Abstract
Background:
The goal of this study was to identify a clinical biomarker signature of brain amyloidosis in the Alzheimer's Disease Neuroimaging Initiative 1 (ADNI1) mild cognitive impairment (MCI) cohort.
Methods:
We developed a multimodal biomarker classifier for predicting brain amyloidosis using cognitive, imaging, and peripheral blood protein ADNI1 MCI data. We used CSF β-amyloid 1–42 (Aβ42) ≤192 pg/mL as proxy measure for Pittsburgh compound B (PiB)-PET standard uptake value ratio ≥1.5. We trained our classifier in the subcohort with CSF Aβ42 but no PiB-PET data and tested its performance in the subcohort with PiB-PET but no CSF Aβ42 data. We also examined the utility of our biomarker signature for predicting disease progression from MCI to Alzheimer dementia.
Results:
The CSF training classifier selected Mini-Mental State Examination, Trails B, Auditory Verbal Learning Test delayed recall, education, APOE genotype, interleukin 6 receptor, clusterin, and ApoE protein, and achieved leave-one-out accuracy of 85% (area under the curve [AUC] = 0.8). The PiB testing classifier achieved an AUC of 0.72, and when classifier self-tuning was allowed, AUC = 0.74. The 36-month disease-progression classifier achieved AUC = 0.75 and accuracy = 71%.
Conclusions:
Automated classifiers based on cognitive and peripheral blood protein variables can identify the presence of brain amyloidosis with a modest level of accuracy. Such methods could have implications for clinical trial design and enrollment in the near future.
Classification of evidence:
This study provides Class II evidence that a classification algorithm based on cognitive, imaging, and peripheral blood protein measures identifies patients with brain amyloid on PiB-PET with moderate accuracy (sensitivity 68%, specificity 78%).
A key breakthrough in Alzheimer dementia (AD) research has been the invention of PET compounds that bind to amyloid deposits in the brain. Randomized secondary prevention trials of anti-amyloid agents that could halt disease progression are presently under way. A vast number of potential participants will need to be screened for these studies. This will expose many amyloid-negative cognitively normal elderly to radiation. Alternatively, blood-based biomarkers would have the important advantage of being safe, affordable, and easy to administer in large cohorts and/or in rural areas and therefore could have an enormous impact on clinical care and clinical trials alike.
The current standard for identifying brain amyloidosis is amyloid PET imaging.1 Recently, one research group proposed that CSF β-amyloid 1–42 (Aβ42) levels could serve as reliable indicator of the presence of brain amyloidosis.2 The pathologically validated cutoff of CSF Aβ42 ≤192 pg/mL for discriminating AD from cognitively normal subjects3 was found to be a reliable surrogate indicator of the presence of brain amyloidosis (defined as Pittsburgh compound B [PiB]-PET standard uptake value ratio [SUVR] ≥1.5).4
We hypothesized that we would identify a clinical biomarker signature of brain amyloidosis composed of highly relevant to AD yet simple to measure cognitive, imaging, and peripheral blood protein markers using the Alzheimer's Disease Neuroimaging Initiative 1 (ADNI1) mild cognitive impairment (MCI) cohort. Using an advanced support vector machine (SVM) approach, we developed a multimodal classifier for predicting brain amyloidosis. Unfortunately, only a small fraction of the ADNI1 MCI cohort received PiB-PET scans. Therefore, we took advantage of the strong agreement between CSF Aβ42 ≤192 pg/mL and PiB SUVR ≥1.5 thresholds and used ADNI1 MCI subjects with CSF Aβ42 data but no PiB-PET biomarker data (n = 151) to develop our classification methodology, which was then tested in the smaller cohort of ADNI1 MCI subjects with PiB-PET data (n = 60). We also assessed the utility of our biomarker signature to predict subsequent clinical progression to AD at 24 and 36 months in all 211 subjects.
METHODS
Subjects.
Data used to prepare this article were obtained from the ADNI database (http://adni.loni.usc.edu). ADNI is the result of efforts of many coinvestigators from a broad range of academic institutions and private corporations; subjects have been recruited from more than 50 sites across the United States and Canada. For up-to-date information, see www.adni-info.org. ADNI1 enrolled approximately 400 subjects with amnestic MCI, 200 with mild AD, and 200 normal control subjects, aged 55 to 90 years, between 2005 and 2008. Written informed consent was obtained from all participants. The clinical description of the ADNI1 cohort was recently published.5 The full list of inclusion/exclusion criteria may be accessed online at http://www.adni-info.org/Scientists/ADNIGrant.aspx.
ADNI1 enrolled 398 subjects with MCI. The 151 subjects with MCI who provided peripheral blood and CSF, but not PiB-PET data, were selected for inclusion in our training sample. Conversely, the 60 subjects with MCI who provided peripheral blood and PiB-PET imaging constituted our testing sample. MCI progressors were defined as subjects with MCI who progressed to AD at any point of their follow-up. Nonprogressors were defined as subjects with MCI who at the respective 24-month and 36-month visit were still diagnosed as MCI.
Standard protocol approvals, registrations, and patient consents.
ADNI study sites received approval from their institutional ethical standards committee on human experimentation. Written informed consent was obtained from all research subjects participating in the study or their surrogates when applicable.
Feature selection.
We selected the following well-established cognitive measures for inclusion in the classifier models: the Mini-Mental State Examination (MMSE),6 the most frequently used brief cognitive screening instrument; Trails B,7 an easy-to-administer measure of executive function; and the Auditory Verbal Learning Test (AVLT) delayed recall,8 a sensitive word list verbal memory measure.
We selected the following peripheral blood proteins for inclusion in our models: ApoE, a protein involved in cholesterol and Aβ metabolism; brain-derived neurotrophic factor (BDNF), whose circulating levels are altered in AD9; tumor necrosis factor α (TNF-α), a proinflammatory and apoptotic marker elevated in AD10; interleukin 6 receptor (IL-6R), the receptor molecule for IL-6—a proinflammatory cytokine; interleukin 13 (IL-13), a cytokine associated with Aβ burden in AD11; and clusterin (ApoJ), a lipoprotein involved in Aβ fibril formation and clearance.12 Many studies have demonstrated aberrant levels of each of these proteins in peripheral blood in AD, and have found associations between their concentrations and subsequent cognitive decline and/or imaging changes.9–25 We applied log10 transformation to normalize the distribution of all ADNI1 plasma protein variables.
We also included APOE ε4 genotype and hippocampal volume. APOE, the most established genetic risk factor for sporadic AD,26 can be reliably determined from peripheral blood samples. Hippocampal atrophy, the most established structural imaging biomarker of AD, is readily detectable in the MCI and even the presymptomatic stages of AD.27–30
PiB biomarker data.
Description of PiB-PET acquisition may be found at http://www.adni-info.org. We downloaded the PiB SUVR measures from the University of Pittsburgh from the ADNI Web site in October 2008. Each subject's PiB SUVR image was sampled with an automated 14 region-of-interest (ROI) template including 9 cortical (anterior cingulate, frontal, sensorimotor, lateral temporal, mesial temporal, parietal and occipital cortex, the occipital pole, and the precuneus) and 3 subcortical (anterior ventral striatum, thalamus, and subcortical white matter) ROIs in addition to the pons and the cerebellum. We derived mean cortical PiB SUVR by averaging the 9 cortical ROI measures. SUVR estimation was done blindly. PiB data were acquired at various time points relative to the baseline MRI. Fourteen subjects had their PiB scan at the time of their ADNI baseline assessments, 40 subjects at the 12-month, one subject at the 18-month, and 5 subjects at the 24-month follow-up assessment. We paired PiB data with the corresponding demographic and hippocampal volumetric variables from the same visit. Plasma protein measures were derived at baseline, and therefore preceded PiB scans in some cases.
Fluid biomarker data.
We downloaded the baseline CSF Aβ42 data from the ADNI Web site (http://adni.loni.usc.edu) in October 2008. The CSF collection and transportation protocols and procedural details on CSF Aβ42 measurements are provided at http://www.adni-info.org and in a recent ADNI publication.3 Plasma collection, proteomic platforms, and assays are described in detail in reference 10.
Hippocampal segmentation.
We downloaded and linearly registered the preprocessed baseline 1.5, 3-dimensional T1-weighted scans to the International Consortium for Brain Mapping 53 brain template31 using a 9-parameter transformation.32 We resampled the images to an isotropic space of 220 voxels along each axis resulting in a final voxel size of 1 mm3. The hippocampi were segmented with our recently developed and validated automated machine-learning hippocampal segmentation technique (AdaBoost), which uses adaptive boosting33 as previously described.34–38 Hippocampal volumes were extracted.
Statistical methods.
Our analyses were performed retrospectively. We examined the baseline differences in subject demographics, cognitive performance, hippocampal volume, and blood protein measures in the CSF training, PiB-PET testing, and MCI progressor/nonprogressor cohorts using 2-tailed t tests for continuous variables and χ2 tests for categorical variables.
Support vector machine classifier.
SVMs are a popular machine-learning algorithm. SVMs have been particularly successful in biological classification, because nonlinear kernels introduced to the algorithm allow for nonplanar, multidimensional surfaces to classify patterns of data. We optimized the radial basis function kernel and the SVM cost parameter through grid search using the e1071 package in R (http://cran.r-project.org). Because noncontributory features degrade the classification, we allowed our classifier to rank all features and to iteratively remove those with lower weights until it finds the set of features that yields maximal classification accuracy.
We trained the SVM algorithm in all ADNI1 subjects with MCI who had available baseline protein and CSF Aβ42 data but no PiB-PET imaging (n = 151). The outcome variable was CSF Aβ42 ≤ or >192 pg/mL. The CSF training classifier also included age, sex, and education. Cross-validation used the leave-one-out (LOOCV) approach. We computed receiver operating characteristic (ROC) curve and area under the curve (AUC) to assess the classifier's prediction performance.
Our PiB classifiers used the subjects with MCI who had available protein and PiB-PET data (n = 60). We ran 2 PiB classifiers: the first one was run with the fixed tuning parameters from the CSF training classifier (PiB testing-without-tuning classifier) and the second where the PiB classifier was allowed to tune its cost and gamma parameters for optimal classification while using LOOCV (PiB testing-with-tuning classifier). The reason for allowing self-tuning while still restricting ourselves to the variables chosen by the CSF training classifier was to further refine the variable list to the ones that are most useful for predicting PiB positivity.
Our primary goal was to identify a clinical biomarker signature of brain amyloidosis in the ADNI1 MCI cohort. The PiB testing-without-tuning classifier results were used assigning level-of-evidence statement because the classifier resulting from PiB testing-with-tuning has incorporation bias. The PiB testing-without-tuning results were assigned as Class II level of evidence, while the PiB testing-with-tuning results were assigned Class IV level of evidence.
Next, we explored how well the CSF training and the PiB testing-with-tuning classifier features performed when predicting future progression from MCI to AD at 24 and 36 months. We ran these classifiers once with fixed parameters and a second time while allowing self-tuning for optimal performance while using LOOCV. Tuning allowed us to detect the optimal classifier performance without restricting ourselves to parameters that worked for predicting closely related but obviously not identical measures (high PiB and low CSF Aβ42).
All classifiers were further subjected to selection bias correction using permutation analyses. The permutation method empirically evaluates the distribution of the test statistic under the null hypothesis. We ran 10,000 permutations of the dependent variable (clinical diagnosis) against the sets of individual biomarker characteristics for each individual classifier and defined a final single corrected p value for each ROC.
We computed sensitivity, specificity, and accuracy for specific biomarker cutpoints and computed normal approximation confidence intervals (CIs) for those parameters.
All classifiers were rerun after excluding APOE ε2 carriers (CSF sample n = 11; PiB sample n = 3). The results can be seen in table e-1 on the Neurology® Web site at Neurology.org.
RESULTS
Demographic and baseline biomarker comparisons.
CSF training sample.
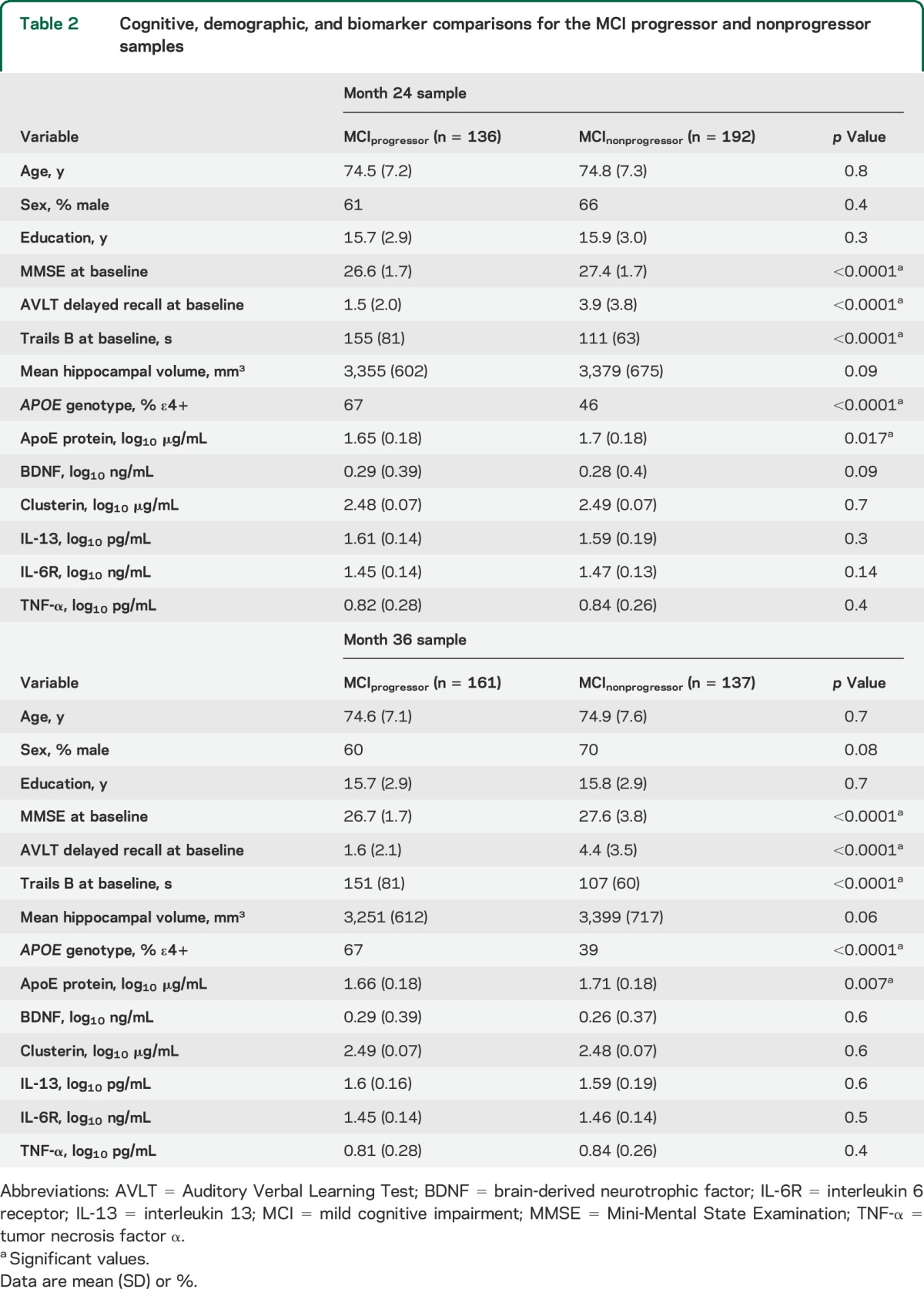
Baseline demographic and biomarker comparisons between the MCI group with CSF Aβ42 ≤192 pg/mL and the MCI group with CSF Aβ42 >192 pg/mL are shown in table 1. The groups showed significant differences in cognitive performance and genotype distribution: the CSF Aβ42 ≤192 pg/mL group had poorer AVLT and higher Trails B scores and higher proportion of APOE ε4+ subjects compared with the CSF Aβ42 >192 pg/mL group.
Table 1.
Cognitive, demographic, and biomarker comparisons for the training and testing samples
PiB testing sample.
Baseline demographic and biomarker comparisons between the MCI group with PiB SUVR ≥1.5 and the MCI group with PiB SUVR <1.5 are shown in table 1. Significantly poorer MMSE scores were seen in the PiB SUVR <1.5 group while AVLT and Trails B were similar in the 2 PiB groups. Plasma ApoE and CSF Aβ42 levels were significantly lower in subjects with MCI who had PiB SUVR <1.5; these subjects were also more likely to be APOE ε4+.
Demographic comparisons of the CSF testing and PiB training samples.
We found no significant differences in age, education, sex, APOE genotype, MMSE score, hippocampal volume, and plasma protein levels between subjects with MCI with CSF Aβ42 ≤192 pg/mL and those with PiB SUVR ≥1.5. Subjects with MCI who had CSF Aβ42 ≤192 pg/mL performed significantly worse on the AVLT and on Trails B relative to subjects with MCI who had PiB SUVR ≥1.5.
Demographic and biomarker variables were similar between subjects with MCI with CSF Aβ42 >192 pg/mL and those with PiB SUVR <1.5 except for marginally lower MMSE scores and plasma IL-6R levels in the subjects with MCI from the CSF sample.
A subset of 29 subjects had both PiB-PET and CSF Aβ42 data, which allowed us to compute the accuracy with which CSF Aβ42 predicted PiB SUVR ≥1.5. Twenty of the 21 subjects with PiB SUVR ≥1.5 also had CSF Aβ42 ≤192 pg/mL (sensitivity 95%). Seven of the 8 subjects with PiB SUVR <1.5 also had CSF Aβ42 >192 pg/mL (specificity 88%). The overall predictive accuracy of PiB SUVR from CSF Aβ42 was 93%.
MCI progression to AD.
The baseline demographic and biomarker comparisons between subjects with MCI who progressed to AD vs those who did not at month 24 and month 36 can be seen in table 2. At baseline, MCI progressors showed significantly lower MMSE and AVLT scores, higher Trails B score, greater proportions of APOE ε4 positivity, and lower plasma ApoE protein concentrations. Hippocampal volumes were marginally smaller in MCI progressors relative to nonprogressors at either time point.
Table 2.
Cognitive, demographic, and biomarker comparisons for the MCI progressor and nonprogressor samples
Classifier results.
CSF training classifier.
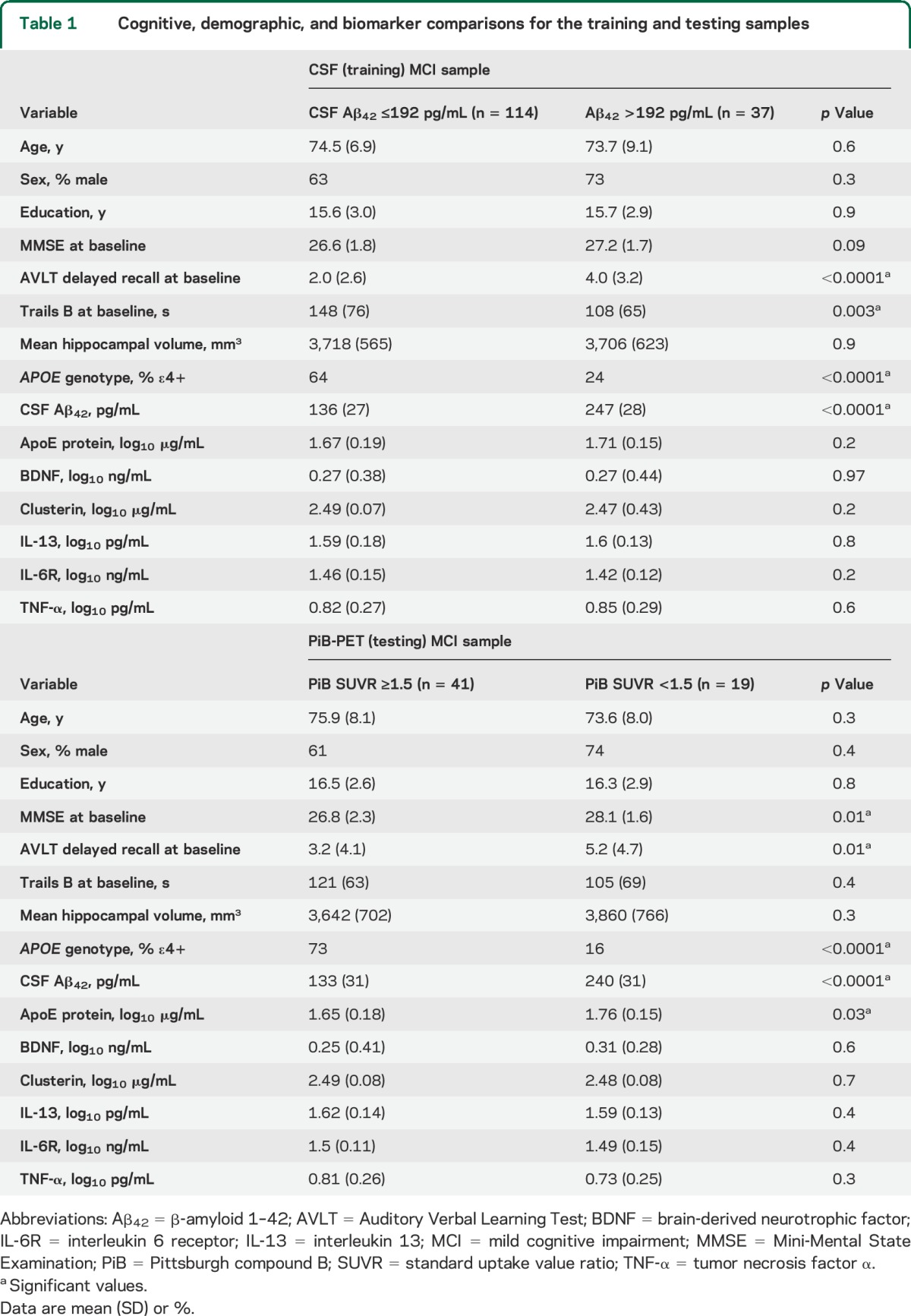
The CSF training classifier used age, sex, education, hippocampal volume, APOE genotype, and all plasma proteins (ApoE, BDNF, clusterin, IL-6R, IL-13, and TNF-α) to predict CSF Aβ42 ≤ or >192 pg/mL. Performance optimization through feature ranking and iterative parameter tuning and LOOCV were used. We achieved the best AUC = 0.8 and accuracy of 85% (95% CI = 79%–91%) with Trails B, APOE genotype, AVLT, education, IL-6R, clusterin, mean hippocampal volume, ApoE protein, and MMSE (listed in rank order). The permutation-corrected classifier significance was p < 0.00001. Taking into account that CSF ≤192 pg/mL is found in 74% of ADNI subjects with MCI, our classifier achieved sensitivity = 92%, specificity = 65%, positive predictive value (PPV) = 88%, and negative predictive value (NPV) = 74%. Results are shown in panel A of the figure and table e-2. APOE genotype alone achieved accuracy of 76% and AUC = 0.7 (95% CI = 69%–83%).
Figure. Classifier results.
AUC = area under the curve; PiB = Pittsburgh compound B.
PiB testing-without-tuning classifier.
We rigidly entered the 9 features and the optimized cost and gamma parameters selected by the CSF training classifier in the PiB testing classifier. The classifier achieved an AUC of 0.78 in predicting PiB-positive (PiB SUVR ≥1.5) and PiB-negative (PiB SUVR <1.5) cases. Taking into account that a positive amyloid PET scan is found in 63% of ADNI subjects with MCI, our classifier achieved sensitivity = 68%, specificity = 78%, PPV = 84%, and NPV = 59%. The ROC curve is shown in panel C of the figure. At fixed 80% sensitivity, the classifier showed 65% sensitivity. The permutation-corrected classifier significance was p = 0.19. APOE genotype alone achieved accuracy of only 28% and AUC = 0.78 (95% CI = 17%–39%).
PiB testing-with-tuning classifier.
Because we developed a classifier model using a proxy measure for brain amyloidosis (CSF Aβ42) that is highly correlated but not identical to the true measure of brain amyloidosis (PiB-PET), we reran the PiB testing classifier using the 9 features selected by the CSF training classifier while allowing the classifier to rank and select from these features and fine-tune its parameters for optimal classification. Using LOOCV, this classifier achieved an accuracy of 83% (95% CI = 74%–93%) and an AUC of 0.78. Taking into account that a positive amyloid PET scan is found in 63% of ADNI subjects with MCI, our classifier achieved sensitivity = 79%, specificity = 83%, PPV = 89%, and NPV = 70%. The ROC curve is shown in panel B of the figure. The features selected by the final model (in rank order) were APOE genotype, MMSE, Trails B, IL-6R, and clusterin (see table e-2). The permutation-corrected classifier significance was p < 0.00001. APOE genotype alone achieved accuracy of 77% and AUC = 0.7 (95% CI = 66%–88%).
Disease-progression classifiers.
We further explored how well the set of 9 variables selected by the CSF training classifier could predict progression from MCI to AD at 24 and 36 months. Running the stringent model using fixed parameters from the CSF training classifier resulted in an AUC = 0.72 at 24 months and AUC = 0.76 at 36 months. Neither classifier produced acceptable accuracy. The ROC curves are shown in panels E and G of the figure.
Once we allowed the disease-progression classifiers to rank and select features and fine-tune their parameters for optimal classification, their performance marginally improved. The 24-month disease-progression classifier achieved an AUC = 0.74 and accuracy of 73% (95% CI = 68%–78%) and the 36-month disease-progression classifier an AUC = 0.77 and accuracy of 74% (95% CI = 69%–79%). The 24-month disease-progression classifier selected (in rank order) AVLT, Trails B, APOE genotype, and IL-6R. The 36-month disease-progression classifier selected (in rank order) AVLT, APOE genotype, and Trails B. The permutation-corrected classifier significance for both classifiers was p < 0.00001. The results are shown in panels D and F of the figure and table e-2.
DISCUSSION
Our data suggest that plasma, imaging, and cognitive measures can be used to potentially predict brain amyloidosis with modest accuracy and confirm the AD relevance of IL-6R and clusterin—the 2 plasma measures that proved useful for this classification.
The LOOCV used by the CSF training classifier is criticized by some for its perceived tendency to inflate the classifier's performance. A true cross-validation should in theory use nonoverlapping samples. Here, we used the PiB sample as our validation sample. Both the rigidly run and the tuned PiB testing classifiers achieved acceptable but modest accuracies.
For identification of subjects with MCI at greatest risk of disease progression to dementia, the self-tuning classifiers achieved reasonable but modest predictive accuracy. Of note, some but not all of the variables that showed between-group differences (see table 2) were selected. MMSE, for instance, was not chosen, but more challenging tests (AVLT and Trails B) were. IL-6R was the only plasma protein measure that was included.
Several strengths and limitations of this study should be acknowledged. ADNI—the premier longitudinal biomarker study in AD—continues to provide researchers with clinical, cognitive, and biomarker samples free of charge. ADNI uses unified subject assessment, MRI, PiB-PET, CSF, and peripheral blood collection protocols and meticulous data quality control across the sites. Because ADNI uses rigorous exclusion criteria typical of clinical trials, our results have direct implications for clinical trials but may be less applicable to outpatient diagnostic assessments. The lack of a sizable MCI cohort with PiB-PET imaging that would have allowed a true classifier development and more extensive cross-validation is a limitation of this study. There is some etiologic/pathologic uncertainty in the MCI stage because at least 30% of subjects with amnestic MCI have been found to harbor non-AD pathology.39 However, it is precisely this etiologic heterogeneity that we are trying to overcome with our analyses. We only assessed the classifier performance with the most widely accepted PiB SUVR cutoff of 1.5. If higher levels of clinical trial cohort enrichment for brain amyloidosis, and hence improved cost-effectiveness, are sought, the use of a higher PiB SUVR cutoff value might prove valuable to further reduce the proportion of false-positive cases. Finally, similar to any other microarray technology, proteomic microarray analyses have inherent technical limitations. An important source of variability for this type of technology is assay time/batch differences between the samples. This could result in future validation issues. Although the biological significance of our plasma analytes has been previously documented, our findings will need to be independently replicated.
Supplementary Material
ACKNOWLEDGMENT
The authors thank the members of the ADNI Imaging Core for their contributions to the image preprocessing, the members of the ADNI Biomarker Core for the CSF biomarker analyses, and the investigators at the University of Pittsburgh for the PiB SUVR analyses.
GLOSSARY
- Aβ42
β-amyloid 1–42
- AD
Alzheimer dementia
- ADNI1
Alzheimer's Disease Neuroimaging Initiative 1
- AUC
area under the curve
- AVLT
Auditory Verbal Learning Test
- BDNF
brain-derived neurotrophic factor
- CI
confidence interval
- IL-6R
interleukin 6 receptor
- IL-13
interleukin 13
- LOOCV
leave-one-out cross-validation
- MCI
mild cognitive impairment
- MMSE
Mini-Mental State Examination
- NPV
negative predictive value
- PiB
Pittsburgh compound B
- PPV
positive predictive value
- ROC
receiver operating characteristic
- ROI
region of interest
- SUVR
standard uptake value ratio
- SVM
support vector machine
- TNF-α
tumor necrosis factor α
Footnotes
Supplemental data at Neurology.org
Contributor Information
Collaborators: Michael Weiner, Paul Aisen, Ronald Petersen, Clifford R. Jack, Jr, Wiliam Jagust, J.Q. Trojanowki, Arthur W. Toga, Laurel Beckett, Robert C. Green, Anthony Gamst, Andrew J. Sakin, John Morris, William Z. Potter, Tom Montine, Michael Donohue, Sarah Walter, Anders Dale, Matthew Bernstein, Joel Felmlee, Nick Fox, Paul Thompson, Norbert Schuff, Gene Alexander, Charles DeCarli, Dan Bandy, Robert A. Koeppe, Norm Foster, Eric M. Reiman, Kewei Chen, Chet Mathis, Nigel J. Cairns, Lisa Taylor-Reinwald, Lee Shaw, Virginia M.-Y. Lee, Magdalena Korecka, Karen Crawford, Scott Neu, Danielle Harvey, John Kornak, Tatiana M. Foroud, Steven Potkin, Li Shen, Zaven Kachaturian, Richard Frank, Peter J. Snyder, Susan Molchan, Jeffrey Kaye, Sara Dolen, Joseph Quinn, Lon Schneider, Sonia Pawluczyk, Bryan M. Spann, James Brewer, Helen Vanderswag, Judith L. Heidebrink, Joanne L Lord, Kris Johnson, Rachelle S. Doody, Javier Villanueva-Meyer, Munir Chowdhury, Yaakov Stern, Lawrence S. Honig, Karen L. Bell, Mark A. Mintun, Stacy Schneider, Daniel Marson, Randall Griffith, David Clark, Hillel Grossman, Cheuk Tang, George Marzloff, Leyla deToledo-Morrell, Raj C. Shah, Ranjan Duara, Daniel Varon, Peggy Robers, Marilyn S. Albert, Nicholas Kozauer, Maria Zerrate, Henry Rusinek, Mony J de Leon, Susan M De Santi, P. Murali Doraiswamy, Jeffrey R. Petrella, Marilyn Aiello, Steve Arnold, Jason H. Karlawish, David Wolk, Charles D. Smith, Curtis A. Given, II, Peter Hardy, Oscar L. Lopez, MaryAnn Oakley, Donna M. Simpson, M. Saleem Ismail, Connie Brand, Jennifer Richard, Ruth A. Mulnard, Gaby Thai, Catherine Mc-Adams-Ortiz, Ramon Diaz-Arrastia, Kristen Martin-Cook, Michael DeVous, Allan I. Levey, James J. Lah, Janet S. Cellar, Jeffrey M. Burns, Heather S. Anderson, Mary M. Laubinger, Liana Apostolova, Daniel H.S. Silverman, Neill R. Graff-Radford, Francine Parfitt, Heather Johnson, Martin Farlow, Scott Herring, Ann M. Hake, Christopher H. van Dyck, Martha G. MacAvoy, Amanda L. Benincasa, Howard Chertkow, Howard Bergman, Chris Hosein, Sandra Black, Bojana Stefanovic, Curtis Caldwell, Ging-Yuek Robin Hsiung, Howard Feldman, Michele Assaly, Andrew Kertesz, John Rogers, Dick Trost, Charles Bernick, Donna Munic, Chuang-Kuo Wu, Nancy Johnson, Marsel Mesulam, Carl Sadowsky, Walter Martinez, Teresa Villena, Raymond Scott Turner, Kathleen Johnson, Brigid Reynolds, Reisa A. Sperling, Dorene M. Rentz, Keith A. Johnson, Allyson Rosen, Jared Tinklenberg, Wes Ashford, Marwan Sabbagh, Donald Connor, Sandra Jacobson, Ronald Killiany, Alexander Norbash, Anil Nair, Thomas O. Obisesan, Annapurni Jayam-Trouth, Paul Wang, Alan Lerner, Leon Hudson, Paula Ogrocki, Evan Fletcher, Owen Carmichael, Smita Kittur, Michael Borrie, T-Y Lee, Rob Bartha, Sterling Johnson, Sanjay Asthana, Cynthia M. Carlsson, Steven G. Potkin, Adrian Preda, Dana Nguyen, Pierre Tariot, Adam Fleisher, Stephanie Reeder, Vernice Bates, Horacio Capote, Michelle Rainka, Barry A. Hendin, Douglas W. Scharre, Maria Kataki, Earl A. Zimmerman, Dzintra Celmins, Alice D. Brown, Godfrey Pearlson, Karen Blank, Karen Anderson, Robert B. Santulli, Jessica Englert, Jeff D. Williamson, Kaycee M. Sing, Franklin Watkins, Brian R. Ott, Edward Stopa, Geoffrey Tremont, Stephen Salloway, Paul Malloy, Stephen Correia, Howard J. Rosen, Bruce L. Miller, Jacobo Mintzer, Crystal Flynn Longmire, and Kenneth Spicer
AUTHOR CONTRIBUTIONS
Liana G. Apostolova: study concept and design, acquisition of data, analysis and interpretation, critical revision of the manuscript for important intellectual content, study supervision. Kristy S. Hwang: analysis and interpretation, critical revision of the manuscript for important intellectual content. David Avila: analysis and interpretation, critical revision of the manuscript for important intellectual content. David Elashoff: study design, analysis and interpretation, critical revision of the manuscript for important intellectual content. Omid Kohannim: study design, critical revision of the manuscript for important intellectual content. Edmond Teng: analysis and interpretation, critical revision of the manuscript for important intellectual content. Sophie Sokolow: analysis and interpretation, critical revision of the manuscript for important intellectual content. Clifford R. Jack: critical revision of the manuscript for important intellectual content, ADNI MRI core supervision. William Jagust: critical revision of the manuscript for important intellectual content, study supervision, ADNI PET core supervision. Leslie Shaw: analysis and interpretation, critical revision of the manuscript for important intellectual content, ADNI Biomarker Core supervision. John Q. Trojanowski: critical revision of the manuscript for important intellectual content, ADNI Biomarker Core supervision. Michael W. Weiner: acquisition of data, critical revision of the manuscript for important intellectual content, overall ADNI supervision. Paul M. Thompson: analysis and interpretation, critical revision of the manuscript for important intellectual content.
STUDY FUNDING
Data collection and sharing for this project were funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (NIH grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Alzheimer's Association; Alzheimer's Drug Discovery Foundation; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; F. Hoffmann-La Roche Ltd. and its affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California. The analyses reported in this manuscript were funded by the Easton Consortium for Alzheimer's Drug Discovery and Biomarker Development, NIA R01 AG040770, NIA P50 AG16570. Algorithm development was also supported, in part, by NIMH R01 MH097268 and NIA R01 AG040060 (to P.T.). O.K. was supported, in part, by a UCLA Dissertation Year Fellowship, and by the UCLA Medical Scientist Training Program.
DISCLOSURE
L. Apostolova has served as a consultant to Lilly and GE Healthcare. K. Hwang, D. Avila, D. Elashoff, and O. Kohannim report no disclosures relevant to the manuscript. E. Teng owns stock in General Electric and Cerner Corporations. S. Sokolow reports no disclosures relevant to the manuscript. C. Jack has provided consulting services for Janssen Research & Development, LLC, and Eli Lilly. W. Jagust has served as a consultant to Banner Alzheimer Institute, Genentech Inc., Synarc, Janssen Alzheimer Immunotherapy, F. Hoffmann-La Roche, and Siemens. L. Shaw was a consultant to Innogenetics and collaborates on quality assessment activities as part of the Alzheimer's Disease Neuroimaging Initiative. He serves as consultant and member of the advisory board, and received speaker fees and travel expenses in 2013 from Eli Lilly & Company. J. Trojanowski may accrue revenue on patents submitted by the University of Pennsylvania wherein he is inventor including: Modified avidin-biotin technique, Method of stabilizing microtubules to treat Alzheimer's disease, Method of detecting abnormally phosphorylated tau, Method of screening for Alzheimer's disease or disease associated with the accumulation of paired helical filaments, Compositions and methods for producing and using homogeneous neuronal cell transplants, Rat comprising straight filaments in its brain, Compositions and methods for producing and using homogeneous neuronal cell transplants to treat neurodegenerative disorders and brain and spinal cord injuries, Diagnostic methods for Alzheimer's disease by detection of multiple MRNAs, Methods and compositions for determining lipid peroxidation levels in oxidant stress syndromes and diseases, Compositions and methods for producing and using homogeneous neuronal cell transplants, Method of identifying, diagnosing, and treating alpha-synuclein positive neurodegenerative disorders, Mutation-specific functional impairments in distinct tau isoforms of hereditary frontotemporal dementia and parkinsonism linked to chromosome-17: genotype predicts phenotype, Microtubule stabilizing therapies for neurodegenerative disorders, and Treatment of Alzheimer's and related diseases with an antibody. He is coinventor on patents submitted by the University of Pennsylvania that have generated income from the sale of Avid to Eli Lilly including: Amyloid plaque aggregation inhibitors and diagnostic imaging agents. M. Weiner has served on the scientific advisory boards for Pfizer, BOLT International, Neurotrope Bioscience, Alzheon, and Eli Lilly. He has provided consulting to Synarc, Pfizer, Janssen, KLJ Associates, Easton Associates, Harvard University, University of California, Los Angeles (UCLA), Alzheimer's Drug Discovery Foundation (ADDF), Avid Radiopharmaceuticals, Clearview Healthcare Partners, Perceptive Informatics, Smartfish AS, Decision Resources, Inc., Araclon, Merck, Defined Health, and Genentech. The following entities have provided funding for travel: Pfizer, Paul Sabatier University, MCI Group France, Travel eDreams, Inc., Neuroscience School of Advanced Studies (NSAS), Danone Trading, BV, CTAD ANT Congrès, Kenes, Intl., ADRC, UCLA, UCSD, Sanofi-Aventis Groupe, University Center Hospital, Toulouse, Araclon, AC Immune, Eli Lilly, New York Academy of Sciences (NYAS), and National Brain Research Center, India, for Johns Hopkins Medicine. He received honoraria from Pfizer, Tohoku University, and Danone Trading, BV. He received research support from Merck and Avid. P. Thompson reports no disclosures relevant to the manuscript. Go to Neurology.org for full disclosures.
REFERENCES
- 1.Johnson KA, Minoshima S, Bohnen NI, et al. Appropriate use criteria for amyloid PET: a report of the Amyloid Imaging Task Force, the Society of Nuclear Medicine and Molecular Imaging, and the Alzheimer's Association. Alzheimers Dement 2013;9:e1–e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weigand SD, Vemuri P, Wiste HJ, et al. Transforming cerebrospinal fluid Aβ42 measures into calculated Pittsburgh Compound B units of brain Aβ amyloid. Alzheimers Dement 2011;7:133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shaw LM, Vanderstichele H, Knapik-Czajka M, et al. Cerebrospinal fluid biomarker signature in Alzheimer's disease neuroimaging initiative subjects. Ann Neurol 2009;65:403–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jack CR, Jr, Wiste HJ, Vemuri P, et al. Brain beta-amyloid measures and magnetic resonance imaging atrophy both predict time-to-progression from mild cognitive impairment to Alzheimer's disease. Brain 2010;133:3336–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petersen RC, Aisen PS, Beckett LA, et al. Alzheimer's disease neuroimaging initiative (ADNI): clinical characterization. Neurology 2010;74:201–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Folstein MF, Folstein SE, McHugh PR. Mini-Mental State: a practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res 1975;12:189–198. [DOI] [PubMed] [Google Scholar]
- 7.Kelland DZ, Lewis R, Gurevitch D. Evaluation of the repeatable cognitive-perceptual-motor battery: reliability, validity and sensitivity to diazepam. J Clin Exp Neuropsychol 1992;14:65 Abstract. [Google Scholar]
- 8.Rey A. L'examen Clinique en Psychologie. Paris: Presses Universitaires de France; 1964. [Google Scholar]
- 9.Doecke JD, Laws SM, Faux NG, et al. Blood-based protein biomarkers for diagnosis of Alzheimer disease. Arch Neurol 2012;69:1318–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soares HD, Potter WZ, Pickering E, et al. Plasma biomarkers associated with the apolipoprotein E genotype and Alzheimer disease. Arch Neurol 2012;69:1310–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ringman JM, Elashoff D, Geschwind DH, et al. Plasma signaling proteins in persons at genetic risk for Alzheimer disease: influence of APOE genotype. Arch Neurol 2012;69:757–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnstone D, Milward EA, Berretta R, Moscato P. Multivariate protein signatures of pre-clinical Alzheimer's disease in the Alzheimer's Disease Neuroimaging Initiative (ADNI) plasma proteome dataset. PLoS One 2012;7:e34341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diniz BS, Teixeira AL. Brain-derived neurotrophic factor and Alzheimer's disease: physiopathology and beyond. Neuromolecular Med 2011;13:217–222. [DOI] [PubMed] [Google Scholar]
- 14.O'Bryant SE, Hobson VL, Hall JR, et al. Serum brain-derived neurotrophic factor levels are specifically associated with memory performance among Alzheimer's disease cases. Dement Geriatr Cogn Disord 2011;31:31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Angelucci F, Spalletta G, di Iulio F, et al. Alzheimer's disease (AD) and mild cognitive impairment (MCI) patients are characterized by increased BDNF serum levels. Curr Alzheimer Res 2010;7:15–20. [DOI] [PubMed] [Google Scholar]
- 16.Holmes C, Cunningham C, Zotova E, et al. Systemic inflammation and disease progression in Alzheimer disease. Neurology 2009;73:768–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim SM, Song J, Kim S, et al. Identification of peripheral inflammatory markers between normal control and Alzheimer's disease. BMC Neurol 2011;11:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rocha de Paula M, Gomez Ravetti M, Berretta R, Moscato P. Differences in abundances of cell-signalling proteins in blood reveal novel biomarkers for early detection of clinical Alzheimer's disease. PLoS One 2011;6:e17481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swardfager W, Lanctot K, Rothenburg L, Wong A, Cappell J, Herrmann N. A meta-analysis of cytokines in Alzheimer's disease. Biol Psychiatry 2010;68:930–941. [DOI] [PubMed] [Google Scholar]
- 20.Helmy AA, Naseer MM, Shafie SE, Nada MA. Role of interleukin 6 and alpha-globulins in differentiating Alzheimer and vascular dementias. Neurodegener Dis 2012;9:81–86. [DOI] [PubMed] [Google Scholar]
- 21.Walker DG, Dalsing-Hernandez JE, Campbell NA, Lue LF. Decreased expression of CD200 and CD200 receptor in Alzheimer's disease: a potential mechanism leading to chronic inflammation. Exp Neurol 2009;215:5–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szczepanik AM, Funes S, Petko W, Ringheim GE. IL-4, IL-10 and IL-13 modulate A beta(1–42)-induced cytokine and chemokine production in primary murine microglia and a human monocyte cell line. J Neuroimmunol 2001;113:49–62. [DOI] [PubMed] [Google Scholar]
- 23.Thambisetty M, Simmons A, Velayudhan L, et al. Association of plasma clusterin concentration with severity, pathology, and progression in Alzheimer disease. Arch Gen Psychiatry 2010;67:739–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schrijvers EM, Koudstaal PJ, Hofman A, Breteler MM. Plasma clusterin and the risk of Alzheimer disease. JAMA 2011;305:1322–1326. [DOI] [PubMed] [Google Scholar]
- 25.Baig S, Palmer LE, Owen MJ, Williams J, Kehoe PG, Love S. Clusterin mRNA and protein in Alzheimer's disease. J Alzheimers Dis 2012;28:337–344. [DOI] [PubMed] [Google Scholar]
- 26.Guerreiro RJ, Gustafson DR, Hardy J. The genetic architecture of Alzheimer's disease: beyond APP, PSENs and APOE. Neurobiol Aging 2012;33:437–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Apostolova LG, Dinov ID, Dutton RA, et al. 3D comparison of hippocampal atrophy in amnestic mild cognitive impairment and Alzheimer's disease. Brain 2006;129:2867–2873. [DOI] [PubMed] [Google Scholar]
- 28.Apostolova LG, Dutton RA, Dinov ID, et al. Conversion of mild cognitive impairment to Alzheimer disease predicted by hippocampal atrophy maps. Arch Neurol 2006;63:693–699. [DOI] [PubMed] [Google Scholar]
- 29.Apostolova LG, Mosconi L, Thompson PM, et al. Subregional hippocampal atrophy predicts Alzheimer's dementia in the cognitively normal. Neurobiol Aging 2010;31:1077–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Apostolova LG, Thompson PM, Green AE, et al. 3D comparison of low, intermediate, and advanced hippocampal atrophy in MCI. Hum Brain Mapp 2010;31:786–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mazziotta J, Toga A, Evans A, et al. A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM). Philos Trans R Soc Lond B Biol Sci 2001;356:1293–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Collins DL, Neelin P, Peters TM, Evans AC. Automatic 3D intersubject registration of MR volumetric data in standardized Talairach space. J Comput Assist Tomogr 1994;18:192–205. [PubMed] [Google Scholar]
- 33.Freund Y, Schapire R. A decision-theoretic generalization of online learning and an application to boosting. J Comp Sys Sci 1997;55:119–139. [Google Scholar]
- 34.Morra JH, Tu Z, Apostolova LG, Green AE, Toga AW, Thompson PM. Automatic subcortical segmentation using a contextual model. Med Image Comput Comput Assist Interv 2008;11:194–201. [DOI] [PubMed] [Google Scholar]
- 35.Morra JH, Tu Z, Apostolova LG, et al. Validation of a fully automated 3D hippocampal segmentation method using subjects with Alzheimer's disease, mild cognitive impairment, and elderly controls. Neuroimage 2008;43:59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morra JH, Tu Z, Apostolova LG, et al. Automated mapping of hippocampal atrophy in 1-year repeat MRI data from 490 subjects with Alzheimer's disease, mild cognitive impairment, and elderly controls. Neuroimage 2009;45:S3–S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Apostolova LG, Morra JH, Green AE, et al. Automated 3D mapping of baseline and 12-month associations between three verbal memory measures and hippocampal atrophy in 490 ADNI subjects. Neuroimage 2010;51:488–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ikonomovic MD, Klunk WE, Abrahamson EE, et al. Post-mortem correlates of in vivo PiB-PET amyloid imaging in a typical case of Alzheimer's disease. Brain 2008;131:1630–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jicha GA, Parisi JE, Dickson DW, et al. Neuropathologic outcome of mild cognitive impairment following progression to clinical dementia. Arch Neurol 2006;63:674–681. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.