Abstract
The blaNDM-1 gene is associated with extensive drug resistance in Gram-negative bacteria. This probably spread to Enterobacteriaceae from Acinetobacter spp., and we characterized plasmids associated with blaNDM-1 in Acinetobacter spp. to gain insight into their role in this dissemination. Four clinical NDM-1-producing Acinetobacter species strains from India and Pakistan were investigated. A plasmid harboring blaNDM-1, pNDM-40-1, was characterized by whole-genome sequencing of Acinetobacter bereziniae CHI-40-1 and comparison with related plasmids. The presence of similar plasmids in strains from Pakistan was sought by PCR and sequencing of amplicons. Conjugation frequency was tested and stability of pNDM-40-1 investigated by real-time PCR of isolates passaged with and without antimicrobial selection pressure. A. bereziniae and Acinetobacter haemolyticus strains contained plasmids similar to the pNDM-BJ01-like plasmids identified in Acinetobacter spp. in China. The backbone of pNDM-40-1 was almost identical to that of pNDM-BJ01-like plasmids, but the transposon harboring blaNDM-1, Tn125, contained two short deletions. Escherichia coli and Acinetobacter pittii transconjugants were readily obtained. Transconjugants retained pNDM-40-1 after a 14-day passage experiment, although stability was greater with meropenem selection. Fragments of pNDM-BJ01-like plasmid backbones are found near blaNDM-1 in some genetic contexts from Enterobacteriaceae, suggesting that cross-genus transfer has occurred. pNDM-BJ01-like plasmids have been described in isolates originating from a wide geographical region in southern Asia. In vitro data on plasmid transfer and stability suggest that these plasmids could have contributed to the spread of blaNDM-1 into Enterobacteriaceae.
INTRODUCTION
Acinetobacter baumannii is a successful nosocomial pathogen, and extensively drug-resistant strains are increasingly prevalent (1, 2). Other Acinetobacter spp. are found in the environment and can cause opportunistic infections (2). There is evidence that the gene encoding New Delhi metallo-β-lactamase-1 (NDM-1) evolved in an Acinetobacter background through fusion between the aminoglycoside resistance gene aphA6 and a β-lactamase-encoding progenitor of blaNDM-1 (3). Subsequently, blaNDM-1 and its closely related variants have spread rapidly among many genera of Gram-negative bacteria (4, 5), and they are found on plasmids of several different incompatibility types and chromosomally (4–6). NDM enzymes hydrolyze all β-lactams except aztreonam and are commonly found with other resistance mechanisms, mediating resistance to almost all clinically available antimicrobials (5, 7).
blaNDM genes are prevalent in clinical Enterobacteriaceae isolates in South Asia. Many cases of infection or colonization with NDM-producing Enterobacteriaceae around the world have been linked to travel to the Indian subcontinent (5, 8). Studies within Indian hospitals have identified NDM-1-producing Acinetobacter spp. causing infections in intensive care units (9–11). Similar cases have been reported in Europe, but most patients probably became colonized during travel to the Balkans or North Africa (12–14). In China, blaNDM-1 has been observed many times in several Acinetobacter spp., including A. baumannii, from clinical, environmental, and farm animal samples but is less commonly reported in Enterobacteriaceae (15–22).
The immediate genetic contexts of blaNDM (the genes flanking blaNDM) in Acinetobacter spp. are well conserved. An intact ISAba125 is normally present upstream (3, 14, 16). Downstream there are usually a conserved set of genes from the bleomycin resistance gene, ble, to ISCR27. Most examples have an intact ISAba125 further downstream from ISCR27, thus capturing the entire context in a Tn125 transposon (13, 14, 16). In Chinese isolates, blaNDM-1 is usually found on plasmids, with all sequenced examples being closely related to pNDM-BJ01 from A. lwoffii WJ10621, despite being reported in many different Acinetobacter spp (15, 16, 19, 23). In most isolates identified outside Asia, blaNDM-1 is found on the chromosome (12–14).
The Acinetobacter plasmids from China share features which suggest that they could have contributed to the acquisition of blaNDM by Enterobacteriaceae. We set out to see if similar plasmids were present in Acinetobacter spp. from India and Pakistan, where some studies have shown a high prevalence of Enterobacteriaceae producing blaNDM. We describe the whole-genome sequence (WGS) of an A. bereziniae isolate from Chennai, India, with such a plasmid. We further show that closely related plasmids carrying blaNDM-1 are present in Acinetobacter spp. isolated in Karachi, Pakistan. We also investigated the conjugation efficiencies and stabilities of these plasmids in different recipients to further explore their potential as vectors in the dissemination of blaNDM-1.
MATERIALS AND METHODS
Bacterial strains studied.
A full list of isolates used in this study is given in Table 1. A. bereziniae CHI-40-1 (11) was investigated in detail in the current study. Other Acinetobacter species isolates were from fecal screening samples collected at the Civil Hospital Karachi, Pakistan, in 2012. Samples from consecutive patients admitted to the hospital were collected at admission and discharge, and isolates growing on selective plates containing ertapenem were further analyzed. The isolates studied here represent all of the blaNDM-1-positive Acinetobacter spp. isolated from 717 fecal swabs processed as of January 2013. Escherichia coli UAB190 (24) and A. pittii AG3528 were used as recipients in mating experiments.
TABLE 1.
Bacterial isolates used in this study
| Isolate | Species | Source | Location where isolated | Yr isolated |
|---|---|---|---|---|
| CHI-40-1 | Acinetobacter berezeniae | Clinical isolate (pus) | Tamil Nadu, India | 2005 |
| 73261-EC | Acinetobacter haemolyticus | Fecal screening | Karachi, Pakistan | 2012 |
| 70114-EC | Acinetobacter haemolyticus | Fecal screening | Karachi, Pakistan | 2012 |
| 69122-EW | Acinetobacter haemolyticus | Fecal screening | Karachi, Pakistan | 2012 |
| 74312-EC | Acinetobacter schindleri | Fecal screening | Karachi, Pakistan | 2012 |
| 73668-ECT | Acinetobacter towneri | Fecal screening | Karachi, Pakistan | 2012 |
Identification and antimicrobial susceptibility testing.
Initial bacterial identification was performed by matrix-assisted laser desorption ionization–time of flight mass spectroscopy (MALDI-TOF) (Bruker, Billerica, MA, USA). Confirmation of the identification was by phylogenetic analysis of 16S rRNA gene sequences for Pakistan isolates, together with ribosomal multilocus sequence typing (rMLST) for CHI-40-1 (see Fig. S1 in the supplemental material). Antimicrobial susceptibility testing was performed by Etest (bioMérieux, LaPlane, France) and MIC test strip (Liofilchem, Roseto degli Abruzzi, Italy). Interpretation was according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoints (version 3.1).
WGS and analysis.
A. bereziniae CHI-40-1 was sequenced using an Illumina HiSeq platform at the Wellcome Trust Sanger Institute, Cambridge. A unique index-tagged insert library was prepared to allow processing of the sample data following multiplex sequencing with other libraries on eight channels of an Illumina Genome Analyzer GAII cell to give 100-bp paired-end reads, as previously described (25). Reads were assembled de novo using the Velvet Assembly Tool (version 1.2.10) (26). Plasmid contigs were identified using Blast searches against the sequence of plasmid pNDM-BJ01 from Acinetobacter lwoffii strain WJ10621 (accession number JQ001791) (16). Links between contigs were confirmed by PCR and sequencing of amplicons. Detailed comparison between pNDM-40-1 and closely related plasmids was performed using nucleotide alignments created in Geneious (27). Antibiotic resistance genes were identified using RESFinder. Annotation was by transfer of annotations for genes with close nucleotide identity from reference sequences in Geneious.
PFGE and in-gel hybridization.
Genomic DNA was prepared in 1% low-melting-point agarose plugs as described previously (28). Briefly, plugs were made using a bacterial cell suspension in Tris-EDTA (TE) buffer at a standard optical density of 1.8 to 2.0 at a wavelength of 600 nm (28). Plugs were treated with ApaI (Thermo Scientific, Waltham, MA, USA) or S1 nuclease (Thermo Scientific) and pulsed-field gel electrophoresis (PFGE) performed as described previously (28, 29). Autoradiographs were prepared by in-gel hybridization of pulsed-field gels with gene probes, made using a random primer method to label blaNDM-1 or traA PCR products with [32P]CTP, as previously described (28).
Conjugation and passage experiments.
Conjugation experiments were performed as described previously using a plate mating assay at 30°C (4), with A. bereziniae CHI-40-1 and A. haemolyticus 69122-EW as donors and E. coli UAB190 and A. pittii AG3528 (both rifampin resistant) as recipients. Selection was performed on Brilliance UTI Clarity agar (Oxoid Ltd., Basingstoke, United Kingdom) supplemented with rifampin (Sigma-Aldrich, St. Louis, MO, USA) for recipient selection, ampicillin (Sigma-Aldrich) and rifampin for UAB190 background transconjugants, or meropenem (AstraZeneca, London, United Kingdom) and rifampin for AG3528 background transconjugants. For each experiment, 5 isolated colonies were subcultured to selective media. Pure growths on subculture were tested by MALDI-TOF to confirm the species background, and the presence of blaNDM-1 was confirmed by PCR. Mating efficiency was calculated as the number of transconjugants per recipient cell. Transconjugants obtained from mating between CHI-40-1 and each recipient background were subjected to S1 PFGE and in-gel hybridization with blaNDM-1 and traA.
A passage experiment was performed on A. bereziniae CHI-40-1 and its transconjugants, E. coli UAB190NDMP2 and A. pittii AG3528NDMP1. Cultures from selective plates were inoculated into Luria-Bertani (LB) broth (Thermo Scientific) with and without antibiotic selection and incubated overnight at 37°C. The following day, cultures were reinoculated into a fresh broth with the same selection as the starting culture. Columbia blood agar plates (E&O Laboratories, Bonnybridge, Scotland) were inoculated daily to check purity, and cultures were stored each day in LB broth with 10% glycerol at −80°C. This procedure was repeated for 14 consecutive days. Antibiotic selection was with meropenem at 10 μg/ml for CHI-40-1 and AG3528NDMP1 and at 1 μg/ml for UAB190NDMP2. Stored cultures were investigated by S1 PFGE and probing for blaNDM-1 and traA as well as real-time quantitative PCR (qPCR) (see below).
PCR.
A full list of PCR and sequencing primers used together with full PCR conditions is given in Table S2 in the supplemental material. The presence of plasmids similar to pNDM-BJ01 was confirmed by PCR with primers described by Hu et al. (16). Sequencing of PCR amplicons was used to resolve gaps in the pNDM-40-1 sequence and to primer walk the blaNDM-1 context in 69122-EW. PCR amplicons for sequencing were purified using the QIAquick gel extraction kit (Qiagen, Limburg, Netherlands) as per the manufacturer's instruction, and products were submitted to Eurofins MWG Operon (Ebersberg, Germany) for sequencing.
Real time-quantitative PCR (qPCR) was performed to quantify changes in blaNDM-1 and traA copy number present in bacterial cells over the course of the passage experiment. The single-copy chromosomal gene rpoB was used as the reference gene. Dual-labeled probes with fluorescent dye and quenchers were synthesized by Eurofins MWG Operon. blaNDM-1 and traA fluorescence cycle threshold (CT) values were compared to rpoB CT values, and quantification was performed by the ΔΔCT method (30). Regression analysis was performed using Excel 2007. A validation experiment showed that ΔCT values were linear over the range of values detected in the passage experiment. All experiments were performed in triplicate.
Nucleotide sequence accession numbers.
Accession numbers for pNDM-40-1 from A. bereziniae CHI-40-1 and the partial sequence for pNDM-69122 from A. haemolyticus 69122-EW are KF702385 and LN611576, respectively. A. bereziniae CHI-40-1 assembly contigs are deposited under study accession number PRJEB7120, contig accession numbers CDEL01000001 to CDEL01000324.
RESULTS
Species identification, antimicrobial susceptibility, and resistance genes present in A. bereziniae CHI-40-1.
The clinical Acinetobacter isolate from Chennai was found to be an A. bereziniae strain by rMLST. Five NDM-1-producing Acinetobacter isolates from the fecal screening study were obtained from five different patients and included three species (Table 1; see Fig. S1 in the supplemental material). Three Acinetobacter haemolyticus isolates were found to be representatives of a single strain by ApaI restriction digestion and PFGE. All strains were extensively drug resistant (Table 2). In keeping with this, genes associated with resistance to β-lactams (blaOXA-58), aminoglycosides (strA, strB, and aacC2), macrolides [msr(E) and mph(E)], trimethoprim (dfrA1), and sulfonamides (sul1 and sul2) were identified in A. bereziniae CHI-40-1.
TABLE 2.
Antimicrobial MICs for Acinetobacter species recipients and transconjugants
| Strain | MIC (mg/liter)a |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ATMb | CAZb | IPM | MEM | TZP | AMK | GEN | TOB | CIP | CST | FOF | SXT | TGCb | |
| CHI-40-1 | 48 (R) | ≥256 (R) | ≥32 (R) | ≥32 (R) | 96 (R) | 48 (R) | ≥256 (R) | 12 (R) | ≥32 (R) | 0.75 (S) | 8 | ≥32 (R) | 0.75 (R) |
| 69122-EW | 3 (S) | ≥256 (R) | ≥32 (R) | ≥32 (R) | 24 (R) | 32 (R) | 2 (S) | 8 (R) | ≥32 (R) | 0.75 (S) | 32 | 0.25 (S) | 1 (R) |
| 73668-ECT | 16 (R) | ≥256 (R) | ≥32 (R) | ≥32 (R) | 48 | 6 (S) | 192 (R) | ND | ≥32 (R) | 0.5 (S) | 4 | 0.25 (S) | 0.5 (I) |
| 74312-EC | 48 (R) | ≥256 (R) | ≥32 (R) | ≥32 (R) | ≥256 (R) | 48 (R) | 96 (R) | 12 (R) | ≥32 (R) | 0.5 (S) | 16 | ≥32 (R) | 0.75 (R) |
| AG3528 | 16 (R) | 3 (S) | 0.75 (S) | 0.75 (S) | 2 | 2 (S) | 0.75 (S) | 0.75 (S) | 4 (R) | 1 (S) | 16 | 0.125 (S) | ND |
| UAB190 | 0.125 (S) | 0.25 (S) | 0.38 (S) | 0.047 (S) | 2 | 2 (S) | 8 (R) | 1.5 (S) | 0.006 (S) | 0.5 (S) | 3 | 0.094 (S) | ND |
| AG3528NDMP1 | 16 (R) | ≥256 (R) | ≥32 (R) | ≥32 (R) | 96 | 3 (S) | 0.75 (S) | 0.75 (S) | 3 (R) | 1 (S) | 16 | 0.125 (S) | ND |
| UAB190NDMP2 | 0.064 (S) | ≥256 (R) | 24 (R) | 4 (I) | 256 | 2 (S) | 8 (R) | 1.5 (S) | 0.006 (S) | 0.5 (S) | 3 | 0.094 (S) | ND |
Antimicrobial susceptibility results based on EUCAST pharmacokinetic/pharmacodynamic (PK/PD) non-species-specific breakpoints. R, resistant; I, intermediate resistance profile; S, sensitive; ATM, aztreonam; CAZ, ceftazidime; IPM, imipenem; MEM, meropenem; TZP, piperacillin-tazobactam; AMK, amikacin; GEN, gentamicin; TOB, tobramycin; CIP, ciprofloxacin; CST, colistin sulfate; FOF, fosfomycin; RIF, rifampin; SXT, co-trimoxazole; TGC, tigecycline; ND, not determined.
Species-specific Acinetobacter species breakpoints not available.
Characterization of plasmid pNDM-40-1.
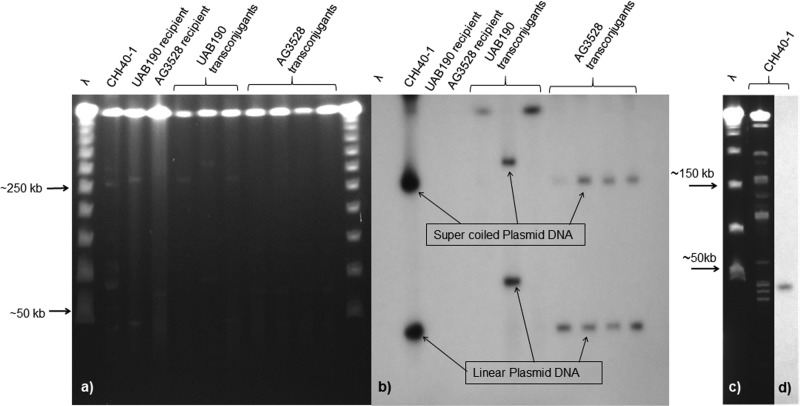
S1 PFGE and blaNDM-1 probing showed that A. bereziniae CHI-40-1 harbored multiple plasmids, with blaNDM-1 present on plasmids corresponding to bands of ∼45 kb and ∼250 kb (Fig. 1a and b). However, blaNDM-1 was present on a single ApaI restriction fragment of ∼45 kb (Fig. 1c and d).
FIG 1.
(a) Pulsed-field gel of S1 nuclease-digested genomic DNA from A. bereziniae CHI-40-1, recipients, and transconjugants; (b) in-gel hybridization with a blaNDM-1 gene probe; (c) pulsed-field gel of ApaI-digested genomic DNA from CHI-40-1; (d) in-gel hybridization with a blaNDM-1 gene probe. The molecular size marker is concatemers of λ of ∼50 to 1,000 kb.
The de novo assembly of the A. bereziniae CHI-40-1 WGS produced 324 contigs, with a mean GC content of 38% and a combined size of 4.78 Mb. A 45,827-bp plasmid harboring blaNDM-1, pNDM-40-1, was closed by PCR and sequencing of amplicons. The GC content of the plasmid backbone is 36.2%, and that of the variable region (from ISAba14 to the end of Tn125, nucleotides [nt] 5427 to 16280) is 52.5%.
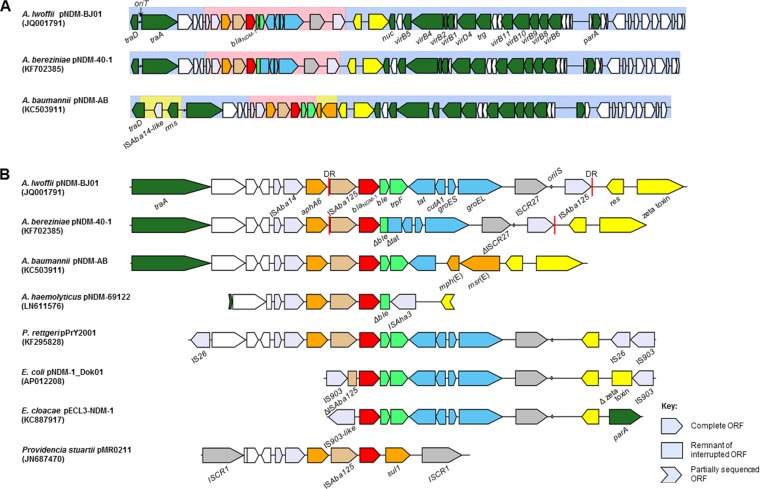
At the time of writing, complete sequences of nine pNDM-BJ01-like plasmids were available in GenBank. The backbone of pNDM-40-1 is 100% identical at the nucleotide level to those of pNDM-BJ01 (bases 1 to 5684 and 17987 to 47274, accession number JQ001791), pNDM-BJ02 (JQ060896), pAbNDM-1 (JN377410), and pXM1 (AMXH01000087). pNDM-AB (KC503911), pM131_NDM-1 (JX072963), and pNDM-Iz4b (KJ547696) exhibit minor differences from the backbone of pNDM-BJ01 and from one another. pNDM-AB differs the most, because of a 3.5-kb insertion containing the genes traD and insB and a putative methyltransferase gene (Fig. 2; see Table S3 in the supplemental material). These plasmids were all identified in isolates from China and were found in five different species, including A. baumannii. Additionally, several blaNDM-1-negative Acinetobacter species sequences contain regions with significant identity to the backbones of pNDM-BJ01-like plasmids (see Fig. S3 in the supplemental material).
FIG 2.
(A) Gene maps of plasmids pNDM-BJ01, pNDM-40-1, and pNDM-AB; (B) immediate blaNDM-1 context from pNDM-40-1, A. haemolyticus 69122-EW, and related sequences in Acinetobacter and Enterobacteriaceae. Open reading frames (ORFs) are color coded with the direction of transcription indicated by arrowheads, and truncated remnants of ORFs are shown as rectangles. Red, blaNDM-1; orange, other antibiotic resistance; lime green, usually immediately downstream of blaNDM-1; blue, from a common context in Xanthomonas and Pseudoxanthomonas; brown, ISAba125; dark gray, ISCR transposases; light gray, other insertion sequence transposases; yellow, resolvase and zeta-toxin from pNDM-BJ01-like plasmids; dark green, named plasmid backbone genes; white, genes for hypothetical proteins. Regions with a light blue shaded background contain plasmid backbone with close identity among pNDM-BJ01-like plasmids. The pink shaded regions represent genes normally found in the blaNDM-1 context in Acinetobacter spp. Regions from pNDM-AB with a yellow background represent genes with no significant identity to those in pNDM-40-1. oriT, origin of transfer; traD, conjugal transfer gene; traA, MobA/L-type relaxase gene; res, resolvase gene; nuc, nuclease homologue; virB1 to -B11 and virD4, putative T4SS genes; trg, putative lytic transglycosylase gene; parA, putative plasmid partition gene; rms, putative type I restriction-modification system methyltransferase subunit gene; aphA6, aminoglycoside resistance gene; ble, bleomycin resistance gene; trpF, phosphoribosylanthranilate isomerase gene; tat, twin-arginine translocation pathway signal sequence domain gene; cutA1, periplasmic divalent cation tolerance gene; groES, cochaperonin gene; groEL, chaperonin gene; oriIS, origin of insertion of ISCR27; res, putative resolvase gene; msr(E) and mph(E), macrolide resistance genes.
The blaNDM-1 gene in pNDM-40-1 is found within a Tn125 transposon (Fig. 2), as with other pNDM-BJ01-like plasmids (12, 13, 16). Tn125 in pNDM-40-1 has two deletions relative to Tn125 in pNDM-BJ01: a 1,298-bp deletion from the 3′ end of ble to tat and a 150-bp deletion within ISCR27 (see Fig. S3 in the supplemental material). pNDM-BJ02 and contig 5 from Acinetobacter soli TCM341 lack the 3′ ISAba125, while in pAB-D499 and pM131_NDM-1, there is an ISAba11 inserted at the 3′ end of the element. In pNDM-AB, a large part of the context from cutA1 to the 3′ ISAba125 is replaced by the macrolide resistance genes msr(E) and mph(E) (23). In all pNDM-BJ01-like plasmids, the aminoglycoside resistance gene aphA6 and an ISAba14 element are found immediately upstream of Tn125 (Fig. 2).
GenBank searches show that blaNDM-1 contexts in Enterobacteriaceae have high degrees of identity with the blaNDM-1 context from pNDM-BJ01-like plasmids. In most cases, this is restricted to genes that make up part of the full Tn125 element harboring blaNDM-1, with at least a fragment of the ISAba125 upstream of blaNDM-1 and the ble and trpF genes being present in almost all cases. Four sequences from Enterobacteriaceae with regions of close identity to the blaNDM-1 context in pNDM-BJ01, which included part of the plasmid backbone, were available at the time of writing (31) (Fig. 2). These were in the plasmids pPrY2001 from Providencia rettgeri (KF295828), pMR0211 from Providencia stuartii (JN687470), pNDM-1_Dok01 from E. coli (AP012208), and pECL3-NDM-1 from Enterobacter cloacae (KC887917). Plasmid pPrY2001 contains the most extensive region of identity. The sequence is nearly identical to that found in pNDM-BJ01 from the far 3′ end of traA to the resolvase gene, with the main difference being the absence of the 3′ ISAba125.
All pNDM-BJ01-like plasmids share a region with genes coding for a type IV secretion system (T4SS) involved in constructing the conjugation machinery and mediating conjugative transfer of plasmid DNA to recipient bacteria (16). In addition, all pNDM-BJ01-like plasmids contain genes proposed to code for a plasmid partition system (parA) and a putative zeta-toxin, which may contribute to plasmid stability through a toxin-antitoxin addiction system. It has not been possible to identify the replicase or the origin of replication of these plasmids. However, the wide range of replication strategies already described means that the lack of an identifiable replicase is not entirely surprising (6, 32).
The blaNDM-1 context in A. haemolyticus.
PCR analysis revealed that the A. haemolyticus strain, but not the other two Acinetobacter species isolates from Karachi, contained several regions of a pNDM-BJ01-like plasmid backbone. The immediate blaNDM-1 context in A. haemolyticus 69122-EW was linked to pNDM-BJ01-like backbone genes traA upstream and the resolvase gene downstream (Fig. 2). The immediate context differed from that described in pNDM-BJ01 in that most of Tn125 was missing.
A previously uncharacterized insertion sequence, ISAha3, most similar to ISAlw1 (95% amino acid [AA] identity between transposases), was inserted between ble and the putative resolvase gene. No direct repeats (DRs) were observed, but this was not uncommon for other closely related ISs deposited in ISFinder. It is possible that transposition of ISAha3 resulted in deletion of the sequence often found between ble and res and also resulted in the loss of one of the DRs (33). S1 PFGE and in-gel hybridization showed that blaNDM-1 was present on ∼45-kb plasmids in A. haemolyticus 69122-EW, pNDM-69122.
Conjugative transfer and stability of plasmids harboring blaNDM-1.
Transconjugants were obtained from mating experiments with A. bereziniae CHI-40-1 and A. haemolyticus 69122-EW donors, in both E. coli UAB190 and A. pittii AG3528 recipients, at rates of 10−4 to 10−5 transconjugants per recipient cell. All putative transconjugants tested were found to be the recipient background species by MALDI-TOF and to be blaNDM-1 positive by PCR. MICs to all β-lactams except aztreonam were elevated in selected transconjugants (Table 2).
PCR analysis showed that the relaxase gene, traA, and other sections of the pNDM-BJ01-like backbone were present in all transconjugant colonies tested. In A. pittii AG3528 transconjugants, in-gel hybridization showed that both traA and blaNDM-1 were present on ∼45-kb plasmids, as expected for pNDM-40-1. However, in E. coli UAB190 transconjugants, these genes were both present on either the chromosome or ∼90-kb plasmids (Fig. 1b) (traA data not shown).
Probing of S1 PFGE gels of CHI-40-1, UAB190NDMP2, and AG3528NDMP1 over the course of a 14-day passage showed that blaNDM-1-positive bands did not alter in size (see Fig. S4 in the supplemental material). The intensity of the blaNDM-1 bands in CHI-40-1 was similar over the course of the experiment with and without meropenem selection. The intensity of the blaNDM-1 bands for both transconjugant strains was stable with antibiotic selection but decreased significantly over the course of the passage without meropenem selection.
Regression analysis of ΔΔCT values from qPCR experiments (see Fig. S5 in the supplemental material) showed statistically significant falls in the quantities of blaNDM-1 and traA template over the course of the passage experiment for transconjugant strains without meropenem selection. For the donor strain, CHI-40-1, there was little change. For all strains tested, blaNDM-1 and traA remained detectable throughout the 14-day passage experiment, even in the absence of antibiotic selection.
DISCUSSION
All the complete NDM-1 plasmid sequences from Acinetobacter spp. that we were able to identify in GenBank were from Chinese isolates and were similar to pNDM-BJ01. pNDM-BJ01 contains a single ApaI restriction site, in keeping with the ∼45-kb bands present in both the S1 and ApaI gels. We conclude that CHI-40-1 harbors just one plasmid containing blaNDM-1 and that the 300-kb band represented residual supercoiled plasmid DNA.
The pNDM-BJ01-like plasmids have a GC content similar to that of most Acinetobacter species WGSs deposited in GenBank (∼40%). All examples of these plasmids harboring blaNDM-1 have very high levels of identity with one another. The small number of related sequences not associated with blaNDM-1 are more distantly related to pNDM-BJ01-like plasmids but are also found exclusively in Acinetobacter spp. We propose that these findings are compatible with this plasmid lineage having evolved within the Acinetobacter genus and with the acquisition of blaNDM-1 being a relatively recent event.
Most descriptions of conjugative transfer of blaNDM-1 from Acinetobacter spp. in vitro are for isolates with pNDM-BJ01-like plasmids (15, 16, 19, 23). Conjugation rates were similar for the pNDM-BJ01-like plasmids studied here into E. coli and A. pittii recipients. The stability of both blaNDM-1 and traA, coding for the pNDM-BJ01-like relaxase, was similar in E. coli and A. pittii transconjugants. However, in the E. coli transconjugants studied in more detail, this apparently required recombination of pNDM-40-1, or part of it, with the chromosome or another plasmid. Although pNDM-40-1 was therefore no longer likely to function as an autonomously replicating plasmid, this still suggests a potential means by which blaNDM-1 contexts could have spread from Acinetobacter spp. to Enterobacteriaceae. This may explain why no complete pNDM-BJ01-like plasmid has yet been described in Enterobacteriaceae. However, sequence data now strongly suggest that at least some blaNDM-1 contexts in Enterobacteriaceae are derived from pNDM-BJ01-like plasmids, since several examples demonstrate high levels of identity with the plasmid backbone sequences which flank ISAba14, aphA6, and Tn125 containing blaNDM-1 (31).
Recent reports suggest that NDM-1-producing Enterobacteriaceae may be more common than originally demonstrated in China (21, 22), and it is possible that the spread of blaNDM-1 into Enterobacteriaceae could have initiated there. However, the high prevalence in some regions of the Indian subcontinent of NDM-producing Enterobacteriaceae has been well documented, and most reports of travel-associated colonization with NDM-producing bacteria involved the subcontinent (5, 8). It is therefore potentially significant that pNDM-BJ01-like plasmids have now been identified from India and Pakistan and from returning travelers to the region (34, 35). Insufficient data are available to establish whether these cases could be linked to recent spread from China or whether these plasmids are prevalent in a wider geographical region including the Indian subcontinent.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to the library construction, sequencing, and core informatics teams at the Wellcome Trust Sanger Institute for assistance, to Mandy Wootton and her team at the Specialist Antimicrobial Chemotherapy Unit, Public Health Wales, for their support and advice, to Keith Jolley and Samuel Sheppard for access to and advice regarding the use of the rMLST database, and to Rosemary Barnes for assistance with editing the manuscript.
This work was supported by the British Society of Antimicrobial Chemotherapy (GA2011-07P to L.S.J. and GA2012-02OS to A.M.), the European Commission Leonardo Da Vinci Lifelong Learning Programme (2012-1-PT1-LEO02-11437 to M.J.C.), the Canadian Institutes of Health Research and Medical Research Council, Canada-UK Partnership on Antibiotic Resistance (G1100135 to M.A.T.), and the UK Clinical Research Collaboration, Translational Infection Research (TIR) Initiative (G1000803 to S.J.P.).
We have no conflicts to declare.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.03242-14.
REFERENCES
- 1.Higgins PG, Dammhayn C, Hackel M, Seifert H. 2010. Global spread of carbapenem-resistant Acinetobacter baumannii. J Antimicrob Chemother 65:233–238. doi: 10.1093/jac/dkp428. [DOI] [PubMed] [Google Scholar]
- 2.Visca P, Seifert H, Towner KJ. 2011. Acinetobacter infection—an emerging threat to human health. IUBMB Life 63:1048–1054. doi: 10.1002/iub.534. [DOI] [PubMed] [Google Scholar]
- 3.Toleman MA, Spencer J, Jones L, Walsh TR. 2012. blaNDM-1 is a chimera likely constructed in Acinetobacter baumannii. Antimicrob Agents Chemother 56:2773–2776. doi: 10.1128/AAC.06297-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walsh TR, Weeks J, Livermore DM, Toleman MA. 2011. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect Dis 11:355–362. doi: 10.1016/S1473-3099(11)70059-7. [DOI] [PubMed] [Google Scholar]
- 5.Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, Chaudhary U, Doumith M, Giske CG, Irfan S, Krishnan P, Kumar AV, Maharjan S, Mushtaq S, Noorie T, Paterson DL, Pearson A, Perry C, Pike R, Rao B, Ray U, Sarma JB, Sharma M, Sheridan E, Thirunarayan MA, Turton J, Upadhyay S, Warner M, Welfare W, Livermore DM, Woodford N. 2010. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis 10:597–602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carattoli A. 2013. Plasmids and the spread of resistance. Int J Med Microbiol 303:298–304. doi: 10.1016/j.ijmm.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 7.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. 2009. Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother 53:5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson AP, Woodford N. 2013. Global spread of antibiotic resistance: the example of New Delhi metallo-β-lactamase (NDM)-mediated carbapenem resistance. J Med Microbiol 62:499–513. doi: 10.1099/jmm.0.052555-0. [DOI] [PubMed] [Google Scholar]
- 9.Karthikeyan K, Thirunarayan MA, Krishnan P. 2010. Coexistence of blaOXA-23 with blaNDM-1 and armA in clinical isolates of Acinetobacter baumannii from India. J Antimicrob Chemother 65:2253–2254. doi: 10.1093/jac/dkq273. [DOI] [PubMed] [Google Scholar]
- 10.Bharadwaj R, Joshi S, Dohe V, Gaikwad V, Kulkarni G, Shouche Y. 2012. Prevalence of New Delhi metallo-β-lactamase (NDM-1)-positive bacteria in a tertiary care centre in Pune, India. Int J Antimicrob Agents 39:265–266. doi: 10.1016/j.ijantimicag.2011.09.027. [DOI] [PubMed] [Google Scholar]
- 11.Jones LS, Toleman MA, Weeks JL, Howe RA, Walsh TR, Kumarasamy KK. 2014. Plasmid carriage of blaNDM-1 in clinical Acinetobacter baumannii isolates from India. Antimicrob Agents Chemother 58:4211–4213. doi: 10.1128/AAC.02500-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonnin RA, Poirel L, Naas T, Pirs M, Seme K, Schrenzel J, Nordmann P. 2012. Dissemination of New Delhi metallo-β-lactamase-1-producing Acinetobacter baumannii in Europe. Clin Microbiol Infect 18:E362–365. doi: 10.1111/j.1469-0691.2012.03928.x. [DOI] [PubMed] [Google Scholar]
- 13.Pfeifer Y, Wilharm G, Zander E, Wichelhaus TA, Gottig S, Hunfeld KP, Seifert H, Witte W, Higgins PG. 2011. Molecular characterization of blaNDM-1 in an Acinetobacter baumannii strain isolated in Germany in 2007. J Antimicrob Chemother 66:1998–2001. doi: 10.1093/jac/dkr256. [DOI] [PubMed] [Google Scholar]
- 14.Poirel L, Bonnin RA, Boulanger A, Schrenzel J, Kaase M, Nordmann P. 2012. Tn125-related acquisition of blaNDM-like genes in Acinetobacter baumannii. Antimicrob Agents Chemother 56:1087–1089. doi: 10.1128/AAC.05620-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Y, Zhou Z, Jiang Y, Yu Y. 2011. Emergence of NDM-1-producing Acinetobacter baumannii in China. J Antimicrob Chemother 66:1255–1259. doi: 10.1093/jac/dkr082. [DOI] [PubMed] [Google Scholar]
- 16.Hu H, Hu Y, Pan Y, Liang H, Wang H, Wang X, Hao Q, Yang X, Xiao X, Luan C, Yang Y, Cui Y, Yang R, Gao GF, Song Y, Zhu B. 2012. Novel plasmid and its variant harboring both a blaNDM-1 gene and type IV secretion system in clinical isolates of Acinetobacter lwoffii. Antimicrob Agents Chemother 56:1698–1702. doi: 10.1128/AAC.06199-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho PL, Li Z, Lai EL, Chiu SS, Cheng VC. 2012. Emergence of NDM-1-producing Enterobacteriaceae in China. J Antimicrob Chemother 67:1553–1555. doi: 10.1093/jac/dks095. [DOI] [PubMed] [Google Scholar]
- 18.Wang X, Liu W, Zou D, Li X, Wei X, Shang W, Wang Y, Li H, Huan Li YW, He X, Huang L, Yuan J. 2013. High rate of New Delhi metallo-β-lactamase 1-producing bacterial infection in China. Clin Infect Dis 56:161–162. doi: 10.1093/cid/cis782. [DOI] [PubMed] [Google Scholar]
- 19.Yang J, Chen Y, Jia X, Luo Y, Song Q, Zhao W, Wang Y, Liu H, Zheng D, Xia Y, Yu R, Han X, Jiang G, Zhou Y, Zhou W, Hu X, Liang L, Han L. 2012. Dissemination and characterization of NDM-1-producing Acinetobacter pittii in an intensive care unit in China. Clin Microbiol Infect 18:E506–E513. doi: 10.1111/1469-0691.12035. [DOI] [PubMed] [Google Scholar]
- 20.Zhang C, Qiu S, Wang Y, Qi L, Hao R, Liu X, Shi Y, Hu X, An D, Li Z, Li P, Wang L, Cui J, Wang P, Huang L, Klena JD, Song H. 2013. Higher isolation of NDM-1 producing Acinetobacter baumannii from the sewage of the hospitals in Beijing. PLoS One 8:e64857. doi: 10.1371/journal.pone.0064857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qin S, Fu Y, Zhang Q, Qi H, Wen JG, Xu H, Xu L, Zeng L, Tian H, Rong L, Li Y, Shan L, Yu Y, Feng X, Liu HM. 2014. High incidence and endemic spread of NDM-1-positive Enterobacteriaceae in Henan Province, China. Antimicrob Agents Chemother 58:4275–4282. doi: 10.1128/AAC.02813-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou G, Guo S, Luo Y, Ye L, Song Y, Sun G, Guo L, Chen Y, Han L, Yang J. 2014. NDM-1-producing strains, family Enterobacteriaceae, in hospital, Beijing, China. Emerg Infect Dis 20:340–342. doi: 10.3201/eid2002.121263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang WJ, Lu Z, Schwarz S, Zhang RM, Wang XM, Si W, Yu S, Chen L, Liu S. 2013. Complete sequence of the blaNDM-1-carrying plasmid pNDM-AB from Acinetobacter baumannii of food animal origin. J Antimicrob Chemother 68:1681–1682. doi: 10.1093/jac/dkt066. [DOI] [PubMed] [Google Scholar]
- 24.Mata C, Navarro F, Miro E, Walsh TR, Mirelis B, Toleman M. 2011. Prevalence of SXT/R391-like integrative and conjugative elements carrying blaCMY-2 in Proteus mirabilis. J Antimicrob Chemother 66:2266–2270. doi: 10.1093/jac/dkr286. [DOI] [PubMed] [Google Scholar]
- 25.Mutreja A, Kim DW, Thomson NR, Connor TR, Lee JH, Kariuki S, Croucher NJ, Choi SY, Harris SR, Lebens M, Niyogi SK, Kim EJ, Ramamurthy T, Chun J, Wood JL, Clemens JD, Czerkinsky C, Nair GB, Holmgren J, Parkhill J, Dougan G. 2011. Evidence for several waves of global transmission in the seventh cholera pandemic. Nature 477:462–465. doi: 10.1038/nature10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drummond A, Ashton B, Buxton S, Cheung M, Cooper A, Duran C, Heled J, Kearse M, Markowitz S, Moir R, Stones-Havas S, Sturrock S, Swidan F, Thierer T, Wilson A. 2012. Geneious v5.6. Biomatters, Auckland, New Zealand. [Google Scholar]
- 28.Patzer JA, Walsh TR, Weeks J, Dzierzanowska D, Toleman MA. 2009. Emergence and persistence of integron structures harbouring VIM genes in the Children's Memorial Health Institute, Warsaw, Poland, 1998-2006. J Antimicrob Chemother 63:269–273. doi: 10.1093/jac/dkn512. [DOI] [PubMed] [Google Scholar]
- 29.ARPAC May 2009, posting date PFGE typing protocol recommended by ARPAC for Acinetobacter baumannii. http://phagetherapylightandshade.blogspot.com/2009/05/pfge-typing-protocol-recommended-by.html.
- 30.Johnson G, Nolan T, Bustin SA. 2013. Real-time quantitative PCR, pathogen detection and MIQE. Methods Mol Biol 943:1–16. doi: 10.1007/978-1-60327-353-4_1. [DOI] [PubMed] [Google Scholar]
- 31.Partridge SR, Iredell JR. 2012. Genetic contexts of blaNDM-1. Antimicrob Agents Chemother 56:6065–6067. (Author reply, 56:6071.) doi: 10.1128/AAC.00117-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Francia MV, Varsaki A, Garcillan-Barcia MP, Latorre A, Drainas C, de la Cruz F. 2004. A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol Rev 28:79–100. doi: 10.1016/j.femsre.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 33.Weinert TA, Schaus NA, Grindley ND. 1983. Insertion sequence duplication in transpositional recombination. Science 222:755–765. doi: 10.1126/science.6314502. [DOI] [PubMed] [Google Scholar]
- 34.Bogaerts P, Huang TD, Rezende de Castro R, Bouchahrouf W, Glupczynski Y. 2013. Could Acinetobacter pittii act as an NDM-1 reservoir for Enterobacteriaceae? J Antimicrob Chemother 68:2414–2415. doi: 10.1093/jac/dkt201. [DOI] [PubMed] [Google Scholar]
- 35.McGann P, Milillo M, Clifford RJ, Snesrud E, Stevenson L, Backlund MG, Viscount HB, Quintero R, Kwak YI, Zapor MJ, Waterman PE, Lesho EP. 2013. Detection of New Delhi metallo-beta-lactamase (encoded by blaNDM-1) in Acinetobacter schindleri during routine surveillance. J Clin Microbiol 51:1942–1944. doi: 10.1128/JCM.00281-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.