Abstract
AmgRS is an envelope stress-responsive two-component system and aminoglycoside resistance determinant in Pseudomonas aeruginosa that is proposed to protect cells from membrane damage caused by aminoglycoside-generated mistranslated polypeptides. Consistent with this, a ΔamgR strain showed increased aminoglycoside-promoted membrane damage, damage that was largely absent in AmgRS-activated amgS-mutant strains. Intriguingly, one such mutation, V121G, while providing for enhanced resistance to aminoglycosides, rendered P. aeruginosa susceptible to several ribosome-targeting nonaminoglycoside antimicrobials that are inducers and presumed substrates of the MexXY-OprM multidrug efflux system. Surprisingly, the amgSV121G mutation increased mexXY expression threefold, suggesting that export of these nonaminoglycosides was compromised in the amgSV121G mutant. Nonetheless, a link was established between AmgRS activation and mexXY expression and this was confirmed in studies showing that aminoglycoside-promoted mexXY expression is dependent on AmgRS. While nonaminoglycosides also induced mexXY expression, this was not AmgRS-dependent, consistent with these agents not generating mistranslated polypeptides and not activating AmgRS. The aminoglycoside inducibility of mexXY was abrogated in a mutant lacking the AmgRS target genes htpX and PA5528, encoding a presumed cytoplasmic membrane-associated protease and a membrane protein of unknown function, respectively. Thus, aminoglycoside induction of mexXY is a response to membrane damage and activation of the AmgRS two-component system.
Keywords: Efflux, envelope stress, multidrug, Pseudomonas aeruginosa, two-component system
Introduction
Pseudomonas aeruginosa is a common nosocomial pathogen (Hidron et al. 2008; Zhanel et al. 2008, 2010) and a major cause of morbidity and mortality in patients with cystic fibrosis (CF) (Govan et al. 2007; de Vrankrijker et al. 2010; Brugha and Davies 2011). Treatment of P. aeruginosa infections is complicated by the microorganism's innate resistance to many antimicrobials, a product of its impressive intrinsic resistome (Olivares et al. 2013), and its access to an array of acquired resistance mechanisms (Breidenstein et al. 2011; Poole 2011). Major contributors to antimicrobial resistance in this organism are multidrug efflux systems of the resistance-nodulation-division (RND) family, including MexAB-OprM, MexCD-OprJ, MexEF-OprN, and MexXY-OprM, which contribute to both intrinsic (MexAB-OprM, MexXY-OprM) and acquired (all) resistance (Poole 2013). MexXY-OprM is somewhat unique in P. aeruginosa in conferring resistance to the aminoglycoside (AG) class of antimicrobials (Sobel et al. 2003; Poole 2005a; Henrichfreise et al. 2007), a class long-used in the management of CF lung infections caused by this microorganism (Prayle and Smyth 2010). While several endogenous AG resistance determinants are present in P. aeruginosa (Schurek et al. 2008; Dötsch et al. 2009; Lee et al. 2009; Krahn et al. 2012), MexXY-OprM is the predominant mechanism of resistance to these agents in CF isolates (Poole 2005a; Henrichfreise et al. 2007; Vettoretti et al. 2009).
The MexXY-OprM efflux system is comprised of a cytoplasmic membrane (CM) drug-proton antiporter (MexY), an outer membrane porin (OprM) and a periplasmic membrane fusion protein that joins the membrane-associated components together (MexX) (Aires et al. 1999). The MexX and MexY components are encoded by a single operon under the control of an adjacent repressor gene, mexZ (Aires et al. 1999; Matsuo et al. 2004), while OprM, which functions as the outer membrane component of several multidrug efflux systems in P. aeruginosa (Poole 2005b), is encoded by the 3rd gene of another multidrug efflux operon, mexAB-oprM (Aires et al. 1999; Mine et al. 1999). The mexXY operon is antimicrobial inducible, with only those agents known to target the ribosome able to promote mexXY expression (Masuda et al. 2000a; Jeannot et al. 2005; Morita et al. 2006). Antimicrobial-inducible mexXY expression is compromised by so-called ribosome protection mechanisms (Jeannot et al. 2005), suggesting that the MexXY efflux system is recruited in response to ribosome disruption or defects in translation. Consistent with this, mutations in fmt (encoding a methionyl-tRNA-formyltransferase) (Caughlan et al. 2009), folD (involved in folate biosynthesis and production of the formyl group added to initiator methionine) (Caughlan et al. 2009), and the ribosomal protein genes rplA (Westbrock-Wadman et al. 1999), rplY (El'Garch et al. 2007), and the rplU-rpmA operon (Lau et al. 2012), all of which are expected to negatively impact protein synthesis, increase the expression of mexXY. Upregulation of mexXY by antimicrobials (Morita et al. 2006) or mutations (fmt/folD [Caughlan et al. 2009], rplY [El'Garch et al. 2007] and rplU-rpmA [Lau et al. 2012]) is dependent upon a gene, armZ (formerly known as PA5471), encoding a MexZ-targeting anti-repressor (Yamamoto et al. 2009; Hay et al. 2013). Expression of armZ is also promoted by ribosome-disrupting antimicrobials (Morita et al. 2006) and fmt/folD (Caughlan et al. 2009) or rplU-rpmA (Lau et al. 2012) mutations. Moreover, armZ expression is governed by a transcriptional attenuation mechanism that directly links ribosome/translation disruption and armZ expression, providing a mechanism whereby ribosome perturbation drives MexXY recruitment (Morita et al. 2009). Still, drug-inducible mexXY expression independent of MexZ (Hay et al. 2013) and ArmZ (Muller et al. 2010) has been reported, an indication that additional regulator(s) influence mexXY expression. Indeed, the ParRS two-component system (TCS) implicated in adaptive resistance to cationic antimicrobial peptides, such as the polymyxins (Fernandez et al. 2010), has been linked to ArmZ-independent mexXY expression (Muller et al. 2010), with mutations in the parRS locus driving mexXY expression and AG resistance (Muller et al. 2010; Guenard et al. 2014).
Although ArmZ is required for mexXY induction in response to ribosome perturbation, it is insufficient for maximal drug-inducible expression of this efflux operon – armZ/PA5471 hyperexpression from a multicopy plasmid or resultant from a mutation in the armZ leader peptide both provide for much more modest mexXY expression as compared with drug-treated cells (Morita et al. 2006). Presumably, additional downstream effects of ribosome perturbation work in concert with ArmZ to effect/promote mexXY derepression. In the case of AGs, which promote mistranslation (Weisblum and Davies 1968), this may relate to the generation of aberrant polypeptides that damage the CM (Davis et al. 1986; Busse et al. 1992). Interestingly, the AmgRS TCS (Lee et al. 2009) in P. aeruginosa that appears to be operationally similar to the CpxRA envelope stress response TCS in Escherichia coli (Ruiz and Silhavy 2005) has been proposed to control an adaptive response to membrane damage caused by AG-generated aberrant polypeptides (Lee et al. 2009). Contributing to intrinsic AG resistance (Lee et al. 2009) this TCS has also been linked to acquired resistance in both laboratories and clinical AG-resistant strains as a result of gain-of-function “activating” mutations in the amgS gene that encodes the sensor kinase component of this TCS (Lau et al. 2013). In the current study we confirm that AmgRS, indeed, provides protection against AG-promoted membrane damage. We further demonstrate that AmgRS activation via an amgS gain-of-function mutation or AG exposure promotes mexXY expression, and that AmgRS mediates mexXY expression in response to AGs, but not to other inducers of this efflux operon.
Experimental Procedures
Bacterial strains and growth conditions
The bacterial strains and plasmids used in this study are described in Table1. Bacterial cells were cultured in Luria broth (L-broth) and on Luria agar (L-agar), with antibiotics as necessary, at 37°C. In E. coli, plasmid pEX18Tc and its derivatives were maintained or selected with 10 μg/mL tetracycline, while plasmid pK18mobsacB and its derivatives were maintained or selected with 50 μg/mL kanamycin.
Table 1.
Bacterial strains and plasmids used in this study.
| Strain or plasmid | Description1 | Reference |
|---|---|---|
| Escherichia coli strains | ||
| DH5α | φ80d lacZΔM15 endA1 recA1 hsdR17 (rK− mK+) supE44 thi- 1 gyrA96 relA1 F−Δ(lacZYA-argF) U169 | Ausubel et al. (1992) |
| S17-1 | thi pro hsdR recA Tra+ | Simon et al. (1983) |
| Pseudomonas aeruginosa strains | ||
| K767 | PAO1 prototroph (wild-type) | Masuda and Ohya (1992) |
| K3249 | K767 derivative carrying the amgSR182C mutation | Lau et al. (2013) |
| K3260 | K767 derivative carrying the amgSV121G mutation | Lau et al. (2013) |
| K3159 | K767ΔamgR | Krahn et al. (2012) |
| K3583 | K767ΔamgS | This study |
| K3584 | K3249ΔamgR | This study |
| K3585 | K3260ΔamgR | This study |
| K1525 | K767ΔmexXY | De Kievit et al. (2001) |
| K3586 | K1525 derivative carrying the amgSV121G mutation | This study |
| K2413 | K767ΔarmZ | Morita et al. (2006) |
| K3587 | K2413 derivative carrying the amgSV121G mutation | This study |
| K2415 | K767ΔmexZ | Morita et al. (2006) |
| K3615 | K2415ΔamgR | This study |
| K3589 | K767ΔyccA | This study |
| K3590 | K767ΔhtpX | This study |
| K3591 | K767ΔPA5528 | This study |
| K3593 | K767ΔhtpXΔyccA | This study |
| K3594 | K767 ΔhtpXΔPA5528 | This study |
| K3595 | K767 ΔyccAΔPA5528 | This study |
| K3596 | K767ΔhtpXΔPA5528ΔyccA | This study |
| Plasmids | ||
| pEX18Tc | Broad-host-range gene replacement vector; sacB Tcr | Hoang et al. (1998) |
| pK18mobsacB | Broad-host-range gene replacement vector; sacB Kanr | Schäfer et al. (1994) |
| pCL6 | pEX18Tc derivative carrying amgSV121G | Lau et al. (2013) |
| pCG005 | pEX18Tc::ΔamgR | Krahn et al. (2012) |
| pCL22 | pK18mobsacB::ΔamgR | This study |
| pCL23 | pEX18Tc::ΔamgS | This study |
| pCL24 | pEX18Tc::ΔyccA | This study |
| pCL25 | pEX18Tc::ΔhtpX | This study |
| pCL26 | pEX18Tc::ΔPA5528 | This study |
Tcr, tetracycline-resistant; Kanr, kanamycin resistant.
DNA methods
Standard protocols were used for restriction endonuclease digestion, ligation, transformation, and agarose gel electrophoresis, as described by Sambrook and Russell (Sambrook and Russell 2001). Plasmid and chromosomal DNA was prepared as before (Lau et al. 2012). DNA fragments used for cloning were extracted from agarose gels using a Wizard® SV gel and PCR clean-up system (Fisher Scientific, Ltd., Nepean, Canada). CaCl2-competent E. coli cells were prepared as described previously (Inoue et al. 1990). Oligonucleotide synthesis was carried out by Integrated DNA Technologies (Coralville, IA). Nucleotide sequencing was carried out by ACGT Corp. (Toronto, Canada) using universal primers.
Construction of P. aeruginosa gene-deletion mutants
Pseudomonas aeruginosa strains carrying various gene deletions were generated by first engineering the deletions in plasmid pEX18Tc (or plasmid pK18mobsacB in the case of the ΔamgR strain K3585) and then mobilizing them into P. aeruginosa from E. coli S17-1 as before (Srikumar et al. 1997; Krahn et al. 2012). Deletions were constructed by amplifying, via PCR, 1-kb fragments upstream and downstream of the sequences being deleted and cloning these individually into plasmid pEX18Tc for sequencing (to ensure that no mutations had been introduced during PCR) and then together into pEX18Tc to generate the deletion construct. PCR fragments were gel purified and digested with restriction enzymes (sites incorporated into the PCR primers) prior to cloning into appropriately digested plasmids. For deletions in amgS, htpX, yccA, and PA5528 the upstream and downstream fragments were amplified using the corresponding UP-F and UP-R, and DN-F and DN-R primers, respectively, (Table2) in separate 50-μL reaction mixtures containing 10 ng of P. aeruginosa K767 chromosomal DNA, 1 U of Phusion high-fidelity DNA polymerase (New England BioLabs, Ltd., Pickering, Ontario, Canada), 1X Phusion HF buffer, 5% (vol/vol) dimethyl sulfoxide (DMSO), primers at a 0.6 μmol/L final concentration, and deoxynucleoside triphosphates (dNTPs) at a 0.2 mmol/L final concentration. The mixtures were heated for 3 min at 98°C, followed by 35 cycles of 0.5 min at 98°C, 0.5 min at 72°C, and 0.5 min at 72°C, before finishing with 10 min at 72°C. For ΔamgR, the previously constructed amgR deletion plasmid pCG005 was employed as described (Krahn et al. 2012) or the ΔamgR gene was excised from pCG005 as a single 2-kb EcoRI-PstI restriction fragment and cloned into plasmid pK18mobsacB to generate an alternat-ive ΔamgR vector. Pseudomonas aeruginosa transconjugants harboring chromosomal inserts of the pEX18Tc- or pK18mobsacB-derived deletion vectors were selected on L-agar plates containing tetracycline (50 μg/mL) and chloramphenicol (5 μg/mL; to counterselect E. coli S17-1), or kanamycin (1500 μg/mL) and chloramphenicol (10 μg/mL; counterselect E. coli S17-1), respectively. These were subsequently streaked onto L-agar containing sucrose (10% [wt/vol]), with sucrose-resistant colonies screened for the appropriate deletion using colony PCR with 2.5 U Taq polymerase in 10% (vol/vol) DMSO (Sheu et al. 2000). Colony PCR was carried out using either the respective UP-F and DN-R primers (ΔamgR, ΔamgS, and ΔPA5528; Table2), or the scr-F and scr-R primer set (ΔyccA and ΔhtpX; Table2) for each deletion. The reaction mixtures were heated for 3 min at 95°C, followed by 30 cycles of 30 sec at 95°C, 30 sec at either 66.8°C (for ΔamgS), 65°C (for ΔamgR), 60°C (for ΔyccA), 55°C (for ΔhtpX) or 62°C (for ΔPA5528), and either 3.5 min (for ΔamgS), 3 min (for ΔamgR and ΔPA5528) or 2 min (for ΔyccA and ΔhtpX) at 72°C, before finishing with 10 min at 75°C.
Table 2.
Oligonucleotides used in this study.
| Primer | Oligonucleotide sequence (5′→3′)1 | Reference |
|---|---|---|
| amgRUp-F | GACTGAATTCCTGTAGAAGTCCTGGCGGT | Krahn et al. (2012) |
| amgRDown-R | GACTCTGCAGCGGCGCTGGAGAAACTGGT | Krahn et al. (2012) |
| amgSUP-F | GGACGTGAATTCGACTTGCTGCGGTTGAAC | This study |
| amgSUP-R | GACCTGTCTAGAACCAGAGCGGCGTTTTC | This study |
| amgSDN-F | GGCAGTTCTAGATGATACCCGACGGGTTTG | This study |
| amgSDN-R | AACGCTAAGCTTGACAGAAGGTCCATGCCAC | This study |
| yccAUP-F | GCAGTTGAGCTCTGGTACTGGTGGACGACGATG | This study |
| yccAUP-R | GCAGTTGGATCCTTGCATGGTGTGGGTACTCC | This study |
| yccADN-F | GAGGCTGGATCCGGCGACGACTGATAGAAAAC | This study |
| yccADN-R | GAGGCATCTAGAGGGTTGAGCAGGTACAGGCA | This study |
| yccA-scr-F | CCGTTGCCTTAAACCACTCG | This study |
| yccA-scr-R | AATCACCTGGTCGTAGCGTTG | This study |
| htpXUP-F | CATCGTGAGCTCGAAGGTCTCGACATCCTCG | This study |
| htpXUP-R | CATCGTGGTACCTAATTCTGGCCGGTGAAG | This study |
| htpXDN-F | CATCGTGGTACCTCCGCTTTCACACTTGGGA | This study |
| htpXDN-R | CGTGAGTCTAGAACGCCAGGCAGTCGTAG | This study |
| htpX-scr-F | CGCCAACACCATTCATTACG | This study |
| htpX-scr-R | TGAACCATTCGGCCATGTG | This study |
| PA5528UP-F | CATCGTGAGCTCGAACTTCGCCACGTAGG | This study |
| PA5528UP-R | GCACTCGGTACCAGAACACGCTGCATTGTAG | This study |
| PA5528DN-F | CATCGTGGTACCGAACTGATCCCGAGTGGTTG | This study |
| PA5528DN-R | GGAGCTTCTAGATGGTGTCGCCATTCAGC | This study |
| qPCR-htpX-F | ATCTCCAAGTGGATGGCGA | Lau et al. (2013) |
| qPCR-htpX-R | CAGCTCTTCGACGGTTTGC | Lau et al. (2013) |
| qPCR-PA5528-F | ATGCAGCGTGTTCTCAGC | Lau et al. (2013) |
| qPCR-PA5528-R | CGCTTGGCATTGGCATCCA | Lau et al. (2013) |
| qPCR-mexX-F | CTATCGGCATCACCAGCG | Lau et al. (2012) |
| qPCR-mexX-R | ATCTGGAACAGCACGGTG | Lau et al. (2012) |
| qPCR-PA5471-F | CATCAAGCCTTTGTCCGC | Lau et al. (2012) |
| qPCR-PA5471-R | CGGTGGTTTGCAGTTGCT | Lau et al. (2012) |
| qPCR-rpoD-F | ATCCTGCGCAACCAGCAGAA | Lau et al. (2012) |
| qPCR-rpoD-R | TCGACATCGCGCGGTTGATT | Lau et al. (2012) |
Restriction endonuclease cleavage sites are underlined.
Construction of P. aeruginosa AmgSV121G mutants
To introduce the amgSV121G mutation into the P. aeruginosa ΔmexXY and ΔarmZ strains K1525 and K2413, respectively, plasmid pCL6, a pEX18Tc-based gene replacement vector carrying the amgSV121G mutation, was mobilized into the P. aeruginosa deletion strains from E. coli S17-1 and amgSV121G derivatives selected as before (Lau et al. 2013).
Membrane depolarization assay
A previously described fluorometric assay (Krahn et al. 2012), involving the membrane potential-sensitive dye bis-(1,3-dibutylbarbituric acid) trimethine oxonol [DiBAC4(3)], was employed to measure the degree of CM depolarization promoted by AG treatment of P. aeruginosa. Briefly, early logarithmic phase (optical density at 600 nm [OD600 nm] = 0.3–0.5) L-broth subcultures of P. aeurginosa were treated with the AGs gentamicin (2 or 5 μg/mL final concentration) or tobramycin (0.5 or 1.25 μg/mL final concentration) or the aminocyclitol, spectinomycin (SPC) (1280 μg/mL). In some experiments P. aeruginosa was pretreated with chloramphenicol (128 μg/mL) for 15 min prior to the addition of gentamicin. Samples (5 mL) of the AG-treated and untreated control cultures were taken immediately and then hourly over 3 h and exposed to DiBAC4(3) (Invitrogen, Burlington, Ontario, Canada) at 37°C for 5 min in the dark at a final concentration of 10 μg/mL. Bacteria were then pelleted and resuspended in phosphate-buffered saline (Nehme et al. 2004) to a final OD600 nm of 0.1. Membrane depolarization-dependent fluorescence emitted by cells was then measured using a Varian (now Agilent, Mississauga, Ontario, Canada) Cary Eclipse fluorescent spectrophotometer with excitation and emission wavelengths of 490 and 518, respectively.
Quantitative real-time PCR
Bacterial RNA was isolated, purified, and reverse transcribed into cDNA as described previously (Lau et al. 2012). The primers used in quantitative real-time PCR (qPCR designation, Table2) were designed to amplify specific gene fragments with lengths of 99 bp (htpX), 74 bp (PA5528), 142 bp (mexX), 89 bp (armZ/PA5471), or 91 bp (rpoD), and were validated and described previously (Lau et al. 2012, 2013). All quantitative real-time PCR primer sets used in the present study had a minimum 4-log10 dynamic range. The expression of htpX, PA5528, mexX, armZ, and rpoD was assessed by quantitative real-time PCR as described previously using a CFX96 real-time PCR detection system (Bio-Rad, Mississauga, Ontario, Canada) (Lau et al. 2012). For each gene studied, at least one control reaction with no cDNA template was included in each experiment to check for contamination of reagent(s) and to identify unintended amplification products (e.g., primer dimers). The levels of expression of the target genes in each strain studied, normalized against that of the reference gene, rpoD, were calculated using the standard analysis feature of the CFX-manager software version 3.0 (Bio-Rad) and were reported herein as fold change relative to that in the P. aeruginosa PAO1 wild-type (WT) strain K767, unless otherwise specified.
Antibiotic susceptibility assay
The susceptibility of P. aeruginosa to antimicrobial agents was assessed using the twofold serial microtiter broth dilution method described previously (Jo et al. 2003), with an inoculum of ∼5 × 105 cells per mL. MICs were recorded as the lowest concentration of antibiotic inhibiting visible growth after 18 h of incubation at 37°C.
Results
AmgRS is a membrane damage-responsive TCS
A recent study showed that treatment of P. aeruginosa with the AG gentamicin-promoted depolarization of the CM and that this was exacerbated in a mutant lacking the AmgR component of the AmgRS TCS (Krahn et al. 2012). These data were interpreted as AG-generated mistranslated polypeptides damaging the CM, and AmgRS playing a role in ameliorating this damage. Consistent with AG-generated mistranslated/polypeptides being responsible for the observed CM depolarization, exposure of wild-type P. aeruginosa K767 to SPC, an aminocyclitol that is related to AGs but does not promote mistranslation (Wallace et al. 1974), failed to promote membrane depolarization (Fig.1A). Moreover, treatment of P. aeruginosa K767 with chloramphenicol, a translation inhibitor (Vazquez 1974), prior to the addition of gentamicin abrogated gentamicin-promoted membrane depolarization (Fig.1A), again consistent with (mis)translated polypeptides being central to the membrane damage being measured in the depolarization assay. In agreement with the previous study, loss of amgR enhanced membrane depolarization promoted by the AGs gentamicin (modestly at 2.5X MIC [data not shown] as seen before [Krahn et al. 2012] and much more strikingly at 1X MIC [Fig.1B, compare WT and ΔamgR]) and, especially, tobramycin (Fig.1C), which was markedly less membrane damaging than gentamicin at a comparable MIC (Fig.1C). In addition, amgS gain-of-function mutants K3249 (amgSR182C) and K3260 (amgSV121G), in which the AmgRS TCS is activated and AG resistance enhanced (Lau et al. 2013), showed reduced membrane depolarization, dependent on AmgR (Fig.1B). Taken together, these data support the conclusion that AmgRS protects the CM from damage(s) caused by AG-generated, mistranslated aberrant polypeptides.
Figure 1.
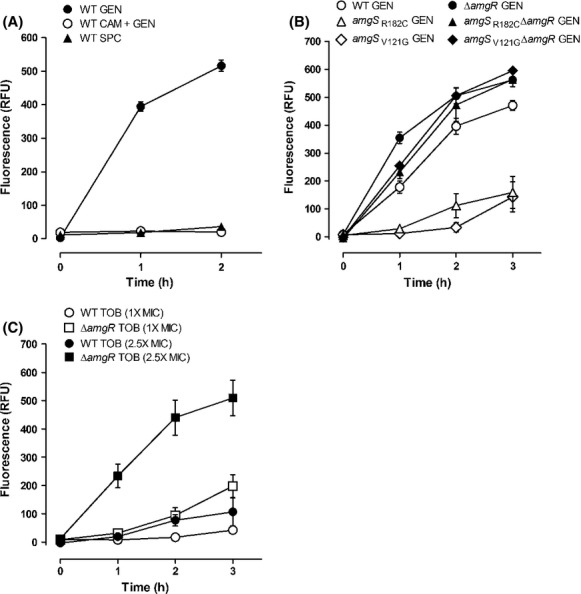
Aminoglycoside-promoted cytoplasmic membrane depolarization. Cytoplasmic membrane depolarization, as assessed by DiBAC4(3) fluorescence, was measured over time following exposure of Pseudomonas aeruginosa to various antimicrobials added at T = 0 h. (A) Wild-type (WT) P. aeruginosa strain K767 exposed to 5 μg/mL (2.5X MIC) of gentamicin (GEN; filled circles), 128 μg/mL chloramphenicol + 5 μg/mL gentamicin (CAM + GEN; open circles) and 1280 μg/mL (2.5X MIC) spectinomycin (SPC; filled triangles). (B) Pseudomonas aeruginosa strains K767 (WT; open circles), K3159 (ΔamgR; filled circles), K3249 (amgSR182C; open triangles), K3584 (amgSR182C ΔamgR; filled triangles), K3260 (amgSV121G; open diamonds), and K3585 (amgSV121G ΔamgR; filled diamonds) exposed to 2 μg/mL (1X MIC) of gentamicin (GEN). (C) Pseudomonas aeruginosa strains K767 (WT; circles) and K3159 (ΔamgR; squares) exposed to 0.5 μg/mL (1X MIC) (open symbols) and 1.25 μg/mL (2.5X MIC) (filled symbols) tobramycin (TOB). The data are means ± standard errors of the means (SEMs) of three to four independent experiments and have been corrected for fluorescence measured in the absence of antimicrobial exposure.
Impact of amgSV121G on expression of the mexXY multidrug efflux operon
While the amgSR182C and amgSV121G mutations provided for a modest increase in resistance to AGs (Lau et al. 2013), evaluation of their impact on susceptibility to non-AGs revealed an unexpected decrease in resistance to several agents for the amgSV121G mutant (Table3). Interestingly, those agents for which resistance was reduced (SPC, chloramphenicol, tetracycline, and erythromycin) are known inducers (Masuda et al. 2000a; Jeannot et al. 2005; Morita et al. 2006) and probable substrates (Aires et al. 1999; Mine et al. 1999; Masuda et al. 2000b) of the MexXY-OprM multidrug efflux system. This suggested that the amgSV121G mutation was adversely affecting the induction of mexXY by these agents. Examination of mexXY expression using quantitative real-time PCR, however, revealed that mexXY expression was actually enhanced ca. threefold in the amgSV121G mutant relative to its parent strain K767 (Fig.2A). Moreover, this enhanced expression was wholly dependent on AmgR (Fig.2A), an indication that the mexXY upregulation was a direct result of AmgRS activation by the V121G mutation present in AmgS. This precisely mirrored the AmgR-dependent increase in the expression of htpX (Fig.2B), an established AmgRS-regulated gene (Lau et al. 2013). Since drug-inducible mexXY expression is invariably dependent on the MexZ anti-repressor, ArmZ (Morita et al. 2006; Hay et al. 2013), the dependence of amgSV121G mutational upregulation of mexXY on ArmZ was also assessed. Surprisingly, while loss of armZ had a modest negative impact on mexXY expression in both the WT and V121G amgS backgrounds relative to their ArmZ+ counterparts, the amgSV121G mutation still provided for a threefold increase in mexXY expression in the absence of armZ (Fig.2A). Thus, mexXY expression promoted by the amgSV121G mutation was independent of this anti-repressor. In agreement with this, and in contrast to drug-inducible mexXY expression where armZ is upregulated in parallel with mexXY (Morita et al. 2006), the amgSV121G mutation had no impact on armZ expression (data not shown).
Table 3.
Impact of the amgSV121G mutation on resistance of Pseudomonas aeruginosa to nonaminoglycosides.
| Strain | AmgS1 | MexXY1 | Minimal inhibitory concentration (μg/mL) for:2 | ||||
|---|---|---|---|---|---|---|---|
| SPC | TET | ERY | CAM | CAR | |||
| K767 | WT | + | 1024 | 16 | 512 | 64 | 64 |
| K3249 | R182C | + | 1024 | 16 | 512 | 64 | 64 |
| K3260 | V121G | + | 256 | 8 | 256 | 32 | 64 |
| K1525 | WT | − | 64 | 16 | 128 | 64 | 64 |
| K3586 | V121G | − | 64 | 16 | 128 | 32 | 64 |
The status of the AmgS (wild-type [WT] or with the indicated amino acid substitution) and MexXY (+, present; −, absent) components of the indicated strains is shown.
SPC, spectinomycin; TET, tetracycline; ERY, erythromycin; CAM, chloramphenicol; CAR, carbenicillin.
Figure 2.
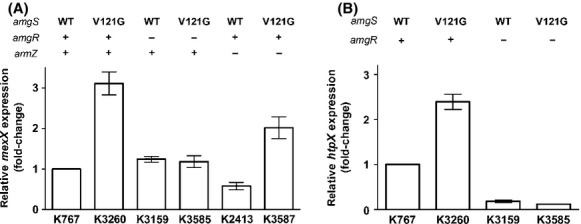
Impact of the amgSV121G mutation on mexXY and htpX expression. Expression of (A) mexXY and (B) htpX was assessed in late-log phase cultures of the indicated strains using real-time quantitative PCR. The status of the amgS (wild-type [WT] or V121G mutation) and the amgR and armZ genes (+, wild-type gene present; −, gene deleted) in each strain is indicated. Expression was normalized to rpoD and is reported relative to the wild-type Pseudomonas aeruginosa strain K767 (fold-change). Values are means ± standard errors of the means (SEMs) from at least three independent determinations, each performed in triplicate.
A possible explanation for the apparent disconnect between the increased susceptibility to non-AG MexXY-OprM substrates and the increase in mexXY expression was that the reduced resistance of the amgSV121G strain K3260 was, in fact, unrelated to mexXY. To assess this, the mexXY genes were deleted in strain K3260 (yielding strain K3586) and susceptibility to these non-AGs reexamined and compared with a ΔmexXY strain carrying a WT amgS gene (strain K1525). As seen in Table3, while the mexXY-mutant strains were both more susceptible to these non-AG mexXY inducers/substrates, there was no longer any difference in susceptibility to these agents between the amgSWT and the amgSV121G derivatives. Thus, the enhanced drug susceptibility seen in the MexXY+ amgSV121G strain K3260 was dependent on the presence of mexXY.
Given the increased mexXY expression seen in strain K3260, it was possible that the enhanced AG resistance of this mutant was attributable to MexXY-OprM and not AmgRS-mediated expression of its other target genes (e.g., htpX, PA5528). To assess this, the impact of the amgSV121G mutation on AG resistance in a ΔmexXY background was examined. As seen in Table4, the amgSV121G mutation promoted a comparable and, in some instances (e.g., for tobramycin and streptomycin), greater increase in AG resistance in the MexXY− derivative K3586 than in the MexXY+ derivative K3260 (although the absolute level of resistance was lower owing to the absence of the AG-accommodating multidrug efflux system in the MexXY− strains). Thus, while MexXY-OprM clearly contributes to AG resistance in the amgSV121G strain K3260, the increase in AG resistance seen in K3260 relative to its wild-type parent strain K767 is not explained by the increased mexXY expression in the mutant.
Table 4.
Impact of the amgSV121G mutation on aminoglycoside resistance of Pseudomonas aeruginosa.
| Strain | AmgS1 | MexXY1 | Minimal inhibitory concentration (μg/mL) for:2 | ||||
|---|---|---|---|---|---|---|---|
| TOB | GEN | STR | PAR | NEO | |||
| K767 | WT | + | 1 | 2 | 32 | 256 | 32 |
| K3260 | V121G | + | 2 | 4 | 64 | 512 | 64 |
| K1525 | WT | − | 0.5 | 1 | 2 | 16 | 8 |
| K3586 | V121G | − | 2 | 2 | 8 | 32 | 16 |
The status of the AmgS (wild-type [WT] or with the indicated amino acid substitution) and MexXY (+, present; −, absent) components of the indicated strains is shown.
TOB, tobramycin; GEN, gentamicin; STR, streptomycin; PAR, paromomycin; NEO, neomycin.
AG induction of mexXY is AmgRS-dependent
Given that mutational activation of the AmgRS TCS upregulated mexXY expression, we questioned whether the previously reported induction of mexXY by AGs (Masuda et al. 2000a; Jeannot et al. 2005; Morita et al. 2006), the only agents known to activate AmgRS (Lau et al. 2013), was similarly AmgRS dependent. Initially, since the original studies of AG activation of AmgRS involved the use of late-log phase cells and 1X MIC of AGs such as paromomycin (PAR) and neomycin (Lau et al. 2013) (vs. mid-log phase cells and ¼ MIC of AGs which was used previously for inducing mexXY expression [Morita et al. 2006]), the potential involvement of AmgRS in AG induction of mexXY was assessed using late-log phase cells and 1X MIC of PAR. In agreement with previous reports of AGs inducing mexXY expression (Masuda et al. 2000a; Jeannot et al. 2005; Morita et al. 2006), PAR-induced mexXY expression in wild-type P. aeruginosa strain K767 (threefold; Fig.3A). PAR-inducible mexXY expression was, however, lacking in amgR and amgS deletion strains (Fig.3A), an indication that AG induction of mexXY expression was dependent on AmgRS. As expected, SPC strongly stimulated mexXY expression in K767 (20-fold; Fig.3A), but this was minimally impacted by the loss of amgR or amgS (Fig.3A), consistent with SPC not activating AmgRS (Lau et al. 2013) and so not stimulating mexXY expression via this TCS. PAR (and SPC) induction of mexXY was, as expected, also dependent on armZ – mexXY expression was lost in the ΔarmZ strain K2413 (Fig.3A). Comparable results were seen when mid-log phase cells were exposed to ¼ MIC of PAR as in previous studies of AG-inducible mexXY expression (Fig.3B), an indication that AG-inducible mexXY expression is generally dependent on AmgRS, regardless of growth phase or inducing drug concentration. To confirm and extend this observation, the AmgRS dependence of AG-inducible mexXY expression was assessed using an additional AG, gentamicin, which was previously shown to induce mexXY expression (Jeannot et al. 2005). As seen in Figure4, gentamicin-induced mexXY expression, and this was dependent on AmgR. Moreover, and consistent with gentamicin activating this TCS, expression of the AmgRS target genes htpX and PA5528 was increased two- to threefold by gentamicin, and this was also dependent on AmgR (Fig.4).
Figure 3.
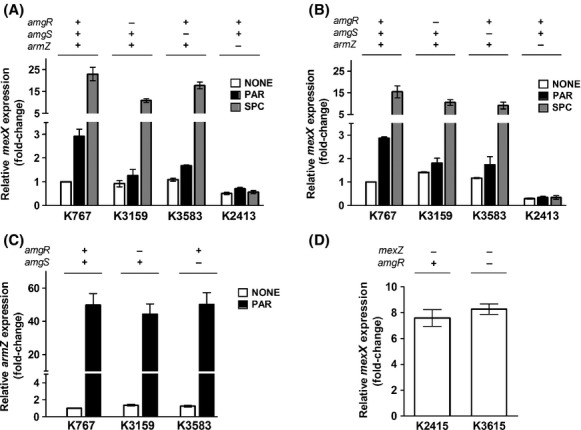
Impact of AmgRS on paromomycin-inducible mexXY expression. Expression of mexXY (A, B, and D) and armZ (C) was assessed in late- (A and C) and mid- (B and D) log phase cultures of the indicated strains without (open bars) or with (filled bars) exposure to paromomycin (PAR; black bars) or spectinomycin (SPC; gray bars) using real-time quantitative PCR. The status of the amgR, amgS, armZ, and mexZ genes (+, wild-type gene present; −, gene deleted) in each strain is indicated. Expression was normalized to rpoD and is reported relative to the wild-type Pseudomonas aeruginosa strain K767 (fold-change). Values are means ± standard errors of the means (SEMs) from at least three independent determinations, each performed in triplicate.
Figure 4.
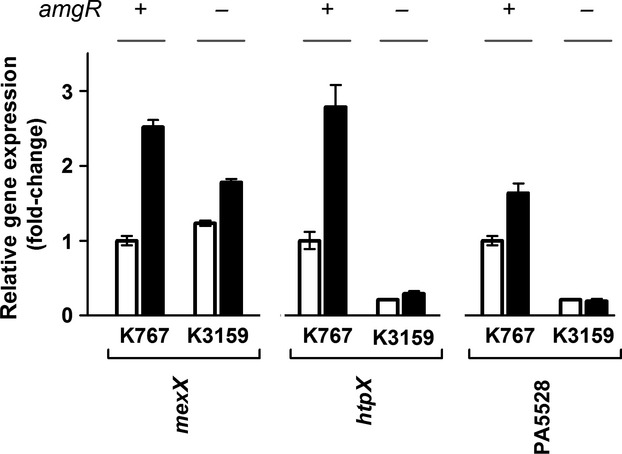
Impact of gentamicin on expression of AmgRS target genes. Expression of mexXY, htpX, and PA5528 was assessed in late-log phase cultures of the indicated strains without (open bars) or with (filled bars) exposure to gentamicin. The status of the amgR gene (+, wild-type gene present; −, gene deleted) in each strain is indicated. Expression was normalized to rpoD and is reported relative to the wild-type Pseudomonas aeruginosa strain K767 (fold-change). Values are means ± standard errors of the means (SEMs) from at least three independent determinations, each performed in triplicate.
AG-inducible mexXY expression is ultimately dependent on AG induction of armZ (Morita et al. 2006). Although not previously studied, we show here that PAR also induces expression of this anti-repressor (Fig.3C). Interestingly, such induction was not compromised by loss of amgR or amgS (Fig.3C), an indication that the AmgRS involvement in AG-inducible mexXY expression is independent of MexZ (i.e., does not promote mexXY expression by influencing ArmZ-modulation of MexZ repressor activity). Consistent with MexZ and AmgRS regulating mexXY expression independently and AmgRS being required only for AG-promoted mexXY expression, the elevated, drug-independent expression of mexXY seen in the ΔmexZ strain K2415 was not lost in the absence of amgR (Fig.3D; see strain K3615). Thus, AG-inducible mexXY expression is not a simple matter of ArmZ-mediated MexZ derepression but also requires activation or operation of the AmgRS TCS.
Involvement of AmgRS-regulated protease genes in AG-inducible mexXY expression
Nine genes show AmgRS-dependent induction by the AG tobramycin (Lee et al. 2009), of which three, htpX, yccA, and PA5528, were primarily responsible for this TCS's contribution to the intrinsic AG resistance of P. aeruginosa (Hinz et al. 2011). The htpX and yccA homologues in E. coli encode a CM-associated protease (Sakoh et al. 2005) and a modulator of the FtsH protease (van Stelten et al. 2009), respectively, with both proteases implicated in membrane protein quality control (Sakoh et al. 2005; van Stelten et al. 2009). PA5528 encodes a predicted CM-associated protein of unknown function. Since MexXY is recruited in response to AmgRS activation (by mutation or AG exposure) and proteases are central to the AmgRS-regulated response to AGs (Hinz et al. 2011), we reasoned that mexXY induction might be dependent on this protease activity, possibly promoted by the degradation products of AG-generated mistranslated polypeptides. These products might, in turn, be the inducers and intended substrates for MexXY. To test this, the aforementioned AmgRS-regulated genes were deleted individually and in combination, and the resultant deletion strains were assessed for AG-inducible mexXY expression. Single knockouts in htpX, PA5528, or yccA had no impact on AG- (i.e., PAR-) inducible expression of mexXY (Fig.5A). Mutants lacking yccA in combination with htpX or PA5528 also retained PAR-inducible mexXY expression (Fig.5B). In contrast, PAR-inducible mexXY expression was absent in the HtpX- PA5528- double-knockout strain K3594, and the further elimination of yccA in the triple knockout stain K3596 had no additional adverse impact on mexXY expression (Fig.5B). Thus, htpX and PA5528 are the sole AmgRS-regulated genes that are required for AmgRS-dependent PAR-inducible mexXY expression. Loss of htpX, PA5528, and yccA, alone or in combination, had no effect on armZ expression (Fig.5C and D), consistent with the AmgRS independence of and, so, lack of a need for AmgRS targets for PAR-inducible armZ expression.
Figure 5.
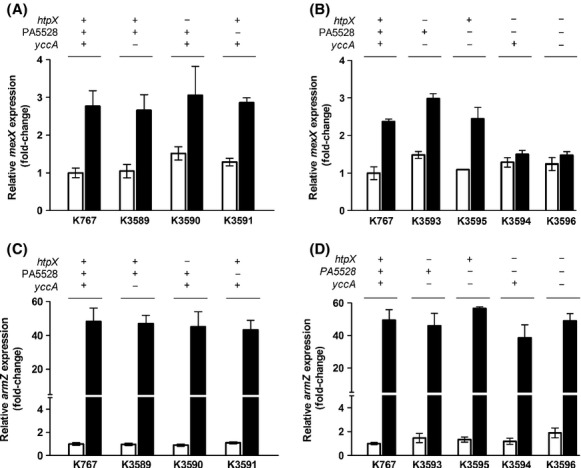
Contribution of AmgRS target genes to aminoglycoside-inducible mexXY expression. Expression of mexXY (A and B) and armZ (C and D) was assessed in late-log phase cultures of the indicated strains without (open bars) and with (filled bars) exposure to paromomycin using real-time quantitative PCR. The status of the htpX, PA5528 and yccA genes (+, wild-type gene present; −, gene deleted) in each strain is indicated. Expression was normalized to rpoD and is reported relative to the wild-type Pseudomonas aeruginosa strain K767 (fold-change). Values are means ± standard errors of the means (SEMs) from at least three independent determinations, each performed in triplicate.
Discussion
Although inducible by ribosome-perturbing agents and mutations, it is clear that mexXY “inducers” are not created equally, with SPC, for example, promoting substantial induction of mexXY (Jeannot et al. 2005; Hay et al. 2013) (this study) while AGs provide for very modest induction of this efflux system (Jeannot et al. 2005) (this study). This speaks to possibly different pathways to the recruitment of the MexXY-OprM efflux system by ribosome-perturbing agents. Indeed, the current study demonstrates that AGs, unique amongst ribosome-targeting antimicrobials in promoting mistranslation and generating membrane-damaging aberrant polypeptides (Davis et al. 1986; Busse et al. 1992), upregulate the mexXY operon by activating the AmgRS TCS that responds to AG-promoted membrane damage and that protects P. aeruginosa from this damage, with AG induction of mexXY dependent on AmgRS. In common with non-AG inducers of mexXY, AG induction of mexXY is, however, also dependent on the ArmZ anti-repressor of MexZ, an indication that AmgR activation of this efflux operon still requires loss of MexZ repression of mexXY. Consistent with these regulators operating independently of one another, armZ expression was not regulated by AmgRS (this study) (loss of armZ also had no impact on the expression of the AmgRS-regulated htpX and PA5528 genes [data not shown]), and the drug-independent expression of mexXY seen in a mexZ mutant was not compromised by the absence of amgR. In addition, loss of amgRS appears only to block AG enhancement of mexXY expression, with basal expression levels of mexXY retained in AmgRS− strains, while loss of armZ has a more significant impact on mexXY expression, eliminating AG-inducible expression of mexXY and reducing basal level expression of the efflux operon. Presumably, additional endogenous ArmZ-dependent mexXY inducers exist in the cell that, as with non-AG antimicrobial inducers of mexXY, operate independent of AmgRS (and likely dependent on other regulators).
Despite differences in AmgRS dependency amongst mexXY inducers it is unclear whether the actual inducing signals and/or efflux substrates that are generated and ultimately accommodated by MexXY-OprM differ in cells treated with AmgRS-dependent (i.e., AGs) versus AmgRS-independent inducers. Since drug-inducible mexXY expression is a response to ribosome perturbation and not the drugs themselves (Jeannot et al. 2005) the actual inducing signal(s) are likely generated as a result of some downstream effect(s) of translation disruption. Whether these are common to both classes of inducers and can be generated with or without membrane perturbation is unknown. Still, it is hard to imagine inducers whose generation is dependent on an envelope stress response (AmgRS) and two of its target genes (htpX, PA5528) being the same as those generated independent of these. It may be, therefore, that other regulators/regulatory systems mediate mexXY induction by non-AGs. The observation, for example, that SPC induction of mexXY greatly exceeds that seen in a mexZ knockout strain and is retained in a MexZ− P. aeruginosa strain (Hay et al. 2013) indicates that MexZ derepression alone does not explain SPC-inducible mexXY expression, and suggests that other factor(s)/regulator(s) (but not AmgRS) mediate this. Still, SPC induction of mexXY expression also requires loss of MexZ repression, just as AmgRS-dependent AG induction of mexXY does, an indication that the downstream-of-ribosome-perturbation signals must similarly be acting through both ArmZ/MexZ as well as the putative additional factor(s)/regulator(s). Thus, the simplest explanation for drug-inducible mexXY expression is that some shared feature or downstream effect of ribosome perturbation by AGs and non-AGs promotes MexZ derepression via ArmZ while unique downstream effects of AG versus non-AG perturbation of the ribosome signals though AmgRS (AGs) and other regulators (non-AG ribosome perturbing agents) (Fig.6). Certainly, ribosome-targeting agents have a myriad of effects on bacterial gene expression and physiology (Ng et al. 2003; Sabina et al. 2003; Shaw et al. 2003; Hutter et al. 2004; Aakra et al. 2005; Lin et al. 2005; Qiu et al. 2005; Nanduri et al. 2009), some common and some agent specific (Ng et al. 2003; Sabina et al. 2003; Hutter et al. 2004). Having mexXY regulators responsive to drug-specific effects would ensure mexXY recruitment in response only to those consequences of translation perturbation where MexXY-OprM function is required. What that function is, and what the intended efflux substrates are, is as yet undetermined, although the observation here that AG-promoted mexXY expression is dependent on AmgRS targets that include a presumed protease (HtpX) is consistent with the AG-generated mexXY inducer and possible MexXY-OprM substrate being a degradation product(s) of AG-generated aberrant polypeptides. As such, MexXY-OprM may function generally as part of an aberrant polypeptide turnover and detoxification process. Still, if this is the case, then there must be some overlap in HtpX and PA5528 function, at least as regards of generation of the mexXY inducer, since loss of either alone has no impact on AG-promoted mexXY expression (i.e., not simply a matter of HtpX cleaving mistranslated proteins to yield the mexXY inducer). Consistent with HtpX and PA5528 having some shared and, thus, mutually compensatory function, PA5528 expression increases threefold in an htpX-mutant strain (data not shown). Although SPC treatment does not yield membrane-damaging aberrant polypeptides, it may well yield truncated/prematurely terminated polypeptides as a result of its interference with the translocation step of translation (Peske et al. 2004). SPC does not appear to fully block translation (Peske et al. 2004), perhaps allowing for partial synthesis of some translation products, with accumulation of these truncated polypeptides possibly having their own deleterious effects on the cell. As such, they may need to be turned over by another (not AmgRS) stress response system and, perhaps, its own set of proteases, with MexXY-OprM again possibly responsible for the export of their degradation products.
Figure 6.
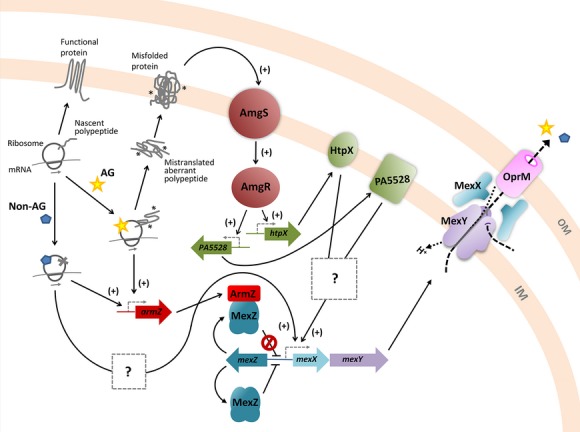
Schematic representation of ArmZ and AmgRS regulation of mexXY in Pseudomonas aeruginosa. In the absence of ribosome-perturbing agents, protein synthesis occurs normally and native, functional proteins are synthesized. In the presence of ribosome-targeting antimicrobials including aminoglycosides (AG), which promote mistranslation and aberrant polypeptide synthesis, and nonaminoglycosides (non-AG), which halt protein synthesis, expression of the amrZ gene is induced. ArmZ, an anti-repressor, modulates the activity of the mexXY repressor, MexZ, leading to expression of mexXY and, ultimately, production and assembly of the MexXY-OprM multidrug efflux system. Additionally, AG-generated aberrant polypeptides disrupt the inner membrane (IM) activating AmgS, the sensor component of the AmgRS two-component system, which in turn activates AmgR to drive expression of the htpX and PA5528 genes. The activities of the htpX and PA5528 gene products in some, as yet unknown way promote expression of the mexXY operon, dependent on ArmZ-mediated loss of MexZ repression of mexXY. Nonaminoglycoside ribosome inhibitors may also promote mexXY expression via additional, as yet unknown regulatory pathway(s), also dependent on ArmZ-mediated loss of MexZ repression. OM, outer membrane.
The observation that the increased mexXY expression of the amgSV121G mutant is ArmZ independent is puzzling, contrasting as it does with the ArmZ dependence of drug- (Morita et al. 2006) and ROS- (Fraud and Poole 2011) inducible mexXY expression. Whether this reflects the presence of a truly ArmZ-independent route to mexXY recruitment in P. aeruginosa is, at present, unknown. It is worth noting, however, that not all amgS gain-of-function mutations impact mexXY expression – the R182C mutation does not – an indication that AmgRS activation alone is insufficient to drive mexXY expression. Presumably, this speaks to some unique feature of the amgSV121G mutation and its impact on AmgRS signaling. Equally puzzling is the observation of decreased resistance to non-AG inducers/substrates of mexXY in the amgSV121G mutant, despite the increase in mexXY expression that results from this amgS mutation. This suggests that the operation of the MexXY-OprM pump is being compromised in some way in the amgSV121G mutant. The failure of the amgSV121G mutation to adversely impact resistance to AGs, which are also mexXY inducers/MexXY-OprM substrates, is presumably due to the positive impact of AmgRS activation on non-MexXY contributors to AG resistance. Consistent with this, the amgSV121G mutation promoted AG resistance in the absence of mexXY. In addition, the observation that the positive impact of the amgSV121G mutation on resistance to some AGs was actually greater in the absence versus the presence of the pump is in agreement with the MexXY-mediated export of/resistance to AGs also being compromised in the amgSV121G mutant (i.e., net impact of the mutation was less in the latter instance owing to the reduced MexXY-OprM contribution to AG resistance). Whatever the explanations for these unique features of the amgSV121G mutant, the mutant provided the crucial link between AmgRS and mexXY, ultimately confirming a role for the TCS in AG-inducible expression of the efflux system.
Acknowledgments
This work was supported by operating grants from Cystic Fibrosis Canada and the Canadian Institutes of Health Research.
Conflict of Interest
None declared.
References
- Aakra A, Vebo H, Snipen L, Hirt H, Aastveit A, Kapur V, et al. Transcriptional response of Enterococcus faecalis V583 to erythromycin. Antimicrob. Agents Chemother. 2005;49:2246–2259. doi: 10.1128/AAC.49.6.2246-2259.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aires JR, Kîhler T, Nikaido H. Plesiat P. Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 1999;43:2624–2628. doi: 10.1128/aac.43.11.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, et al. Short protocols in molecular biology. New York, NY: John Wiley & Sons Inc; 1992. [Google Scholar]
- Breidenstein EB, de la Fuente-Nunez C. Hancock RE. Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol. 2011;19:419–426. doi: 10.1016/j.tim.2011.04.005. [DOI] [PubMed] [Google Scholar]
- Brugha RE. Davies JC. Pseudomonas aeruginosa in cystic fibrosis: pathogenesis and new treatments. Br. J. Hosp. Med. (Lond.) 2011;72:614–619. doi: 10.12968/hmed.2011.72.11.614. [DOI] [PubMed] [Google Scholar]
- Busse HJ, Wostmann C. Bakker EP. The bactericidal action of streptomycin: membrane permeabilization caused by the insertion of mistranslated proteins into the cytoplasmic membrane of Escherichia coli and subsequent caging of the antibiotic inside the cells due to degradation of these proteins. J. Gen. Microbiol. 1992;138:551–561. doi: 10.1099/00221287-138-3-551. [DOI] [PubMed] [Google Scholar]
- Caughlan RE, Sriram S, Daigle DM, Woods AL, Buco J, Peterson RL, et al. Fmt bypass in Pseudomonas aeruginosa causes induction of MexXY efflux pump expression. Antimicrob. Agents Chemother. 2009;53:5015–5021. doi: 10.1128/AAC.00253-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BD, Chen LL. Tai PC. Misread protein creates membrane channels: an essential step in the bactericidal action of aminoglycosides. Proc. Natl. Acad. Sci. USA. 1986;83:6164–6168. doi: 10.1073/pnas.83.16.6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Kievit TR, Parkins MD, Gillis RJ, Srikumar R, Ceri H, Poole K, et al. Multidrug efflux pumps: expression patterns and contribution to antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 2001;45:1761–1770. doi: 10.1128/AAC.45.6.1761-1770.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dötsch A, Becker T, Pommerenke C, Magnowska Z, Jänsch L. Häussler S. Genome-wide identification of genetic determinants of antimicrobial drug resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2009;53:2522–2531. doi: 10.1128/AAC.00035-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El'Garch F, Jeannot K, Hocquet D, Llanes-Barakat C. Plesiat P. Cumulative effects of several nonenzymatic mechanisms on the resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 2007;51:1016–1021. doi: 10.1128/AAC.00704-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez L, Gooderham WJ, Bains M, McPhee JB, Wiegand I. Hancock RE. Adaptive resistance to the “last hope” antibiotics polymyxin B and colistin in Pseudomonas aeruginosa is mediated by the novel two-component regulatory system ParR-ParS. Antimicrob. Agents Chemother. 2010;54:3372–3382. doi: 10.1128/AAC.00242-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraud S. Poole K. Oxidative stress induction of the mexXY multidrug efflux genes and promotion of aminoglycoside resistance development in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2011;55:1068–1074. doi: 10.1128/AAC.01495-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govan JR, Brown AR. Jones AM. Evolving epidemiology of Pseudomonas aeruginosa and the Burkholderia cepacia complex in cystic fibrosis lung infection. Future Microbiol. 2007;2:153–164. doi: 10.2217/17460913.2.2.153. [DOI] [PubMed] [Google Scholar]
- Guenard S, Muller C, Monlezun L, Benas P, Broutin I, Jeannot K, et al. Multiple mutations lead to MexXY-OprM-dependent aminoglycoside resistance in clinical strains of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2014;58:221–228. doi: 10.1128/AAC.01252-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay T, Fraud S, Lau CH, Gilmour C. Poole K. Antibiotic inducibility of the mexXY multidrug efflux operon of Pseudomonas aeruginosa: involvement of the MexZ anti-repressor ArmZ. PLoS One. 2013;8:e56858. doi: 10.1371/journal.pone.0056858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrichfreise B, Wiegand I, Pfister W. Wiedemann B. Resistance mechanisms of multiresistant Pseudomonas aeruginosa strains from Germany and correlation with hypermutation. Antimicrob. Agents Chemother. 2007;51:4062–4070. doi: 10.1128/AAC.00148-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidron AI, Edwards JR, Patel J, Horan TC, Sievert DM, Pollock DA, et al. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006-2007. Infect. Control Hosp. Epidemiol. 2008;29:996–1011. doi: 10.1086/591861. [DOI] [PubMed] [Google Scholar]
- Hinz A, Lee S, Jacoby K. Manoil C. Membrane proteases and aminoglycoside antibiotic resistance. J. Bacteriol. 2011;193:4790–4797. doi: 10.1128/JB.05133-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ. Schweizer HP. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998;212:77–86. doi: 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- Hutter B, Schaab C, Albrecht S, Borgmann M, Brunner NA, Freiberg C, et al. Prediction of mechanisms of action of antibacterial compounds by gene expression profiling. Antimicrob. Agents Chemother. 2004;48:2838–2844. doi: 10.1128/AAC.48.8.2838-2844.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H, Nojima H. Okayama H. High efficiency transformation of Escherichia coli with plasmids. Gene. 1990;96:23–28. doi: 10.1016/0378-1119(90)90336-p. [DOI] [PubMed] [Google Scholar]
- Jeannot K, Sobel ML, El Garch F, Poole K. Plesiat P. Induction of the MexXY efflux pump in Pseudomonas aeruginosa is dependent on drug-ribosome interaction. J. Bacteriol. 2005;187:5341–5346. doi: 10.1128/JB.187.15.5341-5346.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo JTH, Brinkman FSL. Hancock REW. Aminoglycoside efflux in Pseudomonas aeruginosa: involvement of novel outer membrane proteins. Antimicrob. Agents Chemother. 2003;47:1101–1111. doi: 10.1128/AAC.47.3.1101-1111.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahn T, Gilmour C, Tilak J, Fraud S, Kerr N, Lau CH, et al. Determinants of intrinsic aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2012;56:5591–5602. doi: 10.1128/AAC.01446-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau CH-F, Fraud S, Jones M, Peterson SN. Poole K. Reduced expression of the rplU-rpmA ribosomal protein operon in mexXY-expressing pan-aminoglycoside-resistant mutants of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2012;56:5171–5179. doi: 10.1128/AAC.00846-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau CH-F, Fraud S, Jones M, Peterson SN. Poole K. Mutational activation of the AmgRS two-component system in aminoglycoside-resistant Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2013;57:2243–2251. doi: 10.1128/AAC.00170-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Hinz A, Bauerle E, Angermeyer A, Juhaszova K, Kaneko Y, et al. Targeting a bacterial stress response to enhance antibiotic action. Proc. Natl. Acad. Sci. USA. 2009;106:14570–14575. doi: 10.1073/pnas.0903619106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JT, Connelly MB, Amolo C, Otani S. Yaver DS. Global transcriptional response of Bacillus subtilis to treatment with subinhibitory concentrations of antibiotics that inhibit protein synthesis. Antimicrob. Agents Chemother. 2005;49:1915–1926. doi: 10.1128/AAC.49.5.1915-1926.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda N. Ohya S. Cross-resistance to meropenem, cephems, and quinolones in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1992;36:1847–1851. doi: 10.1128/aac.36.9.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda N, Sakagawa E, Ohya S, Gotoh N, Tsujimoto H. Nishino T. Contribution of the MexX-MexY-OprM efflux system to intrinsic resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2000a;44:2242–2246. doi: 10.1128/aac.44.9.2242-2246.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda N, Sakagawa E, Ohya S, Gotoh N, Tsujimoto H. Nishino T. Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-OprM efflux pumps in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2000b;44:3322–3327. doi: 10.1128/aac.44.12.3322-3327.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo Y, Eda S, Gotoh N, Yoshihara E. Nakae T. MexZ-mediated regulation of mexXY multidrug efflux pump expression in Pseudomonas aeruginosa by binding on the mexZ-mexX intergenic DNA. FEMS Microbiol. Lett. 2004;238:23–28. doi: 10.1016/j.femsle.2004.07.010. [DOI] [PubMed] [Google Scholar]
- Mine T, Morita Y, Kataoka A, Mitzushima T. Tsuchiya T. Expression in Escherichia coli of a new multidrug efflux pump, MexXY, from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1999;43:415–417. doi: 10.1128/aac.43.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita Y, Sobel ML. Poole K. Antibiotic inducibility of the MexXY multidrug efflux system of Pseudomonas aeruginosa: involvement of the antibiotic-inducible PA5471 gene product. J. Bacteriol. 2006;188:1847–1855. doi: 10.1128/JB.188.5.1847-1855.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita Y, Gilmour C, Metcalf D. Poole K. Translational control of the antibiotic inducibility of the PA5471 gene required for mexXY multidrug efflux gene expression in Pseudomonas aeruginosa. J. Bacteriol. 2009;191:4966–4975. doi: 10.1128/JB.00073-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller C, Plesiat P. Jeannot K. A two-component regulatory system interconnects resistance to polymyxins, aminoglycosides, fluoroquinolones, and ß-lactams in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2010;55:1211–1221. doi: 10.1128/AAC.01252-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanduri B, Shack LA, Burgess SC. Lawrence ML. The transcriptional response of Pasteurella multocida to three classes of antibiotics. BMC Genom. 2009;10(Suppl. 2):S4. doi: 10.1186/1471-2164-10-S2-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehme D, Li XZ, Elliot R. Poole K. Assembly of the MexAB-OprM multidrug efflux system of Pseudomonas aeruginosa: identification and characterization of mutations in mexA compromising MexA multimerization and interaction with MexB. J. Bacteriol. 2004;186:2973–2983. doi: 10.1128/JB.186.10.2973-2983.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng WL, Kazmierczak KM, Robertson GT, Gilmour R. Winkler ME. Transcriptional regulation and signature patterns revealed by microarray analyses of Streptococcus pneumoniae R6 challenged with sublethal concentrations of translation inhibitors. J. Bacteriol. 2003;185:359–370. doi: 10.1128/JB.185.1.359-370.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivares J, Bernardini A, Garcia-Leon G, Corona F, Sanchez MB. Martinez JL. The intrinsic resistome of bacterial pathogens. Front. Microbiol. 2013;4:103. doi: 10.3389/fmicb.2013.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peske F, Savelsbergh A, Katunin VI, Rodnina MV. Wintermeyer W. Conformational changes of the small ribosomal subunit during elongation factor G-dependent tRNA-mRNA translocation. J. Mol. Biol. 2004;343:1183–1194. doi: 10.1016/j.jmb.2004.08.097. [DOI] [PubMed] [Google Scholar]
- Poole K. Aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2005a;49:479–487. doi: 10.1128/AAC.49.2.479-487.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole K. Efflux-mediated antimicrobial resistance. J. Antimicrob. Chemother. 2005b;56:20–51. doi: 10.1093/jac/dki171. [DOI] [PubMed] [Google Scholar]
- Poole K. Pseudomonas aeruginosa: resistance to the max. Front. Microbiol. 2011;2:65. doi: 10.3389/fmicb.2011.00065. . doi: 10.3389/fmicb.2011.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole K. Pseudomonas aeruginosa efflux pumps. In: Yu EW, Zhang Q, Brown MH, editors. Microbial efflux pumps: current research. Norfolk, U.K: Caister Academic Press; 2013. pp. 175–206. , eds. . [Google Scholar]
- Prayle A. Smyth AR. Aminoglycoside use in cystic fibrosis: therapeutic strategies and toxicity. Curr. Opin. Pulm. Med. 2010;16:604–610. doi: 10.1097/MCP.0b013e32833eebfd. [DOI] [PubMed] [Google Scholar]
- Qiu J, Zhou D, Han Y, Zhang L, Tong Z, Song Y, et al. Global gene expression profile of Yersinia pestis induced by streptomycin. FEMS Microbiol. Lett. 2005;243:489–496. doi: 10.1016/j.femsle.2005.01.018. [DOI] [PubMed] [Google Scholar]
- Ruiz N. Silhavy TJ. Sensing external stress: watchdogs of the Escherichia coli cell envelope. Curr. Opin. Microbiol. 2005;8:122–126. doi: 10.1016/j.mib.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Sabina J, Dover N, Templeton LJ, Smulski DR, Soll D. LaRossa RA. Interfering with different steps of protein synthesis explored by transcriptional profiling of Escherichia coli K-12. J. Bacteriol. 2003;185:6158–6170. doi: 10.1128/JB.185.20.6158-6170.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakoh M, Ito K. Akiyama Y. Proteolytic activity of HtpX, a membrane-bound and stress-controlled protease from Escherichia coli. J. Biol. Chem. 2005;280:33305–33310. doi: 10.1074/jbc.M506180200. [DOI] [PubMed] [Google Scholar]
- Sambrook J. Russell DW. Molecular cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G. Pühler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- Schurek KN, Marr AK, Taylor PK, Wiegand I, Semenec L, Khaira BK, et al. Novel genetic determinants of low-level aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2008;52:4213–4219. doi: 10.1128/AAC.00507-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw KJ, Miller N, Liu X, Lerner D, Wan J, Bittner A, et al. Comparison of the changes in global gene expression of Escherichia coli induced by four bactericidal agents. J. Mol. Microbiol. Biotechnol. 2003;5:105–122. doi: 10.1159/000069981. [DOI] [PubMed] [Google Scholar]
- Sheu DS, Wang YT. Lee CY. Rapid detection of polyhydroxyalkanoate-accumulating bacteria isolated from the environment by colony PCR. Microbiology. 2000;146:2019–2025. doi: 10.1099/00221287-146-8-2019. [DOI] [PubMed] [Google Scholar]
- Simon R, Priefer U. Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Nat. Biotechnol. 1983;1:784–791. [Google Scholar]
- Sobel ML, McKay GA. Poole K. Contribution of the MexXY multidrug transporter to aminoglycoside resistance in Pseudomonas aeruginosa clinical isolates. Antimicrob. Agents Chemother. 2003;47:3202–3207. doi: 10.1128/AAC.47.10.3202-3207.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikumar R, Li XZ. Poole K. Inner membrane efflux components are responsible for beta-lactam specificity of multidrug efflux pumps in Pseudomonas aeruginosa. J. Bacteriol. 1997;179:7875–7881. doi: 10.1128/jb.179.24.7875-7881.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Stelten J, Silva F, Belin D. Silhavy TJ. Effects of antibiotics and a proto-oncogene homolog on destruction of protein translocator SecY. Science. 2009;325:753–756. doi: 10.1126/science.1172221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez D. Inhibitors of protein synthesis. FEBS Lett. 1974;40(Suppl):S63–S84. doi: 10.1016/0014-5793(74)80689-7. [DOI] [PubMed] [Google Scholar]
- Vettoretti L, Plesiat P, Muller C, El Garch F, Phan G, Attree I, et al. Efflux unbalance in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Antimicrob. Agents Chemother. 2009;53:1987–1997. doi: 10.1128/AAC.01024-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vrankrijker AM, Wolfs TF. van der Ent CK. Challenging and emerging pathogens in cystic fibrosis. Paediatr. Respir. Rev. 2010;11:246–254. doi: 10.1016/j.prrv.2010.07.003. [DOI] [PubMed] [Google Scholar]
- Wallace BJ, Tai PC. Davis BD. Selective inhibition of initiating ribosomes by spectinomycin. Proc. Natl. Acad. Sci. USA. 1974;71:1634–1638. doi: 10.1073/pnas.71.5.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisblum B. Davies J. Antibiotic inhibitors of the bacterial ribosome. Bacteriol. Rev. 1968;32:493–528. [PMC free article] [PubMed] [Google Scholar]
- Westbrock-Wadman S, Sherman DR, Hickey MJ, Coulter SN, Zhu YQ, Warrener P, et al. Characterization of a Pseudomonas aeruginosa efflux pump contributing to aminoglycoside impermeability. Antimicrob. Agents Chemother. 1999;43:2975–2983. doi: 10.1128/aac.43.12.2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Ueda A, Kudo M, Matsuo Y, Fukushima J, Nakae T, et al. Role of MexZ and PA5471 in transcriptional regulation of mexXY in Pseudomonas aeruginosa. Microbiology. 2009;155:3312–3321. doi: 10.1099/mic.0.028993-0. [DOI] [PubMed] [Google Scholar]
- Zhanel GG, Decorby M, Laing N, Weshnoweski B, Vashisht R, Tailor F, et al. Antimicrobial-resistant pathogens in intensive care units in Canada: results of the Canadian National Intensive Care Unit (CAN-ICU) study, 2005-2006. Antimicrob. Agents Chemother. 2008;52:1430–1437. doi: 10.1128/AAC.01538-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhanel GG, Decorby M, Adam H, Mulvey MR, McCracken M, Lagace-Wiens P, et al. Prevalence of antimicrobial-resistant pathogens in Canadian hospitals: results of the Canadian Ward Surveillance Study (CANWARD 2008) Antimicrob. Agents Chemother. 2010;54:4684–4693. doi: 10.1128/AAC.00469-10. [DOI] [PMC free article] [PubMed] [Google Scholar]


