Abstract
The translational value of zebrafish high-throughput screens can be improved when more knowledge is available on uptake characteristics of potential drugs. We investigated reference antibiotics and 15 preclinical compounds in a translational zebrafish-rodent screening system for tuberculosis. As a major advance, we have developed a new tool for testing drug uptake in the zebrafish model. This is important, because despite the many applications of assessing drug efficacy in zebrafish research, the current methods for measuring uptake using mass spectrometry do not take into account the possible adherence of drugs to the larval surface. Our approach combines nanoliter sampling from the yolk using a microneedle, followed by mass spectrometric analysis. To date, no single physicochemical property has been identified to accurately predict compound uptake; our method offers a great possibility to monitor how any novel compound behaves within the system. We have correlated the uptake data with high-throughput drug-screening data from Mycobacterium marinum-infected zebrafish larvae. As a result, we present an improved zebrafish larva drug-screening platform which offers new insights into drug efficacy and identifies potential false negatives and drugs that are effective in zebrafish and rodents. We demonstrate that this improved zebrafish drug-screening platform can complement conventional models of in vivo Mycobacterium tuberculosis-infected rodent assays. The detailed comparison of two vertebrate systems, fish and rodent, may give more predictive value for efficacy of drugs in humans.
INTRODUCTION
Studies in zebrafish larvae are used increasingly for modeling human diseases, with the expectation that they will be able to contribute to bridging the gap between in vitro cell-based assays and in vivo mammalian disease models (1–3). Zebrafish larvae have become a popular vertebrate model due to (i) their anatomical, molecular, and genetic similarity to humans, (ii) the availability of a large genetic toolbox, (iii) their easy and low-cost maintenance, (iv) the optical transparency of embryos and larvae, enabling massive phenotype-based screens, (v) very few ethical issues associated with the use of larvae up to the feeding stage, and (vi) the occurrence of many pathological processes resembling those of various human diseases. Among numerous other human diseases, mycobacterial infection caused by Mycobacterium tuberculosis and M. abscessus has been modeled in zebrafish as a potential complement to the currently established in vitro and in vivo models proven to recapitulate the pathology of the human disease progression in zebrafish larvae (4–9). In these systems, the efficacy of a drug can be accurately measured by its effect on the number of bacteria and their presence and survival inside immune cells (10–14). As a result, zebrafish larvae already have yielded significant new insights into the understanding of the pathogenesis of tuberculosis (TB) in humans and contributed to the development of novel strategies for disease treatment (7). Such a gain in understanding is very important, since TB therapy is becoming increasingly difficult with the emergence of new strains of bacteria resistant to currently used antibiotic regimens (multidrug resistant, extensively drug resistant, and totally drug resistant) (15–19). Although new drugs are being discovered, the progress in FDA approval of anti-TB medicines is very limited (20). Moreover, there is also a lack of effective vaccination against TB, pending new approaches and strategies that are under evaluation for further improvement (15). Therefore, the need for more efficient novel drugs, targeting either the pathogen or the host, remains as high and urgent as ever (19, 21). This task can be accomplished only by bringing together academic institutes and pharmaceutical companies with the aim of quickly implementing novel tools that have the potential to accelerate the drug discovery process. As an example of this strategy, GlaxoSmithKline (GSK) recently made publically available the structure and anti-TB properties of a set of 177 potent antitubercular compounds, making samples available to the industrial and academic research communities to stimulate early-stage TB drug discovery activities (22).
Given its previously mentioned attributes, the zebrafish larva potentially can be used at various stages of the drug discovery process, ranging from target identification and lead optimization to preclinical and clinical development (23). The easy availability of large numbers of larvae and their small size makes this model particularly suitable for high-throughput in vivo screening of drugs added to the medium (1, 2, 10, 24–26), significantly reducing material requirements for testing. However, caution must be taken when interpreting the data in the absence of knowledge on drug absorption into the tissues of zebrafish larvae (27).
While the pharmaceutical industry is increasingly gaining confidence in the use of zebrafish larvae in a number of toxicity studies (23, 28–30), progress in the application of zebrafish as a possible alternative or complement to traditional in vitro and in vivo drug efficacy models has been limited (13, 31, 32). Despite the previously mentioned benefits, a possible explanation behind this lack of progress is the limited or altogether missing understanding of the basic pharmacological parameters in zebrafish driving compound efficacy, namely, uptake, distribution, and metabolism (33).
Mass spectrometry (MS) is an analytical technique which is very suitable for the quantification and qualification of small molecules, such as drugs and their metabolites. In the current state-of-the-art zebrafish assays, larvae are treated by adding the compound into their water container, and uptake is assessed by liquid chromatography (LC)-MS analysis of whole-larva lysates (30, 33–35). Here, we demonstrate that the whole-larva lysis method is not always suitable for determining the uptake of drugs. This is due to the capacity of some molecules to persistently adhere to the skin, even after thorough washing, resulting in artificially high background readouts and false-positive results for drug uptake. To remedy this, we present an alternative drug uptake evaluation methodology in zebrafish larvae based on microneedle sampling from the yolk followed by mass spectrometric analysis. Using this novel method, two antitubercular compounds in clinical use, rifampin and moxifloxacin, and 15 preclinical lead compounds developed by GSK were examined. We correlate in vivo uptake levels with the efficacy of the compounds as measured in M. marinum and M. tuberculosis in vitro culture inhibition assays. Furthermore, the data obtained also are correlated with in vivo drug efficacy tests in a murine model of TB infection, as well as with a high-throughput drug screening on M. marinum-infected zebrafish (Fig. 1). The results demonstrate the importance of standardized drug uptake studies in order to be able to understand and correlate the results obtained in zebrafish with other commonly employed efficacy models.
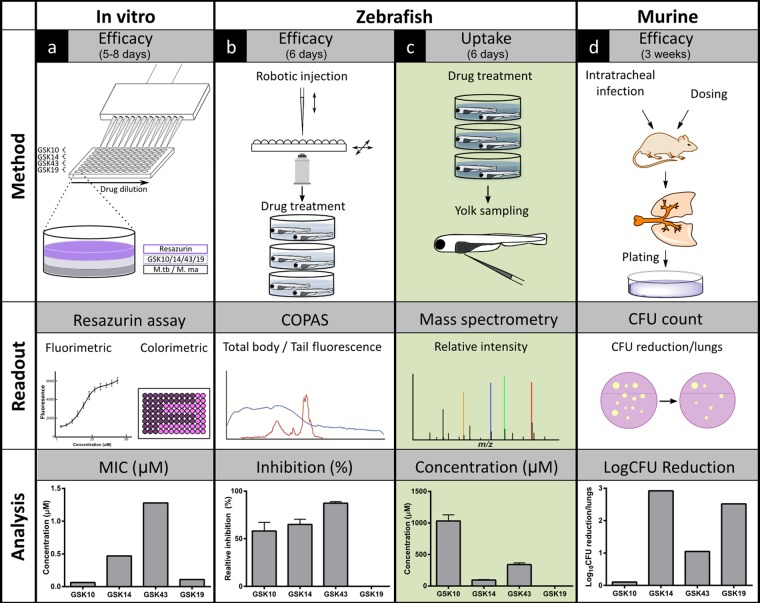
FIG 1.
Scheme of drug efficacy methods using antibacterial activity and uptake assays in the zebrafish model integrated in conventional disease-screening pipeline. Different models are used in our workflow to determine the efficacy of antitubercular compounds (from left to right). (a) Initially, the in vitro efficacy of the tested compounds is determined by their MIC against Mycobacterium marinum and Mycobacterium tuberculosis cultures using fluorimetric and colorimetric readouts from the resazurin assay. (b to d) Biological validation subsequently is performed in in vivo models. (b) First, compound efficacy is screened in M. marinum-infected zebrafish larvae. Embryos are robotically injected, and following compound treatment, the percentage of inhibition is determined using the fluorescence readout of the COPAS system. (c) To unravel whether certain compounds fail to be active in zebrafish larvae due to the lack of antibacterial activity or poor uptake, our microneedle sampling method combined with mass spectrometry is used to assess uptake levels from samples of the yolk. (d) As a gold standard in antitubercular drug development, compound efficacy is established by determining the rate of the CFU reduction in the lungs of M. tuberculosis-infected rodents. After setting (arbitrary) cutoffs in all models, compounds could be categorized into positive or negative groups. By the comparison of these groups along the pipeline, our improved zebrafish platform may give a more predictive value for human efficacy of drugs. Cycle times are indicated for each model.
MATERIALS AND METHODS
Animal experiment ethics statement.
All animal studies were ethically reviewed and carried out in accordance with European Directive 2010/63/EU and the GSK Policy on the Care, Welfare, and Treatment of Animals.
Zebrafish husbandry and compound treatment.
Zebrafish of the AB/TL wild-type strain were handled in compliance with the local animal welfare regulations and maintained according to standard protocols (zfin.org). Embryos were collected from family crosses and grown at 28°C in egg water (60 μg/ml Instant Ocean sea salts; Sera Marin) in the dark. At 3 days postfertilization (dpf), 20 to 30 larvae were transferred to small, 35- by 10-mm petri dishes, each containing 3 to 4 ml of egg water supplemented with the following compounds: rifampin (RIF; Sigma-Aldrich) and moxifloxacin (MOX; Santa Cruz), each at 150 μM, and GSK compounds (GlaxoSmithKline Pharmaceuticals) at 10 μM. Dimethyl sulfoxide (DMSO) at 0.1% was used as a control. Larvae were exposed to the compounds for 17 and 40 h. Controls were exposed for a few seconds (T = 0 h). Compounds were administered separately to the larvae or in combinations. Compounds were refreshed every day.
Assessing compound quantities in zebrafish larvae with lysis method.
Larvae were transferred to 2-ml microcentrifuge tubes one by one; excessive compound was removed, washed three times briefly with 1 ml 50% methanol in water, and then prepared for lysis. Lysis was performed based on snap-freezing in liquid nitrogen followed by sonication, as previously described (36).
Microneedle sampling from the yolk of zebrafish larvae.
Before taking samples, larvae were transferred to 2-ml microcentrifuge tubes and then washed three times briefly with 1 ml 50% methanol in water one by one. After washing, one larva was placed on a 1% agarose plate and excessive methanol was removed. Under a stereomicroscope at 20× magnification, larvae were punctured in the yolk with a microneedle (manually pulled borosilicate glass capillary; Harvard Apparatus) placed in the capillary holder of a CellTram Oil microinjector (Eppendorf) mounted onto a micromanipulator workstation. Vacuum was generated manually via a rotating knob, and the content of the yolk was extracted. When most of the content was removed from the yolk, the suction was balanced and stopped. To recover the sample from the microneedle into a microcentrifuge tube, pressure was generated by turning the knob in the opposite direction.
Determination of the sample volume.
Since individual larvae can differ in their yolk size and volume and the manually pulled microneedles used for sampling were not identical in diameter, the sampled volume was not always the same per larva. In order to quantify the drugs in the sample, the volume has to be known. As a consequence, images of every sample were taken, including the microneedle containing the sample. The volume of the samples was determined by modeling the needle as a cone and measuring the diameter and the length on the image, using the scale bar as a reference. Since the angle for the capillary holder was always adjusted to a 45° angle, we also adjusted the model for this projection. This way the results were normalized with the corresponding sample volume. Sample volumes typically were around 50 nl, varying between 20 and 200 nl.
Sample preparation.
After recovering the sample from the microneedle into the microcentrifuge tube, the yolk proteins were precipitated using a 75% methanol and 25% water solution. Methanol-water solution internal standards (IS) also were added to the sample: rifabutin and levofloxacin were an IS for rifampin and moxifloxacin, respectively, and ampicillin was an IS for the GSK compounds. For the microneedle samples, an IS concentration of 20 nM was used. For the whole-larva samples, the spiked IS resulted in a concentration of 250 nM. After vortexing for 2 min, the precipitated proteins were spun down by centrifugation at 16.1 RCF (relative centrifugal force) for 5 min at 0°C. The supernatant was used for LC-MS analysis.
Mass spectrometry.
A Surveyor Plus ultraperformance liquid chromatography (UPLC) system was used, and a 15-μl sample was injected in an Acquity C18 T3 column (2.1 by 100 mm, 1.8 μm). Mobile phase A was 99% H2O and 1% acetonitrile with 0.3% formic acid. Mobile phase B was 90% acetonitrile and 10% H2O with 0.3% formic acid, and the flow rate was 500 μl/min. An LTQ Orbitrap XL (Thermo Fisher Scientific) was used for detection of the ions in positive electrospray mode. The electrospray voltage was 4.5 kV. The capillary temperature was 375°C, and the capillary and tube lens voltage was 29 V and 120 V, respectively. The sheath gas flow rate and auxiliary gas flow rate was 35 and 10 U, respectively. Resolution was set to 7,500 in order to reduce the scan time and increase the number of data points. For the quantification of the samples, an academic calibration curve was constructed within every batch.
M. marinum infections in zebrafish in vivo model.
Infection experiments were performed on zebrafish embryos at early developmental stages (up to 1,024-cell stage) injected with 40 CFU Mycobacterium marinum M strain into the yolk using an automated robotic injection system as described in Carvalho et al. (10). As a control, an equal volume of carrier solution was injected. After injection, embryos were incubated at 28°C in egg water in a petri dish. Starting at 3 dpf, 200 larvae were treated with the following compounds until 5 dpf by adding the compounds into egg water: 200 μM rifampin (Sigma-Aldrich), 10 μM GSK compounds, and DMSO (0.1%) as a negative control. Water with compounds was refreshed once daily. Bacterial infection was quantified using a complex object parametric analyzer and sorter (COPAS) XL (Union Biometrica) as described previously (10). Fluorescence in the posterior half of the larvae was determined using a custom Perl script that analyzes the COPAS extinction profile to determine the posterior half and then sums the fluorescent values of all data points for that half.
M. tuberculosis H37Rv inhibition assay.
The measurement of the MIC was performed as described in Ballell et al. (22). For each compound, measurements were performed in 96-well flat-bottom polystyrene microtiter plates. Ten 2-fold drug dilutions in neat DMSO starting at 50 mM were performed. These drug solutions (5 μl) were added to 95 μl Middlebrook 7H9 medium (lines A to H, rows 1 to 10 of the plate layout). Isoniazid was used as a positive control. Eight 2-fold dilutions of isoniazid starting at 160 μg ml−1 were prepared, and this control curve (5 μl) was added to 95 μl Middlebrook 7H9 medium (row 11, lines A to H). Neat DMSO (5 μl) was added to row 12 (growth and blank controls). The inoculum was standardized to ∼1 × 107 CFU ml−1 and diluted 1:100 in Middlebrook 7H9 broth (Middlebrook ADC enrichment; a dehydrated culture medium which supports growth of mycobacterial species; catalog no. 211887; available from Becton-Dickinson) to produce the final inoculum of the H37Rv strain (ATCC 25618). This inoculum (100 μl) was added to the entire plate except for wells G-12 and H-12 (blank controls). All plates were placed in a sealed box to prevent drying out of the peripheral wells and were incubated at 37°C without shaking for 6 days. A resazurin solution was prepared by dissolving one tablet of resazurin (resazurin tablets for milk testing; catalog no. 330884Y; VWR International Ltd.) in 30 ml sterile phosphate-buffered saline (PBS). Twenty-five μl of this solution was added to each well. Fluorescence was measured (excitation wavelength [λex], 530 nm; emission wavelength [λem], 590 nm; Spectramax M5; Molecular Devices) after 48 h to determine the MIC value.
M. marinum inhibition assays.
The measurement of the MIC was performed as described above for the M. tuberculosis H37Rv inhibition assay, with minor changes. Each compound measurement was performed in 96-well flat-bottom polystyrene microtiter plates. Ten 2-fold drug dilutions in neat DMSO starting at 50 μM were performed. These drug solutions (2 μl) were added to 98 μl Middlebrook 7H9 medium (lines A to H, rows 1 to 10 of the plate layout). Isoniazid was used as a positive control. Eight 2-fold dilutions of isoniazid starting at 100 μM were prepared, and this control curve (2 μl) was added to 98 μl Middlebrook 7H9 medium (row 11, lines A to H). Neat DMSO (2 μl) was added to row 12 (growth and blank controls). The inoculum was standardized to ∼2 × 105 CFU ml−1 and diluted 1:1 in Middlebrook 7H9 broth (Middlebrook ADC enrichment) to produce the final inoculum. This inoculum (100 μl) was added to the entire plate, except for wells C-12 and D-12 (blank controls). All plates were placed in a sealed box to prevent drying out of the peripheral wells and were incubated at 28°C without shaking for 3 days. A resazurin solution was prepared by dissolving one tablet of resazurin (resazurin tablets for milk testing; VWR International Ltd.) in 30 ml sterile PBS. Of this solution, 25 μl was added to each well. Fluorescence was measured (λex, 535 nm; λem, 590 nm; Tecan Infinite 200Pro) after 48 h to determine the MIC value.
M. tuberculosis infections in mouse in vivo model.
Infections of mice with Mycobacterium tuberculosis was initiated by nonsurgical intratracheal instillation of M. tuberculosis H37Rv as previously described by Rullas et al. (31). In brief, 8- to 10-week-old female mice were anesthetized with 3% isoflurane and intubated with a metal probe (catalog number 27134; Unimed SA, Lausanne, Switzerland). The inoculum (105 CFU/mouse suspended in 50 μl of phosphate-buffered saline) was put into the probe and delivered through forced inhalation with a syringe. Treatment was started 24 h after infection, to allow for phagocytosis of instilled bacteria, and lasted for up to 7 days postinfection (dpi). Finally, an additional 24 h was allowed for clearance of compounds before organ harvesting. To measure infection burden in lungs, all lobes were aseptically removed and homogenized. The homogenates were supplemented with 5% glycerol and stored frozen (−80°C) until plating. After 14 days of culture, colonies were counted using an automatic colony counter (aCOLyte-Supercount; Synoptics Ltd., Cambridge, United Kingdom) and confirmed by visual inspection to correct potential misreadings. Bacterial growth of about 2 logs over the initial inoculum was determined to be a level that would provide enough dynamic range to detect statistically significant growth inhibition.
Statistical analysis.
COPAS data were analyzed (Prism 4.0; GraphPad Software) using nonparametric, two-tailed Wilcoxon signed-rank tests, and the uptake data were analyzed using unpaired, two-tailed t tests. P values shown are P < 0.05 (*), P < 0.01 (**), and P < 0.001 (***).
RESULTS
Assessing rifampin and moxifloxacin quantities in zebrafish by lysis of whole larvae.
We have recently reported an automated high-throughput drug-screening protocol using Mycobacterium marinum-infected zebrafish larvae (10). To better understand the efficacy of antituberculosis compounds used in this test system, we performed uptake experiments using mass spectrometry. This improved zebrafish drug-screening platform then could be integrated into the conventional disease-screening pipeline and give more predictive value for human efficacy of antitubercular compounds (Fig. 1).
To assess the quantity of drugs present in zebrafish, larvae were exposed to rifampin, a first-line anti-TB drug used as a positive control in our drug-screening experiments (10, 24), as well as to moxifloxacin, a gyrase inhibitor currently under exploration for novel antitubercular regimen development to shorten the duration of TB treatment (37).
First, we measured the quantities of the aforementioned compounds using the previously described whole-larva lysis method followed by LC-MS analysis (30). After different exposure times (17 and 40 h), larvae were washed with 50% methanol and lysed individually to determine drug concentration using LC-MS (Fig. 2a). A steady increase was detectable in the level of moxifloxacin during the treatment period, from 3.1 pmol/larva at 17 h of exposure and increasing further to 13.6 pmol/larva at 40 h of exposure. On the other hand, rifampin seemed to have reached a steady-state level already at 17 h of exposure with 6.2 pmol/larva and 6.6 pmol/larva at the 40-h exposure time. As a control, we exposed the larvae to the drugs for only a few seconds, washed them immediately with 50% methanol, and lysed them for analysis (T = 0 h). Surprisingly, the presence of both drugs was detected even after these few seconds of exposure, at which time rifampin was measured at 4.6 pmol/larva and moxifloxacin at 0.8 pmol/larva. Washing after drug exposure also was performed using other solvents (embryo medium, 5% and 50% ethanol), which resulted in the same quantities being detected as those after the 50% methanol wash (data not shown). The detection of rifampin and moxifloxacin in the T = 0 h control lysates presumably was due to the tendency of these drugs to adhere to the skin of the larvae. These results suggest that the whole-larva lysis method is not suitable for monitoring drug uptake.
FIG 2.
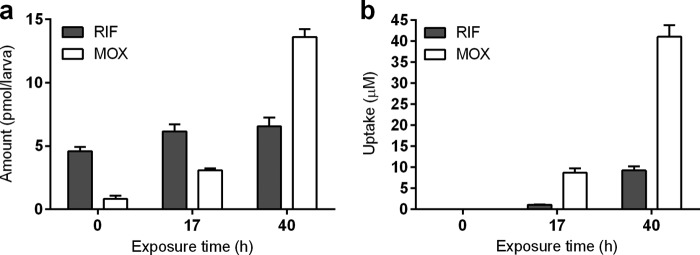
Rifampin and moxifloxacin quantities measured by the whole-zebrafish-larva lysis method (a) and microneedle sampling method from the yolk (b). Three-day-old zebrafish larvae were exposed to a combination of rifampin (RIF; gray bars) and moxifloxacin (MOX; white bars) dosed at a 150 μM concentration for 0, 17, and 40 h (n = 10). Note that as a control, at the zero time point (T = 0 h) the zebrafish larvae were exposed to the compounds (at the same dose) for only a few seconds. This short exposure was followed by three consecutive washing steps with 50% methanol. In the readout, after lysis of the whole larvae the presence of compounds is detected even at T = 0 h (a); however, via our microneedle method the compounds are undetectable in the yolk at T = 0 h (b). All data are expressed as the means ± standard errors of the means.
Determining uptake of rifampin and moxifloxacin in the yolk of zebrafish larvae using a microneedle sampling method.
Since the larva lysis method cannot be used for correctly monitoring the uptake of certain drugs, we aimed to overcome this issue by taking samples (in the nanoliter range) from the yolk of zebrafish larvae using a microneedle attached to an Eppendorf CellTram oil instrument. As detailed in Materials and Methods, we also have developed a method to determine the sample volume needed for quantitative mass spectrometric analysis. This method was used to analyze samples in the same time course experiment as that applied in the lysis method (17 and 40 h of exposure). We observed a gradual increase in the uptake level of both rifampin (17 h of exposure at 1.1 μM, 40 h of exposure at 9.2 μM) and moxifloxacin (17 h of exposure at 8.7 μM, 40 h of exposure at 41 μM) (Fig. 2b). Neither rifampin nor moxifloxacin was detected at the control time point (T = 0 h), indicating that our new method eliminates the issue of drug adherence. The results show that our method can measure drug uptake with high accuracy and sensitivity from samples in the nanoliter range and also is suitable for compounds that persistently stick to the surface of larvae, such as rifampin.
Uptake of preclinical antituberculosis compounds and correlation with their efficacy on bacterial burden in Mycobacterium marinum-infected zebrafish larvae in vivo system.
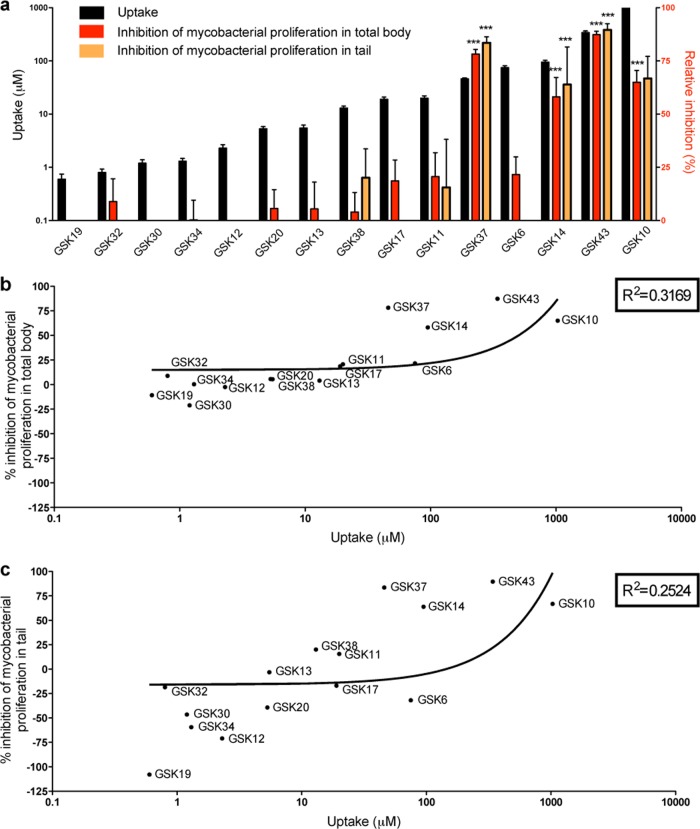
In order to correlate the uptake of a drug with its in vivo effect on microbial infection using drugs that were not previously tested in our zebrafish model, we performed an anti-TB drug screening on M. marinum-infected zebrafish larvae using 15 preclinical compounds provided by GSK. These compounds were prescreened in vitro for their antibacterial activity on M. tuberculosis and M. marinum cultures and showed a large gradient of activities (see Table S1 in the supplemental material). For the in vivo test, zebrafish embryos were robotically injected with fluorescently labeled bacteria at early stages of development, and hatched larvae were treated with compounds from 3 dpi by following the protocol as published by Carvalho et al. (10). The antibacterial efficacy of the compounds was assessed by monitoring the fluorescence intensity correlating with bacterial burden present either in the total body or only in the tail region of 5-day-old zebrafish larvae using COPAS analysis. The efficacy was expressed as the percentage of inhibition of mycobacterial proliferation in the total body and the tail region relative to that of DMSO-treated control groups (Fig. 3). The tail region is representative of a part of the body where mycobacteria are enclosed mainly by immune cells leading to granulomas; therefore, it presents a good measure for disseminated disease (10, 38). The tail measurements compared to the total body are an indicator for potential problems with drug distribution. Compared to the control, we identified compounds reducing bacterial load significantly and others that had moderate or no significant effect on infection (Fig. 3). The results show that at least 3 compounds were significantly active in reducing bacterial burden both in the total body and the tail region. In order to better understand the reason for a compound failing to lower the bacterial load in the zebrafish, we determined GSK compound uptake levels. We tested the uptake of the 15 compounds using the whole-larva lysis method (see Table S2). These experiments showed an adherence to the skin of 8 of these compounds, again making this method unsuitable for correctly measuring the uptake for several compounds. Therefore, samples were taken from yolk of larvae at 5 dpf after 40 h of treatment, and the uptake was measured using the microneedle sampling method (Fig. 3a). The comparison revealed a correlation between the uptake and the antibacterial efficacy of compounds in both the total body and the tail region (Fig. 3a, b, and c).
FIG 3.
Correlation between the uptake level of preclinical antitubercular compounds and their efficacy in M. marinum zebrafish infection model. (a) Uptake levels of 15 preclinical antitubercular GSK compounds were measured from samples taken from the yolk of 5-day-old zebrafish larvae after 40 h of exposure at 10 μM concentration (black bars) (n = 10). The efficacy of the compounds was assessed by monitoring fluorescent bacterial burden in the total body (red bars) or tail region (orange bars) of larvae at 5 days p.i. using the COPAS system after 40 h of treatment (n = 200). Efficacy is expressed as a percentage of inhibition of mycobacterial proliferation relative to the level for DMSO-treated control groups. The bar graphs depict the correlation between the uptake and the relative inhibition, both in the total body and the tail region. Significance in inhibition is indicated with asterisks (***, P < 0.001). (b and c) Correlation between the efficacy in the total body (b) or in the tail (c) and the uptake of the GSK compounds. The efficacy of several compounds shown in panels b and c is below detection limits, as shown in panel a.
Correlation between drug uptake levels and physicochemical properties.
It is important to determine if the uptake level of a compound can be correlated with its physicochemical properties; if such a relationship was identified, it would allow the effective prediction of the uptake of compounds in this model. The compounds used in this study were a selection from three distinct chemical series which represent three separate classes of compounds with very different chemical properties. We compared a number of common metrics used in predictive strategies to the uptake levels measured in our assays. In our case, no relationship was found for the majority of the metrics examined (molecular weight, molecular volume, number of rotatable bonds, hydrogen bond acceptors/donors, number of sp3 carbons, number of aromatic rings, number of Lipinski H-bond acceptors/donors, and polar surface area; see Fig. S1 in the supplemental material). One metric that appeared to have an effect on the measured uptake was chromlogD, which represents the lipophilicity of a compound (Fig. 4). This experimentally determined property is the chromatographic hydrophobicity index (CHI) (39), a retention time of the compound at pH 7.4 on a fast-gradient reverse-phase high-performance liquid chromatography (HPLC) column, modified by a conversion factor which has been shown to be more accurate than classical octanol-water partitioning (40). This perhaps is unsurprising, given the lipophilic nature of the yolk of the larvae. As a result, it may offer the potential for future predictive models. This potential can be tested by the generation of a larger data set from a structurally diverse compound library. Our microneedle sampling method gives valuable additional information on the transportability of particular drugs through epithelial layers and could lead to better theoretical models for use in combination with physicochemical predictions.
FIG 4.
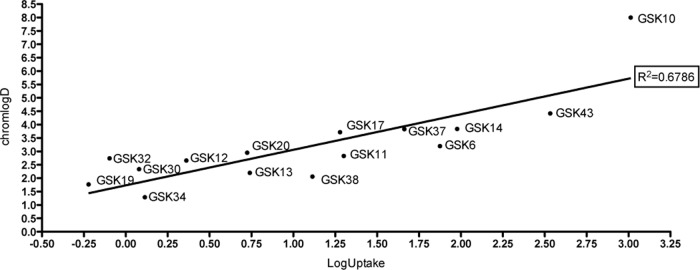
Correlation between uptake levels and compound hydrophobicity. chromlogD is a representation of hydrophobicity; therefore, it represents the solubility/lipophilicity of a compound. To examine the relationship between chromlogD of the compounds and their uptake, chromlogD is plotted against uptake levels measured in 5-day-old zebrafish larvae after 40 h of exposure.
Comparison of uptake levels with in vitro and in vivo antibacterial efficacy models used in our pipeline of anti-TB drug discovery.
We aimed to elucidate if the uptake and antibacterial efficacy results obtained from the zebrafish model translate to other models commonly used in the drug development pipeline. For this reason, we first compared zebrafish data to in vitro M. marinum antibacterial efficacy models.
To determine if there is correlation between uptake and the antibacterial activities, first we normalized the data for uptake with the MIC of compounds against M. marinum (Fig. 5). This log(uptake/MIC) ratio identifies which compounds are present in the larvae at concentrations in excess of the MIC value that is necessary to evoke the minimum inhibition of bacterial growth. A compound with a log(uptake/MIC) value of <0 is not taken up sufficiently to elicit a strong inhibitory effect on the growth of the bacteria. Values of >0 indicate compounds where the measured uptake concentrations exceed the MIC value. Using these data, a plot of the log(uptake/MIC) against the observed percentage of inhibition in the zebrafish assay was produced (Fig. 5). As expected, none of the 3 compounds for which the log(uptake/MIC) was below zero significantly reduced the bacterial burden in the entire body in zebrafish (GSK12, GSK13, and GSK19). However, from the 12 compounds which were present in concentrations well above their MIC values, only 4 were active (indicated with green color in Fig. 5) (GSK 10, GSK14, GSK37, and GSK43).
FIG 5.
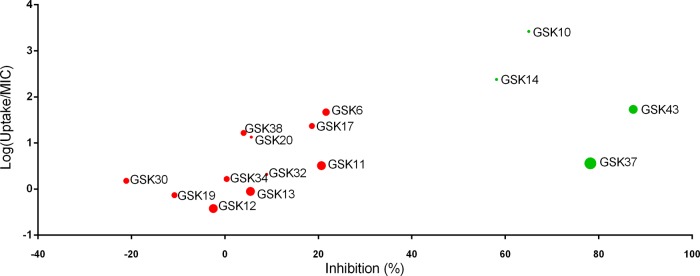
Correlation between uptake, in vitro MIC, and in vivo zebrafish antibacterial activities of compounds. The measured uptake data in zebrafish larvae were normalized with the in vitro MIC of compounds against M. marinum [log(uptake/MIC)]. This ratio is plotted against the observed percentage of inhibition in the zebrafish M. marinum infection assay. The size of the circles indicates their MIC values, meaning smaller circles correspond to lower MIC values with higher potency. Green color represents compounds significantly reducing bacterial burden in the zebrafish infection model, considered active compounds, while red color indicates inactive compounds.
Furthermore, we extended our comparison with in vitro M. tuberculosis antibacterial efficacy data as well as with in vivo M. tuberculosis-infected mouse efficacy data, with the latter being a gold standard in antituberculosis drug development (Fig. 1 and Table 1). In the acute mouse model, the inactive compounds are defined as compounds that produced less than a 2 log reduction in CFU count (31) (see Table S3 in the supplemental material). Conversely, active compounds are those that produced a greater than 2 log reduction in the CFU count in the acute mouse model. Of the 4 active compounds identified via the zebrafish screening, only 2 (GSK14 and GSK37) are considered active in mouse experiments, whereas the other 2 compounds (GSK10 and GSK43) were inactive in the mouse model (Table 1). Of the 11 compounds detected as inactive in the zebrafish assay, 5 were efficacious in mouse (GSK11, GSK17, GSK19, GSK20, and GSK32) and 6 proved to be inactive in mouse (GSK6, GSK12, GSK13, GSK30, GSK34, and GSK38), showing that the in vivo models yield very different results from the in vitro MIC data.
TABLE 1.
Comparison of zebrafish and mouse efficacy data
| Compound | Efficacy in: |
|
|---|---|---|
| Zebrafish | Mouse | |
| GSK14 | + | + |
| GSK37 | + | + |
| GSK10 | + | − |
| GSK43 | + | − |
| GSK11 | − | + |
| GSK17 | − | + |
| GSK19 | − | + |
| GSK20 | − | + |
| GSK32 | − | + |
| GSK6 | − | − |
| GSK12 | − | − |
| GSK13 | − | − |
| GSK30 | − | − |
| GSK34 | − | − |
| GSK38 | − | − |
DISCUSSION
Zebrafish larval screening systems have increasingly been used for testing the effects of drugs. Especially in the context of drug screening against tuberculosis, current methodologies based on bacterial load reduction are accurate and can be automated. In the case of antibiotic screens, this has led to valuable information on the efficacy of existing and new antibiotics using M. marinum infection as a test system (10, 13, 14, 32). However, the reasons behind the failure of some compounds to lower bacterial load in M. marinum-infected zebrafish larvae were unclear, and we wanted to test whether this was caused by poor compound uptake. To date, the mass spectrometric methods currently in use fail to correctly monitor uptake levels in zebrafish. In order to address this problem, we decided to explore an alternative methodology that could measure the uptake of compounds from the yolk of zebrafish larvae by microneedle sampling followed by mass spectrometric analysis. The results show that microneedle sampling of volumes in the nanoliter range from the yolk allows accurate and sensitive analysis of the uptake of drugs by individual larvae.
Our method is based on the assumption that taking samples from the yolk is relevant for predicting limitations of drug efficacy in the rest of the body of zebrafish larvae. This assumption is supported by measured quantities in the yolk and whole-larva lysis experiments for moxifloxacin and the 15 GSK compounds (see Fig. S2 and S3 in the supplemental material). We have compared antimicrobial efficacy in the entire body with that in the tail region in order to check whether spreading of the tested drugs through the body presents a problem (Fig. 3). These results do not show any indication that the effect of the 15 tested compounds is limited by distribution through the body; therefore, they indicate that the yolk is a representative location for measuring drug uptake. However, the concentration measured in the yolk cannot predict concentrations in other tissues. Physicochemical properties of compounds have a great impact on the pharmacokinetics. Caution must be taken when using hydrophobic compounds, as they may accumulate in the yolk due to its lipid-rich environment. Moreover, bioavailability throughout the body can be limited by binding of the compounds to proteins in the yolk. Although several of the tested antitubercular compounds are highly effective in the entire body of the fish, further studies on pharmacokinetic modeling in zebrafish will be needed to understand the importance of differences in local concentrations of antibiotics for combating infectious diseases in an applied medical setting.
Our results show that there is a wide variety of uptake of the various tested compounds. As expected, only compounds that reach uptake levels that are greater than their MICs are active in our antimicrobial test system. In some cases, uptake was shown to be a limiting factor for reaching the MIC as determined in vitro (Fig. 5). The fact that this outcome could not be predicted based solely on physicochemical properties, like hydrophobicity or mass, shows the importance of our test system. At least in one case (compound GSK19), it is clear that the difference from the effect in the rodent test system can be explained by problems with uptake by the zebrafish larva (Fig. 1). By explaining false-negative test results from large screens, our microsampling method opens new possibilities for drug screening using zebrafish larvae in a preclinical test setup. In the future, our screening pipeline also can be the basis for in-depth studies of the mechanisms behind this and other false-negative test results in zebrafish. It is also interesting to study why some drugs that did show high efficacy in the zebrafish larva model were inactive in the mouse model. This could be examined, for instance, by testing other drug administration methods, e.g., drugs can injected directly inside the body or added to the food instead of the medium. This can show whether the negative results in the rodent test model are caused by degradation of drugs added to the food in the acidic environment of the stomach.
In addition to highlighting the importance of identifying potential false-negative and -positive test results, our results show that some drugs that come out as most positive in the combined uptake and efficacy zebrafish test system also are active in the in vivo rodent model. This is of significance, since zebrafish larvae do represent a very different test system compared to rodents with respect to, for instance, (i) the method of application (in the medium versus application in the food in the rodent system), (ii) metabolic rate of these test systems (where zebrafish present a metabolic rate that is much slower than that of rodents based on the fact that zebrafish is an ectothermic model organism and mice are endothermic organisms that are chronically challenged by cold stress under laboratory conditions [41]), and (iii) different levels of genetic polymorphisms (the zebrafish strains are wild types with very high numbers of polymorphisms, whereas the mouse strains are highly inbred and devoid of polymorphisms). Therefore, we believe that the zebrafish model provides added value to the rodent system, since, in comparisons with the human population, test models with a larger number of polymorphisms and a lower metabolic rate are of relevance. From a broader perspective, the observation that compounds that are active in two highly different test systems (in this case a fish versus a rodent) could provide additional confidence that the compound also will provide high efficacy in human subjects with various background genotypes. Some of the compounds tested in this study (such as GSK14, which was shown to have good uptake and high efficacy in the zebrafish system) are members of series of compounds which have progressed to extensive medicinal chemistry programs.
Therefore, we anticipate that the novel uptake measurement technique presented here makes important inroads into our understanding of zebrafish infection model data and opens up new opportunities for zebrafish larval systems to be more widely employed in the field of pharmacology. It should be noted that our uptake method also is highly applicable for toxicity testing in zebrafish larval screening systems, and in this case it also will be of great added value to rodent toxicity test systems. We hope that our results will stimulate future detailed follow-up pharmacokinetic studies in this model system. In conclusion, our results are a good starting point to undertake more detailed pharmacokinetic/pharmacodynamic (PK/PD) studies together with studies of reference compounds. PK/PD studies can help improve the selection procedure during drug discovery, reducing high attrition rates in drug development and reducing costs in future experiments. This eventually can lead to the full acceptance of the zebrafish as a model in pharmacological screening pipelines in industry.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jan de Sonneville (Life Science Methods BV) for support with the automated microinjection system and Rico Bongaarts and Angela Comas (Union Biometrica) for help and advice with COPAS analyses. We thank Peter Racz (ZF-screens B.V.), Rubén Marín Juez (ZF-screens B.V.), and Wouter Veneman (Leiden University) for helpful discussions. We thank Davy de Wit and Ulrike Nehrdich for fish caretaking. From GSK-DDW we thank the Therapeutic Efficacy group for efficacy studies, Antonio Martínez and team for essential animal laboratory upkeep and maintenance, and Esther Pérez-Herrán and Eva López-Román for in vitro biology. In addition, we thank Joaquin Rullas for helpful discussions.
This work was supported by the Leiden University Fund (LUF; A.O.). Additional support was obtained from the EU project ZF-Health (FP7-Health-2009-242048; H.P.S and A.H.M.). R.J.R. was financed by the Netherlands Metabolomics Centre (NMC), which is a part of The Netherlands Genomics Initiative/Netherlands Organization for Scientific Research. F.C. was supported by a Marie Curie fellowship of the European 7th Framework Initial Training Network FishForPharma (PITG-GA-2011-289209). H.J.J, S.J.R., and R.P.D. received funding from the Innovative Medicines Initiative Joint Undertaking under grant agreement no. 115337, resources of which are composed of financial contributions from the European Union's Seventh Framework Programme (FP7/2007-2013) and EFPIA companies in-kind contribution. Some of the research leading to these results received funding from the European Union's Seventh Framework Programme for research, technological development, and demonstration under grant agreement no. 261378. The COPAS system acquisition was supported in part by the Division for Earth and Life Sciences (ALW) with financial aid from the Netherlands Organization for Scientific Research (NWO; 834.10.004).
We have no competing financial interests to declare.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.03588-14.
REFERENCES
- 1.Lieschke GJ, Currie PD. 2007. Animal models of human disease: zebrafish swim into view. Nat Rev Genet 8:353–367. doi: 10.1038/nrg2091. [DOI] [PubMed] [Google Scholar]
- 2.Ali S, Champagne DL, Spaink HP, Richardson MK. 2011. Zebrafish embryos and larvae: a new generation of disease models and drug screens. Birth Defects Res C Embryo Today 93:115–133. doi: 10.1002/bdrc.20206. [DOI] [PubMed] [Google Scholar]
- 3.Zon LI, Peterson RT. 2005. In vivo drug discovery in the zebrafish. Nat Rev Drug Discov 4:35–44. doi: 10.1038/nrd1606. [DOI] [PubMed] [Google Scholar]
- 4.Meijer AH, Spaink HP. 2011. Host-pathogen interactions made transparent with the zebrafish model. Curr Drug Targets 12:1000–1017. doi: 10.2174/138945011795677809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flynn JL. 2006. Lessons from experimental Mycobacterium tuberculosis infections. Microbes Infect 8:1179–1188. doi: 10.1016/j.micinf.2005.10.033. [DOI] [PubMed] [Google Scholar]
- 6.O'Toole R. 2010. Experimental models used to study human tuberculosis. In Laskin A, Gadd G, Sariaslani S (ed), Advances in applied microbiology, 1st ed. Elsevier Inc, New York, NY. [DOI] [PubMed] [Google Scholar]
- 7.Berg RD, Ramakrishnan L. 2012. Insights into tuberculosis from the zebrafish model. Trends Mol Med 18:689–690. doi: 10.1016/j.molmed.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 8.Bernut A, Herrmann J-L, Kissa K, Dubremetz J-F, Gaillard J-L, Lutfalla G, Kremer L. 2014. Mycobacterium abscessus cording prevents phagocytosis and promotes abscess formation. Proc Natl Acad Sci U S A 111:E943–E952. doi: 10.1073/pnas.1321390111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernut A, Le Moigne V, Lesne T, Lutfalla G, Herrmann J-L, Kremer L. 2014. In vivo assessment of drug efficacy against Mycobacterium abscessus using the embryonic zebrafish test system. Antimicrob Agents Chemother 58:4054–4063. doi: 10.1128/AAC.00142-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carvalho R, de Sonneville J, Stockhammer OW, Savage NDL, Veneman WJ, Ottenhoff THM, Dirks RP, Meijer AH, Spaink HP. 2011. A high-throughput screen for tuberculosis progression. PLoS One 6:e16779. doi: 10.1371/journal.pone.0016779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cui C, Benard EL, Kanwal Z, Stockhammer OW, van der Vaart M, Zakrzewska A, Spaink HP, Meijer AH. 2011. Infectious disease modeling and innate immune function in zebrafish embryos. Methods Cell Biol 105:273–308. doi: 10.1016/B978-0-12-381320-6.00012-6. [DOI] [PubMed] [Google Scholar]
- 12.Takaki K, Davis JM, Winglee K, Ramakrishnan L. 2013. Evaluation of the pathogenesis and treatment of Mycobacterium marinum infection in zebrafish. Nat Protoc 8:1114–1124. doi: 10.1038/nprot.2013.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takaki K, Cosma CL, Troll MA, Ramakrishnan L. 2012. An in vivo platform for rapid high-throughput antitubercular drug discovery. Cell Rep 2:175–184. doi: 10.1016/j.celrep.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adams KN, Takaki K, Connolly LE, Wiedenhoft H, Winglee K, Humbert O, Edelstein PH, Cosma CL, Ramakrishnan L. 2011. Drug tolerance in replicating mycobacteria mediated by a macrophage-induced efflux mechanism. Cell 145:39–53. doi: 10.1016/j.cell.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ottenhoff THM, Kaufmann SHE. 2012. Vaccines against tuberculosis: where are we and where do we need to go? PLoS Pathog 8:e1002607. doi: 10.1371/journal.ppat.1002607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gandhi NR, Nunn P, Dheda K, Schaaf HS, Zignol M, van Soolingen D, Jensen P, Bayona J. 2010. Multidrug-resistant and extensively drug-resistant tuberculosis: a threat to global control of tuberculosis. Lancet 375:1830–1843. doi: 10.1016/S0140-6736(10)60410-2. [DOI] [PubMed] [Google Scholar]
- 17.Velayati AA, Masjedi MR, Farnia P, Tabarsi P, Ghanavi J, Ziazarifi AH, Hoffner SE. 2009. Emergence of new forms of totally drug-resistant tuberculosis bacilli: super extensively drug-resistant tuberculosis or totally drug-resistant strains in Iran. Chest 136:420–425. doi: 10.1378/chest.08-2427. [DOI] [PubMed] [Google Scholar]
- 18.Russell DG, Barry CE, Flynn JL. 2010. Tuberculosis: what we don't know can, and does, hurt us. Science 328:852–856. doi: 10.1126/science.1184784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ottenhoff THM. 2009. Overcoming the global crisis: “yes, we can,” but also for TB? Eur J Immunol 39:2014–2020. doi: 10.1002/eji.200939518. [DOI] [PubMed] [Google Scholar]
- 20.Mahajan R. 2013. Bedaquiline: first FDA-approved tuberculosis drug in 40 years. Int J Appl Basic Med Res 3:1–2. doi: 10.4103/2229-516X.112228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuijl C, Savage NDL, Marsman M, Tuin AW, Janssen L, Egan DA, Ketema M, van den Nieuwendijk R, van den Eeden SJF, Geluk A, Poot A, van der Marel G, Beijersbergen RL, Overkleeft H, Ottenhoff THM, Neefjes J. 2007. Intracellular bacterial growth is controlled by a kinase network around PKB/AKT1. Nature 450:725–730. doi: 10.1038/nature06345. [DOI] [PubMed] [Google Scholar]
- 22.Ballell L, Bates RH, Young RJ, Alvarez-Gomez D, Alvarez-Ruiz E, Barroso V, Blanco D, Crespo B, Escribano J, González R, Lozano S, Huss S, Santos-Villarejo A, Martín-Plaza JJ, Mendoza A, Rebollo-Lopez MJ, Remuiñan-Blanco M, Lavandera JL, Pérez-Herran E, Gamo-Benito FJ, García-Bustos JF, Barros D, Castro JP, Cammack N. 2013. Fueling open-source drug discovery: 177 small-molecule leads against tuberculosis. ChemMedChem 8:313–321. doi: 10.1002/cmdc.201200428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fleming A, Alderton WK. 2013. Zebrafish in pharmaceutical industry research: finding the best fit. Drug Discov Today Dis Model 10:e43–e50. doi: 10.1016/j.ddmod.2012.02.006. [DOI] [Google Scholar]
- 24.Spaink HP, Cui C, Wiweger MI, Jansen HJ, Veneman WJ, Marín-Juez R, de Sonneville J, Ordas A, Torraca V, van der Ent W, Leenders WP, Meijer AH, Snaar-Jagalska BE, Dirks RP. 2013. Robotic injection of zebrafish embryos for high-throughput screening in disease models. Methods 62:246–254. doi: 10.1016/j.ymeth.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 25.Gehrig J, Reischl M, Kalmár E, Ferg M, Hadzhiev Y, Zaucker A, Song C, Schindler S, Liebel U, Müller F. 2009. Automated high-throughput mapping of promoter-enhancer interactions in zebrafish embryos. Nat Methods 6:911–916. doi: 10.1038/nmeth.1396. [DOI] [PubMed] [Google Scholar]
- 26.Pardo-Martin C, Chang T-Y, Koo BK, Gilleland CL, Wasserman SC, Yanik MF. 2010. High-throughput in vivo vertebrate screening. Nat Methods 7:634–636. doi: 10.1038/nmeth.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goldsmith P. 2004. Zebrafish as a pharmacological tool: the how, why and when. Curr Opin Pharmacol 4:504–512. doi: 10.1016/j.coph.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 28.Rubinstein AL. 2006. Zebrafish assays for drug. Expert Opin Drug Metab Toxicol 2:231–240. doi: 10.1517/17425255.2.2.231. [DOI] [PubMed] [Google Scholar]
- 29.Barros TP, Alderton WK, Reynolds HM, Roach A, Berghmans GS. 2008. Zebrafish: an emerging technology for in vivo pharmacological assessment to identify potential safety liabilities in early drug discovery. Br J Pharmacol 154:1400–1413. doi: 10.1038/bjp.2008.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berghmans S, Butler P, Goldsmith P, Waldron G, Gardner I, Golder Z, Richards FM, Kimber G, Roach A, Alderton W, Fleming A. 2008. Zebrafish based assays for the assessment of cardiac, visual and gut function–potential safety screens for early drug discovery. J Pharmacol Toxicol Methods 58:59–68. doi: 10.1016/j.vascn.2008.05.130. [DOI] [PubMed] [Google Scholar]
- 31.Rullas J, García JI, Beltrán M, Cardona P-J, Cáceres N, García-Bustos JF, Angulo-Barturen I. 2010. Fast standardized therapeutic-efficacy assay for drug discovery against tuberculosis. Antimicrob Agents Chemother 54:2262–2264. doi: 10.1128/AAC.01423-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Makarov V, Lechartier B, Zhang M, Neres J, van der Sar AM, Raadsen SA, Hartkoorn RC, Ryabova OB, Vocat A, Decosterd LA, Widmer N, Buclin T, Bitter W, Andries K, Pojer F, Dyson PJ, Cole ST. 2014. Towards a new combination therapy for tuberculosis with next generation benzothiazinones. EMBO Mol Med 6:372–383. doi: 10.1002/emmm.201303575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diekmann H, Hill A. 2013. ADMETox in zebrafish. Drug Discov Today Dis Model 10:e31–e35. doi: 10.1016/j.ddmod.2012.02.005. [DOI] [Google Scholar]
- 34.Barreto-Valer K, López-Bellido R, Macho Sánchez-Simón F, Rodríguez RE. 2012. Modulation by cocaine of dopamine receptors through miRNA-133b in zebrafish embryos. PLoS One 7:e52701. doi: 10.1371/journal.pone.0052701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou Y, He M-F, Choi FF-K, He Z-H, Song J-Z, Qiao C-F, Li S-L, Xu H-X. 2011. A high-sensitivity UPLC-MS/MS method for simultaneous determination and confirmation of triptolide in zebrafish embryos. Biomed Chromatogr 25:851–857. doi: 10.1002/bmc.1534. [DOI] [PubMed] [Google Scholar]
- 36.Raterink R-J, Kloet FM, Li J, Wattel NA, Schaaf MJM, Spaink HP, Berger R, Vreeken RJ, Hankemeier T. 2013. Rapid metabolic screening of early zebrafish embryogenesis based on direct infusion-nanoESI-FTMS. Metabolomics 9:864–873. doi: 10.1007/s11306-012-0493-6. [DOI] [Google Scholar]
- 37.Nijland HMJ, Ruslami R, Suroto A, Burger JDM, Alisjahbana B, van Crevel R, Aarnoutse RE. 2007. Rifampicin reduces plasma concentrations of moxifloxacin in patients with tuberculosis. Clin Infect Dis 45:1001–1007. doi: 10.1086/521894. [DOI] [PubMed] [Google Scholar]
- 38.Davis JM, Clay H, Lewis JL, Ghori N, Herbomel P, Ramakrishnan L. 2002. Real-time visualization of mycobacterium-macrophage interactions leading to initiation of granuloma formation in zebrafish embryos. Immunity 17:693–702. doi: 10.1016/S1074-7613(02)00475-2. [DOI] [PubMed] [Google Scholar]
- 39.Valkó K, Bevan C, Reynolds D. 1997. Chromatographic hydrophobicity index by fast-gradient RP-HPLC: a high-throughput alternative to log P/log D. Anal Chem 69:2022–2029. doi: 10.1021/ac961242d. [DOI] [PubMed] [Google Scholar]
- 40.Young RJ, Green DVS, Luscombe CN, Hill AP. 2011. Getting physical in drug discovery II: the impact of chromatographic hydrophobicity measurements and aromaticity. Drug Discov Today 16:822–830. doi: 10.1016/j.drudis.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 41.Seth A, Stemple DL, Barroso I. 2013. The emerging use of zebrafish to model metabolic disease. Dis Model Mech 6:1080–1088. doi: 10.1242/dmm.011346. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.