Abstract
Detailed mutational analysis examines the roles of individual residues of the Vga(A) linker in determining the antibiotic resistance phenotype. It defines a narrowed region of residues 212 to 220 whose composition determines the resistance specificity to lincosamides, pleuromutilins, and/or streptogramins A. From the analogy with the recently described function of the homologous ABC-F protein EttA as a translational factor, we infer that the Vga(A) linker interacts with the ribosome and directly or indirectly affects the binding of the respective antibiotic.
INTRODUCTION
The ABC-F family of ABC transporters comprises soluble proteins with two nucleotide binding domains (NBD) (Pfam accession number PF00005) separated by a flexible linker of approximately 80 amino acid residues. In contrast to typical ABC transporters, ABC-F proteins do not have any transmembrane domain. They usually participate in nontransport cellular functions, including translation regulation and DNA excision, and are also involved in antibiotic resistance. Antibiotic resistance ABC-F proteins are collectively referred to as ARE (antibiotic resistance) proteins (1). The mechanism of resistance conferred by ARE proteins is not well understood.
Vga(A) is one of the most-studied ARE proteins (2–4). Since the first report of Vga(A) as a streptogramin A (SgA) resistance determinant (5), several variants differing in their ability to confer resistance to SgA, lincosamides (L), and/or pleuromutilins (P) were reported (Fig. 1) (4, 6–11). For clarity in this paper, we refer to unspecified Vga(A) variants as Vga(A)*, use Vga(A) only for the originally described Vga(A) protein (NCBI protein database accession number AAA26684) conferring resistance to SgA (5), and use Vga(A)LC (the LC subscript indicates resistance to lincomycin and clindamycin) for the variant with NCBI protein database accession number ABH10964 that confers resistance to both L and SgA (4).
FIG 1.
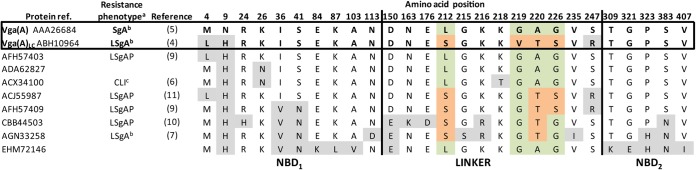
Amino acid sequence variability of Vga(A)* variants mined from the NCBI protein database. Only variable positions are shown. Amino acid residues that differ from those in Vga(A) are indicated by a gray background. Specific residues important for antibiotic specificity of Vga(A) and Vga(A)LC are indicated by light green and light bronze background, respectively (4). In the Resistance phenotypea column, the superscript letter a indicates that the resistance phenotype as published in the relevant reference is shown as follows: SgA, streptogramins A; L, lincosamides; P, pleuromutilins; CLI, clindamycin. The superscript letter b indicates that pleuromutilins were not tested, and the superscript letter c indicates that only CLI was tested.
Comparison of the Vga(A) and Vga(A)LC proteins showed that the shift in resistance between SgA and L is determined by amino acid substitutions L212S, G219V, A220T, and G226S clustered in a sequence only 15 amino acids long within the interdomain linker, which is the main source of polymorphism for all Vga(A)* proteins (Fig. 1) (4). Nothing is known about how these individual variable residues contribute to antibiotic specificity. We set out to decipher the relationship between the linker variability of Vga(A)* variants and the resistance phenotype using detailed mutational study of the Vga(A) and Vga(A)LC linkers. Implications of our results for the proposed resistance mechanism are discussed.
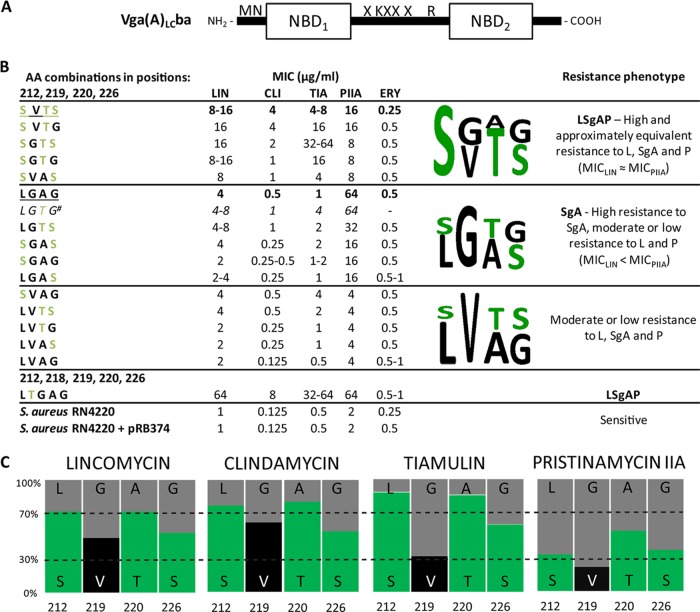
We have mutated amino acid residues of the linker in positions 212, 219, 220, and 226 to create a set of mutant forms of Vga(A)* that contain all but one of the possible combinations of Vga(A)- and Vga(A)LC-specific residues in their respective positions. Site-directed mutagenesis was performed using two complementary primers according to the QuikChange site-directed mutagenesis kit protocol (Stratagene). The construct pBluescript II KS carrying the vga(A)LCba gene encoding a more-efficient hybrid protein, combining the sequences of Vga(A) and Vga(A)LC (Fig. 2A), was used as a template (4). All mutations were verified by resequencing before subcloning of mutant forms into pRB374 shuttle vector as described previously (4). The resulting constructs were transformed to Staphylococcus aureus RN4220, and the MICs of lincomycin (LIN), clindamycin (CLI), pristinamycin IIA (PIIA), tiamulin (TIA), and erythromycin (ERY) were determined by the standard broth microdilution method (Fig. 2B) (12). We repeatedly failed to prepare a mutant form with the combination of amino acids L212, G219, T220, and G226 (LGTG). For the purpose of statistical evaluation, we predicted the MICs for the missing variant using single-layer feed-forward neural network (13). All statistical tests were calculated using R software (14).
FIG 2.
Efficiency of the Vga(A)* variants prepared and analyzed in this study. (A) Scheme of the Vga(A)LCba protein used in preparing the mutant forms of Vga(A). Mutated amino acids (X) at positions 212, 219, 220, and 226 are indicated. (B) MICs, resistance phenotypes, and consensus sequence logos. The amino acid (AA) combinations at positions 212, 219, 220, and 226 are shown. The sequence patterns of the original Vga(A) and Vga(A)LC are underlined. The superscript number indicates that the predicted MIC values for the missing variant LGTG are shown. Polar amino acids are shown in green in the sequence logo. (C) Impact of the individual positions to the level of resistance to a particular antibiotic. Results are expressed as the relative ratio of the mean MIC for variants with a particular residue.
All strains with plasmids bearing mutant forms of the gene showed decreased susceptibility to at least one of the antiobiotics (LIN, CLI, PIIA, or TIA), suggesting that all introduced Vga(A)* variants retained their activities. On the basis of the level of resistance, they can be classified into three groups (Fig. 2B). The first group consists of variants with the same resistance profile as that conferred by Vga(A)LC, i.e., high and approximately equivalent levels of resistance to LIN and PIIA (MIC of LIN [MICLIN] ≈ MICPIIA). These variants also confer substantial resistance to pleuromutilins (P) (LSgAP phenotype). The second group contains variants which, like Vga(A), confer high resistance to PIIA but only moderate or low resistance to L and P (MICLIN < MICPIIA, i.e., SgA phenotype), and the third group contains variants conferring moderate or low resistance to all three groups of antibiotics. Susceptibility to P, represented here by TIA was for the first time tested in the context of antibiotic specificity of Vga(A)* variants. As verified by Spearman's rank correlation coefficient test on a significance level of P ≤ 0.01, susceptibility to P correlates with both lincosamides. Nevertheless, the optimal combination SGTS conferring maximal resistance to P slightly differs from the SVTS one, optimal for lincosamides (Fig. 2B). No significant changes in susceptibility to ERY, which was used as a control, were recorded.
The relative impact of individual positions on the level of resistance to each tested antibiotic is depicted in Fig. 2C. The statistical significance of individual positions and double combinations was evaluated by analysis of variance (ANOVA) for linear model fits (15) performed on ranked MIC values. The analysis indicated that positions 212 and 220 are important for resistance to L (F1,23 = 67.2, P < 0.0001 and F1,23 = 92.5, P < 0.0001, respectively) and to P (F1,13 = 38.9, P < 0.0001 and F1,13 = 24.2, P = 0.0003, respectively) and that position 219 is important for resistance to SgA (F1,12 = 23.5, P = 0.0004), i.e., variants with polar amino acid residues S212 and T220 are the most effective for resistance to L and P and variants with G219 are the most effective for resistance to SgA (Fig. 2B and C). The analysis also showed an interaction effect of positions 212 and 219 on resistance to L (F1,23 = 44.5, P < 0.0001) and on resistance to SgA (F1,12 = 16.3, P < 0.002). Given that position 219 alone is not significant for resistance to L, it modifies the effect of position 212. In the case of SgA, the interaction is reversed. This observed reciprocity of the interaction effect between the two positions on L and SgA resistance probably relates to the observation that increased resistance to L was at the expense of resistance to SgA. Vga(A)* variants exhibiting the LSgAP phenotype hardly achieved maximum levels of resistance to PIIA conferred by variants preferring this compound over L and P and vice versa.
Our data correspond well with published resistance profiles of previously characterized Vga(A)* proteins (Fig. 1), narrowing the variable linker region determining antibiotic specificity to only eight amino acids between positions 212 and 220. Another potentially important position located within this region is position 218. The K218T substitution is the only one in the linker of the variant with NCBI protein database accession number ACX34100, which, unlike Vga(A), confers resistance to clindamycin (11) (Fig. 1). We evaluated the effect of the substitution on the resistance phenotype in the same background protein as before. Surprisingly, the resulting protein with the K218T mutation was the most efficient variant constructed, reaching the maximum levels of resistance to all three groups of antibiotics (Fig. 2B). Remarkably, the K218T substitution altered the antibiotic specificity of Vga(A) toward L and P regardless of the presence of the most efficient SgA specific amino acid combination L212-G219 and without compromising the level of resistance as observed for the set of variants with lysine in position 218.
A Vga(A)* variant (NCBI protein database accession number AFH57403) has linker sequence identical to that of Vga(A); however, it has been shown that it confers resistance to both SgA and L (9) (Fig. 1). This contradicts our previous observations that this variant does not exhibit any preference to lincosamides (MICLIN < MICPIIA) when tested in a heterologous host (4). The observed lincosamide resistance thus may reflect higher expression of this variant from its own promoter in contrast to a moderate expression in our heterologous system using vegII promoter from Bacillus subtilis. It was consistently observed that Vga(A)v cloned under the control of the vegII promoter as well as Vga(A)v encoded by a transposon confers resistance only to SgA; however, the same variant confers resistance to clindamycin when encoded by a plasmid in a clinical isolate (3, 7).
The mechanism of resistance conferred by ARE proteins is not well understood. Because some ARE proteins associate with a membrane even though they do not have a transmembrane domain (3, 16), it is widely accepted that ARE proteins may interact with a membrane partner to constitute a functional transporter (17). Indeed, the first evidence for such cooperation was recently shown between the ARE protein Msr(D) and the major facilitator superfamily Mef(E) transporter, both originating from Streptococcus pneumoniae (18). However, direct involvement of Msr(D) in transport has not yet been demonstrated.
The sequence homology of ARE proteins with translational factors and the fact that these proteins confer resistance exclusively to antibiotics inhibiting translation resulted in the formulation of an alternative hypothesis of ARE protein function. It has been suggested that ARE proteins might interact with the ribosome and affect the binding of antibiotics to their target site (17). Very recently, the function of the first ABC-F protein, EttA, as a translation factor which regulates protein synthesis depending on cell energy status has been characterized (19, 20). The similarity of Vga(A)* to EttA (31% identity) allows us to envisage a mechanism of Vga(A) action based on the EttA one, providing a more-detailed insight into the probable involvement of Vga(A) in antibiotic resistance by ribosome protection than ever before.
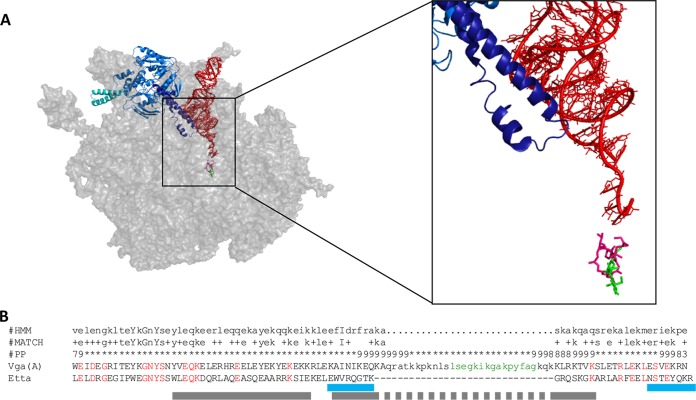
Cryo-electron microscopy (cryo-EM) was used to show that EttA binds to the ribosome at the tRNA exit site (E-site) and interacts with the aminoacyl stem of tRNAfMet bound in the peptidyl-tRNA-binding site (P-site). The contact is mediated through an interdomain linker called the PtlM domain. The PtlM domain forms a distinctive elongated structure, which enters the ribosome in the direction of the peptidyl transferase center (PTC) where the amino acids residues at the tip of the PtlM domain specifically recognize the aminoacyl stem of tRNAfMet (Fig. 3A). The second functionally important structural motif of EttA is a large insertion in NBD2 called arm. The arm subdomain interacts with the L1 stalk of the large ribosomal subunit and restricts the ribosome dynamics in response to availability of ATP. While the arm subdomain is absent in Vga(A)* variants, the interdomain linker is not only conserved but even further enlarged by 30 amino acid residues inserted into its center, thus forming a presumptive tip of the linker structure (Fig. 3B). Moreover, the above-defined variable region from positions 212 to 220 of Vga(A)* determining the antibiotic specificity forms part of the linker extension.
FIG 3.
(A) EttA (blue) bound to the 50S ribosomal subunit in complex with P-tRNAfMet (red) (RCSB Protein Database [PDB] accession numbers 3J5S [19] and 3I9C [24] and binding sites of clindamycin (green) (PDB accession number 3OFZ [25]) and SgA and dalfopristin (purple) (PDB accession number 1SM1 [21]). The arm subdomain of EttA (bright blue) and the PtlM domain (dark blue) are indicated. The enlargement shows a detailed view of the PtlM domain interacting with the stem of P-tRNAfMet in the direction toward bound antibiotics. (B) Significant Pfam-A matches of the Vga(A) linker with ABC_tran_2 domain and sequence alignment with the PtlM domain of EttA. The hidden Markov model sequence (HMM), match between HMM and Vga(A) (MATCH), and posterior probability (PP) (the degree of confidence in each individual aligned residue) are given. Amino acid residues of Vga(A) and EttA that are identical (red) and the antibiotic specificity site of Vga(A) (green) are indicated. The bright blue lines represent the regions of contact of PtlM with P-site tRNA, and the gray lines represent regions of contact of PtlM with 23S rRNA (19).
Both the Vga(A) linker and the PtlM domain of EttA fulfill the requirements for the consensus sequence of ABC_trans_2 domain family (Fig. 3B). Thus, the simplest concept of the Vga(A) function is that the Vga(A) linker interacts, analogously to the PtlM domain, with the ribosome and promotes the displacement of the antibiotic. The observed correlation between Vga(A)* linker amino acid composition and chemical properties of the relevant antibiotic can evoke the concept of direct interaction of the linker tip with the target antibiotic. However, this correlation may only be apparent and may reflect more-complex relationships than direct interaction. In fact, during translation, the P-site is constantly occupied by initiation tRNAfMet or by peptidyl-tRNA, which most probably would prevent direct contact of the Vga(A)* linker with antibiotics. Therefore, an indirect effect of Vga(A)* on antibiotic binding mediated by contact of the linker with P-site tRNAs seems to be more probable. The observed correlation of antibiotic specificity with the linker sequence could reflects differences in ribosome binding sites of L and SgA, which overlap the A-site or A- and P-sites of PTC, respectively (21).
We failed to prove direct interaction of Vga(A) and Vga(A)LC with the relevant antibiotics using surface plasmon resonance (data not shown). This corresponds to the assumption that the linker conformation is context dependent, i.e., induced by interaction of this region with the ribosome. This means that the potential interaction of the protein with an antibiotic could be demonstrated only “in situ” using cryo-EM. On the contrary, considering Vga(A)* as an antibiotic recognition part of a transport system, the protein-antibiotic interaction could be demonstrated with high probability by surface plasmon resonance. In this context, the mechanism of Vga(A) function based on its interaction with the ribosome appears to be more probable than the transport one.
An important aspect for the ribosomal protection resistance is to ensure that the antibiotic will not rebind after it is dislodged from the ribosome. Coupling antibiotic release from the ribosome to antibiotic efflux could be the solution. Indeed, the presence of a macrolide transporter is an essential requirement to manifest the peptide-mediated macrolide resistance in Escherichia coli (22). Similarly, macrolide resistance due to mutations in ribosomal protein L22 relies on the presence of the active macrolide transporter AcrAB-TolC (23). In this context, both considered mechanisms of ARE protein function are likely to be combined rather than exclude each other.
ACKNOWLEDGMENTS
We thank Ondrej Sedivy for helpful comments on statistical analysis and Jan Vojt for neural network computing.
This work was supported by the Grant Agency of the Czech Republic project P302-12-P632 and Ministry of Education, Youth and Sports of the Czech Republic projects CZ.1.07/2.3.00/20.0055 and CZ.1.07/2.3.00/30.0003 and BIOCEV (Biotechnology and Biomedicine Centre of the Academy of Sciences) and Charles University project CZ.1.05/1.1.00/02.0109 from the European Regional Development Fund in the Czech Republic.
REFERENCES
- 1.Dassa E, Bouige P. 2001. The ABC of ABCs: a phylogenetic and functional classification of ABC systems in living organisms. Res Microbiol 152:211–229. doi: 10.1016/S0923-2508(01)01194-9. [DOI] [PubMed] [Google Scholar]
- 2.Jacquet E, Girard J-MM, Ramaen O, Pamlard O, Lévaique H, Betton J-MM, Dassa E, Chesneau O, Levaique H. 2008. ATP hydrolysis and pristinamycin IIA inhibition of the Staphylococcus aureus Vga(A), a dual ABC protein involved in streptogramin A resistance. J Biol Chem 283:25332–25339. doi: 10.1074/jbc.M800418200. [DOI] [PubMed] [Google Scholar]
- 3.Chesneau O, Ligeret H, Hosan-Aghaie N, Morvan A, Dassa E. 2005. Molecular analysis of resistance to streptogramin A compounds conferred by the Vga proteins of staphylococci. Antimicrob Agents Chemother 49:973–980. doi: 10.1128/AAC.49.3.973-980.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Novotna G, Janata J. 2006. A new evolutionary variant of the streptogramin A resistance protein, Vga(A)LC, from Staphylococcus haemolyticus with shifted substrate specificity towards lincosamides. Antimicrob Agents Chemother 50:4070–4076. doi: 10.1128/AAC.00799-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allignet J, Loncle V, El Solh N. 1992. Sequence of a staphylococcal plasmid gene, vga, encoding a putative ATP-binding protein involved in resistance to virginiamycin A-like antibiotics. Gene 117:45–51. doi: 10.1016/0378-1119(92)90488-B. [DOI] [PubMed] [Google Scholar]
- 6.Qin X, Poon B, Kwong J, Niles D, Schmidt BZ, Rajagopal L, Gantt S. 2011. Two paediatric cases of skin and soft-tissue infections due to clindamycin-resistant Staphylococcus aureus carrying a plasmid-encoded vga(A) allelic variant for a putative efflux pump. Int J Antimicrob Agents 38:81–83. doi: 10.1016/j.ijantimicag.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 7.Tessé S, Trueba F, Berthet N, Hot C, Chesneau O. 2013. Resistance genes underlying the LSA phenotype of staphylococcal isolates from France. Antimicrob Agents Chemother 57:4543–4546. doi: 10.1128/AAC.00259-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allignet J, El Solh N. 1999. Comparative analysis of staphylococcal plasmids carrying three streptogramin-resistance genes: vat-vgb-vga. Plasmid 42:134–138. doi: 10.1006/plas.1999.1412. [DOI] [PubMed] [Google Scholar]
- 9.Lozano C, Aspiroz C, Rezusta A, Gómez-Sanz E, Simon C, Gómez P, Ortega C, Revillo MJ, Zarazaga M, Torres C, José M. 2012. Identification of novel vga(A)-carrying plasmids and a Tn5406-like transposon in meticillin-resistant Staphylococcus aureus and Staphylococcus epidermidis of human and animal origin. Int J Antimicrob Agents 40:306–312. doi: 10.1016/j.ijantimicag.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 10.Mendes RE, Smith TC, Deshpande L, Diekema DJ, Sader HS, Jones RN. 2011. Plasmid-borne vga(A)-encoding gene in methicillin-resistant Staphylococcus aureus ST398 recovered from swine and a swine farmer in the United States. Diagn Microbiol Infect Dis 71:177–180. doi: 10.1016/j.diagmicrobio.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 11.Gentry DR, McCloskey L, Gwynn MN, Rittenhouse SF, Scangarella N, Shawar R, Holmes DJ. 2008. Genetic characterization of Vga ABC proteins conferring reduced susceptibility to pleuromutilins in Staphylococcus aureus. Antimicrob Agents Chemother 52:4507–4509. doi: 10.1128/AAC.00915-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Urbaskova P. 1998. Rezistence bakterií k antibiotikům. Vybrané metody. TRIOS, Prague, Czech Republic. [Google Scholar]
- 13.Auer P, Burgsteiner H, Maass W. 2008. A learning rule for very simple universal approximators consisting of a single layer of perceptrons. Neural Netw 21:786–795. doi: 10.1016/j.neunet.2007.12.036. [DOI] [PubMed] [Google Scholar]
- 14.R Core Team. 2013. R: a language and environment for statistical computing. 3.1.1 R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 15.Chambers JM. 1991. Linear models, p 95–138. In Chambers JM, Hastie TJ. (ed), Statistical models in S. Chapman and Hall/CRC, London, United Kingdom. [Google Scholar]
- 16.Olano C, Rodríguez AM, Méndez C, Salas JA. 1995. A second ABC transporter is involved in oleandomycin resistance and its secretion by Streptomyces antibioticus. Mol Microbiol 16:333–343. doi: 10.1111/j.1365-2958.1995.tb02305.x. [DOI] [PubMed] [Google Scholar]
- 17.Kerr ID, Reynolds PE, Cove JH, Reynolds ED. 2005. ABC proteins and antibiotic drug resistance: is it all about transport? Biochem Soc Trans 33:1000–1002. doi: 10.1042/BST20051000. [DOI] [PubMed] [Google Scholar]
- 18.Nunez-Samudio V, Chesneau O. 2013. Functional interplay between the ATP binding cassette Msr(D) protein and the membrane facilitator superfamily Mef(E) transporter for macrolide resistance in Escherichia coli. Res Microbiol 164:226–235. doi: 10.1016/j.resmic.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 19.Chen B, Boël G, Hashem Y, Ning W, Fei J, Wang C, Gonzalez RL Jr, Hunt JF, Frank J. 2014. EttA regulates translation by binding the ribosomal E site and restricting ribosome-tRNA dynamics. Nat Struct Mol Biol 21:152–159. doi: 10.1038/nsmb.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boël G, Smith PC, Ning W, Englander MT, Chen B, Hashem Y, Testa AJ, Fischer JJ, Wieden HJ, Frank J, Gonzalez RL Jr, Hunt JF. 2014. The ABC-F protein EttA gates ribosome entry into the translation elongation cycle. Nat Struct Mol Biol 21:143–151. doi: 10.1038/nsmb.2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harms JM, Schlünzen F, Fucini P, Bartels H, Yonath A. 2004. Alterations at the peptidyl transferase centre of the ribosome induced by the synergistic action of the streptogramins dalfopristin and quinupristin. BMC Biol 2:4. doi: 10.1186/1741-7007-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lovmar M, Nilsson K, Vimberg V, Tenson T, Nervall M, Ehrenberg M. 2006. The molecular mechanism of peptide-mediated erythromycin resistance. J Biol Chem 281:6742–6750. doi: 10.1074/jbc.M511918200. [DOI] [PubMed] [Google Scholar]
- 23.Lovmar M, Nilsson K, Lukk E, Vimberg V, Tenson T, Ehrenberg M. 2009. Erythromycin resistance by L4/L22 mutations and resistance masking by drug efflux pump deficiency. EMBO J 28:736–744. doi: 10.1038/emboj.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jenner LB, Demeshkina N, Yusupova G, Yusupov M. 2010. Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat Struct Mol Biol 17:555–560. doi: 10.1038/nsmb.1790. [DOI] [PubMed] [Google Scholar]
- 25.Dunkle JA, Xiong L, Mankin AS, Cate JHD. 2010. Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action. Proc Natl Acad Sci U S A 107:17152–17157. doi: 10.1073/pnas.1007988107. [DOI] [PMC free article] [PubMed] [Google Scholar]