liver damage, and no evidence of cirrhosis. Exclusion criteria included the following: liver biopsy with a METAVIR fibrosis score of 3 or 4, positive test result for hepatitis B surface antigen or anti-HIV antibodies, history of major depression within the 2 years prior to enrollment, history of disease precluding the use of pegIFN or RBV, and unresolved clinically significant diseases other than HCV.
Patients were randomized to receive 1 of 3 doses of ABT-450/r (50/100 mg, 100/100 mg, or 200/100 mg) or placebo once daily (QD). Following 3 days of monotherapy, pegIFN alfa-2a at 180 μg/week and weight-based RBV at 1,000 to 1,200 mg/day were added, and the same dose of ABT-450/r or placebo was continued to complete a total of 12 weeks. At week 12, ABT-450/r or placebo was discontinued, and patients received pegIFN/RBV alone for up to 36 additional weeks.
Pharmacokinetic evaluations.
ABT-450 and ritonavir concentrations were determined using a liquid chromatography-tandem mass spectrometry (LC-MS/MS) method with lower limits of quantitation of 0.5 ng/ml (ABT-450) and 5 ng/ml (ritonavir). Pharmacokinetic analyses were conducted using WinNonlin Professional version 5.2 (Pharsight Corporation, CA).
Efficacy analysis.
HCV RNA was measured using the COBAS TaqMan HCV Test v2.0 real-time reverse transcriptase PCR (RT-PCR) assay (Roche, Pleasanton, CA), with a lower limit of quantitation of 25 IU/ml and a lower limit of detection of 10 IU/ml. The virologic response was assessed as HCV RNA decrease from baseline in log10 IU/ml.
Sequence analysis of patient samples.
Viral RNA was isolated from plasma samples from HCV-infected patients at baseline (prior to the first dose on day 1) and after 3 days of ABT-450/r dosing (prior to the first dose on day 4) from samples with viral loads of ≥500 IU/ml either by an automated method using the m2000 instrument (Abbott Molecular, Inc., Des Plaines, IL) or with the QIAamp viral RNA minikit (Qiagen, Valencia, CA). RT-PCR was performed with the Superscript III one-step RT-PCR system with Platinum Taq High Fidelity (Invitrogen, Carlsbad, CA) using sense and antisense primers located outside NS3/4A. Nested PCR was performed with Platinum Pfx DNA polymerase (Invitrogen, Carlsbad, CA) using sense and antisense primers specific for the NS3 protease domain. The NS3 protease domain PCR fragment was inserted into the pJET1.2 vector (Fermentas, Glen Burnie, MD), plasmid DNA was isolated from individual colonies, and the plasmid DNA insert was sequenced. An average of 87 clones per sample were sequenced, with a minimum of 75 clones sequenced from each sample.
Phenotypic analysis of patient samples.
The nested PCR products from patient samples described above were digested and cloned into the appropriate genotype 1a or 1b replicon shuttle vector. Plasmid DNA and in vitro-transcribed RNA were prepared and transfected into Huh7-derived cells, and luciferase assays were performed with EC50s calculated as described above.
RESULTS
In vitro activity.
ABT-450 inhibited genotype 1a-H77 and 1b-Con1 HCV subgenomic replicons in cell culture with EC50s of 1.0 and 0.21 nM, respectively. In the presence of 40% human plasma, the EC50s increased by 17- to 24-fold (Table 1). The CC50 of ABT-450 was >37 μM, resulting in an in vitro selectivity index of ≥37,000-fold. ABT-450 demonstrated activity across multiple HCV genotypes, with an EC50 of 5.3 nM against the genotype 2a JFH-1 subgenomic replicon and EC50s of 19, 0.09, and 0.69 nM against replicons containing NS3 protease from genotypes 3a, 4a, and 6a, respectively. As has been observed for other NS3/4A protease inhibitors such as simeprevir and asunaprevir, ABT-450 has reduced potency against genotype 3a, likely caused by the presence of the NS3 polymorphism D168Q, which is commonly found in this genotype (35, 36).
TABLE 1.
Activity of ABT-450 against replicon cell lines with NS3 from different genotypes in the presence or absence of 40% human plasma
| Human plasma concn (%) | EC50 (nM, mean ± SD) with genotype: |
|||||
|---|---|---|---|---|---|---|
| 1a | 1b | 2a | 3a | 4a | 6a | |
| 0 | 1.0 ± 0.33 | 0.21 ± 0.07 | 5.3 ± 1.2 | 19 ± 5.2 | 0.09 ± 0.03 | 0.69 ± 0.09 |
| 40 | 17.3 ± 5.7 | 5.2 ± 2.0 | NDa | ND | ND | ND |
ND, not determined.
Activity of ABT-450 against chimeric replicons containing NS3 protease from genotype 1 patients.
To determine the breadth of coverage in genotype 1, the activity of ABT-450 against chimeric replicons containing sequences derived from 11 genotype 1a- and 9 genotype 1b-infected patients was characterized. EC50s ranged from 0.43 to 1.87 nM against the genotype 1a isolates and from 0.033 to 0.087 nM against the genotype 1b isolates (Table 2), indicating that ABT-450 can inhibit NS3 proteases across a broad range of genotype 1 isolates.
TABLE 2.
Antiviral activity of ABT-450 in transient HCV replicon assays using chimeric replicons containing NS3 protease genes from HCV genotype 1-infected patients
| Genotype | Samplea | EC50, nM |
|---|---|---|
| 1a | 1a-H77 | 0.96 |
| 1 | 0.43 | |
| 2 | 0.69 | |
| 3 | 0.65 | |
| 4 | 1.87 | |
| 5 | 0.60 | |
| 6 | 0.65 | |
| 7 | 0.89 | |
| 8 | 0.68 | |
| 9 | 1.30 | |
| 10 | 0.62 | |
| 11 | 1.03 | |
| Mean | 0.86 | |
| 1b | 1b-Con1 | 0.033 |
| 1 | 0.062 | |
| 2 | 0.046 | |
| 3 | 0.056 | |
| 4 | 0.058 | |
| 5 | 0.087 | |
| 6 | 0.033 | |
| 7 | 0.043 | |
| 8 | 0.074 | |
| 9 | 0.067 | |
| Mean | 0.058 |
Characterization of resistance.
HCV genotype 1a and 1b subgenomic replicon cell lines were passaged in the presence of ABT-450 at concentrations 10-, 100-, or 500-fold over the EC50. No colonies survived selection at a concentration 100- or 500-fold over the EC50 in genotype 1a, and no colonies survived at 500-fold over the EC50 in genotype 1b. The following major variants in HCV NS3 were observed in selected colonies: R155K, D168E, and D168N in 1a-H77 and A156T, D168H, D168V, and D168Y in 1b-Con1 (Table 3). In order to achieve a more complete understanding of the ABT-450 resistance profile, its activity was assessed against HCV replicons containing NS3 variants that have been observed in HCV-infected patients treated with the HCV protease inhibitor telaprevir, boceprevir, simeprevir, asunaprevir, danoprevir, or faldaprevir (14, 37–41). Most of the tested variants conferred less than 100-fold resistance to ABT-450. R155K in genotype 1a, which has been shown to confer resistance to virtually all HCV NS3/4A protease inhibitors, confers 37-fold reduced susceptibility to ABT-450. In both genotypes 1a and 1b, the D168V variant has been shown to confer resistance to the macrocyclic and acyclic noncovalent protease inhibitors such as simeprevir, asunaprevir, danoprevir, and faldaprevir but not to the linear keto amide class of covalent inhibitors such as telaprevir and boceprevir. This variant confers 96- or 159-fold-reduced susceptibility to ABT-450 in genotype 1a or 1b, respectively. The highest level of resistance to ABT-450 among the evaluated variants was observed with the D168Y variant, which conferred 219- or 337-fold reduced susceptibility in genotype 1a or 1b, respectively.
TABLE 3.
Loss in activity for ABT-450 against resistant variants selected by NS3/4A protease inhibitors
| Replicona | NS3 variant | Fold change in EC50 relative to wild-type value | Replication capacity (% of wild-type level) |
|---|---|---|---|
| 1a-H77 | V36A | 3 | 130 |
| V36L | 2 | 82 | |
| V36M | 2 | 81 | |
| F43L | 20 | 17 | |
| T54S | 0.4 | 6 | |
| V55I | 1 | 81 | |
| Q80K | 3 | 91 | |
| Q80L | 2 | 38 | |
| Q80R | 2 | 44 | |
| R155G | 14 | 2 | |
| R155Kb | 37 | 31 | |
| R155S | 7 | 2 | |
| R155T | 7 | 5 | |
| R155W | 11 | 5 | |
| A156T | 17 | 5 | |
| D168A | 50 | 35 | |
| D168Eb | 14 | 34 | |
| D168H | 62 | 24 | |
| D168Nb | 13 | 28 | |
| D168V | 96 | 2 | |
| D168Y | 219 | 4 | |
| V36M + R155K | 79 | 29 | |
| Q80K + R155K | 19 | 77 | |
| 1b-Con1 | T54A | 1 | 59 |
| V55A | 1 | 14 | |
| R155K | 40 | 73 | |
| R155Qb | <0.5 | ||
| A156S | 0.5 | 61 | |
| A156Tb | 7 | 19 | |
| D168A | 27 | 69 | |
| D168E | 4 | 80 | |
| D168Hb | 76 | 108 | |
| D168T | 49 | 129 | |
| D168Vb | 159 | 157 | |
| D168Yb | 337 | 70 | |
| V170A | 1 | 64 |
Three-day monotherapy pharmacokinetics and antiviral efficacy in study M11-602.
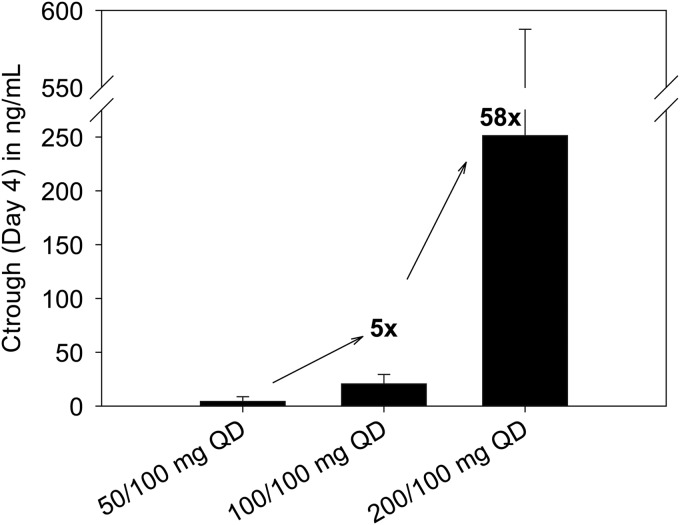
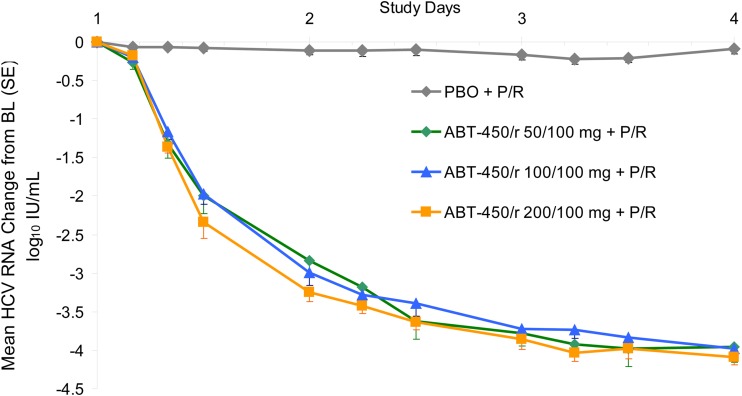
Twenty-four treatment-naive patients infected with HCV genotype 1 (8 per group) were randomized in study M11-602 and administered an ABT-450/r dose of 50/100 mg, 100/100 mg, or 200/100 mg QD for 3 days. Demographic and baseline characteristics were similar between groups. Mean baseline HCV RNA levels were 6.86 log10 IU/ml for patients receiving placebo and 6.74 log10 IU/ml for patients receiving ABT-450/r (34). Pharmacokinetic analysis of samples from the HCV-infected patients administered different doses of ABT-450 showed a greater-than-dose-proportional increase in ABT-450 exposure with increasing dose through 3 days of treatment with ABT-450/r (Fig. 3) (25). The mean trough concentration increased 58-fold when the ABT-450 dose was increased from 50 mg to 200 mg. HCV RNA levels were analyzed during the 3-day monotherapy treatment. The mean maximum HCV RNA decrease from baseline was 4.02 ± 0.43 log10 IU/ml for all patients receiving ABT-450/r, compared with 0.36 ± 0.13 log10 IU/ml for patients receiving placebo (P < 0.001) (34). Similar HCV RNA decreases were seen with all ABT-450/r doses at the end of the 3-day treatment (Fig. 4).
FIG 3.
Mean (± standard deviation [SD]) ABT-450 plasma trough concentration levels after 3-day monotherapy in HCV genotype 1-infected patients following various ABT-450/r doses.
FIG 4.
Mean (± standard error [SE]) HCV RNA change from baseline during 3-day treatment with ABT-450/r in HCV genotype 1-infected patients. BL, baseline; PBO, placebo; P/R, pegylated interferon plus ribavirin.
Phenotypic evaluation of in vivo resistance development.
Phenotypic analyses of viral isolates from baseline (before the first dose was administered) and at the end of 3 days of ABT-450/r monotherapy were performed in order to characterize the selection of resistant variants. The development of phenotypic resistance to ABT-450 during 3 days of dosing was assessed by calculating the fold change in EC50 at the end of the 3-day monotherapy compared to baseline (Table 4). Thirteen of the 24 patients (12 of 19 infected with genotype 1a and 1 of 5 infected with genotype 1b) had a viral load level sufficient (≥500 IU/ml) to allow amplification of the target gene at the end of the 3-day dosing period (5, 3, and 5 patients in the ABT-450/r 200/100-, 100/100-, and 50/100-mg treatment groups, respectively).
TABLE 4.
Phenotypic resistance after 3 days of treatment with ABT-450/r in HCV genotype 1-infected patients
| ABT-450/r treatment (mg QD) | Patient | Genotype | EC50, nM |
Fold change in EC50 | |
|---|---|---|---|---|---|
| Baseline | After 3 days of dosing | ||||
| 200/100 | 1 | 1a | 1.31 | 1.14 | 0.9 |
| 2 | 1a | 1.36 | 1.06 | 0.8 | |
| 3 | 1a | 3.11 | 3.23 | 1.0 | |
| 4 | 1a | 2.42 | 2.57 | 1.1 | |
| 5 | 1a | 0.56 | 0.86 | 1.5 | |
| 100/100 | 6 | 1a | 1.55 | 10.2 | 6.6 |
| 7 | 1b | 0.04 | 15.5 | 386 | |
| 8 | 1a | 2.80 | 8.84 | 3.2 | |
| 50/100 | 9 | 1a | 0.81 | 8.88 | 11 |
| 10 | 1a | 5.97 | 66.7 | 11 | |
| 11 | 1a | 1.52 | 11.5 | 7.6 | |
| 12 | 1a | 1.01 | 1.69 | 1.7 | |
| 13 | 1a | 1.54 | 9.78 | 6.4 | |
Among the genotype 1a-infected patients with amplifiable samples, 4 of 5 (80%) from the ABT-450/r 50/100-mg group, 2 of 2 (100%) from the ABT-450/r 100/100-mg group, and 0 of 5 (0%) from the ABT-450/r 200/100-mg group showed a 3- to 12-fold loss of susceptibility to ABT-450 relative to the baseline value for that patient. The remaining samples had less than a 3-fold loss of susceptibility to ABT-450 relative to the baseline value for that patient. The single genotype 1b-infected patient with amplifiable HCV RNA, who received ABT-450/r at 100/100 mg QD, showed a 386-fold loss of susceptibility to ABT-450 relative to the baseline value for that patient.
Since many of the clinical samples at day 4 contained a mixture of NS3 variants (including the wild type) and since each of these variants may have differing replication fitness and variable susceptibility to ABT-450, the EC50 against clinical isolates is a reflection of the multiple variants present in the quasispecies. Therefore, the EC50s from reference replicons harboring single defined resistant variants differ from those for the clinical isolates.
Sequence analysis at baseline and on day 4 (after 3 days of ABT-450/r monotherapy).
In order to identify known RAVs at baseline and to monitor the emergence of variants at the end of ABT-450/r monotherapy, clonal nucleotide sequence analysis was performed on the NS3 protease gene from all baseline and day 4 samples with HCV viral titers of ≥500 IU/ml. The analysis of the baseline samples focused on changes at amino acid positions 155, 156, and 168 of NS3, since these are the positions at which resistant variants were identified during in vitro selection experiments with ABT-450 and where variants have most commonly emerged in patients treated with other NS3/4A protease inhibitors. The consensus amino acids at these positions, based on the prototypic standards, are arginine (R) at amino acid position 155 (R155), alanine (A) at 156 (A156), and aspartic acid (D) at 168 (D168). For all of the patients in this study, the HCV NS3 sequence at baseline was identical to the prototypic sequence at these 3 amino acid positions.
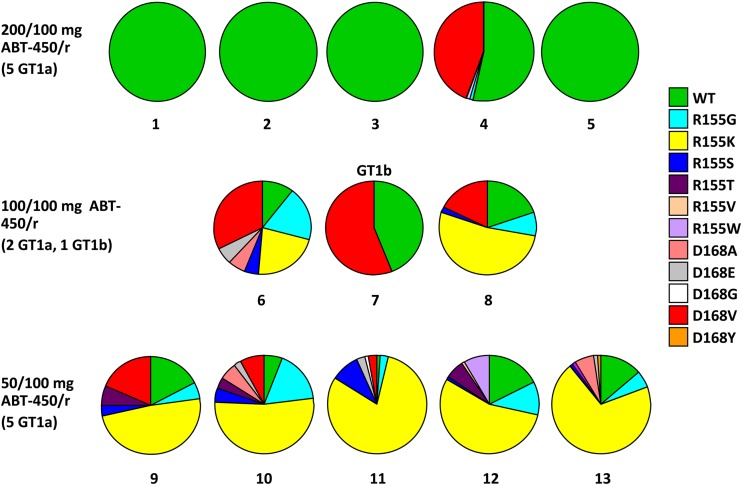
The analysis of the viral isolates obtained at the end of the 3-day monotherapy period also focused on changes at amino acids 155, 156, and 168, as no emergent variants were observed in samples from more than 1 patient at any other amino acid position in NS3/4A. In the ABT-450/r 200/100-mg dose group, clonal sequences from 4 of 5 samples did not contain any known RAVs at the end of 3 days of dosing, while clonal sequences from 1 sample contained D168V (Fig. 5). In contrast, sequences from all patients receiving lower doses of ABT-450 contained RAVs. Samples from the 3 patients from the ABT-450/r 100/100-mg dose group that could be amplified and sequenced had a high percentage of clones containing RAVs, most commonly R155K or D168V. Similarly, samples from the 5 patients in the ABT-450/r 50/100-mg dose group that were sequenced contained clones with known RAVs; R155K was the most common RAV identified. In most of the genotype 1a-infected patients who received the 50- or 100-mg dose of ABT-450, a number of other RAVs which confer modest levels of resistance to ABT-450, such as R155G, R155S, R155T, R155W, D168A, and/or D168E, were present at lower prevalence.
FIG 5.
Distribution of wild-type virus and resistant variants in patient samples with viral loads of ≥500 IU/ml after 3 days of treatment with ABT-450/r. A, alanine; D, aspartic acid; E, glutamic acid; G, glycine; K, lysine; R, arginine; S, serine; T, threonine; V, valine; W, tryptophan; Y, tyrosine.
DISCUSSION
ABT-450 is an inhibitor of HCV NS3/4A protease with nanomolar activity against genotypes 1, 2, 3, 4, and 6. We assessed the resistance-associated variants selected in vitro by ABT-450 since these variants may have clinical implications relative to other HCV protease inhibitors. Resistance selection experiments with ABT-450 in genotype 1a and 1b replicons identified RAVs at amino acid positions 155 and 168 in genotype 1a and at positions 156 and 168 in genotype 1b. When ABT-450 was evaluated against a panel of NS3 variants that have been observed in patients treated with other HCV NS3/4A protease inhibitors, cross-resistance was noted with variants known to confer resistance to other protease inhibitors. The D168Y variant conferred the highest level of resistance to ABT-450 in both genotypes 1a and 1b.
In study M11-602, the mean maximum HCV RNA decrease from baseline was 4.02 ± 0.43 log10 IU/ml for all patients receiving ABT-450/r, and was similar across all 3 ABT-450/r doses during the 3-day treatment (Fig. 4). While the decrease in viral RNA was independent of dose, resistance analysis from the ABT-450/r monotherapy portion of this study demonstrated a relationship between higher ABT-450 doses and a lower likelihood of emergence of RAVs. Among the 13 patients in whom HCV RNA could be amplified and sequenced after 3 days of monotherapy (12 with genotype 1a and 1 with genotype 1b), clonal sequencing of samples from 4 of the 5 patients who received the highest ABT-450 dose (200 mg QD) did not identify RAVs at position 155, 156, or 168, while one sample contained D168V. In contrast, samples from all patients receiving lower doses of ABT-450 contained multiple RAVs, most commonly R155K or D168V. These findings suggest that, despite similar decreases in HCV RNA levels, ABT-450 exposures associated with the 50-mg dose were not sufficient to suppress replication of the R155K and D168V variants, which are associated with approximately 30- to 100-fold loss in ABT-450 susceptibility. In contrast, the higher exposures achieved with the 200-mg dose suppressed emergence of all detectable RAVs at day 4 in a substantial proportion of patients. These findings suggest that the ABT-450 exposures achieved with all tested doses were adequate to suppress replication of wild-type HCV, but only those at the 200-mg dose consistently prevented emergence of virus harboring RAVs during the 3 days of treatment. These 3-day monotherapy findings are consistent with resistance analyses conducted in a dose-ranging phase 2 trial, AVIATOR (M11-652, ClinicalTrials.gov registration no. NCT01464827), in which ABT-450/r was administered for up to 24 weeks. In this study, the most prevalent treatment-emergent variant in NS3 among patients who experienced virologic failure was D168V; treatment-emergent R155K was seen infrequently in patients with virologic failure who received an ABT-450 dose of ≥150 mg (42).
HCV exists as a quasispecies containing all possible preexisting single RAVs even before the onset of drug pressure (16). Thus, sufficient and ongoing drug exposure for the duration of the dosing interval is critical to maintain viral suppression. It has been observed previously that higher plasma concentrations of HCV NS3 protease inhibitors such as telaprevir result in both greater antiviral activity and lower selection of resistant HCV variants (41). Preclinical data suggested that the plasma levels of HCV protease inhibitors might be boosted with ritonavir coadministration (43). Based on these observations, we utilized ritonavir for pharmacokinetic enhancement of ABT-450. Our hypothesis was that a strategy based on achieving consistently high ABT-450 exposures throughout the 24-hour dosing period may reduce the risk of emergence of resistance, improve efficacy, and allow use of a lower dose of ABT-450 while permitting once-daily dosing.
The strategy of ritonavir pharmacokinetic enhancement to increase drug exposures has long been utilized in antiretroviral therapy for the treatment of human immunodeficiency virus (HIV) infection. Coadministration of HIV protease inhibitors with ritonavir improves efficacy by maintaining peak, trough and overall drug exposures of the protease inhibitor at levels necessary to inhibit replication of both wild-type virus and resistant variants (44). High rates of HIV suppression despite less than optimal medication adherence has been attributed to the “forgiveness of ritonavir” and the favorable PK profile of boosted HIV protease inhibitors (45, 46), which have been first-line therapies for more than a decade. In addition, by prolonging the drug's half-life, ritonavir also permits less frequent dosing. These advantages have resulted in the widespread use of low-dose ritonavir to enhance the pharmacokinetics of HIV protease inhibitors (47, 48).
A comparison of pharmacokinetic parameters in healthy volunteers receiving ABT-450 alone or combined with ritonavir at 100 mg demonstrated that ritonavir substantially increases ABT-450 plasma concentration and half-life, with trough concentrations (C24) approximately 340-fold higher in the presence of ritonavir than with ABT-450 alone (25, 49). Based on pharmacokinetic data for healthy patients, the trough ABT-450 concentrations achieved with a daily dose of 200 mg of ABT-450 and 100 mg of ritonavir are predicted to be greater than that seen with 1,000 mg ABT-450 twice daily in the absence of ritonavir.
Thus, coadministration of ABT-450 with low-dose ritonavir enhances ABT-450 exposures considerably. While the relationship between plasma ABT-450 levels and those achieved in hepatocytes is unclear, rapid suppression of HCV RNA and a sustained virologic response after the end of treatment in patients who received ABT-450 in study M11-602 suggest that coadministering an adequate dose of ABT-450 with ritonavir achieves ABT-450 concentrations at the site of action that are well in excess of the EC50 of RAVs (50). As the emergence of RAVs may lead to treatment failure, inhibition of these variants, resulting in more complete HCV suppression, may account for the high response rates seen in large trials of DAA combination therapy that include ABT-450/r, where ABT-450 was administered at a dosage of 150 mg QD with ritonavir at 100 mg QD in a coformulated tablet (19–23).
In conclusion, RAVs in NS3 result in decreased activity of ABT-450 in vitro; however, when ABT-450 exposures are optimized through use of ritonavir for pharmacokinetic enhancement, emergence of these variants is prevented or delayed in vivo. This finding may explain the low rates of virologic breakthrough seen in the phase 3 clinical trials of ABT-450/r, ombitasvir, and dasabuvir with or without RBV, since similarly constructed drug regimens that did not use pharmacokinetic enhancement have been associated with high rates of virologic breakthrough (51, 52). In addition, the lower dosing of ABT-450, facilitated by the boosting effect of ritonavir, may correlate with the low rates of treatment discontinuation and good tolerability seen in these large clinical trials (19–23).
ACKNOWLEDGMENTS
The design, study conduct, and financial support of the M11-602 trial were provided by AbbVie.
T.P.-M., R.T., D.C., I.G., T.D., T.H., L.L., T.R., M.I., T.M., T.N., K.M., Y.S.O., D.K., A.M., and C.C. are current or former Abbott or AbbVie employees and may hold Abbott or AbbVie stock or options. Y.S.O. is an Enanta employee and may hold Enanta stock or options.
We thank the M11-602 study coordinators and nurses, as well as the patients and investigators who participated in the study. We also thank Barbara McGovern for critical reading of the manuscript.
REFERENCES
- 1.Lavanchy D. 2011. Evolving epidemiology of hepatitis C virus. Clin Microbiol Infect 17:107–115. doi: 10.1111/j.1469-0691.2010.03432.x. [DOI] [PubMed] [Google Scholar]
- 2.Mohd Hanafiah K, Groeger J, Flaxman AD, Wiersma ST. 2013. Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. Hepatology 57:1333–1342. doi: 10.1002/hep.26141. [DOI] [PubMed] [Google Scholar]
- 3.Smith DB, Bukh J, Kuiken C, Muerhoff AS, Rice CM, Stapleton JT, Simmonds P. 2014. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment Web resource. Hepatology 59:318–327. doi: 10.1002/hep.26744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zein NN. 2000. Clinical significance of hepatitis C virus genotypes. Clin Microbiol Rev 13:223–235. doi: 10.1128/CMR.13.2.223-235.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bacon BR, Gordon SC, Lawitz E, Marcellin P, Vierling JM, Zeuzem S, Poordad F, Goodman ZD, Sings HL, Boparai N, Burroughs M, Brass CA, Albrecht JK, Esteban R. 2011. Boceprevir for previously treated chronic HCV genotype 1 infection. N Engl J Med 364:1207–1217. doi: 10.1056/NEJMoa1009482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacobson IM, McHutchison JG, Dusheiko G, Di Bisceglie AM, Reddy KR, Bzowej NH, Marcellin P, Muir AJ, Ferenci P, Flisiak R, George J, Rizzetto M, Shouval D, Sola R, Terg RA, Yoshida EM, Adda N, Bengtsson L, Sankoh AJ, Kieffer TL, George S, Kauffman RS, Zeuzem S. 2011. Telaprevir for previously untreated chronic hepatitis C virus infection. N Engl J Med 364:2405–2416. doi: 10.1056/NEJMoa1012912. [DOI] [PubMed] [Google Scholar]
- 7.Poordad F, McCone JJ, Bacon BR, Bruno S, Manns MP, Sulkowski MS, Jacobson IM, Reddy KR, Goodman ZD, Boparai N, DiNubile MJ, Sniukiene V, Brass CA, Albrecht JK, Bronowicki JP. 2011. Boceprevir for untreated chronic HCV genotype 1 infection. N Engl J Med 364:1195–1206. doi: 10.1056/NEJMoa1010494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeuzem S, Andreone P, Pol S, Lawitz E, Diago M, Roberts S, Focaccia R, Younossi Z, Foster GR, Horban A, Ferenci P, Nevens F, Müllhaupt B, Pockros P, Terg R, Shouval D, van Hoek B, Weiland O, Van Heeswijk R, De Meyer S, Luo D, Boogaerts G, Polo R, Picchio G, Beumont M. 2011. Telaprevir for retreatment of HCV infection. N Engl J Med 364:2417–2428. doi: 10.1056/NEJMoa1013086. [DOI] [PubMed] [Google Scholar]
- 9.Jacobson I, Dore G, Foster G, Fried M, Radu M, Rafalskiy V, Moroz I, Craxi A, Peeters M, Lenz O, Ouwerkerk-Mahadevan S, Kalmeijer R, Beumont-Mauviel M. 2013. Simeprevir (TMC435) with peginterferon/ribavirin for chronic HCV genotype-1 infection in treatment-naive parients: results from QUEST-1, a phase III trial. J Hepatol 58:S574. doi: 10.1016/S0168-8278(13)61424-5. [DOI] [Google Scholar]
- 10.Manns MP, Marcellin P, Poordad F, Stanislau Affonso de Araujo E, Buti M, Horsmans Y, Janczewska E, Villamil F, Peeters M, Lenz O, Ouwerkerk-Mahadevan S, Kalmeijer R, Beumont-Mauviel M. 2013. Simeprevir (TMC435) with peginterferon/ribavirin for treatment of chronic HCV genotype-1 infection in treatment-naive patients: results from QUEST-2, a phase III trial. J Hepatol 58:S568. doi: 10.1016/S0168-8278(13)61413-0. [DOI] [Google Scholar]
- 11.Hezode C. 2012. Boceprevir and telaprevir for the treatment of chronic hepatitis C: safety management in clinical practice. Liver Int 32(Suppl 1):S32–S38. doi: 10.1111/j.1478-3231.2011.02707.x. [DOI] [PubMed] [Google Scholar]
- 12.Van Thiel DH, Friedlander L, DeMaria N, Molloy PJ, Kania RJ, Colantoni A. 1998. Treatment of chronic hepatitis C in individuals with pre-existing or confounding neuropsychiatric disease. Hepatogastroenterology 45:328–330. [PubMed] [Google Scholar]
- 13.Kieffer TL, Sarrazin C, Miller JS, Welker MW, Forestier N, Reesink HW, Kwong AD, Zeuzem S. 2007. Telaprevir and pegylated interferon-alpha-2a inhibit wild-type and resistant genotype 1 hepatitis C virus replication in patients. Hepatology 46:631–639. doi: 10.1002/hep.21781. [DOI] [PubMed] [Google Scholar]
- 14.Susser S, Welsch C, Wang Y, Zettler M, Domingues FS, Karey U, Hughes E, Ralston R, Tong X, Herrmann E, Zeuzem S, Sarrazin C. 2009. Characterization of resistance to the protease inhibitor boceprevir in hepatitis C virus-infected patients. Hepatology 50:1709–1718. doi: 10.1002/hep.23192. [DOI] [PubMed] [Google Scholar]
- 15.Lenz O, de Bruijne J, Vijgen L, Verbinnen T, Weegink C, Van Marck H, Vandenbroucke I, Peeters M, Simmen K, Fanning G, Verloes R, Picchio G, Reesink H. 2012. Efficacy of re-treatment with TMC435 as combination therapy in hepatitis C virus-infected patients following TMC435 monotherapy. Gastroenterology 143:1176–1178. doi: 10.1053/j.gastro.2012.07.117. [DOI] [PubMed] [Google Scholar]
- 16.Chatterjee A, Guedj J, Perelson AS. 2012. Mathematical modelling of HCV infection: what can it teach us in the era of direct-acting antiviral agents? Antiviral Ther 17:1171–1182. doi: 10.3851/IMP2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bartenschlager R, Ahlborn-Laake L, Mous J, Jacobsen H. 1994. Kinetic and structural analyses of hepatitis C virus polyprotein processing. J Virol 68:5045–5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krishnan P, Beyer J, Mistry N, Koev G, Reisch T, DeGoey D, Kati W, Campbell A, Williams L, Xie W, Setze C, Molla A, Collins C, Pilot-Matias T. 2015. In vitro and in vivo antiviral activity and resistance profile of ombitasvir, an inhibitor of hepatitis C virus NS5A. Antimicrob Agents Chemother 59:979–987. doi: 10.1128/AAC.04226-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andreone P, Colombo MG, Enejosa JV, Koksal I, Ferenci P, Maieron A, Müllhaupt B, Horsmans Y, Weiland O, Reesink HW, Rodrigues L, Hu YB, Podsadecki T, Bernstein B. 9 May 2014. ABT-450, ritonavir, ombitasvir, and dasabuvir achieves 97% and 100% sustained virologic response with or without ribavirin in treatment-experienced patients with HCV genotype 1b infection. Gastroenterology doi: 10.1053/j.gastro.2014.04.045. [DOI] [PubMed] [Google Scholar]
- 20.Feld JJ, Kowdley KV, Coakley E, Sigal S, Nelson DR, Crawford D, Weiland O, Aguilar H, Xiong J, Pilot-Matias T, DaSilva-Tillmann B, Larsen L, Podsadecki T, Bernstein B. 2014. Treatment of HCV with ABT-450/r-ombitasvir and dasabuvir with ribavirin. N Engl J Med 370:1594–1603. doi: 10.1056/NEJMoa1315722. [DOI] [PubMed] [Google Scholar]
- 21.Ferenci P, Bernstein D, Lalezari J, Cohen D, Luo Y, Cooper C, Tam E, Marinho RT, Tsai N, Nyberg A, Box TD, Younes Z, Enayati P, Green S, Baruch Y, Bhandari BR, Caruntu FA, Sepe T, Chulanov V, Janczewska E, Rizzardini G, Gervain J, Planas R, Moreno C, Hassanein T, Xie W, King M, Podsadecki T, Reddy KR. 2014. ABT-450/r-ombitasvir and dasabuvir with or without ribavirin for HCV. N Engl J Med 370:1983–1992. doi: 10.1056/NEJMoa1402338. [DOI] [PubMed] [Google Scholar]
- 22.Poordad F, Hezode C, Trinh R, Kowdley KV, Zeuzem S, Agarwal K, Shiffman ML, Wedemeyer H, Berg T, Yoshida EM, Forns X, Lovell SS, Da Silva-Tillmann B, Collins CA, Campbell AL, Podsadecki T, Bernstein B. 2014. ABT-450/r-ombitasvir and dasabuvir with ribavirin for hepatitis C with cirrhosis. N Engl J Med 370:1973–1982. doi: 10.1056/NEJMoa1402869. [DOI] [PubMed] [Google Scholar]
- 23.Zeuzem S, Jacobson IM, Baykal T, Marinho RT, Poordad F, Bourliere M, Sulkowski MS, Wedemeyer H, Tam E, Desmond P, Jensen DM, Di Bisceglie AM, Varunok P, Hassanein T, Xiong J, Pilot-Matias T, DaSilva-Tillmann B, Larsen L, Podsadecki T, Bernstein B. 2014. Retreatment of HCV with ABT-450/r-ombitasvir and dasabuvir with ribavirin. N Engl J Med 370:1604–1614. doi: 10.1056/NEJMoa1401561. [DOI] [PubMed] [Google Scholar]
- 24.Kempf DJ, Marsh KC, Kumar G, Rodrigues AD, Denissen JF, McDonald E, Kukulka MJ, Hsu A, Pizzuti D, Plattner JJ, Norbeck DW, Leonard JM. 1997. Pharmacokinetic enhancement of inhibitors of the human immunodeficiency virus protease by coadministation with ritonavir. Antimicrob Agents Chemother 41:654–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Menon RM, Klein CE, Lawal AA, Chiu Y-L, Awni WM, Podsadecki TJ, Nada A, Bernstein BM. 2009. Pharmacokinetics and tolerability of the HCV protease inhibitor ABT-450 following single ascending doses in healthy adult volunteers with and without ritonavir, abstr 57. Abstr HepDART 2009. [Google Scholar]
- 26.Ku Y, McDaniel KF, Chen H-J, Shanley JP, Kempf DJ, Grampovnik DJ, Sun Y, Liu D, Gai Y, Or YS, Wagow SH, Engstrom K, Grieme T, Sheikh A, Mei J. 2010. Preparation of heterocyclic macrocyclic peptides as hepatitis C serine protease inhibitors. PCT WO 2010030359 A2 20100318. [Google Scholar]
- 27.Lohmann V, Korner F, Koch J, Herian U, Theilmann L, Bartenschlager R. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 28.Lohmann V, Hoffmann S, Herian U, Penin F, Bartenschlager R. 2003. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J Virol 77:3007–3019. doi: 10.1128/JVI.77.5.3007-3019.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu L, Pilot-Matias TJ, Stewart KD, Randolph JT, Pithawalla R, He W, Huang PP, Klein LL, Mo H, Molla A. 2004. Mutations conferring resistance to a potent hepatitis C virus serine protease inhibitor in vitro. Antimicrob Agents Chemother 48:2260–2266. doi: 10.1128/AAC.48.6.2260-2266.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kato T, Date T, Miyamoto M, Furusaka A, Tokushige K, Mizokami M, Wakita T. 2003. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 125:1808–1817. doi: 10.1053/j.gastro.2003.09.023. [DOI] [PubMed] [Google Scholar]
- 31.Hopkins S, Scorneaux B, Huang Z, Murray MG, Wring S, Smitley C, Harris R, Erdmann F, Fisher G, Ribeill Y. 2010. A novel nonimmunosuppressive analog of cyclosporin a that exhibits potent inhibition of hepatitis C virus RNA replication in vitro. Antimicrob Agents Chemother 54:660–672. doi: 10.1128/AAC.00660-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Halfman CJ. 1981. Concentrations of binding protein and labeled analyte that are appropriate for measuring at any analyte concentration range in radioimmunoassays. Methods Enzymol 74:481–497. doi: 10.1016/0076-6879(81)74034-5. [DOI] [PubMed] [Google Scholar]
- 33.Pauwels R, Balzarini J, Baba M, Snoeck R, Schols D, Herdewijn P, Desmyter J, De Clercq E. 1988. Rapid and automated tetrazolium-based colorimetric assay for the detection of anti-HIV compounds. J Virol Methods 20:309–321. doi: 10.1016/0166-0934(88)90134-6. [DOI] [PubMed] [Google Scholar]
- 34.Lawitz E, Gaultier I, Poordad F, Cohen DE, Menon R, Larsen LM, Podsadecki TJ, Bernstein B. 2010. Initial antiviral activity of the HCV NS3 protease inhibitor ABT-450 when given with low dose ritonavir as 3-day monotherapy: preliminary results of study M11-602 in genotype-1 (GT1) HCV-infected treatment-naive subjects. Hepatology 52(Suppl 1):1202A. [Google Scholar]
- 35.Lenz O, Vijgen L, Berke JM, Cummings MD, Fevery B, Peeters M, De Smedt G, Moreno C, Picchio G. 2013. Virologic response and characterisation of HCV genotype 2-6 in patients receiving TMC435 monotherapy (study TMC435-C202). J Hepatol 58:445–451. doi: 10.1016/j.jhep.2012.10.028. [DOI] [PubMed] [Google Scholar]
- 36.McPhee F, Sheaffer AK, Friborg J, Hernandez D, Falk P, Zhai G, Levine S, Chaniewski S, Yu F, Barry D, Chen C, Lee MS, Mosure K, Sun LQ, Sinz M, Meanwell NA, Colonno RJ, Knipe J, Scola P. 2012. Preclinical profile and characterization of the hepatitis C virus NS3 protease inhibitor asunaprevir (BMS-650032). Antimicrob Agents Chemother 56:5387–5396. doi: 10.1128/AAC.01186-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berger KL, Lagace L, Triki I, Cartier M, Marquis M, Lawetz C, Bethell R, Scherer J, Kukolj G. 2013. Viral resistance in hepatitis C virus genotype 1-infected patients receiving the NS3 protease inhibitor faldaprevir (BI 201335) in a phase 1b multiple-rising-dose study. Antimicrob Agents Chemother 57:4228–4936. doi: 10.1128/AAC.00822-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lim SR, Qin X, Susser S, Nicholas JB, Lange C, Herrmann E, Hong J, Arfsten A, Hooi L, Bradford W, Nájera I, Smith P, Zeuzem S, Kossen K, Sarrazin C, Seiwert SD. 2012. Virologic escape during danoprevir (ITMN-191/RG7227) monotherapy is hepatitis C virus subtype dependent and associated with R155K substitution. Antimicrob Agents Chemother 56:271–279. doi: 10.1128/AAC.05636-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McPhee F, Friborg J, Levine S, Chen C, Falk P, Yu F, Hernandez D, Lee MS, Chaniewski S, Sheaffer AK, Pasquinelli C. 2012. Resistance analysis of the hepatitis C virus NS3 protease inhibitor asuneprevir. Antimicrob Agents Chemother 56:3670–3681. doi: 10.1128/AAC.00308-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reesink HW, Fanning GC, Farha KA, Weegink C, Van Vliet A, Van 't Klooster G, Lenz O, Aharchi F, Mariën K, Van Remoortere P, de Kock H, Broeckaert F, Meyvisch P, Van Beirendonck E, Simmen K, Verloes R. 2010. Rapid HCV-RNA decline with once daily TMC435: a phase I study in healthy volunteers and hepatitis C patients. Gastroenterology 138:913–921. doi: 10.1053/j.gastro.2009.10.033. [DOI] [PubMed] [Google Scholar]
- 41.Sarrazin C, Kieffer TL, Bartels D, Hanzelka B, Müh U, Welker M, Wincheringer D, Zhou Y, Chu HM, Lin C, Weegink C, Reesink H, Zeuzem S, Kwong AD. 2007. Dynamic hepatitis C virus genotypic and phenotypic changes in patients treated with the protease inhibitor telaprevir. Gastroenterology 132:1767–1777. doi: 10.1053/j.gastro.2007.02.037. [DOI] [PubMed] [Google Scholar]
- 42.Kowdley KS, Lawitz E, Poordad F, Cohen DE, Nelson DR, Zeuzem S, Everson GT, Kwo P, Foster GR, Sulkowsk MS, Xie W, Pilot-Matias T, Liossis G, Larsen L, Khatri A, Podsadecki T, Bernstein B. 2014. Phase 2b trial of interferon-free therapy for hepatitis C virus genotype 1. N Engl J Med 370:222–232. doi: 10.1056/NEJMoa1306227. [DOI] [PubMed] [Google Scholar]
- 43.Kempf DJ, Klein C, Chen H-J, Klein LL, Yeung C, Randolph JT, Lau YY, Chovan LE, Guan Z, Hernandez L, Turner TM, Dandliker PJ, Marsh KC. 2007. Pharmacokinetic enhancement of the hepatitis C virus protease inhibitors VX-950 and SCH 503034 by co-dosing with ritonavir. Antivir Chem Chemother 18:163–167. [DOI] [PubMed] [Google Scholar]
- 44.Moyle GJ, Back D. 2001. Principles and practice of HIV-protease inhibitor pharmacoenhancement. HIV Med 2:105–113. doi: 10.1046/j.1468-1293.2001.00063.x. [DOI] [PubMed] [Google Scholar]
- 45.Shuter J, Sarlo JA, Kanmaz TJ, Rode RA, Zingman BS. 2007. HIV infected patients receiving lopinavir/ritonavir based antiretroviral therapy achieve high rates of virologic suppression despite adherence rates less than 95%. J Acquir Immune Defic Syndr 45:4–7. doi: 10.1097/QAI.0b013e318050d8c2. [DOI] [PubMed] [Google Scholar]
- 46.Shuter J. 2008. Forgiveness of non-adherence to HIV-1 antiretroviral therapy. J Antimicrob Chemother 61:769–773. doi: 10.1093/jac/dkn020. [DOI] [PubMed] [Google Scholar]
- 47.Wensing AMJ, van Maarseveen NM, Nijhuis M. 2010. Fifteen years of HIV protease inhibitors: raising the barrier to resistance. Antiviral Res 85:59–74. doi: 10.1016/j.antiviral.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 48.Hull MW, Montaner JS. 2011. Ritonavir-boosted protease inhibitors in HIV therapy. Ann Med 43:375–388. doi: 10.3109/07853890.2011.572905. [DOI] [PubMed] [Google Scholar]
- 49.Menon R. 2014. ABT-450/ritonavir + ombitasvir + dasabuvir: drug interactions mediated by transporters. 15th International Workshop on Clinical Pharmacology of HIV & Hepatitis Therapy, Washington, DC. [Google Scholar]
- 50.Lawitz E, Poordad F, DeJesus E, Kowdley K, Gaultier I, Cohen D, Xie W, Larsen L, Pilot-Matias T, Menon RM, Podsadecki T, Bernstein B. 2012. ABT-450/ritonavir (ABT-450/r) combined with pegylated interferon alpha-2a/ribavirin after 3-day monotherapy in genotype 1 (GT1) HCV-infected treatment-naïve subjects: 12-week sustained virologic response (SVR12) and safety results. J Hepatol 56:S470. doi: 10.1016/S0168-8278(12)61199-4. [DOI] [Google Scholar]
- 51.Lok AS, Gardiner DF, Hézode C, Lawitz EJ, Bourlière M, Everson GT, Marcellin P, Rodriguez-Torres M, Pol S, Serfaty L, Eley T, Huang SP, Li J, Wind-Rotolo M, Yu F, McPhee F, Grasela DM, Pasquinelli C. 2014. Randomized trial of daclatasvir and asunaprevir with or without PegIFN/RBV for hepatitis C virus genotype 1 null responders. J Hepatol 60:490–499. doi: 10.1016/j.jhep.2013.10.019. [DOI] [PubMed] [Google Scholar]
- 52.Wyles DL, Rodriguez-Torres M, Lawitz E, Shiffman ML, Pol S, Herring RW, Massetto B, Kanwar B, Trenkle JD, Pang PS, Zhu Y, Mo H, Brainard DM, Subramanian GM, McHutchison JG, Habersetzer F, Sulkowski M. 2014. All-oral combination of ledipasvir, vedroprevir, tegobuvir, and ribavirin in treatment-naïve patients with genotype 1 HCV infection. Hepatology 60:56–64. doi: 10.1002/hep.27053. [DOI] [PubMed] [Google Scholar]