Abstract
The anaerobic oxidation of methane (AOM) is an important sink of methane that plays a significant role in global warming. AOM was first found to be coupled with sulfate reduction and mediated by anaerobic methanotrophic archaea (ANME) and sulfate-reducing bacteria (SRB). ANME, often forming consortia with SRB, are phylogenetically related to methanogenic archaea. ANME-1 is even able to produce methane. Subsequently, it has been found that AOM can also be coupled with denitrification. The known microbes responsible for this process are Candidatus Methylomirabilis oxyfera (M. oxyfera) and Candidatus Methanoperedens nitroreducens (M. nitroreducens). Candidatus Methylomirabilis oxyfera belongs to the NC10 bacteria, can catalyze nitrite reduction through an “intra-aerobic” pathway, and may catalyze AOM through an aerobic methane oxidation pathway. However, M. nitroreducens, which is affiliated with ANME-2d archaea, may be able to catalyze AOM through the reverse methanogenesis pathway. Moreover, manganese (Mn4+) and iron (Fe3+) can also be used as electron acceptors of AOM. This review summarizes the mechanisms and associated microbes of AOM. It also discusses recent progress in some unclear key issues about AOM, including ANME-1 in hypersaline environments, the effect of oxygen on M. oxyfera, and the relationship of M. nitroreducens with ANME.
Keywords: Anaerobic methanotrophic archaea, anaerobic oxidation of methane, Candidatus Methanoperedens nitroreducens, Candidatus Methylomirabilis oxyfera, methane
Introduction
Methane is the second most abundant greenhouse gas after carbon dioxide (CO2), which accounts for 14% of global greenhouse gas emissions (EPA 2006). The concentration of methane in the atmosphere has increased ∼2.5 times than the preindustrial level, rising from 720 ppb in 1750 to 1803 ppb in 2011 (Hartmann et al. 2013). Although the methane concentration in the atmosphere is lower than the CO2 concentration (391 ppm), methane is 25-fold more effective in trapping heat in the atmosphere than CO2 on a per-molecule basis (IPCC 2007). Methane contributes to ∼30% of the anthropogenic warming, with the radiative forcing of 0.48 Wm−2 in 2011 (Myhre et al. 2013). After maintaining a relatively stable level for approximately a decade in the 1990s, the atmospheric methane concentration began to grow in 2007 (Hartmann et al. 2013). The concentration of methane in the atmosphere is determined by the balance of sources and sinks. The anaerobic oxidation of methane (AOM) is an important sink of the atmospheric methane concentration (Conrad 2009), which significantly impacts global warming. In marine sediments, the total amount of gas hydrates is up to 150–3000 times the atmospheric methane concentration (500,000–10,000,000 Tg CH4) (Reeburgh 2007). Fortunately, most of the mobilized CH4 is consumed through anaerobic methane oxidation, with a consumption rate of approximately 70–300 Tg CH4 year−1. Without this process, there would be an additional 10–60% of CH4 in the atmosphere (Conrad 2009).
AOM was first discovered in 1976 and is coupled with sulfate reduction in marine sediments (Reeburgh 1976, 1980). However, the responsible microorganisms for this process were actually identified ∼20 years later (Hinrichs et al. 1999; Boetius et al. 2000; Bian et al. 2001). In 2006, a new AOM process named nitrite-dependent anaerobic methane oxidation (N-DAMO) was reported; this process is coupled with denitrification (Raghoebarsing et al. 2006). It was shown that nitrate could also be an electron acceptor of AOM in addition to nitrite (Haroon et al. 2013). Likewise, Beal et al. (2009) suggested that AOM is coupled with the reduction of manganese (Mn4+) and iron (Fe3+) in marine sediments. Overall, there are three different processes of AOM depending on the different electron acceptors: sulfate-dependent anaerobic methane oxidation (S-DAMO) (Fig.1A), nitrate/nitrite-dependent anaerobic methane oxidation (N-DAMO) (Fig.1C and D), and metal ion (Mn4+ and Fe3+)-dependent anaerobic methane oxidation (M-DAMO) (Fig.1B). This review summarizes the biochemistry and microbiology of these three AOM processes, including the mechanisms and distribution of AOM processes, the responsible microbes, and their peculiar properties. Moreover, this review also discusses several key issues about the recent progress of AOM that are still unclear.
Figure 1.
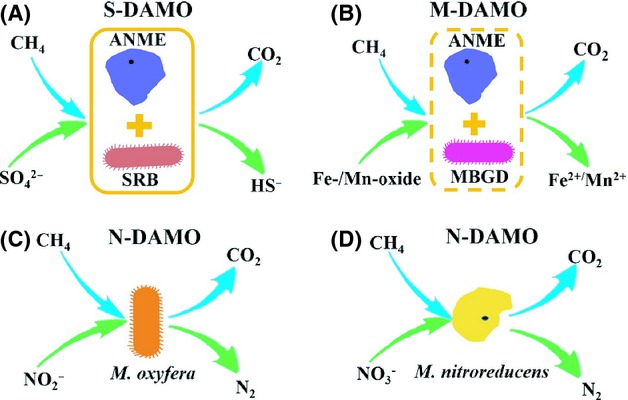
Three different models of anaerobic methane oxidation (AOM) depending on the different electron acceptors: (A) sulfate-dependent anaerobic methane oxidation (S-DAMO); (B) metal ion (Mn4+ and Fe3+)-dependent anaerobic methane oxidation (M-DAMO); and (C, D) nitrate/nitrite-dependent anaerobic methane oxidation (N-DAMO). ANME, an anaerobic methanotrophic archaea; SRB, sulfate-reducing bacteria; M. oxyfera, Candidatus Methylomirabilis oxyfera; M. nitroreducens, Candidatus Methanoperedens nitroreducens; MBGD, marine benthic group D.
Sulfate-Dependent Anaerobic Methane Oxidation
Mechanism and distribution of S-DAMO
S-DAMO (eqs. 1–3) was discovered during geochemical studies conducted ∼40 years ago; these studies first suggested the process of AOM (Reeburgh 1976). In the following decades, more evidences are accumulated indicating that S-DAMO is mainly distributed in marine environments (Reeburgh 1976, 2007; Gal'chenko et al. 2004; Durisch-Kaiser et al. 2005; Orphana et al. 2005; Treude et al. 2007) and in freshwater environments (Murase and Kimura 1994; Grossman et al. 2002; Eller et al. 2005; Alain et al. 2006; Smemo and Yavitt 2007; Miyashita et al. 2009). These studies demonstrated that S-DAMO exists widely in natural ecosystems where it may play an important role in the biogeochemical cycling of carbon and sulfur.
| 1 |
| 2 |
| 3 |
However, the exact metabolic mechanism of S-DAMO still remains unclear. Hoehler et al. (1994) proposed that methane is oxidized via a reversed methanogenesis (a reversal of CO2 reduction) under anoxic conditions. The product of methane oxidation is hydrogen (H2), which is used by sulfate-reducing bacteria (SRB) to yield bicarbonate and sulfide, with methane and sulfate in the ratio of 1:1 (Nauhaus et al. 2002, 2005). Many proteomic and genomic studies have been documented which support for the reverse methanogenesis hypothesis. A new nickel protein (Ni-protein I) was extracted from microbial mats suited for biochemical studies of AOM; this protein may play a catalytic role in AOM and is similar to the nickel cofactor F430 of methyl coenzyme M reductase (MCR), the terminal enzyme of methanogenesis (Krüger et al. 2003; Mayr et al. 2008). Scheller et al. (2010) discovered a purified MCR from a methanogen that could cleave the particularly strong C-H bond of methane to form methyl coenzyme M. The metagenome and mRNA expression analyses of ANME-1 (anaerobic methanotrophic archaea) support the reverse methanogenesis hypothesis (Meyerdierks et al. 2010). Four groups of the mcrA gene (coding for the α-subunit of MCR) were shown to correspond to the ANME community (Hallam et al. 2003). Based on observations of the whole-genome shotgun library and the fosmid library, Hallam et al. (2004) identified many genes associated with methanogenesis. Recently, a complete reverse methanogenesis pathway, including all the mcr subunit genes (mcrABCDG) and the F420-dependent 5,-10-methenyltetrahydromethanopterin reductase (mer) genes, has been identified in the genome of an ANME organism (Haroon et al. 2013).
However, there were repeated attempts that failed to induce reverse methanogenesis using low H2 and high CH4 concentrations (Valentine and Reeburgh 2000). Additionally, the thermodynamic yield of reverse methanogenesis is only −16 kJ mol−1(CH4), which is too low to be shared by an archaea and an SRB. Some bacterial lipids (most likely from SRB) have been found to be isotopically depleted (Thiel et al. 1999; Hinrichs et al. 2000; Pancost et al. 2000), which is difficult to explain without an interspecies carbon transfer. Moreover, instead of H2, many Methanosarcinales use acetate and methylation during methanogenesis, which were shown to be involved in AOM in some environments (Hinrichs et al. 1999). Therefore, reverse methanogenesis may not be the only process of S-DAMO. Subsequently, two new alternative mechanisms of S-DAMO were raised that have greater thermodynamic yields, involve an interspecies carbon transfer, and are identical to the results of phylogenetic analyses (Valentine and Reeburgh 2000).
| 4 |
| 5 |
| 6 |
| 7 |
The first mechanism (eqs. 4–6) involves the formation of acetate and H2 from CH4 and H2O (eq. 4), which would then be consumed with 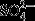 by SRB (eqs. 5 and 6). Acetate, the intermediate, transfers a carbon between CH4 and
by SRB (eqs. 5 and 6). Acetate, the intermediate, transfers a carbon between CH4 and 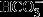 . The net reaction (eq. 7) of this new mechanism is twice the reaction of reverse methanogenesis (eq. 3), which should generate twice as much free energy as the mechanism of reverse methanogenesis.
. The net reaction (eq. 7) of this new mechanism is twice the reaction of reverse methanogenesis (eq. 3), which should generate twice as much free energy as the mechanism of reverse methanogenesis.
| 8 |
| 9 |
In the second mechanism, acetate is produced from CH4 and CO2 (eq. 8) and is consumed with 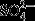 by SRB (eq. 9). Acetate is not shown in the net reaction (eq. 3) because it is an intermediate. This new hypothesis needs to be tested in future studies, but it does explain the documented findings that are inconsistent with reverse methanogenesis.
by SRB (eq. 9). Acetate is not shown in the net reaction (eq. 3) because it is an intermediate. This new hypothesis needs to be tested in future studies, but it does explain the documented findings that are inconsistent with reverse methanogenesis.
CH4 oxidation could not be inhibited by high pressure H2, which suggests that H2 is not an intermediate in AOM (Moran et al. 2008). Hence, Moran proposed a new model for S-DAMO that is named methylogenesis. The methylogenesis model includes two steps: the formation of methyl sulfides from CH4 and CO2 by archaea (eq. 10) and the following consumption of methyl sulfides by SRB (eq. 11). Methanethiol was concluded to play an interspecies role in AOM because the CH4 oxidation rates dropped 68% in the experiments treated with methanethiol (Moran et al. 2008).
| 10 |
| 11 |
However, there is not a definite mechanism to account for S-DAMO in various environments, which may be due to differences in the environmental conditions and the physiological characteristics of the responsible microbes.
The responsible microbes for S-DAMO and their peculiar properties
The type of microbes responsible for S-DAMO are termed anaerobic methanotrophs (ANME) and are represented by three different phylogenetic clusters (ANME-1, ANME-2, and ANME-3) (Hinrichs et al. 1999; Orphan et al. 2001a; Knittel et al. 2005; Schleper et al. 2005; Niemann et al. 2006). ANME-1 and ANME-2 are the most abundant groups of ANME, which are widely distributed in various anaerobic areas and produce methane, while ANME-3 archaea mostly exist in submarine mud volcanoes or occasionally in marine methane seep (Knittel and Boetius 2009; Meulepas et al. 2009). ANME-1 is divided into two subgroups, ANME-1a and ANME-1b (Knittel et al. 2005), while ANME-2 is divided into four distinct subgroups, designated ANME-2a, ANME-2b, ANME-2c, and ANME-2d (Orphan et al. 2001a,b; Mills et al. 2003). The features of these three ANME groups are summarized in Table1.
Table 1.
Features of the three ANME groups
| ANME-1 | ANME-2 | ANME-3 | |
|---|---|---|---|
| Common features | |||
| Habitat | Various anaerobic areas (marine sediments, cold seep, lake sediments, soils, oil field sediments, etc.) | Various anaerobic areas (marine sediments, cold seep, lake sediments, soils, oil field sediments, etc.) | Submarine mud volcanoes and marine methane seep |
| Subgroup | a, b | a, b, c, d | ND |
| Pure culture | No | No | No |
| Features associated with SRB | |||
| Associated SRB | Desulfosarcina and Desulfococcus | Desulfosarcina and Desulfococcus | Desulfobulbus |
| Associated form | Often loose | Often form structured consortia | Often form structured consortia |
| Associated necessity | No | No | No |
| Single-cell form | Often | Yes | Yes |
| Features related to methanogens | |||
| Related methanogen | Methanosarcinales and Methanomicrobiales | Methanosarcinales | Methanococcoides |
| Shape | Often rod shaped (like Methanobacteriales and Methanomicrobiales) | Often coccoid shaped (like Methanosarcinales) | Often coccoid shaped (like Methanosarcinales) |
| Harbour mcrA | Yes | Yes | Yes |
| mcrA subgroup | a, b (identified) | c, d (identified) | f (identified) |
| e (possible) | |||
| Produce methane | Yes | ND | ND |
| Autofluorescent under UV light (like methanogens) | Yes | Yes | Yes |
ANME, anaerobic methanotrophic archaea; ND, not determined; SRB, sulfate-reducing bacteria.
These three archaeal groups of ANME are phylogenetically related to different methanogenic archaea. ANME-1 are distantly related to the orders Methanosarcinales and Methanomicrobiales (Michaelis et al. 2002; Orphan et al. 2002; Knittel et al. 2005), ANME-2 are affiliated with the order Methanosarcinales (Orphan et al. 2001b; Knittel et al. 2005), and ANME-3 are related to the genera Methanococcoides (Niemann et al. 2006; Lösekann et al. 2007; Lazar et al. 2011). The lipid structures of ANME are quite similar to those of methanogens (Elvert and Suess 1999; Hinrichs et al. 1999), and the shapes of ANME are also similar to methanogenic archaea. ANME-1 often appear as rod-shaped cells (Orphan et al. 2002), as do methanogens of Methanobacteriales and Methanomicrobiales (Lloyd et al. 2011); ANME-2 and ANME-3 often exist as coccoid-shaped cells and form clusters (Orphan et al. 2002), as do methanogens of Methanosarcinales (Lloyd et al. 2011). In addition, there are other remarkable similarities between ANME and methanogens. MCR, which is present in all known methanogens (Luton et al. 2002), has also been found in microbial mats that are dominated by ANME (Krüger et al. 2003); additionally, its evolutionary path in ANME mirrors that of methanogens (Hallam et al. 2003). In addition, six subgroups of mcrA (a, b, c, d, e, and f) among ANME archaea have been defined (Lösekann et al. 2007). Due to the presence of an F420 flavin-derived coenzyme, ANME fluoresce blue green under ultraviolet (UV) light, which is a notable characteristic of methanogens (Michaelis et al. 2002; Knittel et al. 2005; Lösekann et al. 2007). The genomes of ANME-1 and ANME-2 contain all homologous genes for enzymes associated with the canonical seven-step methanogenic pathway, although one enzyme (N5,N10-methenyl-tetrahydromethanopterin reductase) encoded by mer was not found in ANME-1 (Hallam et al. 2004; Meyerdierks et al. 2010). The genes, encoding the same carbon fixation pathway as methanogens, were also found in ANME-1 (Meyerdierks et al. 2010). ANME-2 have been shown to be cable to fix N2 (Dekas et al. 2009), as are methanogens in Methanosarcinales (Murray and Zinder 1984; Leigh 2000). Furthermore, ANME-1 have been shown to function as a methanogen in the methane production zone (Lloyd et al. 2011).
ANME often form consortia with SRB to catalyze S-DAMO (Fig.1A). ANME-1 and ANME-2 are associated with SRB of the Desulfosarcina–Desulfococcus (DSS) branch of Deltaproteobacteria (Boetius et al. 2000; Orphan et al. 2002), while ANME-3 are associated with SRB of the Desulfobulbus (DBB) branch (Niemann et al. 2006), also belonging to Deltaproteobacteria. ANME-1 are always loosely associated with SRB (Knittel et al. 2005), while ANME-2 and ANME-3 are usually associated with SRB forming structured consortia (Orphan et al. 2002; Niemann et al. 2006). The typical observed ANME/DSS ratio is 1:1 to 1:3 in a shell-type consortia (Boetius et al. 2000; Orcutt and Meile 2008); however, a very different ANME/DSS ratio of 7:1 was observed in hypersaline environments (Maignien et al. 2013). The ratio of ANME-3 cells to DBB cells is »1 (Lösekann et al. 2007), which differs strongly from the ANME/DSS ratio. However, a physical association with SRB is not obligatory for all three clades of ANME archaea. Most ANME-1 archaea exist as single cells or form monospecific chains without any attached partner (Orphan et al. 2002; Maignien et al. 2013). ANME-2 (Treude et al. 2005) and ANME-3 (Lösekann et al. 2007) have also been found to exist without sulfate-reducing partners. In addition, the syntrophical partners of ANME are not limited to SRB. ANME-2 are able to live syntrophically with various bacteria of Deltaproteobacteria as well as with Sphingomonas spp. of Alphaproteobacteria and Burkholderia of Betaproteobacteria (Knittel and Boetius 2009). ANME-3 have been found to occur with yet unidentified bacteria, forming mixed-type aggregates (Lösekann et al. 2007).
Nitrate/Nitrite-Dependent Anaerobic Methane Oxidation
Mechanism and distribution of N-DAMO
Although there were documented lines of environmental and experimental evidence of N-DAMO years ago (Smith et al. 1991; Islas-Lima et al. 2004), N-DAMO was first proposed in 2006 by Raghoebarsing et al. (2006), who discovered an n-damo enrichment culture obtained from an anoxic freshwater sediment rich in nitrate. The mechanism of N-DAMO (eq. 12) was initially hypothesized to be similar to the reverse methanogenesis of S-DAMO, which was mediated by an “ANME archaeon” (Fig.1C) with electron shuttling to denitrification (Raghoebarsing et al. 2006).
| 12 |
NC10, a new bacterial candidate division, was discovered to be prevalent in the n-damo enrichment (Ettwig et al. 2008). At the same time, an archaeon affiliated with Methanosarcinales that was distantly related to ANME-2 (86–87%) and to methanogens (86–88%) was observed to be associated with NC10 members forming consortia. However, this archaeon was not detected in the later stages of incubation. After the apparent disappearance of the archaeon, the rate of N-DAMO did not decrease. This suggested that the archaeon was not obligatory for N-DAMO and that the process of N-DAMO was performed exclusively by members of NC10 (Ettwig et al. 2008). Then, a new “intra-aerobic” pathway of nitrite reduction was discovered based on the complete genome analysis of Candidatus Methylomirabilis oxyfera, the dominant bacterium affiliated with NC10, and based on isotopic labeling experiments (Ettwig et al. 2010). The new mechanism of N-DAMO suggested that 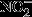 decomposes into NO and O2, which are mainly used to oxidize CH4 (Ettwig et al. 2010). The remaining O2 is consumed in normal respiration by terminal respiratory oxidases (Wu et al. 2011). The whole process might be exclusively mediated by M. oxyfera, the genome of which includes genes encoding the complete pathway for aerobic methane oxidation (Ettwig et al. 2010). The culture, including the NC10 group and the archaea partner, displayed ∼30 times higher nitrate reduction rates than the culture just including the NC10 group (Hu et al. 2009). It was suggested that the archaea might contribute significantly to the reduction of nitrate to nitrite and that the NC10 bacteria might play an important role in the reduction of nitrite (Hu et al. 2009). It is possible that M. oxyfera prefer to use
decomposes into NO and O2, which are mainly used to oxidize CH4 (Ettwig et al. 2010). The remaining O2 is consumed in normal respiration by terminal respiratory oxidases (Wu et al. 2011). The whole process might be exclusively mediated by M. oxyfera, the genome of which includes genes encoding the complete pathway for aerobic methane oxidation (Ettwig et al. 2010). The culture, including the NC10 group and the archaea partner, displayed ∼30 times higher nitrate reduction rates than the culture just including the NC10 group (Hu et al. 2009). It was suggested that the archaea might contribute significantly to the reduction of nitrate to nitrite and that the NC10 bacteria might play an important role in the reduction of nitrite (Hu et al. 2009). It is possible that M. oxyfera prefer to use 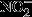 as a substrate rather than
as a substrate rather than 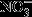 . However, a high concentration of nitrite, an inhibitor to a wide range of microorganisms (Yarbrough et al. 1980), showed a toxic effect on M. oxyfera (Hu et al. 2011). Recently, Haroon et al. (2013) demonstrated that ANME-2d were able to independently achieve AOM (Fig.1D) via reverse methanogenesis (eq. 13) (Raghoebarsing et al. 2006) using nitrate as the terminal electron acceptor; he named the ANME-2d population Candidatus Methanoperedens nitroreducens and the ANME-2d lineage Candidatus Methanoperedenaceae.
. However, a high concentration of nitrite, an inhibitor to a wide range of microorganisms (Yarbrough et al. 1980), showed a toxic effect on M. oxyfera (Hu et al. 2011). Recently, Haroon et al. (2013) demonstrated that ANME-2d were able to independently achieve AOM (Fig.1D) via reverse methanogenesis (eq. 13) (Raghoebarsing et al. 2006) using nitrate as the terminal electron acceptor; he named the ANME-2d population Candidatus Methanoperedens nitroreducens and the ANME-2d lineage Candidatus Methanoperedenaceae.
| 13 |
Even though the mechanism of N-DAMO still remains unclear, the N-DAMO process has been found to occur in different natural freshwater habitats (Raghoebarsing et al. 2006; Ettwig et al. 2008, 2009; Hu et al. 2009; Deutzmann and Schink 2011; Luesken et al. 2011a; Kampman et al. 2012; Wang et al. 2012; Yang et al. 2012; Zhu et al. 2013) where it may play an important role in the biogeochemical cycling of carbon and nitrogen.
The microbes responsible for N-DAMO and their peculiar properties
The microbe responsible for independently coupling AOM to nitrite reduction is called Candidatus Methylomirabilis oxyfera; this microbe is able to reduce nitrite to dinitrogen gas without a nitrous oxide reductase (Ettwig et al. 2010). According to the ultrastructural study of M. oxyfera (Wu et al. 2012), there are three special aspects of M. oxyfera. First, the shape of M. oxyfera is typically polygonal (Wu et al. 2012), which is different from other bacterial shapes described (Hanson and Hanson 1996). Second, the outermost layer of the M. oxyfera cell consists of a putative protein surface layer (S-layer) (Wu et al. 2012) that is known to contribute significantly to mechanical cell stabilization (Engelhardt 2007). Finally, under the growth conditions used in the ultrastructural study, M. oxyfera did not develop intracytoplasmic membranes (ICMs), which are an ultrastructural feature shared by most methanotrophs (Wu et al. 2012). In addition, the M. oxyfera genome contains a complete pmo gene cluster for aerobic methane oxidation (Ettwig et al. 2010), but the genetic analyses of different M. oxyfera enrichment cultures showed that they formed a distinct group affiliated with the pmoA genes of aerobic methanotrophs (Luesken et al. 2011b).
In the N-DAMO enrichment culture, the archaea partner of the NC10 bacteria was subsequently named Candidatus Methanoperedens nitroreducens (Haroon et al. 2013). Candidatus Methanoperedens nitroreducens is able to use nitrate instead of nitrite as the terminal electron acceptor, which is different from M. oxyfera (see Table2) (Ettwig et al. 2010). The N-DAMO pathway of M. nitroreducens is proposed as the reverse of methanogenesis because the genome of M. nitroreducens includes all mcr subunit genes (mcrABCDG) and F420-dependent mer genes for a full reverse methanogenesis (Haroon et al. 2013). In addition, owing to the existence of a full reductive acetyl-CoA (carbon fixation) pathway and acetyl-CoA synthetase in M. nitroreducens (Haroon et al. 2013), it was predicted that M. nitroreducens might be capable of producing acetate, as was suggested for ANME-1 (Meyerdierks et al. 2010). Candidatus Methanoperedens nitroreducens is a new species responsible for N-DAMO, so more studies are needed regarding this new member of N-DAMO in the future.
Table 2.
Comparisons between Candidatus Methylomirabilis oxyfera (M. oxyfera) and Candidatus Methanoperedens nitroreducens (M. nitroreducens), the two known species responsible for N-DAMO
| Features | M. oxyfera | M. nitroreducens |
|---|---|---|
| Habitat | Freshwater environments | Freshwater environments |
| Pure culture | No | No |
| Kingdom | Bacteria | Archaea |
| Affiliated microbes | NC10 | ANME-2d |
| Shape | Atypical polygonal | Irregular coccus |
| Growth conditions | Anaerobic | Anaerobic |
| Produce O2 | Yes | No |
| Electron acceptor (N-DAMO) | Nitrite | Nitrate |
| Mechanism hypothesis (N-DAMO) | Aerobic methane oxidation | Reverse methanogenesis |
| Related gene (N-DAMO) | pmo gene cluster | mcr gene cluster |
ANME, anaerobic methanotrophic archaea; N-DAMO, nitrate/nitrite-dependent anaerobic methane oxidation.
Metal Ion (Mn4+ and Fe3+)-Dependent Anaerobic Methane Oxidation
Similar to the sulfate-dependent mode, manganese (Mn4+) (eq. 14) and iron (Fe3+) (eq. 15) can be used as electron acceptors of AOM in marine methane-seep sediments (Beal et al. 2009). This new pathway that involves coupling AOM with metal ion reduction is called M-DAMO.
| 14 |
| 15 |
An uncultivated group, affiliated with the marine benthic group D (MBGD) (Fig.1B), was found to be the most abundant microorganisms in the sediment of M-DAMO (Beal et al. 2009). ANME-1 and ANME-2 were also identified, while a small percentage of ANME-3 were observed only in the subsequent manganese incubations (Beal et al. 2009). Although the mechanism of M-DAMO and the responsible microbes involved still remains unclear, M-DAMO may play an important role in global marine AOM because of the large amounts of manganese and iron provided to continental margins from rivers (Beal et al. 2009).
Discussion of Key Issues
In the light of recent progress regarding AOM, several unclear issues need to be further elucidated. These issues are discussed below.
Is ANME-1 a hypersaline anaerobic methanotroph ecotype?
The ANME population consisting only of ANME-1 was first found in a natural sedimentary that was high in salt (Lloyd et al. 2006). Then, ANME-1 was reported in other hypersaline environments (Yakimov et al. 2007; Cono et al. 2011; Maignien et al. 2013). Therefore, ANME-1 may be a hypersaline anaerobic methanotroph ecotype, which was suggested to be related to the comparatively low effect of ionic strength on ANME-1 (Maignien et al. 2013). ANME-1 cell membranes contain high contents of glycerol dialkyl glycerol tetraethers (GDGTs) (Niemann and Elvert 2008), which are characterized by a lower permeability compared with typical membrane lipids (Yamauchi et al. 1993; Valentine 2007). ANME-2 and ANME-3 cell membranes contain less or no GDGTs; the dominant component of ANME-2 and ANME-3 cell membranes is diethers exhibiting a higher permeability (Rossel et al. 2011). In addition, the ANME-1 genome was shown to contain genes coding for mannosylglycerate and di-myo-inositol-phosphate synthesis pathways (Meyerdierks et al. 2010), which are widely used to increase the turgor pressure by halophilic microorganisms (Roberts 2004; Empadinhas and da Costa 2008). Recently, proteins involved in gas vesicle synthesis have been identified in the proteome of ANME-1 (Stokke et al. 2012). Gas vesicles have also been observed in halophilic archaea (Walsby 1994), which might function in a salt stress response (Hechler and Pfeifer 2009). The above may contribute to the domination of ANME-1 in hypersaline environments. More needs to be investigated on this topic in the future.
The effect of oxygen on M. oxyfera
Candidatus Methylomirabilis oxyfera has the ability to conduct methane oxidation through a strictly O2-dependent reaction catalyzed by particulate methane monooxygenase (pMMO) (Ettwig et al. 2010). However, it was found that the addition of either 2% or 8% of O2 had an overall detrimental effect on M. oxyfera, and the ability of this bacterial species did not resume the original level (Luesken et al. 2012). These observations suggest that the O2 production and consumption of M. oxyfera is tightly controlled process, and the detrimental effect of O2 on M. oxyfera may be unrecoverable. However, most M. oxyfera and M. oxyfera-like bacteria have been observed in the oxic/anoxic interface of freshwater habitats (Raghoebarsing et al. 2006; Ettwig et al. 2008; Hu et al. 2011; Luesken et al. 2011a,b). Additionally, it is possible that the applied oxygen concentration was too high. In consideration of the above information, it is controversial whether M. oxyfera could use external O2 to oxidize methane. The effect of oxygen on M. oxyfera still remains unclear.
The relationship of M. nitroreducens with ANME
Candidatus Methanoperedens nitroreducens, a new species responsible for N-DAMO (Haroon et al. 2013), which is affiliated with ANME-2d, is the fourth subgroup of ANME-2 for S-DAMO (Mills et al. 2003). In addition, a full reductive acetyl-CoA (carbon fixation) pathway and acetyl-CoA synthetase have been identified in M. nitroreducens. It was predicted that M. nitroreducens might be able to produce acetate (Haroon et al. 2013), as can ANME-1 (Meyerdierks et al. 2010). The reported relationship of M. nitroreducens with ANME suggests that N-DAMO may be associated with S-DAMO, which warrants further investigation.
Conclusion
The microbes responsible for AOM are difficult to cultivate because of their low growth rates (Jagersma et al. 2009). AOM is an “active” process in microbial studies that contributes significantly to the global methane cycle. Currently, three different processes (Table3) are thought to be responsible for AOM, with sulfate, nitrite/nitrate, and metal ions (Mn4+ and Fe3+) serving as electron acceptors. However, the specific mechanism of AOM is not fully known, and the exact features of the responsible microbes require further study.
Table 3.
Comparisons between the three processes of AOM: S-DAMO, N-DAMO, and M-DAMO
| Features | S-DAMO | N-DAMO | M-DAMO |
|---|---|---|---|
| Habitat | Marine environments and freshwater environments | Freshwater environments | Marine environments |
| Mechanism hypothesis | Reverse methanogenesis, acetogenesis, and methylogenesis | Aerobic methane oxidation and reverse methanogenesis | ND |
| Electron acceptor | 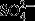 |
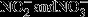 |
Mn4+ and Fe3+ |
| Responsible microbes | ANME | M. oxyfera and M. nitroreducens | MBGD (possible) |
| Reaction (AOM) |
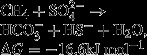 (eq. 3) (eq. 3) |
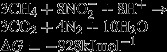 (eq. 12) and (eq. 12) and |
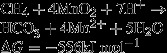 (eq. 14) and (eq. 14) and |
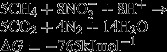 (eq. 13) (eq. 13) |
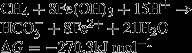 (eq. 15) (eq. 15) |
S-DAMO, sulfate-dependent anaerobic methane oxidation; N-DAMO, nitrate/nitrite-dependent anaerobic methane oxidation; M-DAMO, metal ion (Mn4+ and Fe3+)-dependent anaerobic methane oxidation; ANME, anaerobic methanotrophic archaea; M. oxyfera, Candidatus Methylomirabilis oxyfera; M. nitroreducens, Candidatus Methanoperedens nitroreducens; MBGD, marine benthic group D; ND, not determined; AOM, anaerobic oxidation of methane.
Acknowledgments
This research was supported by the National Natural Science Foundation of China (41473079, 41001151, and 41173089), the National Key Technology R&D Program (2013BAD11B03-3), and the Young Scientists Fund of RCEES (RCEES-QN-20130059F). We are grateful to the anonymous reviewers for their helpful comments regarding the manuscript.
Conflict of Interest
None declared.
References
- Alain K, Holler T, Musat F, Elvert M, Treude T. Krüger M. Microbiological investigation of methane- and hydrocarbon-discharging mud volcanoes in the Carpathian Mountains, Romania. Environ. Microbiol. 2006;8:574–590. doi: 10.1111/j.1462-2920.2005.00922.x. [DOI] [PubMed] [Google Scholar]
- Beal EJ, House CH. Orphan VJ. Manganese- and iron-dependent marine methane oxidation. Science. 2009;325:184–187. doi: 10.1126/science.1169984. [DOI] [PubMed] [Google Scholar]
- Bian L, Hinrichs K-U, Xie T, Brassell SC, Iversen N, Fossing H, et al. Algal and archaeal polyisoprenoids in a recent marine sediment: molecular isotopic evidence for anaerobic oxidation of methane. Geochem. Geophy. Geosy. 2001;2:1–23. [Google Scholar]
- Boetius A, Ravenschlag K, Schubert CJ, Rickert D, Widdel F, Gieseke A, et al. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature. 2000;407:623–626. doi: 10.1038/35036572. [DOI] [PubMed] [Google Scholar]
- La Cono V, Smedile F, Bortoluzzi G, Arcadi E, Maimone G, Messina E, et al. Unveiling microbial life in new deep-sea hypersaline Lake Thetis. Part I: Prokaryotes and environmental settingsemi. Environ. Microbiol. 2011;13:2250–2268. doi: 10.1111/j.1462-2920.2011.02478.x. [DOI] [PubMed] [Google Scholar]
- Conrad R. The global methane cycle: recent advances in understanding the microbial processes involved. Env. Microbiol. Rep. 2009;1:285–292. doi: 10.1111/j.1758-2229.2009.00038.x. [DOI] [PubMed] [Google Scholar]
- Dekas AE, Poretsky RS. Orphan VJ. Deep-sea archaea fix and share nitrogen in methane-consuming microbial consortia. Science. 2009;326:422–426. doi: 10.1126/science.1178223. [DOI] [PubMed] [Google Scholar]
- Deutzmann JS. Schink B. Anaerobic oxidation of methane in sediments of Lake Constance, an oligotrophic freshwater Lake. Appl. Environ. Microbiol. 2011;77:4429–4436. doi: 10.1128/AEM.00340-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durisch-Kaiser E, Klauser L, Wehrli B. Schubert C. Evidence of intense archaeal and bacterial methanotrophic activity in the Black Sea water column. Appl. Environ. Microbiol. 2005;71:8099–8106. doi: 10.1128/AEM.71.12.8099-8106.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eller G, Känel L. Krüger M. Cooccurrence of aerobic and anaerobic methane oxidation in the water column of Lake Plußsee. Appl. Environ. Microbiol. 2005;71:8925–8928. doi: 10.1128/AEM.71.12.8925-8928.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvert M. Suess E. Anaerobic methane oxidation associated with marine gas hydrates: superlight C-isotopes from saturated and unsaturated C20 and C25 irregular isoprenoids. Naturwissenschaften. 1999;86:295–300. [Google Scholar]
- Empadinhas N. da Costa MS. Osmoadaptation mechanisms in prokaryotes: distribution of compatible solutes. Int. Microbiol. 2008;11:151–161. [PubMed] [Google Scholar]
- Engelhardt H. Are S-layers exoskeletons? The basic function of protein surface layers revisited. J. Struct. Biol. 2007;160:115–124. doi: 10.1016/j.jsb.2007.08.003. [DOI] [PubMed] [Google Scholar]
- EPA. 2006. Global anthropogenic emissions of Non-CO2 greenhouse gases: 1990–2020 (EPA Report 430-R-06-003). Available at www.epa.gov/climatechange/economics/international.html (accessed June 2006)
- Ettwig KF, Shima S, de van Pas-Schoonen KT, Kahnt J, Medema MH, Op den Camp HJ, Jetten MS, Strous M. Denitrifying bacteria anaerobically oxidize methane in the absence of archaea. Environ. Microbiol. 2008;10:3164–3173. doi: 10.1111/j.1462-2920.2008.01724.x. [DOI] [PubMed] [Google Scholar]
- Ettwig KF, van Alen T, van de Pas-Schoonen KT, Jetten MS. Strous M. Enrichment and molecular detection of denitrifying methanotrophic bacteria of the NC10 phylum. Appl. Environ. Microbiol. 2009;75:3656–3662. doi: 10.1128/AEM.00067-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettwig KF, Butler MK, Le Paslier D, Pelletier E, Mangenot S, Kuypers MM, et al. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature. 2010;464:543–550. doi: 10.1038/nature08883. [DOI] [PubMed] [Google Scholar]
- Gal'chenko VF, Lein AY. Ivanov MV. Rates of microbial production and oxidation of methane in the bottom sediments and water column of the Black Sea. Microbiology. 2004;73:224–236. [PubMed] [Google Scholar]
- Grossman EL, Cifuentes LA. Cozzarelli IM. Anaerobic methane oxidation in a landfill-leachate plume. Environ. Sci. Technol. 2002;36:2436–2442. doi: 10.1021/es015695y. [DOI] [PubMed] [Google Scholar]
- Hallam SJ, Girguis PR, Preston CM, Richardson PM. DeLong EF. Identification of methyl coenzyme M reductase A (mcrA) genes associated with methane-oxidizing archaea. Appl. Environ. Microbiol. 2003;69:5483–5491. doi: 10.1128/AEM.69.9.5483-5491.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallam SJ, Putnam N, Preston CM, Detter JC, Rokhsar D, Richardson PM, et al. Reverse methanogenesis: testing the hypothesis with environmental genomics. Science. 2004;305:1457–1462. doi: 10.1126/science.1100025. [DOI] [PubMed] [Google Scholar]
- Hanson RS. Hanson TE. Methanotrophic bacteria. Microbiol. Rev. 1996;60:439–471. doi: 10.1128/mr.60.2.439-471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haroon MF, Hu S, Shi Y, Imelfort M, Keller J, Hugenholtz P, Yuan Z, Tyson GW. Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature. 2013;501:1–7. doi: 10.1038/nature12375. [DOI] [PubMed] [Google Scholar]
- Hartmann DL, Klein Tank AMG, Rusticucci M, Alexander L V, Brönnimann S, Charabi Y, et al. Observations: atmosphere and surface. In: Stocker TF, Qin D, Plattner G-K, Tignor M, Allen SK, Boschung J, Nauels A, Xia Y, Bex V, Midgley PM, editors. Climate change 2013: the physical science basis. Cambridge, United Kingdom and New York, NY, USA: Cambridge University Press; 2013. p. 1535. , and, eds., et al. . . Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change, [Google Scholar]
- Hechler T. Pfeifer F. Anaerobiosis inhibits gas vesicle formation in halophilic archaea. Mol. Microbiol. 2009;71:132–145. doi: 10.1111/j.1365-2958.2008.06517.x. [DOI] [PubMed] [Google Scholar]
- Hinrichs K-U, Hayes JM, Sylva SP, Brewer PG. DeLong E F. Methane-consuming archaebacteria in marine sediments. Nature. 1999;398:802–805. doi: 10.1038/19751. [DOI] [PubMed] [Google Scholar]
- Hinrichs K-U, Summons RE, Orphanc V, Sylvaa SP. Hayes JM. Molecular and isotopic analysis of anaerobic methane oxidizing communities in marine sediments. Org. Geochem. 2000;31:1685–1701. [Google Scholar]
- Hoehler TM, Alperin MJ, Albert DB. Martens CS. Field and laboratory studies of methane oxidation in an anoxic marine sediment: evidence for a methanogen-sulfate reducer consortium. Global Biogeochem. Cy. 1994;8:451–463. [Google Scholar]
- Hu S, Zeng RJ, Burow LC, Lant P, Keller J. Yuan Z. Enrichment of denitrifying anaerobic methane oxidizing microorganisms. Env. Microbiol. Rep. 2009;1:377–384. doi: 10.1111/j.1758-2229.2009.00083.x. , and . [DOI] [PubMed] [Google Scholar]
- Hu S, Zeng RJ, Keller J, Lant PA. Yuan Z. Effect of nitrate and nitrite on the selection of microorganisms in the denitrifying anaerobic methane oxidation process. Env. Microbiol. Rep. 2011;3:315–319. doi: 10.1111/j.1758-2229.2010.00227.x. [DOI] [PubMed] [Google Scholar]
- IPCC. Observations: surface and atmospheric climate change. In: Solomon S, Qin D, Manning M, Chen Z, Marquis M, Averyt KB, Tignor M, editors; Miller HL, editor. Climate change 2007: the physical science basis. Cambridge, U.K. and NY, U.S.A: Cambridge University Press; 2007. p. 996. Contribution of Working Group I to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change. [Google Scholar]
- Islas-Lima S, Thalasso F. Gómez-Hernandez J. Evidence of anoxic methane oxidation coupled to denitrification. Water Res. 2004;38:13–16. doi: 10.1016/j.watres.2003.08.024. [DOI] [PubMed] [Google Scholar]
- Jagersma GC, Meulepas RJ, de Heikamp- Jong I, Gieteling J, Klimiuk A, Schouten S, et al. Microbial diversity and community structure of a highly active anaerobic methane-oxidizing sulfate-reducing enrichment. Environ. Microbiol. 2009;11:3223–3232. doi: 10.1111/j.1462-2920.2009.02036.x. [DOI] [PubMed] [Google Scholar]
- Kampman C, Hendrickx TL, Luesken FA, van Alen TA, Op den Camp HJ, Jetten MS, et al. Enrichment of denitrifying methanotrophic bacteria for application after direct low-temperature anaerobic sewage treatment. J. Hazard. Mater. 2012;227:164–171. doi: 10.1016/j.jhazmat.2012.05.032. [DOI] [PubMed] [Google Scholar]
- Knittel K. Boetius A. Anaerobic oxidation of methane: progress with an unknown process. Annu. Rev. Microbiol. 2009;63:311–334. doi: 10.1146/annurev.micro.61.080706.093130. [DOI] [PubMed] [Google Scholar]
- Knittel K, Lösekann T, Boetius A, Kort R. Amann R. Diversity and distribution of methanotrophic archaea at cold seeps. Appl. Environ. Microbiol. 2005;71:467–479. doi: 10.1128/AEM.71.1.467-479.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger M, Meyerdierks A, Glöckner FO, Amann R, Widdel F, Kube M, et al. A conspicuous nickel protein in microbial mats that oxidize methane anaerobically. Nature. 2003;426:878–881. doi: 10.1038/nature02207. [DOI] [PubMed] [Google Scholar]
- Lazar CS, L'Haridon S, Pignet P. Toffin L. Archaeal populations in hypersaline sediments underlying orange microbial mats in the Napoli Mud Volcano. Appl. Environ. Microbiol. 2011;77:3120–3131. doi: 10.1128/AEM.01296-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh JA. Nitrogen fixation in methanogens: the archaeal perspective. Curr. Issues Mol. Biol. 2000;2:125–131. [PubMed] [Google Scholar]
- Lloyd KG, Lapham L. Teske A. An anaerobic methane-oxidizing community of ANME-1b archaea in hypersaline Gulf of Mexico sediments. Appl. Environ. Microbiol. 2006;72:7218–7230. doi: 10.1128/AEM.00886-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd KG, Alperin MJ. Teske A. Environmental evidence for net methane production and oxidation in putative anaerobic methanotrophic (ANME) archaea. Environ. Microbiol. 2011;13:2548–2564. doi: 10.1111/j.1462-2920.2011.02526.x. [DOI] [PubMed] [Google Scholar]
- Lösekann T, Knittel K, Nadalig T, Fuchs B, Niemann H, Boetius A, et al. Diversity and abundance of aerobic and anaerobic methane oxidizers at the Haakon Mosby Mud Volcano, Barents Sea. Appl. Environ. Microbiol. 2007;73:3348–3362. doi: 10.1128/AEM.00016-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luesken FA, Zhu B, van Alen TA, Butler MK, Diaz MR, Song B, et al. pmoA primers for detection of anaerobic methanotrophs. Appl. Environ. Microbiol. 2011a;77:3877–3880. doi: 10.1128/AEM.02960-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luesken FA, van Alen TA, der van Biezen E, Frijters C, Toonen G, Kampman C, et al. Diversity and enrichment of nitrite-dependent anaerobic methane oxidizing bacteria from wastewater sludge. Appl. Microbiol. Biotechnol. 2011b;92:845–854. doi: 10.1007/s00253-011-3361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luesken FA, Wu ML, Op den Camp HJ, Keltjens JT, Stunnenberg H, Francoijs KJ, et al. Effect of oxygen on the anaerobic methanotroph ‘Candidatus Methylomirabilis oxyfera’: kinetic and transcriptional analysis. Environ. Microbiol. 2012;14:1024–1034. doi: 10.1111/j.1462-2920.2011.02682.x. [DOI] [PubMed] [Google Scholar]
- Luton PE, Wayne JM, Sharp RJ. Riley PW. The mcrA gene as an alternative to 16S rRNA in the phylogenetic analysis of methanogen populations in landfill. Microbiology. 2002;148:3521–3530. doi: 10.1099/00221287-148-11-3521. [DOI] [PubMed] [Google Scholar]
- Maignien L, Parkes RJ, Cragg B, Niemann H, Knittel K, Coulon S, Akhmetzhanov A, Boon N. Anaerobic oxidation of methane in hypersaline cold seep sediments. FEMS Microbiol. Ecol. 2013;83:214–231. doi: 10.1111/j.1574-6941.2012.01466.x. [DOI] [PubMed] [Google Scholar]
- Mayr S, Latkoczy C, Krüger M, Günther D, Shima S, Thauer RK, Widdel F, Jaun B. Structure of an F430 variant from archaea associated with anaerobic oxidation of methane. J. Am. Chem. Soc. 2008;130:10758–10767. doi: 10.1021/ja802929z. [DOI] [PubMed] [Google Scholar]
- Meulepas RJW, Jagersma CG, Gieteling J, Buisman CJN, Stams AJM. Lens PNL. Enrichment of anaerobic methanotrophs in sulfate-reducing membrane bioreactors. Biotechnol. Bioeng. 2009;104:458–460. doi: 10.1002/bit.22412. [DOI] [PubMed] [Google Scholar]
- Meyerdierks A, Kube M, Kostadinov I, Teeling H, Glöckner FO, Reinhardt R, et al. Metagenome and mRNA expression analyses of anaerobic methanotrophic archaea of the ANME-1 group. Environ. Microbiol. 2010;12:422–439. doi: 10.1111/j.1462-2920.2009.02083.x. [DOI] [PubMed] [Google Scholar]
- Michaelis W, Seifert R, Nauhaus K, Treude T, Thiel V, Blumenberg M, et al. Microbial reefs in the Black Sea fueled by anaerobic oxidation of methane. Science. 2002;297:1013–1015. doi: 10.1126/science.1072502. [DOI] [PubMed] [Google Scholar]
- Mills HJ, Hodges C, Wilson K, MacDonald IR. Sobecky PA. Microbial diversity in sediments associated with surface-breaching gas hydrate mounds in the Gulf of Mexico. FEMS Microbiol. Ecol. 2003;46:39–52. doi: 10.1016/S0168-6496(03)00191-0. [DOI] [PubMed] [Google Scholar]
- Miyashita A, Mochimaru H, Kazama H, Ohashi A, Yamaguchi T, Nunoura T, et al. Development of 16S rRNA gene-targeted primers for detection of archaeal anaerobic methanotrophs (ANMEs) FEMS Microbiol. Lett. 2009;297:31–37. doi: 10.1111/j.1574-6968.2009.01648.x. [DOI] [PubMed] [Google Scholar]
- Moran JJ, Beal EJ, Vrentas JM, Orphan VJ, Freeman KH. House CH. Methyl sulfides as intermediates in the anaerobic oxidation of methane. Environ. Microbiol. 2008;10:162–173. doi: 10.1111/j.1462-2920.2007.01441.x. [DOI] [PubMed] [Google Scholar]
- Murase J. Kimura M. Methane production and its fate in paddy fields. 6. Anaerobic oxidation of methane in plow layer soil. Soil Sci. Plant Nutr. 1994;40:505–514. [Google Scholar]
- Murray PA. Zinder SH. Nitrogen fixation by a methanogenic archaebacterium. Nature. 1984;312:284–286. [Google Scholar]
- Myhre G, Shindell D, Bréon F-M, Collins W, Fuglestvedt J, Huang J, Koch D, Lamarque J-F, Lee D, Mendoza B, Nakajima T, Robock A, Stephens G, Takemura T. Zhang H. Anthropogenic and natural radiative forcing. In: Midgley PM, editor; Stocker TF, Qin D, Plattner G-K, Tignor M, Allen SK, Boschung J, Nauels A, Xia Y, Bex V, editors. Climate change 2013: the physical science basis. Cambridge, United Kingdom and New York, NY, USA: Cambridge University Press; 2013. pp. 659–740. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change, [Google Scholar]
- Nauhaus K, Boetius A, Krüger M. Widdel F. In vitro demonstration of anaerobic oxidation of methane coupled to sulphate reduction in sediment from a marine gas hydrate area. Environ. Microbiol. 2002;4:296–305. doi: 10.1046/j.1462-2920.2002.00299.x. [DOI] [PubMed] [Google Scholar]
- Nauhaus K, Treude T, Boetius A. Krüger M. Environmental regulation of the anaerobic oxidation of methane: a comparison of ANME-I and ANME-II communities. Environ. Microbiol. 2005;7:98–106. doi: 10.1111/j.1462-2920.2004.00669.x. [DOI] [PubMed] [Google Scholar]
- Niemann H. Elvert M. Diagnostic lipid biomarker and stable carbon isotope signatures of microbial communities mediating the anaerobic oxidation of methane with sulphate. Org. Geochem. 2008;39:1668–1677. [Google Scholar]
- Niemann H, Lösekann T, de Beer D, et al. Novel microbial communities of the Haakon Mosby mud volcano and their role as a methane sink. Nature. 2006;443:854–858. doi: 10.1038/nature05227. [DOI] [PubMed] [Google Scholar]
- Orcutt B. Meile C. Constraints on mechanisms and rates of anaerobic oxidation of methane by microbial consortia: process-based modeling of ANME-2 archaea and sulfate reducing bacteria interactions. Biogeosciences. 2008;5:1587–1599. [Google Scholar]
- Orphan VJ, House CH, Hinrichs K-U, McKeegan KD. DeLong EF. Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis. Science. 2001a;293:484–487. doi: 10.1126/science.1061338. [DOI] [PubMed] [Google Scholar]
- Orphan VJ, Hinrichs KU, Ussler W, 3rd, Paull CK, Taylor LT, Sylva SP, Hayes JM, et al. Comparative analysis of methane-oxidizing archaea and sulfate-reducing bacteria in anoxic marine sediments. Appl. Environ. Microbiol. 2001b;67:1922–1934. doi: 10.1128/AEM.67.4.1922-1934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphan VJ, House CH, Hinrichs K-U, McKeegan KD. DeLong EF. Multiple archaeal groups mediate methane oxidation in anoxic cold seep sediments. Proc. Natl Acad. Sci. USA. 2002;99:7663–7668. doi: 10.1073/pnas.072210299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphana VJ, Ussler W, III, Naehr TH. Geological, geochemical, and microbiological heterogeneity of the seafloor around methane vents in the Eel River Basin, offshore California. Chem. Geol. 2005;205:265–289. [Google Scholar]
- Pancost RD, Damsté JSS, de Lint S, Van der Maarel MJEC, Gottschal JC. Party TMSS. Biomarker evidence for widespread anaerobic methane oxidation in Mediterranean sediments by a consortium of methanogenic archaea and bacteria. Appl. Environ. Microbiol. 2000;66:1126–1132. doi: 10.1128/aem.66.3.1126-1132.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghoebarsing AA, Pol A A, de van Pas-Schoonen KT, Smolders A J, Ettwig KF, Rijpstra WT, et al. A microbial consortium couples anaerobic methane oxidation to denitrification. Nature. 2006;440:918–921. doi: 10.1038/nature04617. [DOI] [PubMed] [Google Scholar]
- Reeburgh WS. Methane consumption in Cariaco Trench waters and sediments. Earth Planet. Sci. Lett. 1976;28:337–344. [Google Scholar]
- Reeburgh WS. Anaerobic methane oxidation: rate depth distributions in Skan Bay sediments. Earth Planet. Sci. Lett. 1980;47:345–352. [Google Scholar]
- Reeburgh WS. Oceanic methane biogeochemistry. Chem. Rev. 2007;107:486–513. doi: 10.1021/cr050362v. [DOI] [PubMed] [Google Scholar]
- Roberts MF. Osmoadaptation and osmoregulation in archaea: update 2004. Front. Biosci. 2004;9:1999–2019. doi: 10.2741/1366. [DOI] [PubMed] [Google Scholar]
- Rossel PE, Elvert M, Ramette A, Boetius A. Hinrichs K-U. Factors controlling the distribution of anaerobic methanotrophic communities in marine environments: evidence from intact polar membrane lipids. Geochim. Cosmochim. Acta. 2011;75:164–184. [Google Scholar]
- Scheller S, Goenrich M, Boecher R, Thauer RK. Jaun B. The key nickel enzyme of methanogenesis catalyses the anaerobic oxidation of methane. Nature. 2010;465:606–609. doi: 10.1038/nature09015. [DOI] [PubMed] [Google Scholar]
- Schleper C, Jurgens G. Jonuscheit M. Genomic studies of uncultivated archaea. Nat. Rev. Microbiol. 2005;3:479–488. doi: 10.1038/nrmicro1159. [DOI] [PubMed] [Google Scholar]
- Smemo KA. Yavitt JB. Evidence for anaerobic CH4 oxidation in freshwater peatlands. Geomicrobiol. J. 2007;24:583–597. [Google Scholar]
- Smith RL, Howes BL. Garabedian SP. In situ measurement of methane oxidation in groundwater by using natural-gradient tracer tests. Appl. Environ. Microbiol. 1991;57:1997–2004. doi: 10.1128/aem.57.7.1997-2004.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokke R, Roalkvam I, Lanzen A, Haflidason H. Steen IH. Integrated metagenomic and metaproteomic analyses of an ANME-1-dominated community in marine cold seep sediments. Environ. Microbiol. 2012;14:1333–1346. doi: 10.1111/j.1462-2920.2012.02716.x. [DOI] [PubMed] [Google Scholar]
- Thiel V, Peckmann J, Seifert R, Wehrung P, Reitner J. Michaells W. Highly isotopically depleted isoprenoids: molecular markers for ancient methane venting. Geochim. Cosmochim. Acta. 1999;63:3959–3966. [Google Scholar]
- Treude T, Krüger M, Boetius A. Jørgensen BB. Environmental control on anaerobic oxidation of methane in the gassy sediments of Eckernförde Bay (German Baltic) Limnol. Oceanogr. 2005;50:1771–1786. [Google Scholar]
- Treude T, Orphan V, Knittel K, Gieseke A, House CH. Boetius A. Consumption of methane and CO2 by methanotrophic microbial mats from gas seeps of the anoxic Black Sea. Appl. Environ. Microbiol. 2007;73:2271–2283. doi: 10.1128/AEM.02685-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine DL. Adaptations to energy stress dictate the ecology and evolution of the archaea. Nat. Rev. Microbiol. 2007;5:316–323. doi: 10.1038/nrmicro1619. [DOI] [PubMed] [Google Scholar]
- Valentine DL. Reeburgh WS. New perspectives on anaerobic methane oxidation. Environ. Microbiol. 2000;2:477–484. doi: 10.1046/j.1462-2920.2000.00135.x. [DOI] [PubMed] [Google Scholar]
- Walsby AE. Gas vesicles. Microbiol. Rev. 1994;58:94–148. doi: 10.1128/mr.58.1.94-144.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhu G, Harhangi HR, Zhu B, Jetten MSM, Yin C, et al. Co-occurrence and distribution of nitrite-dependent anaerobic ammonium and methane-oxidizing bacteria in a paddy soil. FEMS Microbiol. Lett. 2012;336:79–88. doi: 10.1111/j.1574-6968.2012.02654.x. [DOI] [PubMed] [Google Scholar]
- Wu ML, de Vries S, Van Alen TA, Butler MK, Op den Camp HJ, Keltjens JT, et al. Physiological role of the respiratory quinol oxidase in the anaerobic nitrite-reducing methanotroph ‘Candidatus Methylomirabilis oxyfera’. Microbiology. 2011;157:890–898. doi: 10.1099/mic.0.045187-0. [DOI] [PubMed] [Google Scholar]
- Wu ML, van Teeseling MC, Willems MJ, van Donselaar EG, Klingl A, Rachel R, Geerts WJ, Jetten MS, Strous M, van Niftrik L. Ultrastructure of the denitrifying methanotroph “Candidatus Methylomirabilis oxyfera”, a novel polygon-shaped bacterium. J. Bacteriol. 2012;194:284–291. doi: 10.1128/JB.05816-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakimov MM, La Cono V, Denaro R, D'Auria G, Decembrini F, Timmis KN, Golyshin PN, Giuliano L. Primary producing prokaryotic communities of brine, interface and seawater above the halocline of deep anoxic lake L'Atalante, Eastern Mediterranean Sea. ISME J. 2007;1:743–755. doi: 10.1038/ismej.2007.83. [DOI] [PubMed] [Google Scholar]
- Yamauchi K, Doi K, Yoshida Y. Kinoshita M. Archaebacterial lipids: highly proton-impermeable membranes from 1,2-diphytanyl-sn-glycero-3-phosphocholine. Biochim. Biophys. Acta. 1993;2:178–182. doi: 10.1016/0005-2736(93)90353-2. [DOI] [PubMed] [Google Scholar]
- Yang J, Jiang H, Wu G, Hou W, Sun Y, Lai Z, et al. Co-occurrence of nitrite-dependent anaerobic methane oxidizing and anaerobic ammonia oxidizing bacteria in two Qinghai-Tibetan saline lakes. Front. Earth Sci. 2012;6:383–391. [Google Scholar]
- Yarbrough JM, Rake JB. Eagon RG. Bacterial inhibitory effects of nitrite: inhibition of active transport, but not of group translocation, and of intracellular enzymes. Appl. Environ. Microbiol. 1980;39:831–834. doi: 10.1128/aem.39.4.831-834.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Shen L, Hu B, Lou L. Cheng D. Molecular detection of denitrifying anaerobic methane oxidizing bacteria in the sediment of West Lake. Acta Sci. Circumst. 2013;33:1321–1325. [Google Scholar]


