Abstract
Extracellular vesicles (EVs), including ‘microvesicles’ and ‘exosomes’, are highly abundant in bodily fluids. Recent years have witnessed a tremendous increase in interest in EVs. EVs have been shown to play important roles in various physiological and pathological processes, including coagulation, immune responses, and cancer. In addition, EVs have potential as therapeutic agents, for instance as drug delivery vehicles or as regenerative medicine. Because of their small size (50 to 1,000 nm) accurate quantification and size profiling of EVs is technically challenging.
This protocol describes how tunable resistive pulse sensing (tRPS) technology, using the qNano system, can be used to determine the concentration and size of EVs. The method, which relies on the detection of EVs upon their transfer through a nano sized pore, is relatively fast, suffices the use of small sample volumes and does not require the purification and concentration of EVs. Next to the regular operation protocol an alternative approach is described using samples spiked with polystyrene beads of known size and concentration. This real-time calibration technique can be used to overcome technical hurdles encountered when measuring EVs directly in biological fluids.
Keywords: Bioengineering, Issue 92, exosomes, microvesicles, extracellular vesicles, quantification, characterization, Tunable Resistive Pulse Sensing, qNano
Introduction
Vesicles from cellular origin are highly abundant in bodily fluids1. These so called extracellular vesicles (EVs) (50 - 1,000 nm in size) are formed by either fusion of multi-vesicular bodies with the cellular membrane or by direct outward budding of the cellular membrane. In recent years, scientific interest in EVs has greatly increased, resulting in a plethora of EV-focused publications, in which new functions and characteristics of EVs are described1. EVs are now believed to be involved in a broad array of physiological and pathological processes such as signal transduction, immune regulation, and blood coagulation1-4. In cancer, EVs seem to play a role in the formation of premetastatic niches5,6, transfer of pro-cancerous content7,8 and stimulation of angiogenesis8. Besides this, EVs are explored as delivery agents of therapeutic agents9.
Despite these developments, reliable quantification of EVs remains challenging. Traditionally, indirect quantification methods are used, which rely on the quantification of total protein content or specific proteins. Although broadly used, these techniques do not account for protein-per-EV differences, and do not discriminate between contaminating protein aggregates and proteins in EVs. Moreover, these techniques require isolation of EVs, which in many cases makes comparison of EV concentrations in biological samples impossible.
Therefore, efforts are undertaken to develop novel methods that allow for more precise and direct EV measurement10. This report describes the use of tunable resistive pulse sensing (tRPS) for reliable quantification and size profiling of EVs.
Currently, the qNano instrument (Figure 1a) is the only commercially available platform for tRPS. In tRPS, a non-conductive elastic membrane punctuated with a nano-sized pore is separating two fluid cells. One of the fluid cells is filled with the sample of interest, whereas the other cell is filled with particle-free electrolyte. By applying a voltage, an ionic flow/electric current is established, which is altered upon the transfer of particles through the pore (Figure 1b). The magnitude of this current blockade (‘resistive pulse’) is proportional to the volume of the particle11 (Figure 1c). The blockade duration can be used to assess the zeta-potential of particles, which relies on particle characteristics such as charge or shape12. Size profiling of unknown particles can be performed by comparing the resistive pulses caused by the unknown particles with the resistive pulses caused by calibration particles with a known diameter. Besides the magnitude of a blockade event, the rate of which these occur is measured. This count rate relies on the particle concentration. Since the concentration and rate of blockades are linearly proportional13, using a single calibration sample with particles of known concentration and particle size allows for the measurement of concentration14 and size distribution11 of an unknown sample.
The movement of particles through the nanopore is determined by electro kinetic- (electrophoretic and electro-osmotic) and fluidic forces15. By using the variable pressure module (VPM) a pressure difference between the fluid cells can be induced as an additional force. Applying positive pressure increases the flow rate of particles, which may be of benefit when the particle concentration is low. Also, pressure can be applied to reduce the effect of electro-kinetic forces. This is especially important when using nanopores with a relative small pore diameter (NP100, NP150 and possibly NP200) as often used for the detection of EVs. For these nanopores, even when applying significant pressure, the electro-kinetic forces can, depending on particle surface charge, remain nonnegligible16. By measuring the particle rate at multiple pressures, an electro- kinetically corrected, and thus more accurate, EV concentration can be calculated.
Here, detailed protocols are provided to determine the size distribution and concentration of EVs. Next to the regular operation protocol, an alternative approach is described where samples are spiked with polystyrene beads of known size and concentration17. This real-time calibration technique can be used to overcome some of the technical challenges encountered when measuring EVs directly in biological fluids, such as urine, plasma and cell culture supernatant, or when stability of the nanopore over a long period of measurement time cannot be ensured.
Protocol
1. Standard Operating Protocol
1.1 Instrument Setup and Sample Preparation
Connect the instrument to a computer with the Izon Control Suite software installed.
Choose the nanopore size to use: for size and concentration measurement of EVs a NP150 (target size range 85 - 300 nm) or NP200 (target size range 100 - 400 nm) are most often used. When working with EVs that have been isolated using a protocol that involves removal of larger EVs, for instance by passing the sample through a 220 nm filter, a NP100 pore (target size range 70 - 200 nm) can be used. When working with EVs in a biological sample or EVs isolated using a different protocol, the NP200 can be used, as it will get clogged less often. Other nanopores such as the NP300 (target size range 150 - 600 nm) or the NP400 (target size range 200 - 800 nm), or even larger nanopores, can be used for larger types of EVs.
Choose the polystyrene calibration particles that complement the nanopore selected in step 1.1.2. For a NP100, NP150 and NP200 nanopore use the CPC100, CP100 and CPC200 particles, respectively. For accurate size estimation, ensure that the calibration particles have a similar size as the unknown particles.
To ensure homogeneity of the calibration particles, vortex briefly (30 sec). Optionally, apply sonication to remove aggregates.
Dilute the calibration particles in PBS to the target concentration in a volume of at least 40 µl. Note: The target concentration varies based on the nanopore selected in step 1.1.2. Target concentrations are supplied with the nanopores.
Apply and directly remove 78 µl of PBS on the lower fluid cell; this wetting of the lower fluid cell reduces the risk of air bubble formation under the nanopore when applying an electrolyte to the lower fluid cell when the nanopore is in position.
Place the nanopore onto the 4 arms of the instrument. Use the digital calipers to measure the distance between two opposite arms and enter the distance in mm into the “Stretch” input field and click “calibrate stretch” to calibrate the nanopore stretch.
Stretch the nanopore to 47 mm, by turning the side wheel and thus increasing the distance between the opposing arms of the instrument, before reapplying 78 µl PBS to the lower fluid cell. Note: Electrical interference can substantially influence the quality of measurements. When using a laptop to run the Izon Control Suite software, make sure the laptop is connected to the power grid using a grounded socket and plug. Mobile phones kept close to the instrument can also be a source of electrical interference. Electrical interference is observed as constantly repeated peaks in the baseline current, often with root mean square (RMS) noise >10 pA. Note: Almost any buffer can be used to dilute calibration particles and EVs for tRPS characterization. The presence of salts is a prerequisite for establishment of an electric current. For EV measurements use PBS as a buffer. Calibration particles should always be diluted in the same buffer as the EVs to ensure accurate measurements.
1.2. Determine the Optimal Settings for Measurement
Note: Before recording, it is important to establish optimal measurement settings. The blockade magnitude caused by a particle passing through the nanopore is dependent on the applied stretch and the voltage applied. For reliable measurements the RMS noise should be <10 pA and the mode blockade magnitude should be >0.1 nA.
Place the upper fluid cell and shielding cage on the nanopore and introduce 40 µl of diluted calibration particles in the upper fluid cell. Use the VPM to apply ≥0.8 kPa positive pressure.
Reduce the applied stretch slowly towards 44 mm whilst analyzing the blockade events caused by the calibration particles. Note: When reducing the pore diameter the movement of particles through the nanopore will be less likely and thus the particle rate will decrease. However, due to increased relative blockade of the pore a larger blockade event will occur resulting in improved signal-to-noise ratio. Increasing the voltage may further increase the blockade magnitude but can also increase the RMS noise.
Reduce the stretch until appropriate blockade events (Figure 1c) are observed in the “Signal Trace” panel. (mode >0.1 nA) and the corresponding particle rate is >100/min. Note: The particle rate is a less strict cut-off, however as measurements of at least 500 particles are ideal, particle rates of <100/min will cause recording durations of at least 5 min. Particle rates higher than 2,000/min can result in less accurate measurements (if present, sample dilution should be performed).
1.3 Measurement of Calibration Particles, Washing of the Uper Fluid Cell and Sample Measurement
In this section EVs from the cell culture supernatant of the glioblastoma multiforme cell line U87-MG/EGFRvIII are characterized. The isolation and preparation of these EVs has previously been described and visualized18.
Place the calibration particles in the upper fluid cell. Apply pressure (for example 0.8 kPa) using the VPM and record >500 particles.
If performing a multi-pressure measurement, increase the applied pressure (for example to 1.0 kPa) and record a second calibration file. Note: A minimum of 0.2 kPa difference is required.
Remove the calibration sample from the upper fluid cell. Wash the upper fluid cell 3 times with 100 µl PBS to remove residual particles. Before introduction of the sample into the upper fluid cell, use lint-free tissue to remove any residual PBS from the upper fluid cell.
Introduce the sample to the upper fluid cell. Make sure the baseline current is within 3% of the baseline current observed when measuring the calibration particles. If not within 3%, apply the strategy described below to stabilize the baseline current. Apply the exact pressures as applied to the calibration particles and record the sample files. Note: The particle rate plot should display constant particle detection (Figure 2a). In case of sudden interruption of particle detection, a sudden drop in baseline current, or a sudden increase in RMS noise, the pore may be clogged; thus, pause the recording. In order to restore the baseline, tap or twist the shielding cap, apply the plunger, or completely remove the nanopore and wash it with deionized water and replace it on the instrument. Note: Alternatively, increase the nanopore stretch to 47 mm in combination with maximum pressure from the VPM for approximately 5 min.
1.4 Data Analysis
Click on the “Analyze Data” tab to enter the analysis section of the software. Process the calibration and sample files by right-clicking the “Unprocessed Files” and selecting “Process Files”.
Click the checkbox next to the sample in the “Calibrated” column to couple the sample files to the calibration recordings. Select corresponding sample and calibration files and click “OK”. Note: when using the multi-pressure calibration option select the “Multi-pressure Calibration” tab on the left to couple multiple samples to multiple calibration files.
Once successfully coupled, the Izon Control Suite software will display different sample characteristics such as a size-distribution (Figure 2b), baseline durations, full width half maximum (FWHM) and a concentration analysis. Optionally: For each sample, individual data points can be exported as a comma separated file.
2. Alternative Protocol – Spiking Samples with Calibration Beads
Note: In general, the standard operating procedure can be used when working with isolated EVs. When working with non-isolated EVs in biological samples, or isolated EV preparations contaminated with large protein aggregates, operating the instrument can be challenging. These challenges consist mainly of a high rate of nanopore blocking (sudden drop in baseline current), inability to recover baseline currents within 3% of calibration measurement or significant differences in particle rates between identical samples (Figure 3a). For samples displaying these difficulties an alternative protocol to quantify EVs was developed17. This methodology relies on the introduction of larger polystyrene calibration beads into the sample of interest (Figure 3b). A detailed procedure for this alternative protocol is discussed below.
2.1. Sample Preparation
Note: When preparing samples using the alternative method, it is desired to establish an EV-to-bead ratio of around 1. Also, it is essential to include a ‘calibration bead only’ sample, to allow for accurate ‘gating’ of the calibration beads and to determine the number of background particles (for example protein aggregates) present in the buffer.
Centrifuge 100 µl of cell culture supernatant for 7 min at 300 x g
Add 20 µl of the supernatant to 20 µl of PBS and 10 µl of 75 times diluted 335 nm polystyrene beads (stock 7e10/ml).
2.2. Sample Measurement
Use the strategy described in section 1.2 to determine the optimal instrument settings.
Measure the ‘calibration bead only’ sample first. Ensure that the background detection of small non-bead particles is as low as possible (<10% of the beads).
Measure each individual sample once before recording replicates in order to distribute fluctuation in nanopore conditions equally over the different samples. Measure at least 3 replicates of each sample.
Remeasure the ‘calibration bead only’ sample after finishing the recording of all samples.
2.3. Data Analysis
Note: When using the alternative protocol, exclusive use of the Izon Control Suite software does not suffice for concentration calculation. Additional spreadsheet software is required. Table 1 indicates an example of the concentration calculation of the samples depicted in Figure 3.
Open the ‘calibration bead only’ sample and one or more sample files.
Determine which blockade event size (in nA) can be used as a cut-off for distinction between EVs and polystyrene beads. Determining the blockade value (nA) corresponding to the left base of the polystyrene beads population (Figure 3b). Note: ensure equal setting of the bin-sizes of all measurements (can be adjusted in ‘ViewSettings’ which is accessed by clicking the “pop-up” button below “Individual Blockade Trace”).
Retrieve the values of the total particle count for each sample by clicking the “Particle Analysis Summary” tab of the sample.
Filter the data sets using the cut-off level determined in step 2.3.2. by selecting the “Data Filtering” pop-up. Display only particles smaller than the cut-off.
Retrieve the values of the EV count for each sample from the “Particle Analysis Summary”.
Subtract the amount of EVs from the total particles to determine the amount of calibration beads.
Determine the EV-to-bead ratio by dividing EV count by calibration bead count.
Determine the average background ratio by averaging the ratios determined for each “bead only” calibration sample. Subtract this value from each individual sample.
Multiply the adjusted EV-to-bead ratio by the concentration of calibration beads to determine the concentration of EVs for each sample.
Multiply the concentration found in step 2.3.9 by the EV dilution factor introduced by the addition of calibration beads to the EV sample. Note: In the example sample setup, the total dilution of sample in PBS and calibration beads is 2.5 times and thus the concentration found in step 2.3.9 should be multiplied by 2.5 to determine the raw EV sample concentration.
Calculate statistics such as the averages, standard deviation and standard error of the mean for each group of replicates. Note: in some cases overlap between EVs and spiked polystyrene beads is observed. If correction for the underestimation of EV concentration is required, samples without spiked polystyrene beads should also be measured. Use the same cut-off as determined in step 2.3.2 to determine a “bead-to-EV” ratio, to calculate the ratio of EVs that fall within the range of the spiked polystyrene beads. This bead-to-EV ratio should be added to the EV-to-bead ratio determined in step 2.3.8.
2.4. Optional: EV Size Distribution Using the Alternative Method.
Open a sample recording twice in the Control Suite Software.
Set the filter options of one of the samples to only display particles bigger than the cut-off determined above. This will display the calibration particles only.
Set the filtered sample to “calibration file” and enter the mode size of the calibration beads.
Couple the sample file and the “calibration file” created in step 2.4.3 as described in 1.4.2. The sample file will now display a size distribution of both EVs and calibration beads based on the spiked calibration beads. Note: The standard operating protocol will most often suffice for the determination of size-distributions of EVs. Sometimes however, exact buffer components are unknown (for example in plasma or urine) which makes it impossible to prepare a sample of calibration beads in the same buffer as the EVs of interest. An EV sample spiked with calibration particles can be used for EV size-estimation in these specific conditions.
Representative Results
To use the tRPS instrument, a non-conductive nanopore has to be placed on the 4 arms of the machine (Figure 1a) and a voltage (Figure 1b) has to be applied. Once an electric baseline current is established, resistive pulses caused by particles passing through the pore will be detected as illustrated in Figure 1c.
EVs were purified from the cell culture supernatant of the glioblastoma cell line U87-MG/EGFRvIII by ultracentrifugation. A stable particle rate-plot is observed when measuring the isolated EVs (Figure 2a) on a NP100 nanopore. This stable particle rate-plot is required for a reliable EV concentration measurement. After pairing the EV-sample recording to a recording of 115 nm polystyrene calibration beads, a size distribution (Figure 2b) and concentration estimate of the EV-sample can be obtained (data not shown).
EVs were also quantified directly in glioblastoma cell culture supernatant. When measuring EVs in biological samples, nanopore clogging often results in interruptions and/or fluctuations in particle rate plots (Figrue 3a). This results in inaccurate EV concentration estimations. By spiking the sample with polystyrene beads of known concentration and size, an EV-to-bead ratio can be determined. Figure 3b illustrates the results obtained after spiking cell culture supernatant with polystyrene beads of 335 nm in size. Two clear populations are observed. The particles inducing a blockade of less then 0.46 nA are determined EVs, the larger particles are determined polystyrene beads. The ratio of EVs to polystyrene beads is used to calculate the raw concentration of EVs (Table 1). Figure 3c illustrates the size estimation of the two populations based on the spiked polystyrene beads. The nanopore setup used resulted in the detection of EVs >140 nm in size. This can be lowered by reducing the nanopore opening, however this will also result in more clogging events.
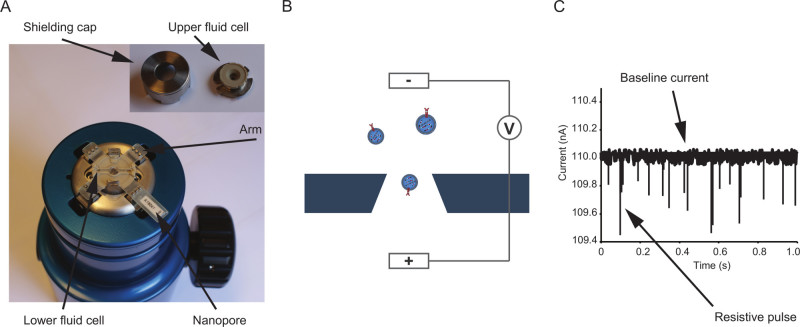 Figure 1: qNano instrument and mode of operation. (A) Photograph of the instrument. A nanopore is positioned on the instrument, separating a lower fluid cell from an upper fluid cell. The fluid cells are protected from environmental electrical interference by the shielding cap. (B) Illustration outlining tunable resistive pulse sensing (tRPS). A non-conductive elastic nanopore is separating two fluid cells. By applying a voltage an electric current is established through the pore punctured in the nanopore. As extracellular vesicles move through the nanopore, the ionic flow is altered and detected as a resistive pulse. In tRPS the opening size of the nanopore can be tuned (reduced or increased) by stretching the nanopore by increasing the distance between the opposing arms of the instrument, or reducing this distance. (C) Illustrative example of resistive pulses. The magnitude of a single resistive pulse is proportional to the volume of the particle: larger pulses indicate larger particles. Please click here to view a larger version of this figure.
Figure 1: qNano instrument and mode of operation. (A) Photograph of the instrument. A nanopore is positioned on the instrument, separating a lower fluid cell from an upper fluid cell. The fluid cells are protected from environmental electrical interference by the shielding cap. (B) Illustration outlining tunable resistive pulse sensing (tRPS). A non-conductive elastic nanopore is separating two fluid cells. By applying a voltage an electric current is established through the pore punctured in the nanopore. As extracellular vesicles move through the nanopore, the ionic flow is altered and detected as a resistive pulse. In tRPS the opening size of the nanopore can be tuned (reduced or increased) by stretching the nanopore by increasing the distance between the opposing arms of the instrument, or reducing this distance. (C) Illustrative example of resistive pulses. The magnitude of a single resistive pulse is proportional to the volume of the particle: larger pulses indicate larger particles. Please click here to view a larger version of this figure.
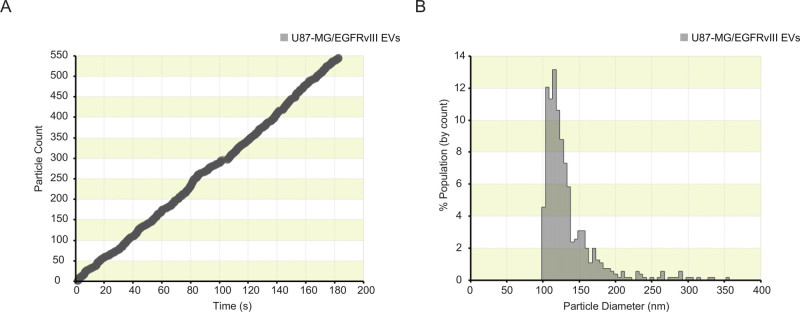 Figure 2: Particle count-plot and size-distribution obtained from measuring isolated EVs from U87-MG/EGFRvIII cell culture supernatant.(A) Particle count-plot indicating overall constant particle detection. Brief reduction of particle detection was observed between 80 and 100 sec of recording. After pausing the recording and tapping the shielding cap, the particle rate stabilized after which the recording was resumed. (B) The size distribution of isolated EVs is plotted after calibrating the unknown sample (EVs) to 115 nm polystyrene calibration beads. (5 nm bin size). Please click here to view a larger version of this figure.
Figure 2: Particle count-plot and size-distribution obtained from measuring isolated EVs from U87-MG/EGFRvIII cell culture supernatant.(A) Particle count-plot indicating overall constant particle detection. Brief reduction of particle detection was observed between 80 and 100 sec of recording. After pausing the recording and tapping the shielding cap, the particle rate stabilized after which the recording was resumed. (B) The size distribution of isolated EVs is plotted after calibrating the unknown sample (EVs) to 115 nm polystyrene calibration beads. (5 nm bin size). Please click here to view a larger version of this figure.
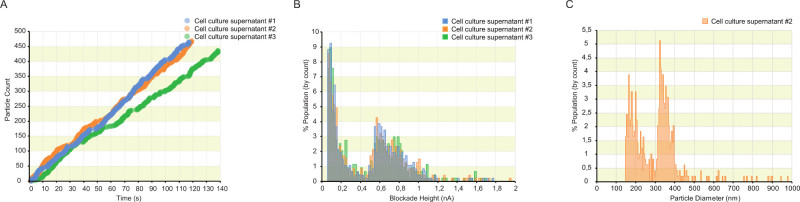 Figure 3: tRPS quantification of EVs in cell culture supernatant using the alternative protocol. (A) Typical particle-rate plots obtained when measuring EVs directly in a biological fluid. Pore clogging causes brief interruptions and fluctuations in the rate of particle detection. Each plot represents a replicate measurement of the same sample. (B) Three replicate size-distribution graphs obtained after spiking cell culture supernatant with 335 nm polystyrene calibration beads. All particles inducing a resistive pulse of less than 0.46 nA are selected as EVs. (C) The spiked polystyrene beads can be used to obtain a size-distribution of the sample. (5 nm bin size). Please click here to view a larger version of this figure.
Figure 3: tRPS quantification of EVs in cell culture supernatant using the alternative protocol. (A) Typical particle-rate plots obtained when measuring EVs directly in a biological fluid. Pore clogging causes brief interruptions and fluctuations in the rate of particle detection. Each plot represents a replicate measurement of the same sample. (B) Three replicate size-distribution graphs obtained after spiking cell culture supernatant with 335 nm polystyrene calibration beads. All particles inducing a resistive pulse of less than 0.46 nA are selected as EVs. (C) The spiked polystyrene beads can be used to obtain a size-distribution of the sample. (5 nm bin size). Please click here to view a larger version of this figure.
| Measurement | Calibration only #1 | Calibration only #2 | Supernatant #1 | Supernatant #2 | Supernatant #3 |
| Average current (nA) | 117 | 120 | 116 | 118 | 120 |
| Particle rate | 172 | 194 | 250 | 246 | 196 |
| cutoff used (nA) | 0.46 | 0.46 | 0.46 | 0.46 | 0.46 |
| Total particles | 303 | 317 | 489 | 488 | 454 |
| Extracellular vesicles | 3 | 1 | 213 | 215 | 213 |
| Spiked calibration beads | 300 | 316 | 276 | 273 | 241 |
| EVs/calibration beads | 0.01 | 0.003 | 0.772 | 0.788 | 0.884 |
| Sample - background | 0.765 | 0.781 | 0.877 | ||
| Extracelullar vesicles (107)/ml | 7.14 | 7.29 | 8.18 | ||
| Sample 2.5x diluted | |||||
| Raw concentration EVs (107)/ml | 17.85 | 18.22 | 20.46 |
Table 1: Example calculation of EV concentration using the alternative protocol. A cut-off value is determined to distinguish EVs from calibration beads. Subsequently, the total number of EVs and beads can be retrieved. For each measurement the EV-to-bead ratio is calculated. The amount of background particles in the electrolyte (for example protein aggregates) is calculated by averaging the EV-to-bead ratio for the individual measurements of the ‘calibration beads only’ sample. For each sample the background ratio is subtracted from the obtained ratio. This adjusted ratio is multiplied by the concentration of the calibration beads in the sample (in this example: 9.33e7/ml). To determine the raw concentration of EVs, the obtained concentration is multiplied by the total EVs dilution factor (in this example: 2.5).
Discussion
The protocols described in this manuscript offer methodologies for quantification and size characterization of EVs using tRPS. The major advantages of the tRPS platform are the small sample size, relative short measurement duration and the absence of required sample manipulation.
Prerequisite for accurate tRPS measurement is to keep conditions identical between calibration and sample measurements. This encompasses the usage of identical buffers as well as identical instrument settings, such as nanopore size, voltage and applied pressure. The original VPM lacks a mechanism for exact setting of the applied pressure, thereby causing minor differences in applied pressure between samples. Also, evaporation of priming fluid in the VPM can induce minor pressure differences when measuring at different time points and the VPM should therefore often be re-primed. These limitations have potentially been solved by introduction of the VPM2, which has a click-based scaling and is air pressure based.
The alternative protocol described in this manuscript is particularly suited for measurement of EVs in non purified biological samples17. We believe that buffer components, such as sugars, lipids, proteins and other larger debris, can in some cases influence the measurement conditions too much for the standard protocol to be applicable. Addition of calibration beads to the sample rather than comparing two separate measurements introduces ‘real time calibration’. This method is especially suitable when comparing samples (e.g. blood plasma of different donors) that have different and/or unknown fluidic background contents. Although differences exist between EVs and polystyrene particles (e.g. particle density and surface charge), theoretical models as well as experimental data underscore the usability of polystyrene beads for quantification and size profiling of EVs, under the prerequisite that significant pressure is applied15,19. To minimize the influence of electrokinetic forces, usage of the relatively larger NP150/NP200 nanopore and significant positive pressure is advised.
EVs and calibration beads are distinguished by size. Consequently, the nanopore has to be opened by applying stretch, to a diameter where detection of both EVs and the larger calibration particles is observed. Since opening of the pore will decrease the sensitivity towards smaller particles, only EVs larger than a certain size are recorded (often EVs >120 nm when using a 335 nm calibration bead). The minimum detection limit for EVs can be decreased to approximately 90 nm, using 203 nm calibration beads on a NP150 nanopore. However, this setup may be unviable when larger EVs induce frequent clogging of the nanopore. The presence of these obstructing EVs may force the utilization of a setup where a population of EVs, too small to reach the detection threshold, will not be detected.
The difficulty to operate the system increases when trying to measure particles smaller than 100 nm in size. In such cases, detection may be improved by increasing the salt concentration of the electrolyte. An increased ion concentration will induce relatively increased blockade magnitudes for small particles (larger signal-to-noise ratio). The viability of this technique for measurements of EVs has to be validated though, as increased salt concentrations may influence the volume of EVs.
In conclusion, the tRPS platform can be used for direct quantification and size characterization of EVs. Since no isolation or EV manipulation (antibody binding or fluorescent labeling) is required, the platform is suitable for direct EV quantification in biological fluids. An alternative protocol is provided that can be beneficial for samples where buffer components induce significant pore clogging events, making reliable utilization of the standard protocol unviable.
Disclosures
The development of the outlined protocol and the writing of this manuscript has been financially supported, in part, by the Dutch Brain Foundation, Schumacher Kramer Foundation, and Bohnenn Fund. Production of this video-article was partially sponsorted by Izon.
References
- Andaloussi S, Mager I, Breakefield XO, Wood MJ. Extracellular vesicles: biology and emerging therapeutic opportunities. Nature reviews. Drug discovery. 2013;12:347–357. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- Lacroix R, Dubois C, Leroyer AS, Sabatier F, Dignat-George F. Revisited role of microparticles in arterial and venous thrombosis. Journal of thrombosis and haemostasis : JTH. 2013;11:24–35. doi: 10.1111/jth.12268. [DOI] [PubMed] [Google Scholar]
- Lee TH, et al. Microvesicles as mediators of intercellular communication in cancer--the emerging science of cellular 'debris'. Seminars in immunopathology. 2011;33:455–467. doi: 10.1007/s00281-011-0250-3. [DOI] [PubMed] [Google Scholar]
- Schorey JS, Bhatnagar S. Exosome function: from tumor immunology to pathogen biology. Traffic. 2008;9:871–881. doi: 10.1111/j.1600-0854.2008.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrie A, et al. Rab27a supports exosome-dependent and -independent mechanisms that modify the tumor microenvironment and can promote tumor progression. Cancer research. 2012;72:4920–4930. doi: 10.1158/0008-5472.CAN-12-0925. [DOI] [PubMed] [Google Scholar]
- Peinado H, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nature medicine. 2012;18:883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Nedawi K, et al. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nature cell biology. 2008;10:619–624. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- Skog J, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nature cell biology. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dommelen SM, et al. Microvesicles and exosomes: opportunities for cell-derived membrane vesicles in drug delivery. Journal of controlled release : official journal of the Controlled Release Society. 2012;161:635–644. doi: 10.1016/j.jconrel.2011.11.021. [DOI] [PubMed] [Google Scholar]
- Pol E, Coumans F, Varga Z, Krumrey M, Nieuwland R. Innovation in detection of microparticles and exosomes. Journal of thrombosis and haemostasis : JTH. 2013;11:36–45. doi: 10.1111/jth.12254. [DOI] [PubMed] [Google Scholar]
- Vogel R, et al. Quantitative sizing of nano/microparticles with a tunable elastomeric pore sensor. Analytical chemistry. 2011;83:3499–3506. doi: 10.1021/ac200195n. [DOI] [PubMed] [Google Scholar]
- Kozak D, et al. Simultaneous size and zeta-potential measurements of individual nanoparticles in dispersion using size-tunable pore sensors. ACS. 2012;6:6990–6997. doi: 10.1021/nn3020322. [DOI] [PubMed] [Google Scholar]
- Willmott GR, et al. Use of tunable nanopore blockade rates to investigate colloidal dispersions. Journal of physics. Condensed matter : an Institute of Physics journal. 2010;22:454116–4510. doi: 10.1088/0953-8984/22/45/454116. [DOI] [PubMed] [Google Scholar]
- Roberts GS, et al. Tunable pores for measuring concentrations of synthetic and biological nanoparticle dispersions. Biosensor. 2012;31:17–25. doi: 10.1016/j.bios.2011.09.040. [DOI] [PubMed] [Google Scholar]
- Vogel R, Anderson W, Eldridge J, Glossop B, Willmott G. A variable pressure method for characterizing nanoparticle surface charge using pore sensors. Analytical chemistry. 2012;84:3125–3131. doi: 10.1021/ac2030915. [DOI] [PubMed] [Google Scholar]
- Kozak D, Anderson W, Trau M. Tuning Particle Velocity and Measurement Sensitivity by Changing Pore Sensor Dimensions. Chemistry Letters. 2012;41:1134–1136. [Google Scholar]
- Vrij J, et al. Quantification of nanosized extracellular membrane vesicles with scanning ion occlusion sensing. Nanomedicine. 2013;8:1443–1458. doi: 10.2217/nnm.12.173. [DOI] [PubMed] [Google Scholar]
- Lasser C, Eldh M, Lotvall J. Isolation and characterization of RNA-containing exosomes. J. Vis. Exp. 2012. [DOI] [PMC free article] [PubMed]
- Yang L, Broom MF, Tucker IG. Characterization of a nanoparticulate drug delivery system using scanning ion occlusion sensing. Pharmaceutical research. 2012;29:2578–2586. doi: 10.1007/s11095-012-0788-3. [DOI] [PubMed] [Google Scholar]


