Abstract
Jen proteins in yeast are involved in the uptake of mono/dicarboxylic acids. The Jen1 subfamily transports lactate and pyruvate, while the Jen2 subfamily transports fumarate, malate, and succinate. Yarrowia lipolytica has six JEN genes: YALI0B19470g, YALI0C15488g, YALI0C21406g, YALI0D20108g, YALI0D24607g, and YALI0E32901g. Through phylogenetic analyses, we found that these genes represent a new subfamily, Jen3 and that these three Jen subfamilies derivate from three putative ancestral genes. Reverse transcription-PCR. revealed that only four YLJEN genes are expressed and they are upregulated in the presence of lactate, pyruvate, fumarate, malate, and/or succinate, suggesting that they are able to transport these substrates. Analysis of deletion mutant strains revealed that Jen3 subfamily proteins transport fumarate, malate, and succinate. We found evidence that YALI0C15488 encodes the main transporter because its deletion was sufficient to strongly reduce or suppress growth in media containing fumarate, malate, or succinate. It appears that the other YLJEN genes play a minor role, with the exception of YALI0E32901g, which is important for malate uptake. However, the overexpression of each YLJEN gene in the sextuple-deletion mutant strain ΔYLjen1-6 revealed that all six genes are functional and have evolved to transport different substrates with varying degrees of efficacy. In addition, we found that YALI0E32901p transported succinate more efficiently in the presence of lactate or fumarate.
Keywords: Fumarate, lactate, malate, succinate, transport, yeast
Introduction
Jen proteins are transporters of mono- and dicarboxylic acids, which are localized in the plasma membrane (Paiva et al. 2002; Vieira et al. 2010). Their functions were first described in Saccharomyces cerevisiae (Casal et al. 1999), in which the expression and activity of SCJen1p (encoded by YKL217W) are (1) regulated through a complex pathway which involves different stages of expression (transcriptional, translational, and posttranslational); (2) strain-dependent; and (3) strongly affected by the carbon source present, especially in the case of glucose (Andrade and Casal 2001; Paiva et al. 2002; Andrade et al. 2005). Interestingly, glucose, fructose, sucrose, and mannose have been shown to inhibit the expression of SCJEN1, whereas galactose, raffinose, ethanol, glycerol, and lactate have the opposite effect (Andrade and Casal 2001; Paiva et al. 2002; Tsuboi et al. 2003; Chambers et al. 2004). SCJen1p has been shown to be responsible for the active transport of lactate and pyruvate in S. cerevisiae (Casal et al. 1999; Akita et al. 2000), while in Candida albicans, CAJen1p can actively transport lactate, pyruvate, and propionate (Soares-Silva et al. 2004). Moreover, a recent study showed that SCJen1p is involved in selenite transport (McDermott et al. 2010).
Some species, like C. albicans and Kluyveromyces lactis, have two types of Jen proteins. In addition to JEN1, they also possess a JEN2 gene, which is repressed in the presence of glucose and upregulated when exposed to succinate or malate (Lodi et al. 2004; Vieira et al. 2010). As mentioned above, CAJen1p transports lactate, pyruvate, and propionate (Soares-Silva et al. 2004), while Jen2p is a fumarate, succinate, and malate transporter in both C. albicans and K. lactis (Lodi et al. 2004; Queirós et al. 2007; Vieira et al. 2010). In species with two types of Jenp, the disruption of both JEN1 and JEN2 affects growth on mono- and dicarboxylic acids (Lodi et al. 2004; Vieira et al. 2010). For example, a ΔCAjen1ΔCAjen2 mutant displayed a growth defect on single-carbon-source media containing lactic, succinic, malic, or pyruvic acids (Vieira et al. 2010).
The genome of Yarrowia lipolytica encodes six proteins that are homologous to Jen proteins: YALI0B19470p (YLJen5p), YALI0C15488p (YLJen1p), YALI0C21406p (YL-Jen2p), YALI0D20108p (YLJen3p), YALI0D24607p (YLJen4p), and YALI0E32901p (YLJen6p). Phylogenetic analyses by Lodi et al. (2007) revealed that Jen proteins in Y. lipolytica form a group separate from those in S. cerevisiae, K. lactis, C. albicans, and other Hemiascomycetes and Euascomycetes. In the same study, a comparison of gene synteny assigned YLJEN2, YLJEN3, YLJEN4, and YLJEN6 to the Jen2 cluster, and YLJEN5 and YLJEN1 to the Jen1 cluster (Lodi et al. 2007). The authors also identified three molecular motifs that distinguished Jen2 proteins from Jen1 proteins: the absence of a C-terminal helix, TM11, usually found in Jen1 proteins; a C-terminal proline, rather than histidine, residue; and a sequence gap between transmembrane domains (TMs) 6 and 7 (Lodi et al. 2007). However, they also noted that YLJen1p and YLJen5p did not present the motifs typical of the Jen1 subfamily, and thus classified these two proteins in a “preJen1” cluster. They hypothesized that preJEN1 represented an intermediate evolutionary step between JEN2 (the ancestral gene) and JEN1.
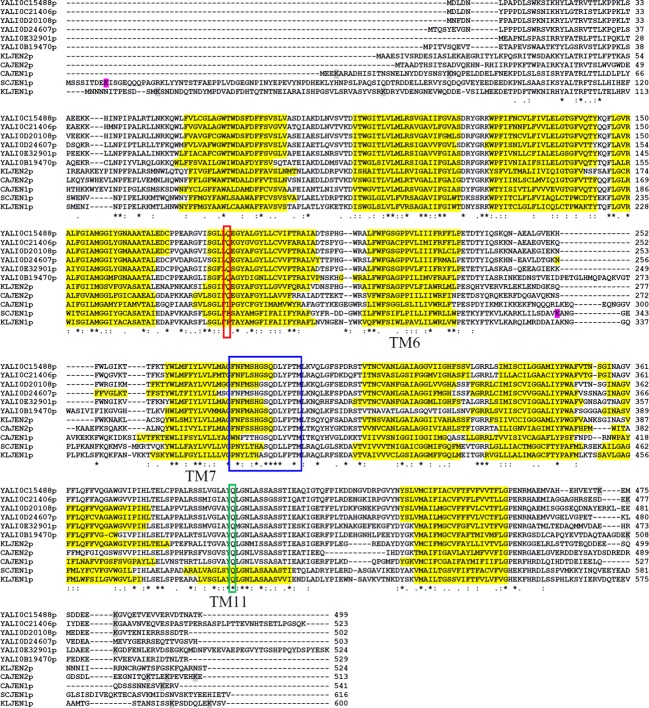
All six Jen proteins in Y. lipolytica share a conserved motif in the seventh TM, NXX(S/T)HX(S/T)QDXXXT, which is involved in transport ability and substrate affinity (Soares-Silva et al. 2007; Fig.1). Indeed, substitution at the N379, H383, or D387 residues of SCJen1p resulted in a major reduction in lactate and pyruvate uptake, but did not affect acetate transport. Instead, mutations at the Q386 and T391 residues resulted in no or only moderate changes in Jen1p's ability to transport lactate, pyruvate, and acetate, but did result in modifications in its respective affinity for each acid. The same study also presented evidence for the existence of a charged interaction between amino acids H383 and D387 which contributes to Jen1p's protein structure. Interestingly, lactate and pyruvate transport were completely suppressed in cells with the double mutation H383D/D387H, while the kinetics of acetate transport were similar to those found with wild-type Jen1p (Soares-Silva et al. 2007).
Figure 1.
Alignment of the six YLJen protein sequences with Jen1p and/or Jen2p sequences of Saccharomyces cerevisiae, Kluyveromyces lactis, and Candida albicans, constructed with ClustalW. SC, S. cerevisiae (YKL217W for SCJen1p), CA, C. albicans (CaO19.7447 for CAJen1p; CaO19.12767 for CAJen2p) and KL, K. lactis (KLLA0E16259 for KLJen1p; KLLA0F10043 for KLJen2p). The blue frame contains the NXX(S/T)HX(S/T)QDXXXT motif and the F270 and Q498 residues of S. cerevisiae are indicated by red and green frames, respectively. Yellow-highlighted amino acids represent transmembrane domains predicted by TMHMM. Gray-highlighted amino acids represent putative ubiquitination sites predicted with high confidence by www.ubpred.org. Purple-highlighted amino acids indicate K9 and K338, the experimentally confirmed ubiquitination sites of SCJen1p (Paiva et al. 2009).
As the synteny analysis of Lodi et al. (2007) was poorly supported and that the phylogeny placed the six Y. lipolytica genes in a monophyletic group, we investigated the possibility that these genes derive from a species-specific gene expansion event. Such an expansion could have conferred unique characteristics on the Jen proteins in Y. lipolytica that would distinguish them from the Jen proteins in other species, we also explored the substrate specificity of each protein. Through comparative genomics analyses involving species closely related to Y. lipolytica, we found that this protein family has a dynamic evolutionary history dating from the emergence of the Yarrowia clade and constitutes a new subfamily of Jen proteins, Jen3. To analyze the specificity and function of each YLJenp, we first constructed multiple mutant strains and then expressed each YLJEN gene in a ΔYLjen1-6 background. We found that the most important proteins, that is, the ones whose absence had the largest effects on cell function, were YLJen1p and YLJen6p. We also show that Jen3p proteins are fumarate, malate, and succinate transporters.
Material and Methods
Yeast growth and culture conditions
The Y. lipolytica strains used in this study were derived from the wild-type Y. lipolytica W29 (ATCC20460) strain (Table1). The auxotrophic strain Po1d (Leu− Ura−) has been previously described by Barth and Gaillardin (1996). The prototrophic strain JMY2900 was obtained by transformation of JMY330, a po1d Ura+ derivative (Beopoulos et al. 2008), with a fragment carrying the LEU2 gene and verification that the strain have recovered the wild-type LEU2 locus as described by Dulermo et al. (2013). All of the strains used in this study are listed in Table1. Media and growth conditions for Escherichia coli have been previously described by Sambrook et al. (1989), and those for Y. lipolytica have been described by Barth and Gaillardin (1996). Rich medium (Yeast extract, Peptone, Dextrose (YPD)) and minimal glucose medium (YNB) were prepared as described previously (Mlícková et al. 2004). Minimal medium (Yeast Nitrogen Base (YNB)) contained 0.17% (wt/vol) yeast nitrogen base (without amino acids and ammonium sulfate, YNBww; Difco, Paris, France), 0.5% (wt/vol) NH4Cl, and 50 mmol/L phosphate buffer (pH 6.8). During the experiment, this minimal medium was supplemented with uracil (0.1 g/L) and/or leucine (0.1 g/L) if necessary. To test growth on different carbon sources, we used the following medium: 0.17% (wt/vol) yeast nitrogen base (without amino acids and ammonium sulfate, YNBww; Difco), 0.5% (wt/vol) NH4Cl, (Difco), and 50 mmol/L phosphate buffer (pH 6.8). For growth and consumption tests, a carbon source was added at 0.15–0.3% (wt/vol). Solid media were obtained by the addition of 1.6% agar. Y. lipolytica precultures were grown in minimal medium to avoid stress on strains that would be used in phenotype tests.
Table 1.
Strains and plasmids
| Strain or plasmid | Genotype or other relevant characteristics (name in figures) | Source or reference |
|---|---|---|
| Escherichia coli | ||
| DH5α | Φ80dlacZΔm15, recA1, endA1, gyrA96, thi-1, hsdR17 (rk−, mk+), supE44, relA1, deoR, Δ(lacZYA-argF)U169 | Promega |
| Yarrowia lipolytica | ||
| W29 | MatA, wild-type | Barth and Gaillardin (1996) |
| Po1d | MatA ura3-302 leu2-270 xpr2-322 | Barth and Gaillardin (1996) |
| JMY330 | MatA ura3-302 leu2-270 xpr2-322, URA3ex; Ura+ Leu− | Beopoulos et al. (2008) |
| JMY2900 | MatA ura3-302 xpr2-322, URA3ex; Ura+ Leu+ | F. Brunel, (unpubl. data) |
| JMY2946 | Po1d ΔYALI0B19470::URA3ex Leu− | This study |
| JMY2949 | Po1d YALI0C21406::URA3ex YALI0C15488::LEU2ex (ΔYLjen1,2) | This study |
| JMY2956 | Po1d ΔYALI0C15488::URA3ex Leu− | This study |
| JMY3007 | Po1d ΔYALI0C21406::URA3ex Leu− | This study |
| JMY3012 | Po1d ΔYALI0D20108::URA3ex Leu− | This study |
| JMY3014 | Po1d YALI0C15488::URA3ex YALI0B19470::LEU2ex (ΔYLjen1,5) | This study |
| JMY3018 | Po1d YALI0C21406::URA3ex YALI0B19470::LEU2ex (ΔYLjen2,5) | This study |
| JMY3040 | Po1d YALI0E32901::URA3ex Leu− | This study |
| JMY3075 | Po1d YALI0D20108::URA3ex YALI0B19470::LEU2ex (ΔYLjen3,5) | This study |
| JMY3084 | Po1d ΔYALI0C21406 ΔYALI0C15488 Ura− Leu− | This study |
| JMY3086 | Po1d ΔYALI0C15488 ΔYALI0B19470 Ura− Leu− | This study |
| JMY3088 | Po1d ΔYALI0C21406 ΔYALI0B19470 Ura− Leu− | This study |
| JMY3090 | Po1d YALI0B19470::URA3ex YALI0E32901::LEU2ex (ΔYLjen5,6) | This study |
| JMY3092 | Po1d YALI0C15488::URA3ex YALI0E32901::LEU2ex (ΔYLjen1,6) | This study |
| JMY3093 | Po1d YALI0D20108::URA3ex YALI0E32901::LEU2ex (ΔYLjen3,6) | This study |
| JMY3094 | Po1d YALI0D20108::URA3ex YALI0C15488::LEU2ex (ΔYLjen1,3) | This study |
| JMY3101 | Po1d YALI0B19470::URA3ex + LEU2ex (ΔYLjen5) | This study |
| JMY3104 | Po1d YALI0C15488::URA3ex + LEU2ex (ΔYLjen1) | This study |
| JMY3106 | Po1d YALI0C21406::URA3ex + LEU2ex (ΔYLjen2) | This study |
| JMY3109 | Po1d YALI0D20108::URA3ex + LEU2ex (ΔYLjen3) | This study |
| JMY3112 | Po1d YALI0E32901::URA3ex + LEU2ex (ΔYLjen6) | This study |
| JMY3146 | JMY3084 + YALI0D20108::URA3ex Leu− | This study |
| JMY3147 | JMY3086 + YALI0D20108::URA3ex Leu− | This study |
| JMY3151 | JMY3084 + YALI0B19470::URA3ex Leu− | This study |
| JMY3152 | JMY3084 + YALI0B19470::URA3ex Leu− | This study |
| JMY3156 | JMY3088 + YALI0D20108::URA3ex Leu− | This study |
| JMY3171 | JMY3147 + YALI0E32901::LEU2ex (ΔYLjen1,3,5,6) | This study |
| JMY3209 | JMY3151 + LEU2ex (ΔYLjen1,2,5) | This study |
| JMY3210 | JMY3156 + LEU2ex (ΔYLjen2,3,5) | This study |
| JMY3213 | JMY3156 + YALI0E32901::LEU2ex (ΔYLjen2,3,5,6) | This study |
| JMY3215 | JMY3171 Ura− Leu− | This study |
| JMY3249 | JMY3156 + YALI0D24607::LEU2ex (ΔYLjen2,3,4,5) | This study |
| JMY3250 | JMY3151 + YALI0E32901::LEU2ex (ΔYLjen1,2,5,6) | This study |
| JMY3252 | Y3147 + YALI0D24607::LEU2ex (ΔYLjen1,3,4,5) | This study |
| JMY3257 | Po1d + YALI0D24607::LEU2ex Ura− | This study |
| JMY3278 | JMY3257 + URA3ex (ΔYLjen4) | This study |
| JMY3282 | JMY3215 + YALI0D24607::LEU2ex Ura− | This study |
| JMY3333 | JMY3282 + URA3ex (ΔYLjen1,3,4,5,6) | This study |
| JMY3598 | JMY3282 + YALI0C21406::URA3ex (ΔYLjen1-6) | This study |
| JMY3702 | JMY3598 Ura− Leu− | This study |
| JMY3722 | JMY3598 LEU2ex Ura− | This study |
| JMY3751 | JMY3722 + pTEF-YALI0B19470-URA3ex (YLJEN5++) | This study |
| JMY3754 | JMY3722 + pTEF-YALI0C15488-URA3ex (YLJEN1++) | This study |
| JMY3757 | JMY3722 + pTEF-YALI0D20108-URA3ex (YLJEN3++) | This study |
| JMY3760 | JMY3722 + pTEF-YALI0D24607-URA3ex (YLJEN4++) | This study |
| JMY3763 | JMY3722 + pTEF-YALI0E32901-URA3ex (YLJEN6++) | This study |
| JMY4125 | JMY3702 + pTEF-C21406-LEU2ex Ura− | This study |
| JMY4157 | JMY4125 + URA3ex (YLJEN2++) | This study |
| Plasmids | ||
| pCR4Blunt-TOPO | Cloning vector | Invitrogen |
| JMP802 | JMP62-pTEF-LEU2ex | |
| JMP803 | JMP62-pTEF-URA3ex | (Müller et al. 1998; Nicaud et al. 2002) |
| JMP547 | pUB4-CRE | Fickers et al. (2003) |
| JMP1392 | JMP62-pTEF-RedStar2SKL-LEU2ex | Kabran et al. (2012) |
| JMP1647 | pCR4Blunt-TOPO YALI0B19470UpDn | This study |
| JMP1649 | pCR4Blunt-TOPO YALI0C15488UpDn | This study |
| JMP1683 | pCR4Blunt-TOPO YALI0C21406UpDn | This study |
| JMP1684 | pCR4Blunt-TOPO YALI0D20108UpDn | This study |
| JMP1689 | pCR4Blunt-TOPO YALI0E32901UpDn | This study |
| JMP1667 | pCR4Blunt-TOPO YALI0B19470UpDn-URA3ex | This study |
| JMP1671 | pCR4Blunt-TOPO YALI0C15488UpDn-URA3ex | This study |
| JMP1679 | pCR4Blunt-TOPO YALI0B19470UpDn-LEU2ex | This study |
| JMP1681 | pCR4Blunt-TOPO YALI0C15488UpDn-LEU2ex | This study |
| JMP1698 | pCR4Blunt-TOPO YALI0C21406UpDn-URA3ex | This study |
| JMP1702 | pCR4Blunt-TOPO YALI0D20108UpDn-URA3ex | This study |
| JMP1713 | pCR4Blunt-TOPO YALI0E32901UpDn-URA3ex | This study |
| JMP1715 | pCR4Blunt-TOPO YALI0E32901UpDn-LEU2ex | This study |
| JMP 1792 | JMP62 pTEF-YALI0B19470-URA3ex | This study |
| JMP1794 | JMP62 pTEF-YALI0C15488-URA3ex | This study |
| JMP1796 | JMP62 pTEF-YALI0D24607-URA3ex | This study |
| JMP1798 | JMP62 pTEF-YALI0E32901-URA3ex | This study |
| JMP1809 | JMP62 pTEF-YALI0D20108-URA3ex | This study |
| JMP1813 | pCR4Blunt-TOPO YALI0D24607UpDn-LEU2ex | This study |
| JMP2366 | JMP62 pTEF-YALI0C21406-LEU2ex | This study |
General genetic techniques
Standard molecular genetic techniques were used throughout this study (Sambrook et al. 1989). Restriction enzymes were obtained from OZYME (Saint-Quentin-en-Yvelines, France). Yeast cells were transformed using the lithium acetate technique (Le Dall et al. 1994). Genomic DNA from yeast transformants was prepared as described by Querol et al. (1992). PCR amplifications were performed in an Eppendorf 2720 thermal cycler with GoTaq DNA polymerases (Promega, Charbonniére-les-Bains, France) for verification PCR and PyroBest DNA polymerases (Takara, Saint-Germain-en-laye, France) for cloning. Table2 gives the primers that were used during this study. PCR fragments were purified with the QIAgen Purification Kit (Qiagen, Courtaboeuf, France), and DNA fragments were recovered from agarose gels with the QIAquick Gel Extraction Kit (Qiagen). The Clone Manager software package was used for gene sequence analysis Sci-Ed Software, Scientific and Education Softwara, Morrisville, NC, USA.
Table 2.
Primer list
| Genes | Primers | Sequences | Utilization |
|---|---|---|---|
| YALI0B19470g (YLJEN5) | B19470UpNotI | GAATGCGGCCGCCAGTTGGTTCAACTGGGTCC | Upstream fragment of YALI0B19470 |
| B19470UpIsceI | CGATTACCCTGTTATCCCTACCGGCATGAGAGTGGTGTTTGG | ||
| B19470DnNotI | GAATGCGGCCGCCAGCCAGGTCAGCTACTATT | Downstream fragment of YALI0B19470 | |
| B19470DnIsceIIceuI | GGTAGGGATAACAGGGTAATCGTAACTATAACGGTCCTAAGGTAGCGACCAATCTCACTCGTTAATTAG | ||
| Ver1B19470 | CAAGTATCATGGTTTGTGGG | Verification of disruption | |
| Ver2B19470 | GTGTCACAACAGTGTCACCC | ||
| StartB19470 | ATCGGATCCCACAATGCCCATCACAGTTTCACAAGAAGTG | Overexpression of YALI0B19470 | |
| EndB19470 | CATCCTAGGTTAACGAGTGAGATTGGTGTCGATTCG | ||
| B19470F | TCTTCTTCTTGCAGTTCTTTG | Expression of YALI0B19470 by RT-PCR | |
| B19470R | GTGCTCATCGAGAATAGGG | ||
| YALI0C15488g (YLJEN1) | C15488UpNotI | GAATGCGGCCGCTTTGTCTGTCTTCCCGTTGC | Upstream fragment of YALI0C15488 |
| C15488UpIsceI | CGATTACCCTGTTATCCCTACCGGTGAATGAACGAACGAAGAAGG | ||
| C15488DnNotI | GAATGCGGCCGCTGAGAGTCCTTCTGTCTACC | Downstream fragment of YALI0C15488 | |
| C15488DnIsceIIceuI | GGTAGGGATAACAGGGTAATCGTAACTATAACGGTCCTAAGGTAGCGAGTGTAGCAACTAACGTATTG | ||
| Ver1-1C15488 | CCACTCCTCCTCTCCTTAGACCG | Verification of disruption | |
| Ver2-2C15488 | CCTCTCCATCGATTTCGAGGTGC | ||
| StartC15488 | ATCGGATCCCACAATGGATTTGGACAACCTCCCTGCCCC | Overexpression of YALI0C15488 | |
| EndC15488 | CATCCTAGGCTACTTAGTAGCATTGGTGTCAACTC | ||
| C15488F | TTCTTCTTGCAGTTCTTCGT | Expression of YALI0C15488 by RT-PCR | |
| C15488R | CGACCCCATTATCATCTTT | ||
| YALI0C21406g (YLJEN2) | C21406UpNotI | GAATGCGGCCGCGGAGAAAATGGACGTGTGAGACGC | Upstream fragment of YALI0C21406 |
| C21406UpIsceI | CGATTACCCTGTTATCCCTACCGAGATAGAGCAAGTAGAAGCAGCG | ||
| C21406DnNotI | GAATGCGGCCGCGGACTTGTAACAGCACACGTTCGC | Downstream fragment of YALI0C21406 | |
| C21406Dn2IsceIIceuI | GGTAGGGATAACAGGGTAATCGTAACTATAACGGTCCTAAGGTAGCGACTCTGAGACCCTCCCCGGATCCC | ||
| Ver1C21406 | CATTAGCGTGGTTTCCATGCG | Verification of disruption | |
| Ver2C21406 | AAGAGAGTTACTTTTGCGGGAGG | ||
| StartC21406 | ATCGGATCCCACAATGGATCTCGACAACTACCCTCCTC | Overexpression of YALI0C21406 | |
| EndC21406 | CATCCTAGGTCACTTTTGGGATCCGGGGAGGGTCTC | ||
| C21406F | GATCTTCCATGATTGGTATTG | Expression of YALI0C21406 by RT-PCR | |
| C21406R | CAGGGTGACGACAAAGAG | ||
| YALI0D20108g (YLJEN3) | D20108UpNotI | GAATGCGGCCGCGCACAAGTAGGTGGGTCCTTCCG | Upstream fragment of YALI0D20108 |
| D20108UpIsceI | CGATTACCCTGTTATCCCTACCGACAGATTGAGCGAGTAGACAGG | ||
| D20108DnNotI | GAATGCGGCCGCGAGAATCTGGAAGCATCCTGCTGCC | Downstream fragment of YALI0D20108 | |
| D20108Dn2IsceIIceuI | GGTAGGGATAACAGGGTAATCGTAACTATAACGGTCCTAAGGTAGCGATAATGTAGGGTGTGTGTGTGACG | ||
| Ver1-2D20108 | CCAAGGGTGGCCATTAGGAGTCG | Verification of disruption | |
| Ver2-2D20108 | GGTGTCATATTCGAGTCCTCGC | ||
| StartD20108-2 | ATCGGATCCCACAATGAATTTTGACAACTTCCCAGCACCAGATCTG | Overexpression of YALI0D20108 | |
| EndD20108-2 | CATCCTAGGTTATCGAGTATCGCTCGAAGAACGTCTTTCAATGTTC | ||
| D20108F | CTTCTTCTGCTTCTTCAACC | Expression of YALI0D20108 by RT-PCR | |
| D20108R | CATCTCTGCTTGTCTGTTTTC | ||
| YALI0D24607g (YLJEN4) | D24607UpNotI | GAATGCGGCCGCGATGCAGATCTTTCCGAGCGCTGG | Upstream fragment of YALI0D24607 |
| D24607UpIsceI | CGATTACCCTGTTATCCCTACCGGAAGGAGTCTTCCTGTTTATGG | ||
| D24607DnNotI | GAATGCGGCCGCCAAGTGGTTCCTGGACCGAATGGC | Downstream fragment of YALI0D24607 | |
| D24607Dn2IsceIIceuI | GGTAGGGATAACAGGGTAATCGTAACTATAACGGTCCTAAGGTAGCGAGGCAGGTATTAGATTTATATGAGTAGACG | ||
| Ver1D24607 | CGACATTTGAAGGAGATGACGG | Verification of disruption | |
| Ver2D24607 | GGACATTGTGCCCTGGGCCACC | ||
| StartD24607 | ATCGGATCCCACAATGACCCAGTCGTACGAAGTCGGAAAC | Overexpression of YALI0D24607 | |
| EndD24607 | CATCCTAGGCTAATGAACACTTCCAACAGTGGTCTG | ||
| HG-RT-D24607F2 | CCGGAGGAATCTTTATGG | Expression of YALI0D24607 by RT-PCR | |
| HG-RT-D24607R2 | GTGAGATGGATGGGGATG | ||
| YALI0E32901g (YLJEN6) | E32901UpNotI | GAATGCGGCCGCAGAGAGTTCTTTATCCACCCCACGG | Upstream fragment of YALI0E32901 |
| E32901UpIsceI | CGATTACCCTGTTATCCCTACCGGACTAGTGAGTGCTTGCCACGAG | ||
| E32901DnNotI | GAATGCGGCCGCCAGTACTCATTACCAGGCAATACGG | Downstream fragment of YALI0E32901 | |
| E32901Dn2IsceIIceuI | GGTAGGGATAACAGGGTAATCGTAACTATAACGGTCCTAAGGTAGCGACCAATATGACTCCCCCTACGAGTCC | ||
| Ver1 E32901 | CTCAACAAAGAGATGATAAGCC | Verification of disruption | |
| Ver2 E32901 | CTGTATTGAATATTTGACTGCTCC | ||
| StartE32901 | ATCGGATCCCACAATGGAAGCTCCTAATCTCTCGCCAGC | Overexpression of YALI0E32901 | |
| EndE32901 | CATCCTAGGTTACTTGGACTCGTAGGGGGAGTC | ||
| E32901F | CGGCTCTCAAGACCTCTAC | Expression of YALI0E32901 by RT-PCR | |
| E32901R | GAGCACCTCCAATAACACAA | ||
| pTEF-Start | GGGTATAAAAGACCACCGTCC | Verification of insertion of overexpression cassettes | |
| 61 Stop | GTAGATAGTTGAGGTAGAAGTTG | ||
| Actin | ACT-A1 | TCCAGGCCGTCCTCTCCC | Expression of Actin by RT-PCR |
| ACT-A2 | GGCCAGCCATATCGAGTCGCA |
Construction of the disruptant strains in Y. lipolytica
The deletion cassettes were generated by PCR amplification as described by Fickers et al. (2003) using the primer pairs described in Table2. The Up-Dn cassettes, which contained an IsceI site, were then inserted into the pCR®4Blunt-TOPO® vector (Invitrogen, Saint-Aubin, France). The auxotrophic marker (either URA3ex or LEU2ex) was then inserted into the vector IsceI site to generate the corresponding JMP (UpDnURA3ex/LEU2ex) vectors (Table1). Transformants were selected on YNB and Leu or YNB and Ura, respectively. The corresponding ver1 and ver2 primers (Table2) were used to check gene disruption by PCR amplification of the genomic loci.
For YALI0B19470g disruption, the primer pairs B19470UpNotI/B19470UpIsceI and B19470DnNotI/B19470DnIsceIIceuI were used (Table2). The Up and Dn regions were purified and used for the PCR fusion. The resulting UpDn fragment was ligated into pCR4Blunt-TOPO, yielding the construct JMP1647. The URA3ex and LEU2ex markers (from JMP802 and JMP803) were then introduced at the I-sceI site, yielding the constructs JMP1667 and JME1680, which respectively contained the YALI0B19470g::URA3ex and YALI0B19470g::LEU2ex cassettes. The deletion cassettes used in this study were obtained by digestion of these plasmids by NotI.
For YALI0C15488g disruption, primer pairs C15488UpNotI/C15488UpIsceI and C15488DnNotI/C15488DnIsceIIceuI were used (Table2). The Up and Dn regions were purified and used for the PCR fusion. The resulting UpDn fragment was ligated into pCR4Blunt-TOPO, yielding the construct JMP1649. The URA3ex and LEU2ex markers (from JMP802 and JMP803) were then introduced at the I-sceI site, yielding the constructs JMP1671 and JME1682, which respectively contained the YALI0C15488g::URA3ex and YALI0C15488g::LEU2ex cassettes. Deletion cassettes were obtained by digestion of these plasmids by NotI.
For YALI0C21406g disruption, primer pairs C21406UpNotI/C21406UpIsceI and C21406DnNotI/C21406Dn2IsceIIceuI were used (Table2). The Up and Dn regions were purified and used for the PCR fusion. The resulting UpDn fragment was ligated into pCR4Blunt-TOPO, yielding the construct JMP1683. The URA3ex marker (from JMP803) was then introduced at the I-sceI site, yielding the construct JMP1698, which contained the YALI0C21406g::URA3ex cassette. Deletion cassettes were obtained by digestion of these plasmids by NotI.
For YALI0D20108g disruption, primer pairs D20108UpNotI/D20108UpIsceI and D20108DnNotI/D20108Dn2IsceIIceuI were used (Table2). The Up and Dn regions were purified and used for the PCR fusion. The resulting UpDn fragment was ligated into pCR4Blunt-TOPO, yielding the construct JMP1684. The URA3ex marker (from JMP803) was then introduced at the I-sceI site, yielding the construct JMP1702, which contained the YALI0D20108g::URA3ex cassette. Deletion cassettes were obtained by digestion of these plasmids by NotI.
For YALI0D24607g disruption, primer pairs D24607UpNotI/D24607UpIsceI and D24607DnNotI/D24607Dn2IsceIIceuI were used (Table2). The Up and Dn regions were purified and used for the PCR fusion. The resulting UpDn fragment was ligated into pCR4Blunt-TOPO, yielding the construct JMP1777. The LEU2ex marker (from JMP802) was then introduced at the I-sceI site, yielding the construct JMP1813, which contained the YALI0D24607g::LEU2ex cassette. Deletion cassettes were obtained by digestion of these plasmids by NotI.
For YALI0E32901g disruption, primer pairs E32901UpNotI/E32901UpIsceI and E32901DnNotI/E32901Dn2IsceIIceuI were used (Table2). The Up and Dn regions were purified and used for the PCR fusion. The resulting UpDn fragment was ligated into pCR4Blunt-TOPO, yielding the construct JMP1689. The URA3ex and LEU2ex markers (from JMP802 and JMP803) were then introduced at the I-sceI site, yielding the constructs JMP1713 and JME1715, which respectively contained the YALI0E32901g::URA3ex and YALI0E32901g::LEU2ex cassettes. Deletion cassettes were obtained by digestion of these plasmids by NotI.
Complementation of the sextuple mutant with individual YLJEN1-6 genes was performed using the respective primer pairs StartB19470/EndB19470, StartC15488/EndC15488, StartC21406/EndC21406, StartD20108/EndD20108, StartD24607/EndD24607, and StartE32901/EndE32901. All complementation genes were cloned into pCR4Blunt-TOPO and then digested by AvrII and BamHI. They were then cloned into JMP1392 that had been previously digested by BamHI and AvrII; this generated JMP1792, JMP1794, JMP1821, JMP1809, JMP1796, and JMP1798, respectively.
Construction of deletion strains was controlled by PCR using Ver1 and Ver2 primers (Table2). Strain complementation was verified by PCR using pTEF-Start and 61 Stop primers.
Growth microtiter plate analysis
Y. lipolytica strain precultures were grown overnight, then centrifuged and washed two times with YNB supplemented with NH4Cl and phosphate buffer (YNB N and P). They were then resuspended into 1 mL of YNB N and P. Washed cells were then sown into 96-well microplates with adapted medium at OD600 = 0.1 in 200 μL of final volume. Growth was observed using a microtiter plate reader (Biotek, Colmar, France), following the manufacturer's instructions.
Analysis of YLJEN gene expression
To determine if carbon sources induced the expression of YLJEN genes, precultures of the reference strain JMY2900 were established in liquid YNB supplemented with 1% glucose and 0.5% yeast extract; cultures grew for 15 h at 28°C. Cells were then washed with distilled water and transferred into fresh liquid YNB media supplemented with one of the following: 1% glucose, 0.3% lactate, 0.3% fumarate, 0.3% malate, 0.3% pyruvate, or 0.3% succinate. Cultures were incubated in baffled Erlenmeyer flasks at 28°C and 160 rpm, then harvested at 2 and 6 h postinoculation, frozen in liquid nitrogen, and stored at −80°C. RNA was extracted from cells using the RNeasy Mini kit (Qiagen), and 2 μg were treated with DNAse (Ambion; Life Technologies, Saint-Aubain, France). cDNA was synthesized using the Maxima First Strand cDNA synthesis kit (Thermo Fisher Scientific, Villebon sur Yvette, France). PCR was then performed using the GoTaq DNA polymerase kit (Promega) with specific primers designed by the Primer3 program and listed in Table2. PCR reactions were performed with 1 μL of cDNA.
HPLC analysis
Y. lipolytica strain precultures were grown overnight, then centrifuged and washed two times with YNB supplemented with NH4Cl and phosphate buffer (YNB N and P). They were then resuspended in 1 mL of YNB N and P. Cultures were inoculated at OD600 nm = 0.1 in 50 mL of YNB medium supplemented with 50, 100, or 200 mmol/L of phosphate buffer (pH = 6.8) depending of the quantity of substrate (1, 2, or 4) and 170 mmol/L each of fumarate, lactate, malate, and succinate, then incubated at 28°C with agitation (160 rpm). To analyze the consumption of carbon sources, a sample of each culture was centrifuged for 1 min at 13 000 rpm; the resulting supernatant was analyzed with HPLC (UltiMate 3000; Dionex-Thermo Fisher Scientific, UK) using a HyperREZ XP carbohydrate H+ 8-μm column (Thermo Fisher Scientific, Villebon sur Yvette, France) coupled to a UV (210 nm) detector. The column was eluted with 0.01 N H2SO4 at room temperature and a flow rate of 0.6 mL min−1. Standard samples were used to determine the quantity of each compound. Prior to HPLC analysis, samples were filtered on 0.45-μm pore-size membranes.
Identification of Jen proteins in ascomycetous yeasts
Twenty-three species were chosen based on their positions in the phylogenetic tree of hemiascomycetes (see Kurtzman 2011; for reference tree). For each species, genomes were retrieved from the indicated database: S. cerevisiae – SGD (http://www.yeastgenome.org/); C. albicans – CGD (http://www.candidagenome.org/); Schizosaccharomyces pombe – Pombase (http://www.pombase.org/); Komagataella (Pichia) pastoris – Gent University (https://bioinformatics.psb.ugent.be/gdb/pichia/); Wickerhamomyces ciferrii, Kazachstania naganishii, and Naumovozyma castellii – the NCBI database (http://www.ncbi.nlm.nih.gov/); Candida tropicalis – the Broad Institute (http://www.broadinstitute.org/); Candida glabrata, Zygosaccharomyces rouxii, Eremothecium (Ashbya) gossypii, K. lactis, Lachancea kluyveri, Lachancea thermotolerans, and Debaryomyces hansenii – Génolevures (http://www.genolevures.org/); Blastobotrys (Arxula) adeninivorans, Cyberlindnera fabianii, and Y. lipolytica – GRYC (http://gryc.inra.fr). Species closely related to Y. lipolytica, that is, Y. yakushimensis CBS10253, Yarrowia galli CBS9722, Y. phangngensis CBS10407, Y. alimentaria CBS10151, and C. hispaniensis CBS9996, were sequenced and annotated in our laboratory at INRA Thiverval-Grignon (data not shown). Homologues of YLJEN genes were found via a homology search based on a two-step reciprocal approach using BLASTp (Altschul et al. 1990). In the first step, each YLJen protein was used as a query for a BLASTp search of the other species, with a cutoff E-value of 1e-10. Then, we used the top hit of each search to perform a reciprocal BLASTp search; if the top hit returned a Y. lipolytica gene different from a JEN gene, it was discarded. Amino acid sequences of all Jen proteins are given in Table S1. Pairwise comparisons of both amino acid identity and similarity were calculated using aligncopypair (EMBOSS; Rice et al. 2000).
Jen gene phylogeny
The JEN gene tree was constructed based on an alignment of 61 Jen proteins, representing 18 yeast species. The alignment was performed with MultAlin (Corpet 1988) and manually corrected with GeneDoc v2.7.0 (Nicholas et al. 1997). The final 405-aa alignment was used to reconstruct trees with both Neighbor-Joining and Maximum-Likelihood algorithms, with three different amino acid substitution matrices (WAG, Dayhoff, and JTT). The models were corrected by a Γ-law distribution with four categories of evolution rates; both invariable sites and the α-parameter of the Γ-law distribution were optimized according to the data. Seaview v4.3.3 was used for these analyses (Gouy et al. 2010).
Reconstruction of an evolutionary scenario
To reconstruct the evolutionary scenario of the Jen protein family in the Yarrowia clade, a species tree based on the concatenation of 912 proteins (data not shown) and a Jenp tree built as described above were reconciled with synteny data. A parsimonious approach was used to minimize the number of duplications and losses of JEN genes. For the phylogeny of JEN genes within the Yarrowia clade, KLJen2p was used as an outgroup. Protein sequences were aligned with MAFFT (Katoh et al. 2002) and edited with Gblocks (Castresana 2000). The phylogenetic tree constructed from the Jen protein alignment was constructed with Seaview v4.3.3 using the BioNJ distance method (Gouy et al. 2010). Bootstrap values were calculated with 100 replicates.
Results and Discussion
The six Y. lipolytica Jen1 proteins are more similar to members of the Jen2 subfamily
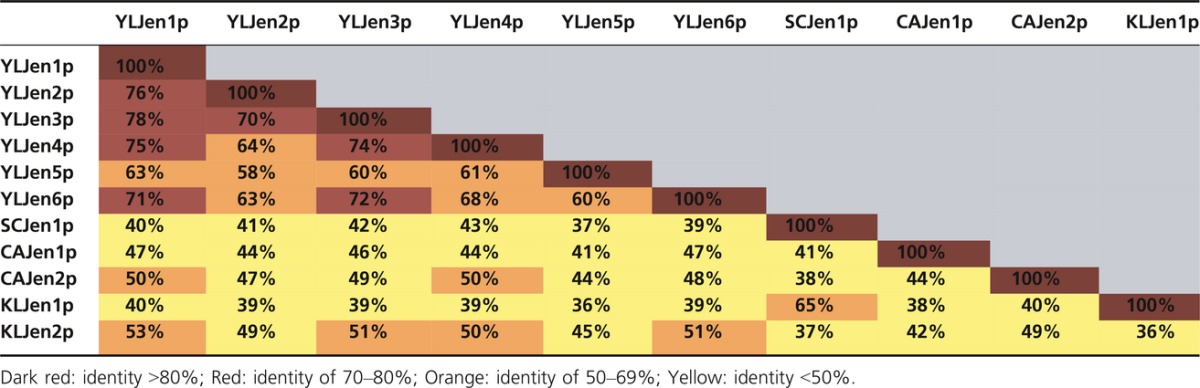
The six YLJen proteins were much more similar to each other (60–78% shared identity; Table3) than they were to Jen proteins from other species (37–47% identity with Jen1p, 44–51% identity with Jen2p). Additionally, all YLJen proteins presented 10 TM domains (Fig. S1). Among the six YLJenp, YALI0B19470p (YLJen5p) seemed to be the most divergent, as it shared the smallest percentage of identity both with the other YLJenp and with sequences from other species, and it presented an additional, unique sequence between TM6 and TM7 at residues 258–279 (PETGLHMQPAQKVGTWASIVI; Fig.1). The six YLJenp were shorter than SCJen1p and KLJen1p, both of which have an N-terminal extension that the YLJenp lacked. The alignment of the six YLJenp sequences with those of Jen1p and/or Jen2p from S. cerevisiae, K. lactis, and C. albicans was consistent with the results of Lodi et al. (2007) in revealing a conserved motif in TM7, NXX(S/T)HX(S/T)QDXXXT. One exception to this was found, however, in YALI0B19470p, in which the first S/T residue was replaced by an alanine (Fig.1, blue frame). In S. cerevisiae, F270 and Q498 residues are highly important for the determination of mono- or dicarboxylate transport function (Soares-Silva et al. 2011). Here, we found that the Q498 residue of SCJen1p was perfectly conserved in all Jenp (Fig.1, green frame), but this was not the case for F270, which was conserved only in KLJen1p. Instead, in all six YLJenp this phenylalanine was replaced by glutamine, as found in Jen2p of K. lactis and C. albicans (Fig.1, red frame). This observation suggests that the molecules transported by the YLJenp transporters may be different from those of the Jen1p of S. cerevisiae or K. lactis, and may potentially be more similar to the Jen2p in K. lactis and C. albicans in their target specificity (i.e., dicarboxylic acids). We also note that the F270 residue was replaced by a leucine in CAJen1p, but as both F and L are nonpolar amino acids, this change is unlikely to have serious consequences for protein specificity.
Table 3.
Percentage of shared amino acids between YLJenp and the Jen1p and Jen2p of Saccharomyces cerevisiae, Candida albicans, and Kluyveromyces lactis
SCJen1p has two ubiquitination sites, K9 and K338, but only the latter is involved in the degradation of the protein (Paiva et al. 2009). Whereas all Jen1p had at least one predicted ubiquitination site in the N-terminal domain (K9/K338 for SCJen1p, K4/K28/K538 for CAJen1p, and K14/K589/K597 for KLJen1p), the potential ubiquitination sites of YLJenp and CAJen2p were generally predicted to be in the proteins C-terminal domains: K508 for YALI0B19470p, K473/K481 for YALI0C15488p, K477/K483 for YALI0C21406p, K487 for YALI0D20108p, K479 for YALI0E32901p, and K502/K506 for CAJen2p (Fig.1, highlighted in gray). No ubiquitination site was predicted with high confidence for YALI0D24607p and KLJen2p.
Together, these three results – the observation that YLJenp sequences shared a higher degree of identity with Jen2 proteins than with Jen1p; the replacement of the F240 residue, found in SCJen1p, with a glutamine, as found in the Jen2p sequences; and the difference between YLJenp and Jen1p sequences in the distribution of ubiquitination sites – suggests that the YLJenp are more closely related to the Jen2 protein family than to the Jen1 protein family, in contrast with what has been previously reported by Lodi et al. (2007). To test this hypothesis, we investigated the evolution of these genes by reconstructing the evolutionary history of the Jen family in the Yarrowia clade as well as in a broader group of hemiascomycetes.
Reconstruction of the evolutionary scenario of the JEN family in hemiascomycetes
To better understand the evolution of JEN genes, we chose 23 species based on their phylogenetic position on the hemiascomycete tree, with the goal of including a broad representation of the genetic diversity within this taxonomic group. We selected nine species of Saccharomycetaceae (four that emerged after the whole genome duplication event (“duplicated”) and five that emerged prior), two species of Wickerhamomycetaceae, and three CTG species, in addition to Komagataella (Pichia) pastoris, Blastobotrys (Arxula) adeninivorans, six species closely related to Y. lipolytica, and Schizosaccharomyces pombe, which served as an outgroup species (Material and Methods). We then retrieved the genomes of these yeasts from genetic databases and attempted to identify the JEN genes by BLAST searches; this effort was successful in 18 of the 23 species. With the exception of S. cerevisiae, none of the species that diverged after the whole genome duplication event has a JEN gene, nor S. pombe and Zygosaccharomyces rouxii. In total, 61 genes representing 18 species were included in the phylogenetic analysis (Fig.2A). The tree generated from the protein sequences contained three monophylogenetic groups, and the branching patterns within these groups were consistent with the species tree topology (see Kurtzman 2011 for reference tree). Each group corresponded to one of three putative ancestral genes, which then underwent numerous further evolutionary events. This hypothesis is supported by the fact that some species, such as D. hansenii and L. kluyveri, had genes in each group. The first group included the known genes SCJEN1, KLJEN1, and CAJEN1, as well as genes from B. adeninivorans, the CTG, and the Wickerhamomycetaceae. It appears that at least seven duplications occurred in this group, with the most ancestral predating the emergence of the Wickerhamomycetaceae genus and the six others being species-specific duplications. The KLJEN2 and CAJEN2 genes belonged to the second monophyletic group, which also contained members of the CTG, the Saccharomycetaceae, the Wickerhamomycetaceae, and K. pastoris. The proteins in this group were highly conserved, with pairwise amino acid similarity ranging from 61% to 89%. The branching pattern of the tree indicated that the JEN2 ancestral gene has been lost in S. cerevisiae and, in contrast to our initial hypothesis, none of the YLJEN genes clustered with JEN2 homologues. All branches of the hemiascomycete tree were represented in the third group, which may derive from an ancestral gene that we called JEN3. All genes from Y. lipolytica, as well as their homologues in closely related species, clustered together within this group, indicating a monophyletic origin. A detailed analysis of their evolutionary history, based on sequence similarity and synteny conservation, revealed an ancestral duplication event which occurred prior to the speciation of Candida hispaniensis, a species closely related to Y. lipolytica, at the base of the Yarrowia clade (Kurtzman 2005; Michely et al. 2013). This duplication led to the evolution of a highly conserved group which contains YLJEN5, and a more dynamically evolving group, whose ancestral member has been lost in C. hispaniensis (Fig.2B). Three successive duplications occurred in this latter group after the divergence of C. hispaniensis and led to the creation of four subgroups. The subgroup represented by YLJEN6 is highly conserved without any subsequent duplications or losses. In contrast, numerous events (duplications and losses) occurred in the three remaining subgroups. In the group containing the YLJEN3 and YLJEN4 genes, an additional duplication occurred, but this duplication could not be dated, as all other species, with the exception of Y. galli, have lost these two genes. In total, 12 duplications and losses were predicted in the Yarrowia clade, indicating that the Jen3 family has a highly dynamic history.
Figure 2.
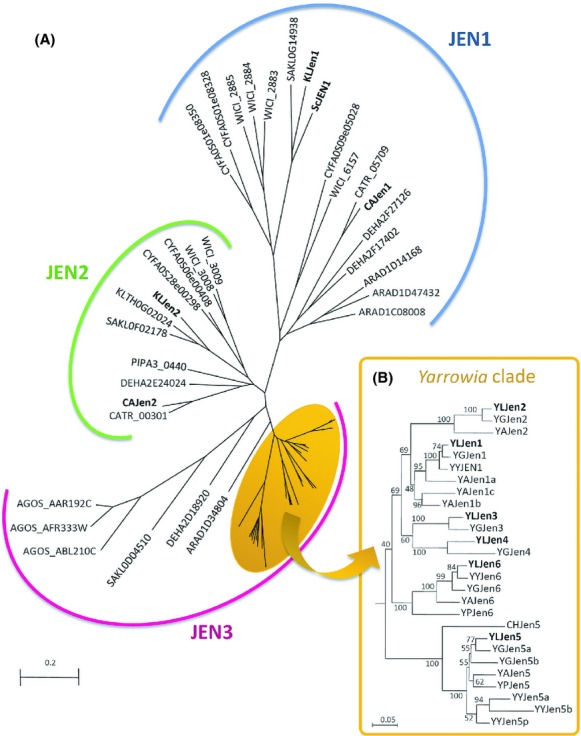
Phylogenetic trees of the Jen protein family. Trees are based on an alignment of Jen proteins and reconstructed with the Neighbor-Joining algorithm. A. Phylogeny of 61 Jen proteins from 18 hemiascomycetous species as inferred from 405 amino acid residues. Each subfamily of Jen proteins is surrounded by a different color: blue for Jen1p, green for Jen2p, and pink for Jen3p. The orange oval indicates the 28 Jen proteins found in the Yarrowia clade. Protein names in bold refer to protein from which experimental data have been published. B. Detailed phylogeny of 28 Jen proteins from the Yarrowia clade as inferred from an alignment of 423 amino acid residues. KLJen2p was used as an outgroup.
Transcription of YLJEN is substrate-specific and regulated by mono- and dicarboxylic acids
SCJEN1 is tightly regulated by various carbon sources; for example, it is repressed by glucose but activated by lactate (Chambers et al. 2004). KLJEN2 and CAJEN2 are likewise repressed by glucose and upregulated by succinate and malate (Lodi et al. 2004; Vieira et al. 2010). To determine how JEN genes are regulated in Y. lipolytica, RT-PCR was performed in media which contained one of the following carbon sources: glucose, fumarate, lactate, malate, pyruvate, and succinate (Fig.3). Surprisingly, the six YLJEN genes differed dramatically in their respective expression profiles. YLJEN1 and YLJEN2 were expressed under all tested conditions (Fig.3), with increased expression in the presence of lactate and pyruvate. YLJEN1 was also upregulated in fumarate and malate media, and slightly upregulated in the presence of succinate (Fig.3). YLJEN1 and YLJEN2 are closely related in the phylogenetic tree (Fig.2B), suggesting that their common ancestor was also expressed under similar conditions. It is possible that, following the divergence of the two genes, either the promoter of YLJEN1 evolved so that it became induced by fumarate, malate, and succinate, or, alternatively, this represented an ancestral state, which the promoter of YLJEN2 subsequently lost.
Figure 3.
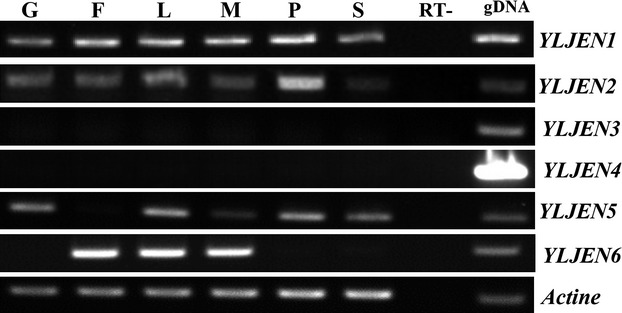
Expression profiles of YLJEN genes of reference strain JMY2900 in the presence of glucose, fumarate, lactate, malate, pyruvate, or succinate. Strain precultures were grown in liquid YNB supplemented with 1% glucose and 0.5% yeast extract for 15 h at 28°C (T0) and then transferred into fresh liquid YNB media supplemented with 1% glucose (G), 0.3% fumarate (F), 0.3% lactate (L), 0.3% malate (M), 0.3% pyruvate (P), or 0.3% succinate (S). RT-PCR was performed from cells incubated after 2 h post-inoculation. Actin was used as an endogenous control for all conditions tested. RT- and gDNA represent negative and positive controls, respectively.
YLJEN3 and YLJEN4 were not expressed in any medium. One potential explanation for this could be that these genes have lost, or are in the process of changing, their ability to be regulated (Fig.3). Interestingly, these genes derivate from the same ancestor (Fig.2B), suggesting that the lack of expression may derive from a common ancestor. Because the lineages YLJEN1/YLJEN2 and YLJEN3/YLJEN4 share a common ancestor, it is tempting to think that the promoter of the YLJEN3/YLJEN4 ancestor was lost during the duplication event that created these two lineages, which would explain the absence of expression of both modern genes. However, we cannot exclude the hypotheses that YLJEN3 and YLJEN4 are in fact expressed, but too weakly to be detected by RT-PCR, or that their promoters are activated under conditions other than those tested here. Alternatively, these two genes may be in the process of becoming pseudogenes, similar to YAGA0D19768g in Y. galli (a close relative of YLJEN4), a pseudogene that has been lost in all other species.
YLJEN5 was slightly upregulated in the presence of lactate and pyruvate, repressed by fumarate and, to a lesser extent, malate, and expressed in succinate as in glucose (Fig.3). YLJEN6 was only expressed when exposed to fumarate, lactate, and malate (Fig.3). In contrast to what has been reported for SCJEN1, KLJEN2, and CAJEN2, glucose did not repress the expression of YLJEN1, YLJEN2, YLJEN5, or YLJEN6. In this regard the YLJEN genes are similar to other genes involved in the metabolic pathways of Y. lipolytica, such as PEX11 or FAT1 (Dulermo et al. 2014 and unpublished data), a pattern which suggests that glucose catabolite repression does not occur in Y. lipolytica.
These results indicate that the regulation of individual YLJEN genes differs depending on the carbon source present but, as for SCJEN1, most are induced by lactate. The YLJEN gene family has thus probably evolved to enable the uptake of several carbon sources under different environmental conditions. In accordance with the expression patterns generated via RT-PCR, we hypothesized that YLJen1p and YLJen2p are involved in the transport of fumarate, lactate, malate, pyruvate and succinate, while YLJen5p transports lactate, pyruvate and probably succinate, and YLJen6p transports fumarate, lactate, and malate.
YLJen1p is involved in fumarate, malate, and succinate transport
We generated a deletion mutant strain for each YLJEN gene (Fig. S2, Table1) and studied their growth in media that each contained one of the following carbon sources: 0.3% glucose, 0.3% acetate, 0.3% butyrate, 0.3% citrate, 0.3% fumarate, 0.3% glutarate, 0.3% DL-lactate, 0.3% malate, 0.3% oxaloacetate, 0.3% pyruvate, and, 0.3% succinate. Surprisingly, we found that Y. lipolytica did not grow on glutarate-containing medium (data not shown), suggesting one of three potential scenarios: (1) this yeast does not possess a transporter for this compound, (2) the glutarate concentration was too low to support observable growth, or (3) Y. lipolytica is not able to metabolize glutarate. The single-deletion strains grew in the presence of all other carbon sources tested, with the exception of ΔYLjen1 when fumarate, malate, or succinate are added into the medium (Fig.4A–C and data not shown). Interestingly, this mutant did not grow at all in succinate-based medium, a result which suggests that YLJen1p may be the only, or at least the main, succinate transporter in Y. lipolytica (Fig.4C). Moreover, the growth of this strain was strongly reduced in media that contained fumarate or malate as the sole carbon sources (Fig.4A and B), a finding which indicates that YLJen1p is also involved in the transport of these compounds. However, the fact that the strain grew at all demonstrates that other YLJenp proteins are also involved in the transport of fumarate and malate.
Figure 4.
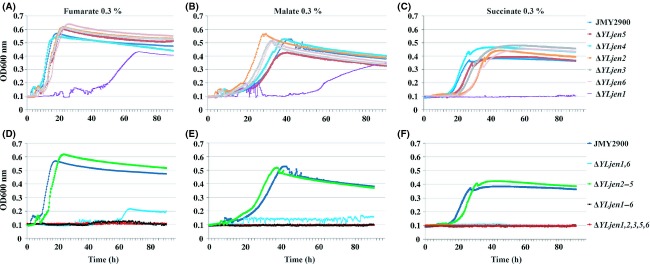
Growth of various YLJEN-deletion strains in media which contained 0.3% fumarate, malate, or succinate. (A and D) Growth in fumarate medium. (B and E) Growth in malate medium. (C and F) Growth in succinate medium. Growth curves are representative of three independent tests.
YLJen6p and YLJen1p are the main transporters of malate and fumarate
To determine the proteins involved in the transport of malate and fumarate, several deletion mutant strains were created in which different combinations of Jen proteins were absent (Fig. S2, Table3). We then evaluated their ability to grow on media that contained different carbon sources using the same procedure as for the single-deletion mutants. Interestingly, the quadruple-deletion strains ΔYLjen2-5 and ΔYLjen2,3,5,6 grew as well as the reference strain JMY2900 on all the carbon sources tested (Fig.4D–F and data not shown), a result showing that YLJEN2, YLJEN3, YLJEN4, YLJEN5, and YLJEN6 are not necessary for growth on the substrates tested here. Moreover, mutants in which YLJEN2, YLJEN3, YLJEN4, and/or YLJEN5 were deleted together with YLJEN1 exhibited the same reduction in growth on malate- and fumarate-based media as the ΔYLjen1 mutant (data not shown). This result confirms that YLJEN2, YLJEN3, YLJEN4, and YLJEN5 are not involved, or are only weakly involved, in the transport of fumarate and malate. This finding was not surprising with regard to YLJEN3 and YLJEN4 because they were not expressed even in the wild-type strain, but it was unexpected for YLJEN2 and YLJEN5 because both are expressed in the wild-type under the conditions tested here. Nevertheless, the deletion of YLJEN6 together with YLJEN1 was sufficient to prevent growth on malate medium and strongly reduce growth on fumarate medium (Fig.4D and E). This result reveals that these two genes serve as the main transporters of malate and fumarate in Y. lipolytica. An absence of growth on fumarate medium was observed when, in addition to YLJEN1 and YLJEN6, YLJEN2, and YLJEN5 or YLJEN3 and YLJEN5 were deleted (data not shown). This shows that YLJen5p is also able, at least weakly, to transport fumarate. Analysis of the quintuple mutant ΔYLjen1,2,3,5,6 and sextuple mutant ΔYLjen1-6 also reveal an absence of growth in media containing fumarate, malate, or succinate (Fig.4D–F). Surprisingly, ΔYLjen1-6 grew as well as JMY2900 in media containing 0.3% acetate, 0.3% butyrate, 0.3% citrate, 0.3% DL-lactate, 0.3% pyruvate, and 0.3% oxaloacetate (Fig. S3). It thus seems that YLJen proteins are not involved in the transport of these substrates, although we cannot exclude the possibility, especially for oxaloacetate, that other transporter(s) also take up these kinds of substrates and therefore compensate for the absence of the six YLJEN genes.
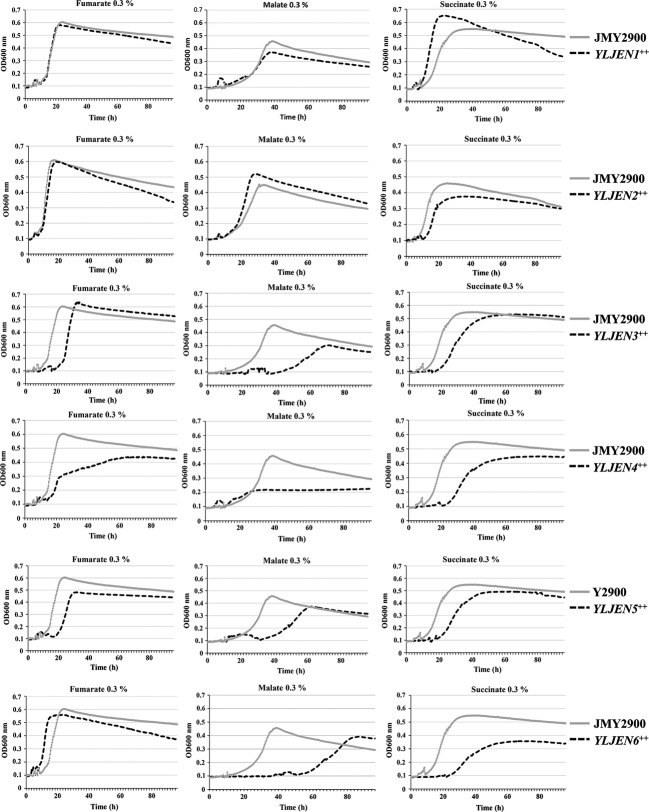
Overexpression of YLJen proteins rescues the transport of fumarate, malate, and succinate in the sextuple-deletion mutant ΔYLjen1-6
To better understand the specificity of the YLJenp, we analyzed the ability of each protein to rescue substrate-transport functions in the sextuple-deletion mutant ΔYLjen1-6. Each gene was expressed under the strong and constitutive promoter pTEF. We then evaluated the ability of each constructed strain to grow in media containing fumarate, malate, or succinate (Fig.5). As expected, the overexpression of YLJEN1 completely restored growth in the three types of media (Fig.5). However, the overexpression of YLJEN2 also had similar effects (Fig.5), which was surprising given that its deletion had not any observable effect on the growth of Y. lipolytica in these media. Unexpectedly, strains that overexpressed YLJEN5 or YLJEN3 grew almost as well as the YLJEN1-overexpression mutant in media containing fumarate or succinate (Fig.5), whereas, under the same conditions in the JMY2900 wild-type strain, these genes were poorly or not expressed (Fig.3). However, their overexpression only partially rescued the growth of ΔYLjen1-6 in malate medium (Fig.5), as would be expected from the RT-PCR results (no or weak expression in malate-based medium, Fig.3). Although YLJEN4 was not expressed in JMY2900, its overexpression partially restored growth in succinate and fumarate media and, to a much lesser degree, also in malate medium (Fig.5). YLJEN6 restored the growth of ΔYLjen1-6 completely in fumarate-based medium but only weakly in malate and succinate media (Fig.5). This result was surprising given the RT-PCR results, which had shown that YLJEN6 was strongly induced in the presence of fumarate and malate but not expressed in succinate medium (Fig.3).
Figure 5.
Growth of YLJEN-overexpression ΔYLjen1-6 mutants on media containing 0.3% fumarate, malate, or succinate. Growth curves are representative of three independent tests.
Taken together, these results indicate that (1) YLJen1p and YLJen2p are generalist transporters with no specific substrate affinity; (2) YLJen3p, YLJen4p, and YLJen5p are more specialized for the transport of fumarate and succinate; and (3) YLJen6p displays a high degree of affinity for fumarate. The best transporters, both in terms of transport efficiency and the diversity of substrates transported, are therefore YLJen1p and YLJen2p, which derivate from a common ancestor in the Jen3 phylogenetic tree (Fig.2B). It is possible, then, that their malate-transporting capabilities may derive from a common ancestor. To test this hypothesis, we attempted to investigate Jen protein from C. hispaniensis, a species closely related to Y. lipolytica, at the base of the Yarrowia clade (Kurtzman 2005; Michely et al. 2013). This species successfully grew in glucose-based medium, but was unable to grow in media that contained, alone or in mixture, lactate, malate, succinate, or fumarate (data not shown), suggesting that CHJenp is nonfunctional. We then overexpressed CHJEN in the ΔYLjen1-6 mutant, and found that this strain did not grow in media containing lactate, malate, succinate, and/or fumarate (data not shown), a result which lent support to this hypothesis.
In showing that multiple-YLJEN genes participate in fumarate transport, our results provide an explanation for the diversity of phenotypes found in the different deletion mutants grown with this substrate. However, it is very curious that, in vivo, the deletion of YLJEN1 completely inhibited growth in succinate-based medium, because our overexpression analysis revealed that YLJen2p and YLJen5p can also transport this compound (YLJen3p and YLJen4p can also transport succinate but they are not expressed in JMY2900). It is therefore possible that the transport of succinate is highly regulated. The same observation can be made about malate transport, as the deletion of YLJEN1 with YLJEN6 prevented growth, even though YLJEN5 and more specially YLJEN2 are also able to transport malate. One explanation could be that YLJen1p and/or YLJen6p control expression of the other YLJEN genes. However, RT-PCR performed in ΔYLjen1 and ΔYLjen1ΔYLjen6 deletion mutants revealed that the remaining JEN genes had the same expression profile (data not shown) as seen in the wild-type (Fig.3). This result indicates that YLJen1p and YLJen6p do not regulate the expression of the other YLJEN genes, at least at a transcriptional level. Posttranslational regulation may exist and could explain our results. It therefore seems that, as has been shown in S. cerevisiae (Andrade and Casal 2001; Paiva et al. 2002; Andrade et al. 2005), the regulation of JEN genes in Y. lipolytica has the potential to be very complex.
Patterns of carbon-source consumption in YLJEN-overexpression strains confirm substrate specificities of most YLJenp transporters
To better characterize the specificity and transport capacity of each YLJen protein, we used HPLC analysis to analyze the consumption of fumarate, malate, and succinate in the wild-type strain JMY2900, the deletion mutants ΔYLjen1, ΔYLjen6, ΔYLjen1,6, ΔYLjen1-6, and the six YLJEN-overexpression mutants. Strains were cultivated in YNB medium supplemented with 170 mmol/L of fumarate, malate, succinate, and lactate; lactate was added to enable the survival of strains, such as ΔYLjen1-6, that are not able to use other carbon sources.
Interestingly, three general patterns of strain behavior were observed. In the first, strains did not grow and did not consume fumarate, malate, and succinate; this group included ΔYLjen1-6, ΔYLjen1, and ΔYLjen1,6 (Fig.6A). However, the lactate concentration in the medium decreased slowly, at a rate of about 0.008 g L−1 h−1. This rate was 26-fold lower that that observed for the reference strain JMY2900 (0.21 g L−1 h−1), suggesting that these strains consumed lactate solely for the purpose of cell maintenance. Based on the results of our previous experiments, we expected that ΔYLjen1,6 and ΔYLjen1-6 would not grow in this medium; however, it was surprising to see that the ΔYLjen1 mutant did not grow either. One potential explanation for this could be that the culture conditions in this experiment were different from those used to generate the results presented in Figure4. Indeed, our initial growth experiments were conducted with a Biotek apparatus using media that contained only one carbon source, while the consumption tests were performed in flasks using a medium that contained four different carbon sources. Moreover, we note that the lag phase for JMY2900 in the growth experiments lasted around 10 h (Fig.4) compared to 20 h in the consumption experiments (Fig.6), an observation which suggests that cells may have been more stressed in the latter conditions.
Figure 6.
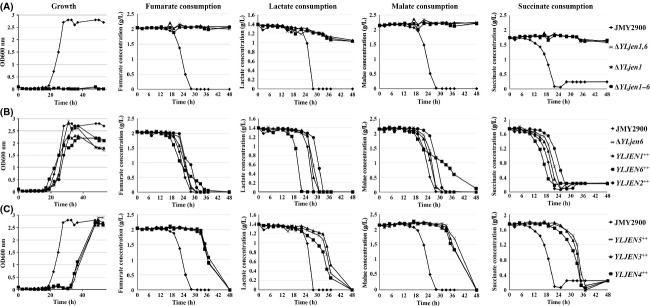
Growth and carbon-source consumption of reference strain JMY2900; YLJEN1-, YLJEN6-, and YLJEN1,6-deletion mutants; and YLJEN-overexpression strains. Strains were grown on media which contained fumarate, lactate, malate, or succinate, and three patterns of strain behavior were observed, presented here in A, B, and C, respectively. Growth and consumption curves are representative of three independent tests.
The second pattern of strain behavior involved growth that began at approximately 20 h of culture and substrate consumption that was as efficient as or better than that found in the JMY2900 wild-type, at 86–129% of the WT rate of consumption. This group was composed of ΔYLjen6 and ΔYLjen1-6 strains in which YLJEN1 or YLJEN2 were overexpressed (Fig.6B and C4). Finally, there was a third identifiable pattern: in this group, strains began to grow after approximately 30 h in culture and consumption rates of at least one substrate was lower than those found in the case of JMY2900. This group contained ΔYLjen1-6 strains that overexpressed YLJEN5 (consumption rate of malate reduced 2.2-fold compared to WT), YLJEN3 (consumption rate of malate and lactate reduced 1.4- and 1.3-fold, respectively), and YLJEN4 (consumption rate of malate and lactate reduced 1.5- and 1.75-fold, respectively) (Fig.6C and D4). In general, the results of the consumption tests were consistent with the growth curves shown in Figures4 and 5 in showing that members of group 2 (i.e., strains that overexpressed YLJEN1 or YLJEN2) grew similarly to JMY2900 in the different substrates while members of group 3 (i.e., strains that overexpressed YLJEN3, YLJEN4, or YLJEN5) grew more slowly in malate.
Table 4.
Substrate consumption rates of the different strains
| Fumarate | Malate | Succinate | Lactate | |||||
|---|---|---|---|---|---|---|---|---|
| Rate (g L−1 h−1) | R 2 | Rate (g L−1 h−1) | R 2 | Rate (g L−1 h−1) | R 2 | Rate (g L−1 h−1) | R 2 | |
| Reference strain JMY2900 | 0.22 (1) | 0.98 | 0.24 (1) | 0.96 | 0.19 (1) | 0.97 | 0.21 (1) | 0.91 |
| ΔYLjen6 | 0.28 (1.2) | 0.99 | 0.29 (1.2) | 0.95 | 0.18 (0.95) | 0.99 | 0.20 (0.94) | 0.99 |
| YLJEN1++ | 0.24 (1.1) | 0.99 | 0.27 (1.13) | 0.99 | 0.21 (1.1) | 0.97 | 0.20 (1.1) | 0.92 |
| YLJEN2++ | 0.19 (0.88) | 0.96 | 0.29 (1.2) | 0.99 | 0.17 (0.86) | 0.99 | 0.24 (0.86) | 0.99 |
| YLJEN3++ | 0.26 (1.2) | 0.99 | 0.18 (0.74) | 1 | 0.29 (1.52) | 0.94 | 0.16 (0.77) | 0.98 |
| YLJEN4++ | 0.24 (1.1) | 0.99 | 0.16 (0.65) | 0.96 | 0.23 (1.2) | 0.97 | 0.12 (0.57) | 0.96 |
| YLJEN5++ | 0.22 (1) | 1 | 0.10 (0.44) | 0.97 | 0.27 (1.4) | 0.94 | 0.18 (0.85) | 0.99 |
| YLJEN6++ | 0.16 (0.74)* 0.057 (0.26)** | 0.990.99 | 0.12 (0.49)* 0045 (0.19)** | 10.99 | 0.2 (1.05) | 1 | 0.25 (1.14) | 0.96 |
Values in parenthesis indicate ratio of rate of mutant to rate of reference strain, that is rate of substrate consumption before (*) and after (**) 22 h of growth. The results represent the mean values of at least two independent experiments and SD did not exceed 7%. R2 corresponds to the coefficient of linear correlation.
Interestingly, we observed a pattern of growth in the YLJEN6-overexpression strain of ΔYLjen1-6 which displayed characteristics of both groups 2 and 3. It began to grow after approximately 20 h in culture (more slowly than the other strains in group 2) and it consumed fumarate and malate more slowly as well (consumption rate decreased 1.35- and 2-fold, respectively, until 22 h of growth; Fig.6B and C4). Moreover, this strain showed some substrate specificity. Indeed, at 22 h, a point at which the supply of succinate and lactate in the medium had been exhausted, the consumption rate of malate and fumarate decreased drastically and simultaneously, 2.6- and 2.9-fold respectively, but was not followed by an immediate decrease in growth rate. The decrease in growth rate that happened at 28 h of culture could be caused by this change in substrate consumption (Fig.6 and Table4). In our gene-rescue experiments, we found that overexpression of YLJEN6 did not completely restore growth in succinate- and malate-based media (Fig.5), a finding which suggests that YLJen6p is not able to efficiently transport these compounds. The strong consumption rate of succinate in the YLJEN6-overexpression strain, then, is surprising, and could be explained by the presence of lactate, which may potentially be co-transported with succinate, and thus increase the rate of uptake. Similarly, the observation that the rate of fumarate consumption in the YLJEN6-overexpression strain was much weaker here compared to the efficient utilization of fumarate by this strain in the gene-rescue experiment (Fig.5) could be due to transport inhibition by malate.
Lactate and fumarate facilitate succinate consumption in the YLJEN6-overexpression strain
To test these hypotheses, we cultivated YLJEN6-overexpression ΔYLjen1-6 mutants in various media, containing several combinations of mono- and di-acids (fumarate only; lactate only; malate only; succinate only; fumarate and lactate; malate and lactate; succinate and lactate; fumarate and malate; fumarate and succinate). Growth and substrate consumption were quantified by spectroscopy and HPLC, respectively (Fig.7). Growth in media which contained only fumarate, malate, or succinate was similar to that shown in Figure5; however, this strain did not grow in lactate-based medium (Fig.7A). This was explained by the too low concentration of lactate in this experiment (170 mmol/L vs. 340 mmol/L in the growth experiment above; data not shown). Because of this lack of growth, lactate was consumed very slowly (rate = 0.013 g L−1 h−1; R = 0.99; Fig.7B), similarly to what had been observed in the previous experiment for the ΔYLjen1-6 strain (Fig.6), and indicating that it was only used for cell maintenance and survival. As expected, the patterns of consumption of fumarate, malate, and succinate reflected those found in the growth curves: fumarate was consumed rapidly (rate = 0.11 g L−1 h−1; R = 0.99), while malate and succinate were consumed slowly (at respective rates of 0.04 g L−1 h−1; R = 0.98 and 0.07 g L−1 h−1; R = 0.99; Fig.7B).
Figure 7.
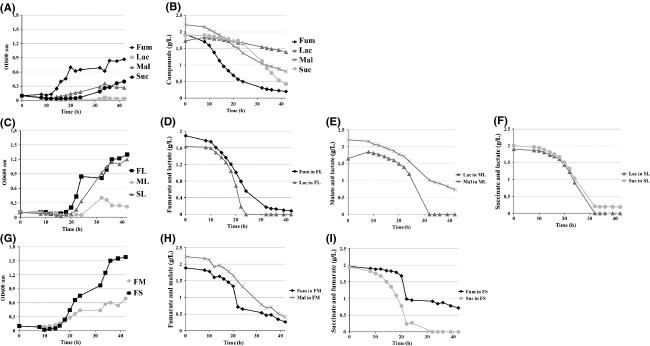
Growth and carbon-source consumption of the YLJEN6-overexpression strain when grown on media which contained, alone or in mixture, fumarate, lactate, malate, and/or succinate. (A) Growth in media containing fumarate (Fum), lactate (Lac), malate (Mal), or succinate (Suc). (B) Substrate consumption in media containing fumarate (Fum), lactate (Lac), malate (Mal), or succinate (Suc). (C) Growth in media containing lactate and fumarate (FL), lactate and malate (ML), or lactate and succinate (SL). (D) Substrate consumption in media containing lactate and fumarate. (E) Substrate consumption in media containing lactate and malate. (F) Substrate consumption in media containing lactate and succinate. (G) Growth in media containing fumarate and malate (FM) or fumarate and succinate (FS). (H) Substrate consumption in media containing fumarate and malate (I) Substrate consumption in media containing fumarate and succinate.
Interestingly, the addition of lactate to media containing fumarate or succinate improved both the growth rate and the amount of growth achieved by the YLJEN6-overexpression strain (Fig.7C); however, its addition to malate medium had no effect (Fig.7C). In these media, lactate was consumed more rapidly than either fumarate (rate = 0.21 g L−1 lactate h−1; R = 0.98 vs. rate = 0.1 g L−1 fumarate h−1; R = 0.99) or malate (rate = 0.14 g lactate L−1 h−1; R = 0.99 vs. rate = 0.05 g malate L−1 h−1; R = 0.98), an observation which suggests that the transport of lactate is not coupled with that of either fumarate or malate (Fig.7D and E). Conversely, the consumption curves of succinate and lactate overlapped (both at a rate of around 0.11 g L−1 h−1 with R = 0.99), providing evidence that they are co-transported (Fig.7F) and supporting our previous hypothesis.
The combination of malate and fumarate in the medium resulted in growth that was weaker than that observed in fumarate-only medium (Fig.7G). This was likely the result of low malate and fumarate consumption rates (rate = 0.06 g.L−1 h−1; R = 0.98 and rate = 0.05 g L−1 h−1; R = 0.98, respectively; Fig.7H). Interestingly, the consumption curves of these two substrates were quite similar. However, in this mixture, the fumarate consumption rate was twofold weaker that than found in the fumarate-only medium (rate = 0.11 g L−1 h−1). Instead, malate consumption was quite similar to that observed in other malate-containing media. Together, these results suggest that malate somehow inhibited fumarate consumption. We observed better growth on the fumarate and succinate medium. Under these conditions, fumarate was consumed 15-fold more slowly than succinate (with the exception of the period between 20 and 22 h, the consumption rate of fumarate was around 0.01 g L−1 h−1; R = 0.90 vs. 0.16 g L−1 h−1; R = 0.96 for succinate) and after 42 h of culture, there was still approximately 0.6 g L−1 fumarate remaining in the medium (Fig.7I). Additionally, the rate of succinate consumption was much higher in this medium than in the succinate-only medium. It seems, therefore, that the presence of fumarate somehow promotes succinate uptake, while succinate inhibits the transport of fumarate.
By evaluating the consumption profiles of fumarate, lactate, malate, and succinate both individually and when mixed together, we can observe that the consumption profiles presented in Figure6 are influenced by the competitive or facilitative roles of the different carbon sources with regard to each other. Although YLJen6p displays some preference for fumarate when only one substrate is present in the medium, its affinity for succinate changes strongly in the presence of lactate or fumarate, compounds which appear to be effectors of succinate uptake.
Conclusions
Lodi et al. (2007) showed that the Jen proteins in Y. lipolytica form a distinct group comparable to those found in S. cerevisiae, K. lactis, C. albicans, and other Hemiascomycetes and Euascomycetes. They assigned YLJEN2, YLJEN3, YLJEN4, and YLJEN6 to the Jen2 cluster and YLJEN5 and YLJEN1 to the Jen1 cluster, and furthermore identified YLJen1p and YLJen5p as “preJen1p” proteins (Lodi et al. 2007). However, through the use of a comparative genomics approach involving species closely related to Y. lipolytica, we found that this protein family has a dynamic evolutionary history dating from the emergence of the Yarrowia clade and represents a distinct subfamily of Jen proteins, Jen3p. In analyses of single-YLJEN and multiple-YLJEN-deletion mutants, we observed that growth was unaffected in acetate-, butyrate-, citrate-, DL-lactate-, oxaloacetate-, and pyruvate-based media, suggesting that either YLJen proteins are not involved in the transport of these substrates or there are other transporters that can compensate for their absence. In general, we found that the substrate specificities of the YLJen proteins are similar to members of the Jen2 subfamily, but that the different YLJenp vary a great deal in their expression level and transport targets. Indeed, it seems that YLJen1p is the main, most efficient transporter, followed by YLJen6p, although this will require specific transport essay to determine the kinetic parameters. Interestingly, we found a strong correlation between phylogeny, expression level, and transport targets. For example, YLJen1p and YlJen2p are derived from a common ancestor, transport the same substrates when they are overexpressed (fumarate, malate, and succinate), and demonstrate similar expression patterns, even if YLJEN1 is up-regulated under more conditions than YLJEN2 is. We were unable to determine if the YLJenp can transport pyruvate or lactate. However, some YLJEN genes, for example, YLJEN6, are upregulated by lactate. YLJen6p is also unusual in that exposure to lactate, but also to fumarate, increased succinate uptake in a mutant strain in which YLJen6p was the only functioning Jen protein, an observation which could explain the divergent results we obtained in experiments that involved YLJEN6 (Figs.5, 6, or 7).
Since Y. lipolytica is a an important organism in the biotechnology sector for its ability to produce both lipids (Beopoulos et al. 2011) as well as also other compounds such as succinate (Kamzolova et al. 2009, 2014; Yuzbashev et al. 2010), more investigations on succinate production with a strain devoid of the six YLJEN genes could be a promising avenue for future research since the absence of succinate consumption by the mutant strain could increase its production.
Acknowledgments
We thank FIDOP/FASO (fonds d'action stratégique des oléagineux) of the French vegetable oil and protein sector for financial support. Stéphanie Michely has a PhD fellowship funded by the French Ministry of Research via the ABIES doctoral school. We would also like to thank Jessica Pearce and Lindsay Higgins for their language editing services.
Conflict of Interest
None declared.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Table S1. Jen proteins found in the Hemiascomycetes.
Figure S1. TMHMM profiles of different Jen proteins.
Figure S2. Strain construction of the mutants used in this study.
Figure S3. Growth of ΔYLjen1-6 and JMY2900 in media which contained 0.3% acetate, butyrate, citrate, lactate, pyruvate, or oxaloacetate. ΔYLjen1-6 and JMY2900 growth curves are represented in gray and black, respectively. Growth curves are representative of three independent tests.
References
- Akita O, Nishimori C, Shimamoto T, Fujii T. Iefuji H. Transport of pyruvate in Saccharomyces cerevisiae and cloning of the gene encoded pyruvate permease. Biosci. Biotechnol. Biochem. 2000;64:980–984. doi: 10.1271/bbb.64.980. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW. Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Andrade RP. Casal M. Expression of the lactate permease gene JEN1 from the yeast Saccharomyces cerevisiae. Fungal Genet. Biol. 2001;32:105–111. doi: 10.1006/fgbi.2001.1254. [DOI] [PubMed] [Google Scholar]
- Andrade RP, Kötter P, Entian KD. Casal M. Multiple transcripts regulate glucose-triggered mRNA decay of the lactate transporter JEN1 from Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 2005;332:254–262. doi: 10.1016/j.bbrc.2005.04.119. [DOI] [PubMed] [Google Scholar]
- Barth G. Gaillardin C. Non conventional yeasts in biotechnology, a handbook. In: Wolf K, editor; Yarrowia lipolytica. Berlin: Springer; 1996. pp. 313–388. , ed., and . [Google Scholar]
- Beopoulos A, Mrozova Z, Thevenieau F, Le Dall M-T, Hapala I, Papanikolaou S, et al. Control of lipid accumulation in the yeast Yarrowia lipolytica. Appl. Environ. Microbiol. 2008;74:7779–7789. doi: 10.1128/AEM.01412-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beopoulos A, Nicaud JM. Gaillardin C. An overview of lipid metabolism in yeasts and its impact on biotechnological processes. Appl. Microbiol. Biotechnol. 2011;90:1193–1206. doi: 10.1007/s00253-011-3212-8. [DOI] [PubMed] [Google Scholar]
- Casal M, Paiva S, Andrade RP, Gancedo C. Leão C. The lactate-proton symport of Saccharomyces cerevisiae is encoded by JEN1. J. Bacteriol. 1999;181:2620–2623. doi: 10.1128/jb.181.8.2620-2623.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Chambers P, Issaka A. Palecek SP. Saccharomyces cerevisiae JEN1 promoter activity is inversely related to concentration of repressing sugar. Appl. Environ. Microbiol. 2004;70:8–17. doi: 10.1128/AEM.70.1.8-17.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulermo T, Treton B, Beopoulos A, Kabran Gnankon AP, Haddouche R. Nicaud JM. Characterization of the two intracellular lipases of Y. lipolytica encoded by TGL3 and TGL4 genes: new insights into the role of intracellular lipases and lipid body organisation. Biochim. Biophys. Acta. 2013;1831:1486–1495. doi: 10.1016/j.bbalip.2013.07.001. [DOI] [PubMed] [Google Scholar]
- Dulermo R, Gamboa-Meléndez H, Dulermo T, Thevenieau F. Nicaud JM. The fatty acid transport protein Fat1p is involved in the export of fatty acids from lipid bodies in Yarrowia lipolytica. FEMS Yeast Res. 2014;14:883–896. doi: 10.1111/1567-1364.12177. [DOI] [PubMed] [Google Scholar]
- Fickers P, Le Dall MT, Gaillardin C, Thonart P. Nicaud JM. New disruption cassettes for rapid gene disruption and marker rescue in the yeast Yarrowia lipolytica. J. Microbiol. Methods. 2003;55:727–737. doi: 10.1016/j.mimet.2003.07.003. [DOI] [PubMed] [Google Scholar]
- Gouy M, Guindon S. Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- Kabran P, Rossignol T, Gaillardin C, Nicaud JM. Neuvéglise C. Alternative splicing regulates targeting of malate dehydrogenase in Yarrowia lipolytica. DNA Res. 2012;19:231–244. doi: 10.1093/dnares/dss007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamzolova SV, Yusupova AI, Vinokurova NG, Fedotcheva NI, Kondrashova MN, Finogenova TV, et al. Chemically assisted microbial production of succinic acid by the yeast Yarrowia lipolytica grown on ethanol. Appl. Microbiol. Biotechnol. 2009;83:1027–1034. doi: 10.1007/s00253-009-1948-1. [DOI] [PubMed] [Google Scholar]
- Kamzolova SV, Vinokurova NG, Dedyukhina EG, Samoilenko VA, Lunina JN, Mironov AA, et al. The peculiarities of succinic acid production from rapeseed oil by Yarrowia lipolytica yeast. Appl. Microbiol. Biotechnol. 2014;98:4149–4157. doi: 10.1007/s00253-014-5585-y. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K. Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtzman CP. New species and a new combination in the Hyphopichia and Yarrowia yeast clades. Antonie Van Leeuwenhoek. 2005;88:121–130. doi: 10.1007/s10482-005-2495-0. [DOI] [PubMed] [Google Scholar]
- Kurtzman CP. Discussion of teleomorphic and ascomycetous yeasts and yeast-like taxa. In: Kurtzman CP, Fell JW, Boekhout T, editors. The Yeasts A Taxonomic Study. London: Elsevier; 2011. pp. 293–310. , eds. . [Google Scholar]
- Le Dall MT, Nicaud JM. Gaillardin C. Multiple-copy integration in the yeast Yarrowia lipolytica. Curr. Genet. 1994;26:38–44. doi: 10.1007/BF00326302. [DOI] [PubMed] [Google Scholar]
- Lodi T, Fontanesi F, Ferrero I. Donnini C. Carboxylic acids permeases in yeast: two genes in Kluyveromyces lactis. Gene. 2004;339:111–119. doi: 10.1016/j.gene.2004.06.019. [DOI] [PubMed] [Google Scholar]
- Lodi T, Diffels J, Goffeau A. Baret PV. Evolution of the carboxylate Jen transporters in fungi. FEMS Yeast Res. 2007;7:646–656. doi: 10.1111/j.1567-1364.2007.00245.x. [DOI] [PubMed] [Google Scholar]
- McDermott JR, Rosen BP. Liu Z. Jen1p: a high affinity selenite transporter in yeast. Mol. Biol. Cell. 2010;15:3934–3941. doi: 10.1091/mbc.E10-06-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michely S, Gaillardin C, Nicaud JM. Neuvéglise C. Comparative physiology of oleaginous species from the Yarrowia clade. PLoS ONE. 2013;8:e63356. doi: 10.1371/journal.pone.0063356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlícková K, Roux E, Athenstaedt K, Andrea S, Daum G, Chardot T, et al. Lipid accumulation, lipid body formation, and acyl coenzyme A oxidases of the yeast Yarrowia lipolytica. Appl. Environ. Microbiol. 2004;70:3918–3924. doi: 10.1128/AEM.70.7.3918-3924.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller S, Sandal T, Kamp-Hansen P. Dalbøge H. Comparison of expression systems in the yeasts Saccharomyces cerevisiae Hansenula polymorpha Klyveromyces lactis Schizosaccharomyces pombe and Yarrowia lipolytica. Cloning of two novel promoters from Yarrowia lipolytica. Yeast. 1998;14:1267–1283. doi: 10.1002/(SICI)1097-0061(1998100)14:14<1267::AID-YEA327>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Nicaud JM, Madzak C, van den Broek P, Gysler C, Duboc P, Niederberger P, et al. Protein expression and secretion in the yeast Yarrowia lipolytica. FEMS Yeast Res. 2002;2:371–379. doi: 10.1016/S1567-1356(02)00082-X. [DOI] [PubMed] [Google Scholar]
- Nicholas KB, Nicholas HB. Deerfield DW., II GeneDoc: analysis and visualisation of genetic variation. EMBNEW NEWS. 1997;4:14. [Google Scholar]
- Paiva S, Kruckeberg AL. Casal M. Utilization of green fluorescent protein as a marker for studying the expression and turnover of the monocarboxylate permease Jen1p of Saccharomyces cerevisiae. Biochem. J. 2002;363:737–744. doi: 10.1042/0264-6021:3630737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiva S, Vieira N, Nondier I, Haguenauer-Tsapis R, Casal M. Urban-Grimal D. Glucose-induced ubiquitylation and endocytosis of the yeast Jen1 transporter: role of lysine 63-linked ubiquitin chains. J. Biol. Chem. 2009;284:19228–192236. doi: 10.1074/jbc.M109.008318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queirós O, Pereira L, Paiva S, Moradas-Ferreira P. Casal M. Functional analysis of Kluyveromyces lactis carboxylic acids permeases: heterologous expression of KlJEN1 and KlJEN2 genes. Curr. Genet. 2007;51:161–169. doi: 10.1007/s00294-006-0107-9. [DOI] [PubMed] [Google Scholar]
- Querol A, Barrio E, Huerta T. Ramón D. Molecular monitoring of wine fermentations conducted by active dry yeast strains. Appl. Environ. Microbiol. 1992;58:2948–2953. doi: 10.1128/aem.58.9.2948-2953.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I. Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Maniatis T. Fritsch EF. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Soares-Silva I, Paiva S, Kötter P, Entian KD. Casal M. The disruption of JEN1 from Candida albicans impairs the transport of lactate. Mol. Membr. Biol. 2004;21:403–411. doi: 10.1080/09687860400011373. [DOI] [PubMed] [Google Scholar]
- Soares-Silva I, Paiva S, Diallinas G. Casal M. The conserved sequence NXX[S/T]HX[S/T]QDXXXT of the lactate/pyruvate: H(+) symporter subfamily defines the function of the substrate translocation pathway. Mol. Membr. Biol. 2007;24:464–474. doi: 10.1080/09687680701342669. [DOI] [PubMed] [Google Scholar]
- Soares-Silva I, Sá-Pessoa J, Myrianthopoulos V, Mikros E, Casal M. Diallinas G. A substrate translocation trajectory in a cytoplasm-facing topological model of the monocarboxylate/H symporter Jen1p. Mol. Microbiol. 2011;81:805–817. doi: 10.1111/j.1365-2958.2011.07729.x. [DOI] [PubMed] [Google Scholar]
- Tsuboi H, Wakisaka Y, Hirotsune M, Akao T, Yamada O. Akita O. Analysis of the pyruvate permease gene (JEN1) in glucose derepression yeast (Saccharomyces cerevisiae) Isolated from a 2-deoxyglucose-tolerant mutant, and its application to sake making. Biosci. Biotechnol. Biochem. 2003;67:765–771. doi: 10.1271/bbb.67.765. [DOI] [PubMed] [Google Scholar]
- Vieira N, Casal M, Johansson B, MacCallum DM, Brown AJ. Paiva S. Functional specialization and differential regulation of short-chain carboxylic acid transporters in the pathogen Candida albicans. Mol. Microbiol. 2010;75:1337–1354. doi: 10.1111/j.1365-2958.2009.07003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzbashev TV, Yuzbasheva EY, Sobolevskaya TI, Laptev IA, Vybornaya TV, Larina AS, et al. Production of succinic acid at low pH by a recombinant strain of the aerobic yeast Yarrowia lipolytica. Biotechnol. Bioeng. 2010;107:673–682. doi: 10.1002/bit.22859. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Jen proteins found in the Hemiascomycetes.
Figure S1. TMHMM profiles of different Jen proteins.
Figure S2. Strain construction of the mutants used in this study.
Figure S3. Growth of ΔYLjen1-6 and JMY2900 in media which contained 0.3% acetate, butyrate, citrate, lactate, pyruvate, or oxaloacetate. ΔYLjen1-6 and JMY2900 growth curves are represented in gray and black, respectively. Growth curves are representative of three independent tests.