Abstract
Knowledge of the methylation profile of genes allow for the identification of biomarkers that may guide diagnosis and effective treatment of disease. Human surfactant protein A (SP-A) plays an important role in lung homeostasis and immunity, and is encoded by two genes (SFTPA1 and SFTPA2). The goal of this study was to identify differentially methylated CpG sites in the promoter region of the SFTPA2 gene in lung cancer tissue, and to determine the correlation between the promoter’s methylation profile and gene expression. For this, we collected 28 pairs of cancerous human lung tissue and adjacent non-cancerous (NC) lung tissue: 17 adenocarcinoma (AC), 9 squamous cell carcinoma (SCC), and 2 AC with SCC features, and we evaluated DNA methylation of the SFTPA2 promoter region by bisulfite conversion. Our results identified a higher methylation ratio in one CpG site of the SFTPA2 gene in cancerous tissue vs. NC tissue (0.36 vs. 0.11, p=0.001). When assessing AC samples, we also found cancerous tissues associated with a higher methylation ratio (0.43 vs. 0.10, p=0.02). In the SCC group, although cancerous tissue showed a higher methylation ratio (0.22 vs. 0.11), this difference was not statistically significant (p=0.35). Expression of SFTPA2 mRNA and total SP-A protein was significantly lower in cancer tissue when compared to adjacent NC tissue (p<0.001), and correlated with the hypermethylated status of a SFTPA2 CpG site in AC samples. The findings of this pilot study may hold promise for future use of SFTPA2 as a biomarker for the diagnosis of lung cancer.
Keywords: Methylation, SP-A, adenocarcinoma, squamous cell carcinoma
Introduction
Epigenetics is the study of heritable changes in gene expression or cellular phenotypes caused by mechanisms other than changes in the underlying sequence. Epigenetic processes in lung cells include histone modifications, methylation of CpG sites in gene promoters, and miRNA modulation (1). These molecular events are affected by environmental factors, such as diet, air pollution, smoking, and others, and they can play an important role in gene expression regulation (2, 3). Therefore, epigenetic mechanisms may represent a link between genetics, the environment, and disease development (1, 4).
DNA methylation at deoxycytosines (C) of CG pairs in gene promoter regions has been shown to have a profound effect on mRNA expression. This process is mainly regulated by DNA methyltransferases (DNMT) that catalyze the addition of a methyl group to C bases in CpG sites, resulting in 5-methyl-cytosines (5). In particular, DNMT1 and DNMT3 have been shown to cooperate to silence genes in human cancer cells (6). Hypermethylation has been shown to mediate gene silencing of tumor suppressor genes in a number of cancers (7–9). On the other hand, DNA hypomethylation can also affect cancer development by enhancing expression of oncogenic genes. Recent work has demonstrated that hypermethylation and hypomethylation of specific genes, as well as overexpression of DNMT are major contributors to the development of various cancers, particularly at early stages (10, 11). Together, these findings indicate that early detection of altered methylation in gene promoters may represent a powerful diagnostic tool. Moreover, knowledge of DNA methylation states may also complete the pathophysiologic picture of disease entities, and allow the development of novel therapeutics (12–15).
In general, genes that are highly expressed have lower methylation content in the promoter region than inactive genes, whose promoters are usually hypermethylated (12, 16–18). We have previously shown alterations in the DNA methylation profile of genes that encode innate immunity molecules, the surfactant proteins in lung cancer (19). We concluded that methylation/demethylation of CpG sites of the SFTPA1 and SP-D promoters results in altered gene expression and this in turn may contribute to the development of lung inflammatory disease and lung cancer (19).
SP-A, the most abundant protein of surfactant, is involved in both host defense and surfactant-related functions (20). The role of SP-A in innate immunity is primarily mediated by its ability to bind several pathogens, enhance phagocytosis and chemotaxis of alveolar macrophages, induce proliferation of immune cells, and stimulate pro-inflammatory cytokine production, as well as modulate the generation of reactive oxygen species (21–23). Furthermore, SP-A participates in several other functions, including serving as a hormone in parturition, and maintaining the structure of the extracellular form of tubular myelin (24–26).
Two genes encode human SP-A: SFTPA1 (or SP-A1), and SFTPA2 (or SP-A2). Both SFTPA1 and SFTPA2 gene products have been shown previously to differ in structure and function, phagocytic activity, and pro-inflammatory cytokine production (27–30). In the genome, these genes are found in opposite transcriptional orientation, but show the same gene organization. The promoter regions of the two genes, although only 25% similar in sequence composition, both contain abundant CpG sites, and thus represent an ideal region to study DNA methylation patterns (Supplementary Figure 1). CpG sites (or islands) are short stretches of DNA with a high cytosine and guanine content (i.e., CG content), with a phosphodiester bond connecting the two residues. Significant differences in the methylation status of two CpG sites of the SFTPA1 gene in lung cancer tissue compared to adjacent non-cancerous (NC) lung tissue have already been reported by us (19). These observations pointed to the possibility that SFTPA1 is a potential biomarker that could be used for diagnosis and treatment of lung cancer (8).
Lung cancer, the leading cause of cancer-related death, is clinically divided into 2 types: small cell lung carcinoma (SCLC) and non-small cell lung carcinoma (NSCLC). The subtypes for NSCLC include adenocarcinoma (AC), squamous cell carcinoma (SCC) and large cell carcinoma. Non-small cell lung carcinoma accounts for 85–90% of lung cancers and is the less aggressive subtype. Its early detection thus holds the most promise for saving lives. There are numerous studies on DNA methylation studies in NSCLC (31–33). The methylation profiles of the SFTPA2 gene in NC and cancer lung tissue, however, remain undetermined.
The hypothesis of the present study is that differences exist in the DNA methylation status of the SFTPA2 gene promoter between NC and cancer samples of human lungs, and that these differences may affect transcription, resulting in altered levels of SFTPA2. To test this hypothesis, we determined the methylation status of 14 CpG sites at the SFTPA2 promoter (Figure 1) and investigated correlations with SFTPA2 and DNMT (DNMT1 and DNMT3) mRNA expression in a group of paired samples of cancerous and adjacent NC lung tissues. We found: a) a hypermethylated CpG site at the SFTPA2 promoter in lung AC samples that correlated with repression of SFTPA2 mRNA and SP-A protein expression, and b) overexpression of DNMT1 and DNMT3 in AC and SCC tumor samples. Our results indicate that SFTPA2 methylation represents a potential biomarker for lung cancer diagnosis.
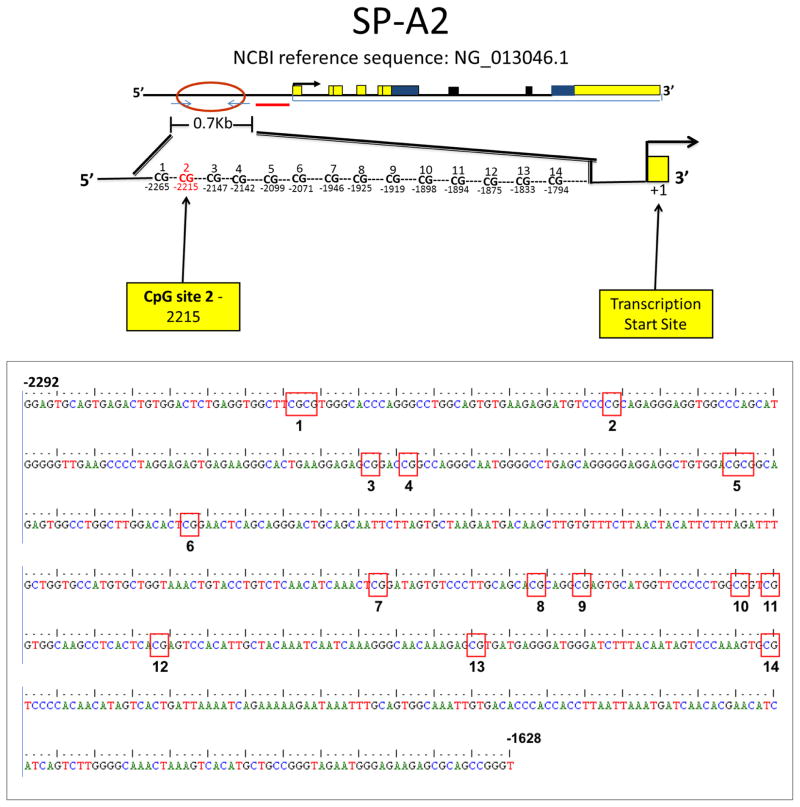
Figure 1. SFTPA2 promoter CpG sites analyzed in this study.
A schematic diagram of the region studied and the 14 CpG sites is shown (top panel). The sequence shown on the bottom panel (664 bp) corresponds to the region between nucleotides −1628 and −2292 upstream of the SFTPA2 transcription start site (NCBI reference sequence: NG_013046.1).
Materials and Methods
Lung samples
A total of 45 pairs of cancerous and adjacent NC lung tissues were obtained from the Penn State Hershey Tumor Bank, Penn State College of Medicine Department of Pathology, and University of Kansas Biospecimen Shared Resource. Based on histopathological analysis, the cancerous tissue was identified as adenocarcinoma (n=25), squamous cell carcinoma (n=17), or adenocarcinoma with squamous cell features (n=3). All protocols were reviewed and approved by the Penn State Hershey College of Medicine Institutional Review Board.
DNA Extraction
Lung tissue was homogenized, and DNA was purified using the QIAmp DNA Mini Kit (QIAGEN, Valencia, CA) following the manufacturer’s protocol. Both DNA quality and concentration were verified by Nanodrop.
Bisulfite Conversion, Polymerase Chain Reaction (PCR), and Sequencing
Approximately 200–500ng of purified DNA were analyzed for methylation by bisulfite conversion using the EZ DNA Methylation Kit (Zymo Research, Irvine, CA), with slight modifications. In brief, after in-column desulfonation and clean-up, the column was spun for 1 min, and then allowed to dry for 10 min before proceeding to elution. An amplicon of 664 nucleotides of the SFTPA2 gene promoter region (SFTPA2, Gene ID: 729238), covering nucleotides −2292/−1628 upstream of the transcription initiation site (Figure 1), was amplified by PCR with the following specific primers: MV1611 (Sense, GGAGTGTAGTGAGATTGTGGATTTTGA, covering nucleotides −2292/−2266 upstream of the transcription initiation site) and MV1613 (Anti-sense, GGTAGAATGGGAGAAGAGTGTAGTTGGGT, covering nucleotides −1656/−1628 upstream of the transcription initiation site). PCR products were purified with the QIAquick DNA Purification Kit (QIAGEN, Valencia, CA) and checked on 2% agarose gels prior to sequencing analysis at the Penn State Hershey Molecular Genetics Core Facility. Due to low quality of sequencing post bisulfite conversion, we were able to obtain data from only 28 out of the 45 paired samples: 17 pairs of AC, 9 pairs of SCC, and 2 pairs of AC with SCC features.
Methylation Analysis
The methylation status of all 14 CpG sites of the amplified region of the SFTPA2 promoter (Figure 1) was determined by sequence analysis of the converted products with the Bioedit software. The data are shown as the ratio of the methylated allele to the total (methylated + unmethylated alleles) (34).
Gene expression analysis
Total RNA was extracted from cancerous and adjacent NC lung tissues using the PureLink RNA mini kit (Life Technologies, Carlsbad, CA). Both RNA concentration and quality were determined by Bioanalyzer 2100 at the Penn State Hershey Functional Genomics Core Facility. Only 25 pairs of tumor/non-tumor pairs showed acceptable RNA quality (RIN>7) after purification: 12 AC, 12 SCC, and 1 AC with SCC features. A total of 600ng of these were retrotranscribed using the High Capacity cDNA reverse transcription kit (Life Technologies, Carlsbad, CA). Expression of SFTPA2, DNMT1, and DNMT3 was measured using real time PCR with TaqMan assays (Life Technologies, Carlsbad, CA) and normalized to 18s rRNA expression (Human Euk 18s rRNA Taqman Assay, Applied Biosystems) using the relative quantification method (35).
Protein purification and Western Blot
RIPA buffer (Thermo, Rockford, IL) was used to extract protein from 8 cancerous tissues (4 AC, and 4 SCC) and 8 non-cancerous adjacent NC lung tissue, following the manufacturer’s protocol. Protein concentration was determined by BCA assay (Thermo, Rockford, IL), and 30 μg were used for Western Blot analysis with an antibody that recognizes both SP-A1 and SP-A2 proteins (41), and cyclophilin B (Abcam, Cambridge, MA) as a loading control.
Prediction of transcription factor binding sites
The pattern-based online software Patch (36) was used to predict transcription factor binding sites in the SFTPA2 promoter. The DNA sequence interrogated for factor binding corresponded to the region surrounding the differentially methylated CpG site. Only human identifiers were included, and no mismatches were allowed.
Statistical Analysis
Methylation ratios of NC and cancer groups were compared using the paired t-test. Data on the adenocarcinoma group were also analyzed separately from the squamous cell carcinoma group. Differences in gene and protein expression between NC and cancer groups were analyzed by t-test. Statistically significant differences were considered when p<0.05.
Results
A CpG site at the SFTPA2 promoter is hypermethylated in lung adenocarcinoma
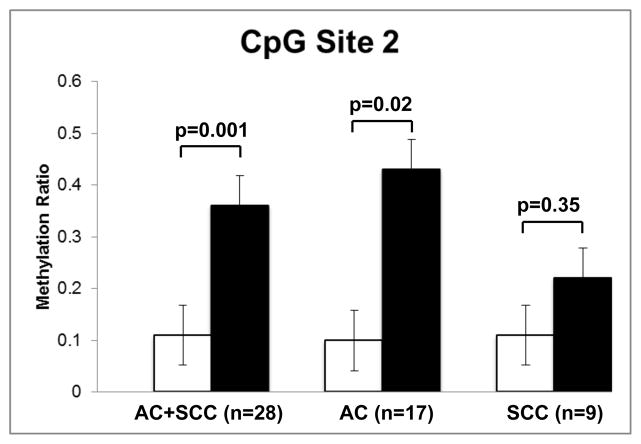
The DNA product amplified by primers MV1611 and MV1613 (664 nucleotides) contained 14 CpG sites (Figure 1). The methylation ratio of these was analyzed by bisulfite conversion, and results are shown in Table 1. Due to low quality of sequencing post bisulfite conversion, we were able to obtain data from only 28 out of the 45 paired samples: 17 pairs of AC, 9 pairs of SCC, and 2 pairs of AC with SCC features. In these, we found that one CpG site (site 2, located −2215 upstream of the SFTPA2 transcription start site) had a higher methylation ratio in cancerous tissue (n=28) when compared to adjacent NC lung tissue (0.36 vs. 0.11, p=0.001). When assessing only the AC group and its corresponding adjacent NC control samples (n=17), we found that the cancerous tissue samples were associated with a higher methylation ratio compared to adjacent NC lung tissue from the same patient (0.43 vs. 0.1, p=0.02) (Figure 2). However, although in the SCC group (n=9), the cancerous tissue had a higher methylation ratio (0.22 versus 0.11 in NC), this difference was not statistically significant (p=0.35) (Figure 2).
Table 1.
Comparison of methylation of CpG Sites in the promoter region of the SFTPA2 gene in lung cancer tissue vs. adjacent non-cancerous lung tissue
| CpG Site | Position* | Methylation ratio (mean ± SD) | p value | |
|---|---|---|---|---|
| Tumor | Normal | |||
| 1 | −2257 | 0.99±0.06 | 1±0 | 0.33 |
| 2 | −2215 | 0.36±0.47 | 0.11±0.29 | 0.001 |
| 3 | −2147 | 0.90±0.28 | 0.90±0.28 | 1.0 |
| 4 | −2142 | 0.73±0.44 | 0.82±0.37 | 0.43 |
| 5 | −2099 | 0.32±0.43 | 0.24±0.41 | 0.65 |
| 6 | −2071 | 0 | 0.09±0.27 | 0.1 |
| 7 | −1946 | 0 | 0 | n/a |
| 8 | −1925 | 0 | 0 | n/a |
| 9 | −1919 | 0 | 0.04±0.19 | 0.33 |
| 10 | −1898 | 0 | 0 | n/a |
| 11 | −1894 | 0 | 0 | n/a |
| 12 | −1875 | 0 | 0 | n/a |
| 13 | −1833 | 0.07±0.04 | 0.06±0.04 | 0.87 |
| 14 | −1794 | 0.10±0.28 | 0.12±0.32 | 0.83 |
relative to the SFTPA2 transcription start site (NCBI reference sequence: NG_013046.1)
Figure 2. DNA Methylation ratios of CpG Site 2 in cancer and adjacent NC lung samples.
Statistical differences were observed when all lung cancer tissues (AC+SCC) were compared to adjacent NC lung tissue (p=0.001, n=28), and the AC group to adjacent NC lung tissue (p=0.02, n=17). No statistically significant differences (p=0.35, n=9) were observed when the SCC and NC were compared. Results are expressed as mean ± SEM.
SFTPA2 mRNA and protein expression levels are significantly reduced in cancer vs. adjacent NC tissue
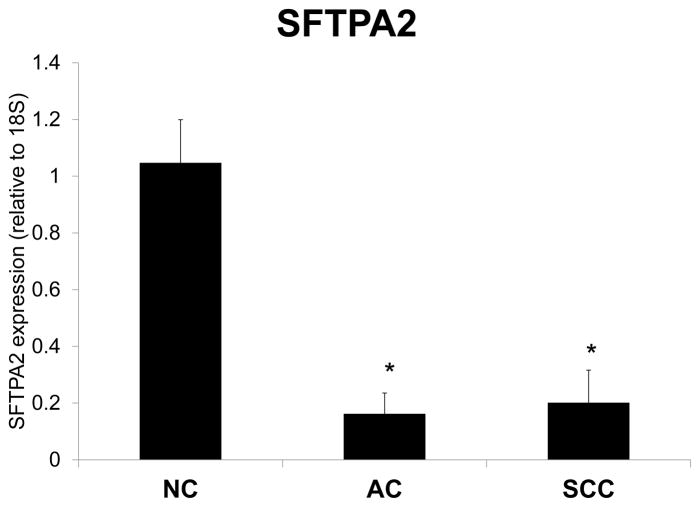
To evaluate the effects of hypermethylation at the SFTPA2 promoter, we measured the expression of SFTPA2 mRNA levels in a subset of cancer and adjacent NC tissues (12 AC/NC pairs, and 12 SCC/NC pairs) by Real Time PCR. Figure 3 shows the relative SFTPA2 mRNA expression in lung cancer (AC and SCC) vs. adjacent NC samples, normalized to 18s. SFTPA2 expression was significantly lower in cancer tissue when compared to NC tissue (p<0.001). Moreover, total SP-A protein levels measured by Western Blot in a subset of paired samples (4 AC/NC pairs, and 4 SCC/NC pairs) were also significantly reduced (Figure 4).
Figure 3. SFTPA2 Gene Expression in NC vs. Lung Cancer Samples.
Expression of SFTPA2 mRNA in NC and lung cancer tissue was measured by Real Time PCR, and expressed as mean ± SEM. Both AC (n=12) and SCC (n=12) cancer samples expressed significantly lower levels of SFTPA2 (p<0.001) compared to NC adjacent tissues.
Figure 4. Total SP-A protein expression in NC vs. Lung Cancer Samples.
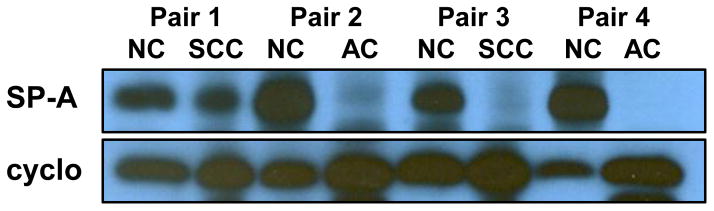
Western Blot analysis of total SP-A and cyclophilin B (loading control) in NC and lung cancer tissue. A representative gel is shown for 4 NC/tumor pairs (2 AC and 2 SCC) of the total 8 pairs (4 AC and 4 SCC) analyzed.
DNMT1 and DNMT3 mRNA levels are increased in cancer vs. adjacent NC tissue
To determine whether the hypermethylated status of SFTPA2 in cancer samples was associated with an increase in DNA methylases, expression of DNMT1 and DNMT3 were measured by Real Time PCR, and normalized to 18s in a subset of samples (12 AC/NC pairs, and 12 SCC/NC pairs). Figure 5 shows that both DNMT1 and DNMT3 were significantly increased in cancer vs. NC (p<0.05). In a subset of samples (9 AC and 5 SCC) for which both SFTPA2 methylation and DNMT expression data were obtained we found that decreased SFTPA2 was correlated with increased DNMT1 expression in AC but not in SCC. However DNMT3 expression was significantly increased in the SCC samples as well (Supplementary table 2).
Figure 5. DNMT gene expression in lung cancer and NC tissue.
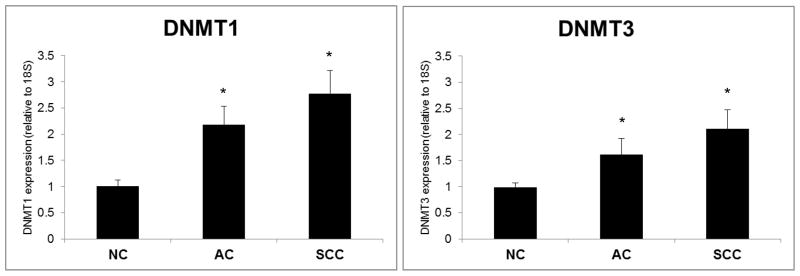
Expression of DNMT1 and DNMT3 mRNA in NC and lung cancer tissue was measured by Real Time PCR, and expressed as mean + SEM. Both AC (n=12) and SCC (n=12) tumor samples had significantly higher levels of DNMT1 and DNMT3 (p<0.05).
Differential binding of transcription factors as a result of DNA methylation
By using the pattern-based online software Patch (36), we identified regulatory sequences for human transcription factors in DNA sequences adjacent to the CpG site 2 (Table 2). The software identified the following transcription factors with predicted binding sites in the sequences surrounding the differentially methylated CpG site: vitamin D receptor (VDR), adenovirus enhancer binding factor / polyomavirus enhancer activator 3 protein (E1AF/PEA3), CCAAT Enhancer binding protein beta (C/EBP beta), sterol regulatory element binding protein (SRE BP), serum response factor (SRF), specificity protein 1 (SP1), c-Ets-1 transcription factor, lymphoid enhancer binding factor 1 (LEF-1), thyroid hormone receptor (T3R), and retinoid X receptor alpha (RXR alpha) (Table 2).
Table 2.
Predicted binding of transcription factors to the SFTPA2 promoter
| Position * | Sequence | Binding Factors** |
|---|---|---|
| −2224 | GGATG | PEA3 |
| −2223 | GATGTCC | C/EBP beta SRE BP SRF |
| −2218 | CCCC | SP1 |
| −2214 | CCGCA | c-Ets-1 LEF-1 |
| −2210 | AGGGAG | T3R-alpha RXR-alpha VDR |
relative to the SFTPA2 transcription start site
Adenovirus enhancer binding factor / polyomavirus enhancer activator 3 protein (E1AF/PEA3), CCAAT Enhancer binding protein beta (C/EBP beta), sterol regulatory element binding protein (SRE BP), serum response factor (SRF), specificity protein 1 (SP1), c-Ets-1 transcription factor, lymphoid enhancer binding protein 1 (LEF-1), thyroid hormone receptor (T3R), and retinoid X receptor alpha (RXR alpha), vitamin D receptor (VDR),
Discussion
Lung cancer is the leading cause of cancer-related death, killing more patients than breast, prostate, and colon cancer combined. Currently, more than 50% of lung cancer patients die within one year after being diagnosed, and the efficiency of treatment has been proven to be highly dependent on early diagnosis (37). Thus, identification of novel biomarkers for lung cancer diagnosis may help improve patient survival. Epigenetic changes, including DNA methylation, occur during the development of many cancers, and these result in alterations of gene expression that contribute to the cancer pathogenesis. Thus, identification of epigenetic signatures not only can contribute to the development of additional diagnostic tools, but also can help identify molecular mechanisms that contribute to the cancer progression.
Human SP-A plays an important role in lung innate immunity, and in the maintenance of normal lung function. Expression of the two SP-A genes is mediated by numerous cellular and molecular factors, and correlations of SP-A1 and SP-A2 genetic variation, and/or expression with lung disease susceptibility has been reported (38–42). Decreased surfactant protein A levels have been reported in pulmonary diseases such as idiopathic pulmonary fibrosis and respiratory distress syndrome of the newborn (43). In addition, rare, heterozygous mutations of SFTPA2 have been associated with pulmonary fibrosis and lung adenocarcinoma (44, 45). In a previous study, differences in the methylation status of the SFTPA1 gene have been found in lung cancer samples (19). Here, we have studied DNA methylation in a CpG dense region of the SFTPA2 promoter located 1600–2300 bp upstream of the transcription start site that contains 14 CpG sites (Figure 1). This region was chosen based on the high density of CpG sites found in preliminary sequence analyses. We found that one of these CpG sites had a significantly higher methylation ratio in AC lung samples, when compared to adjacent NC lung tissue, and a higher methylation, although not significant, in the SCC group vs. NC (Figure 2). In both cases, this pattern correlated with decreased expression of the SFTPA2 gene (Figures 3 and 4) in 9 AC/NC pairs and 5 SCC/NC sample pairs, for which both methylation of CpG site 2 and SFTPA2 mRNA expression results were obtained (Supplementary Table 1). Based on these findings, we speculate that hypermethylation of the SFTPA2 promoter, and decreased expression of the SFTPA2 gene may contribute to a cascade of events that contribute to carcinogenesis (46).
Aberrant DNA methylation of gene promoters has previously been observed in various lung cancers (10, 13–15, 47, 48). Maintenance of methylation patterns is controlled by DNA methyltransferases (DNMT1, DNMT3), in response to various insults and environmental exposures (1, 6, 49). Because overexpression of this enzyme has been previously found in cancer cells of various tumors and stages (50–52), DNMT1 and DNMT3 expression was analyzed in our sample set. Consistent with previous findings, in the current study, both AC and SCC samples showed higher DNMT1 and DNMT3 expression than non-cancerous tissue (Figure 5). However, in a subset where the same samples were analyzed for both methylation and DNMT expression the DNMT1 expression was significantly higher in AC only but the DNMT3 was increased in both AC and SCC (Supplementary tables 1 and 2). This indicates that the hypermethylated status of the SFTPA2 promoter could potentially contribute to AC and/or SCC pathogenesis and perhaps other lung cancers, but other explanations could certainly exist to explain the present findings.
As mentioned above, hypermethylation of the SFTPA2 promoter region may affect binding of essential transcription factors that can promote carcinogenesis by altering SFTPA2 expression (18). To test this hypothesis, we performed in silico analysis of the DNA surrounding sequence of the CpG site 2, with an online tool that allows prediction of transcription factor binding sites, and identified potential binding sites for at least 10 factors (Table 2). Figure 6 shows a diagrammatic representation of the predicted binding sites of the identified transcription factors in the DNA region containing the CpG site 2 (positions −2200/−2230 upstream of the SFTPA2 transcription start site). We speculate that one of the mechanisms that may control the observed SFTPA2 decreased gene expression in lung carcinoma is mediated by impaired binding of one or more transcription factors to hypermethylated CpG sites. Future investigations will focus on characterizing these interactions, as well as on the study of the effects of altered SFTPA2 levels in lung function in patients with lung cancer, including decreased compliance with surfactant deficiency, and increased risk for immune host dysfunction.
Figure 6. Predicted binding of transcription factors to CpG site 2.
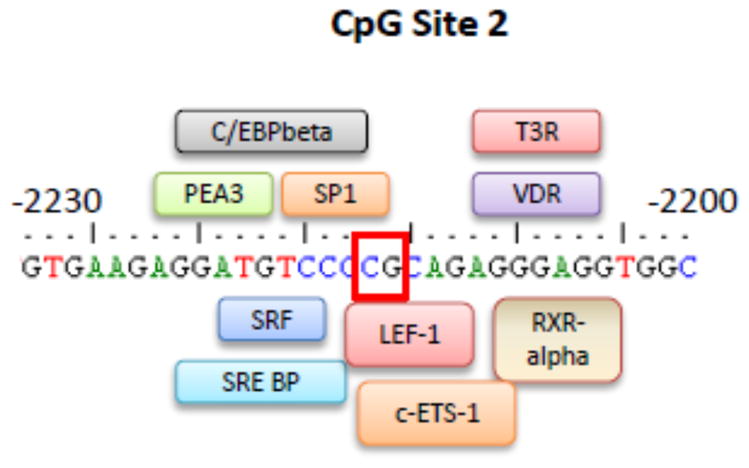
Binding of transcription factors was predicted in the SFTPA2 promoter sequence by the online software Patch (www.gene-regulation.com/cgi-bin/pub/programs/patch/bin/patch.cgi). The figure shows predictions for the region surrounding CpG site 2 (positions −2200/−2230 upstream of the SFTPA2 transcription start site). The box indicates the position of CpG site 2 (−2215).
Of the transcription factors identified in the surrounding region of the hypermethylated CpG site (Table 2), PEA3 and VDR have been most studied and associated with lung malignancies (53–57). While PEA3 plays a key role in metastasis of lung cancer cells, an increase in VDR expression in lung cancer has been associated with improved survival in patients with AC (58, 59). Moreover, associations between VDR and surfactant physiology have been previously described. A natural metabolite of vitamin D3 (1α, 25-dihydroxy-3-epi-vitamin D3) was previously found to play a significant role in stimulating surfactant synthesis (57). In addition, VDR plays a role in the expression of surfactant proteins in the neonate (60). We speculate that methylation of the SFTPA2 promoter region can significantly affect PEA3 and/or VDR binding to this region (Figure 6).
In summary, we have identified a methylation signature for lung cancer in the SFTPA2 promoter that represents a potential biomarker for lung cancer diagnosis. We speculate that, in the future, addition of SFTPA2 methylation profiling to a diagnostic panel for adenocarcinoma may increase diagnostic specificity, and represent a novel adjunct to current diagnostic methods. Furthermore, the SFTPA2 DNA methylation profile could be used as a potential tool to monitor progression of disease and immunity (i.e., host defense). With regards to lung cancer prevention, knowledge of the DNA methylation status of individuals may help identify those who may be high-risk for developing adenocarcinoma and associated dysfunction or decreased production of SFTPA2.
In conclusion, there is a significant difference in the methylation status of the SFTPA2 gene promoter between samples from human lung adenocarcinoma, and adjacent non-cancerous lung tissue. The hypermethylated status of the SFTPA2 gene promoter in cancerous tissue samples was associated with decreased SP-A gene expression. These findings may hold promise for future use of SFTPA2 as a biomarker for the diagnosis and/or therapies of lung cancer.
Supplementary Material
Comparison of CpG sites in the promoter regions (5000bp) of SFTPA2 (upper panel, NCBI Reference Sequence: NG_013046.1) and SFTPA1 (lower panel, NCBI Reference Sequence NG_021189.1) sequences using the online software CpG Island Searcher (http://cpgislands.usc.edu/) (61). The sequence of interest for the present study is indicated between arrows, and contains a region with dense CpG sites, shown in blue (CpG island 1).
Acknowledgments
Funding
This study was supported by the National Institutes of Health Grant HL-34788 and the Barsumian Trust Grant #126717.
The authors thank Sanmei Hu and David Stanford for technical support, and Dr. David Mu for providing the lung samples from the Penn State Department of Pathology.
Footnotes
Declaration of Interests
The authors report no conflicts of interest.
References
- 1.Silveyra P, Floros J. Air pollution and epigenetics: effects on SP-A and innate host defence in the lung. Swiss Med Wkly. 2012;142:w13579. doi: 10.4414/smw.2012.13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ogino S, Lochhead P, Chan AT, Nishihara R, Cho E, Wolpin BM, et al. Molecular pathological epidemiology of epigenetics: emerging integrative science to analyze environment, host, and disease. Mod Pathol. 2013 Apr;26(4):465–84. doi: 10.1038/modpathol.2012.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Y, Li H, Xiao T, Lu Q. Epigenetics in immune-mediated pulmonary diseases. Clin Rev Allergy Immunol. 2013 Dec;45(3):314–30. doi: 10.1007/s12016-013-8398-3. [DOI] [PubMed] [Google Scholar]
- 4.Herceg Z, Vaissière T. Epigenetic mechanisms and cancer: an interface between the environment and the genome. Epigenetics. 2011 Jul;6(7):804–19. doi: 10.4161/epi.6.7.16262. [DOI] [PubMed] [Google Scholar]
- 5.Song J, Teplova M, Ishibe-Murakami S, Patel DJ. Structure-based mechanistic insights into DNMT1-mediated maintenance DNA methylation. Science. 2012 Feb;335(6069):709–12. doi: 10.1126/science.1214453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rhee I, Bachman KE, Park BH, Jair KW, Yen RW, Schuebel KE, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002 Apr;416(6880):552–6. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- 7.Jakopovic M, Thomas A, Balasubramaniam S, Schrump D, Giaccone G, Bates SE. Targeting the Epigenome in Lung Cancer: Expanding Approaches to Epigenetic Therapy. Front Oncol. 2013;3:261. doi: 10.3389/fonc.2013.00261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vaid M, Floros J. Surfactant protein DNA methylation: a new entrant in the field of lung cancer diagnostics? (Review) Oncol Rep. 2009 Jan;21(1):3–11. [PMC free article] [PubMed] [Google Scholar]
- 9.Risch A, Plass C. Lung cancer epigenetics and genetics. Int J Cancer. 2008 Jul;123(1):1–7. doi: 10.1002/ijc.23605. [DOI] [PubMed] [Google Scholar]
- 10.Chen C, Yin N, Yin B, Lu Q. DNA methylation in thoracic neoplasms. Cancer Lett. 2011 Feb;301(1):7–16. doi: 10.1016/j.canlet.2010.10.017. [DOI] [PubMed] [Google Scholar]
- 11.Pogribny IP, Beland FA. DNA hypomethylation in the origin and pathogenesis of human diseases. Cell Mol Life Sci. 2009 Jul;66(14):2249–61. doi: 10.1007/s00018-009-0015-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferguson LR, Tatham AL, Lin Z, Denny WA. Epigenetic regulation of gene expression as an anticancer drug target. Curr Cancer Drug Targets. 2011 Feb;11(2):199–212. doi: 10.2174/156800911794328510. [DOI] [PubMed] [Google Scholar]
- 13.Carvalho RH, Hou J, Haberle V, Aerts J, Grosveld F, Lenhard B, et al. Genomewide DNA methylation analysis identifies novel methylated genes in non-small-cell lung carcinomas. J Thorac Oncol. 2013 May;8(5):562–73. doi: 10.1097/JTO.0b013e3182863ed2. [DOI] [PubMed] [Google Scholar]
- 14.Nikolaidis G, Raji OY, Markopoulou S, Gosney JR, Bryan J, Warburton C, et al. DNA methylation biomarkers offer improved diagnostic efficiency in lung cancer. Cancer Res. 2012 Nov;72(22):5692–701. doi: 10.1158/0008-5472.CAN-12-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suzuki M, Yoshino I. Aberrant methylation in non-small cell lung cancer. Surg Today. 2010 Jul;40(7):602–7. doi: 10.1007/s00595-009-4094-6. [DOI] [PubMed] [Google Scholar]
- 16.Bayarsaihan D. Epigenetic mechanisms in inflammation. J Dent Res. 2011 Jan;90(1):9–17. doi: 10.1177/0022034510378683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanada M, Delia D, Aiello A, Stadtmauer E, Reed JC. bcl-2 gene hypomethylation and high-level expression in B-cell chronic lymphocytic leukemia. Blood. 1993 Sep;82(6):1820–8. [PubMed] [Google Scholar]
- 18.Medvedeva YA, Khamis AM, Kulakovskiy IV, Ba-Alawi W, Bhuyan MS, Kawaji H, et al. Effects of cytosine methylation on transcription factor binding sites. BMC Genomics. 2014;15:119. doi: 10.1186/1471-2164-15-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin Z, Thomas NJ, Bibikova M, Seifart C, Wang Y, Guo X, et al. DNA methylation markers of surfactant proteins in lung cancer. Int J Oncol. 2007 Jul;31(1):181–91. [PubMed] [Google Scholar]
- 20.Floros J, Wang G, Mikerov AN. Genetic complexity of the human innate host defense molecules, surfactant protein A1 (SP-A1) and SP-A2--impact on function. Crit Rev Eukaryot Gene Expr. 2009;19(2):125–37. doi: 10.1615/critreveukargeneexpr.v19.i2.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Phelps DS. Surfactant regulation of host defense function in the lung: a question of balance. Pediatr Pathol Mol Med. 2001 Jul-Aug;20(4):269–92. [PubMed] [Google Scholar]
- 22.Wright JR. Immunoregulatory functions of surfactant proteins. Nat Rev Immunol. 2005 Jan;5(1):58–68. doi: 10.1038/nri1528. [DOI] [PubMed] [Google Scholar]
- 23.Wang J, Reid K. The immunoregulatory roles of lung surfactant collectins SP-A, and SP-D, in allergen-induced airway inflammation. Immunobiology. 2007;212(4–5):417–25. doi: 10.1016/j.imbio.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 24.Williams MC, Hawgood S, Hamilton RL. Changes in lipid structure produced by surfactant proteins SP-A, SP-B, and SP-C. Am J Respir Cell Mol Biol. 1991 Jul;5(1):41–50. doi: 10.1165/ajrcmb/5.1.41. [DOI] [PubMed] [Google Scholar]
- 25.Condon J, Jeyasuria P, Faust J, Mendelson CR. Surfactant protein secreted by the maturing mouse fetal lung acts as a hormone that signals the initiation of parturition. Proc Natl Acad Sci U S A. 2004;101(14):4978–83. doi: 10.1073/pnas.0401124101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang G, Guo X, Diangelo S, Thomas N, Floros J. Humanized SFTPA1 and SFTPA2 transgenic mice reveal functional divergence of SP-A1 and SP-A2: Formation of tubular myelin in vivo requires both gene products. J Biol Chem. 2010 Jan; doi: 10.1074/jbc.M109.046243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang G, Myers C, Mikerov A, Floros J. Effect of cysteine 85 on biochemical properties and biological function of human surfactant protein A variants. Biochemistry. 2007 Jul;46(28):8425–35. doi: 10.1021/bi7004569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Floros J, Wang G, Lin Z. Genetic Diversity of Human SP-A, a Molecule with Innate host Defense and Surfactant-Related Functions; Characteristics, Primary Function, and Significance. Current Pharmacogenomics. 2005;3:87–95. [Google Scholar]
- 29.Mikerov A, Wang G, Umstead T, Zacharatos M, Thomas N, Phelps D, et al. Surfactant protein A2 (SP-A2) variants expressed in CHO cells stimulate phagocytosis of Pseudomonas aeruginosa more than do SP-A1 variants. Infect Immun. 2007 Mar;75(3):1403–12. doi: 10.1128/IAI.01341-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mikerov A, Umstead T, Gan X, Huang W, Guo X, Wang G, et al. Impact of ozone exposure on the phagocytic activity of human surfactant protein A (SP-A) and SP-A variants. Am J Physiol Lung Cell Mol Physiol. 2008 Jan;294(1):L121–30. doi: 10.1152/ajplung.00288.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anglim PP, Alonzo TA, Laird-Offringa IA. DNA methylation-based biomarkers for early detection of non-small cell lung cancer: an update. Mol Cancer. 2008;7:81. doi: 10.1186/1476-4598-7-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haroun RA, Zakhary NI, Mohamed MR, Abdelrahman AM, Kandil EI, Shalaby KA. Assessment of the prognostic value of methylation status and expression levels of FHIT, GSTP1 and p16 in non-small cell lung cancer in Egyptian patients. Asian Pac J Cancer Prev. 2014;15(10):4281–7. doi: 10.7314/apjcp.2014.15.10.4281. [DOI] [PubMed] [Google Scholar]
- 33.Topaloglu O, Hoque MO, Tokumaru Y, Lee J, Ratovitski E, Sidransky D, et al. Detection of promoter hypermethylation of multiple genes in the tumor and bronchoalveolar lavage of patients with lung cancer. Clin Cancer Res. 2004 Apr;10(7):2284–8. doi: 10.1158/1078-0432.ccr-1111-3. [DOI] [PubMed] [Google Scholar]
- 34.Laird PW. Principles and challenges of genomewide DNA methylation analysis. Nat Rev Genet. 2010 Mar;11(3):191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- 35.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001 Dec;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 36.Matys V, Fricke E, Geffers R, Gössling E, Haubrock M, Hehl R, et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003 Jan;31(1):374–8. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rivera MP. Lung cancer in women: differences in epidemiology, biology, histology, and treatment outcomes. Semin Respir Crit Care Med. 2013 Dec;34(6):792–801. doi: 10.1055/s-0033-1358550. [DOI] [PubMed] [Google Scholar]
- 38.Floros J, Thomas N. Research Signpost K, India, editor. Genetic variations of surfactant proteins and lung injury. In: Nakos G, Papathanasiou A, editors. Surfactant Pathogenesis and Treatment of Lung Diasease. 2009. pp. 25–48. [Google Scholar]
- 39.Silveyra P, Floros J. Genetic variant associations of human SP-A and SP-D with acute and chronic lung injury. Front Biosci. 2012;17:407–29. doi: 10.2741/3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.D’Ovidio F, Kaneda H, Chaparro C, Mura M, Lederer D, Di Angelo S, et al. Pilot Study Exploring Lung Allograft Surfactant Protein A (SP-A) Expression in Association With Lung Transplant Outcome. Am J Transplant. 2013 Sep; doi: 10.1111/ajt.12407. [DOI] [PubMed] [Google Scholar]
- 41.Tagaram HR, Wang G, Umstead TM, Mikerov AN, Thomas NJ, Graff GR, et al. Characterization of a human surfactant protein A1 (SP-A1) gene-specific antibody; SP-A1 content variation among individuals of varying age and pulmonary health. Am J Physiol Lung Cell Mol Physiol. 2007 May;292(5):L1052–63. doi: 10.1152/ajplung.00249.2006. Epub 2006/12/26. eng. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y, Voelker DR, Lugogo NL, Wang G, Floros J, Ingram JL, et al. Surfactant Protein-A is Defective in Abrogating Inflammation in Asthma. Am J Physiol Lung Cell Mol Physiol. 2011 Jul; doi: 10.1152/ajplung.00381.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuroki Y, Takahashi H, Chiba H, Akino T. Surfactant proteins A and D: disease markers. Biochim Biophys Acta. 1998 Nov;1408(2–3):334–45. doi: 10.1016/s0925-4439(98)00079-9. [DOI] [PubMed] [Google Scholar]
- 44.Maitra M, Wang Y, Gerard RD, Mendelson CR, Garcia CK. Surfactant protein A2 mutations associated with pulmonary fibrosis lead to protein instability and endoplasmic reticulum stress. J Biol Chem. 2010 Jul;285(29):22103–13. doi: 10.1074/jbc.M110.121467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y, Kuan PJ, Xing C, Cronkhite JT, Torres F, Rosenblatt RL, et al. Genetic defects in surfactant protein A2 are associated with pulmonary fibrosis and lung cancer. Am J Hum Genet. 2009 Jan;84(1):52–9. doi: 10.1016/j.ajhg.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rountree MR, Bachman KE, Herman JG, Baylin SB. DNA methylation, chromatin inheritance, and cancer. Oncogene. 2001 May;20(24):3156–65. doi: 10.1038/sj.onc.1204339. [DOI] [PubMed] [Google Scholar]
- 47.Gomes A, Reis-Silva M, Alarcão A, Couceiro P, Sousa V, Carvalho L. Promoter hypermethylation of DNA repair genes MLH1 and MSH2 in adenocarcinomas and squamous cell carcinomas of the lung. Rev Port Pneumol. 2013 Dec; doi: 10.1016/j.rppneu.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 48.Li L, Shen Y, Wang M, Tang D, Luo Y, Jiao W, et al. Identification of the methylation of p14ARF promoter as a novel non-invasive biomarker for early detection of lung cancer. Clin Transl Oncol. 2013 Oct; doi: 10.1007/s12094-013-1122-1. [DOI] [PubMed] [Google Scholar]
- 49.Baccarelli A, Wright RO, Bollati V, Tarantini L, Litonjua AA, Suh HH, et al. Rapid DNA methylation changes after exposure to traffic particles. Am J Respir Crit Care Med. 2009 Apr;179(7):572–8. doi: 10.1164/rccm.200807-1097OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim H, Kwon YM, Kim JS, Han J, Shim YM, Park J, et al. Elevated mRNA levels of DNA methyltransferase-1 as an independent prognostic factor in primary nonsmall cell lung cancer. Cancer. 2006 Sep;107(5):1042–9. doi: 10.1002/cncr.22087. [DOI] [PubMed] [Google Scholar]
- 51.Lin RK, Hsu HS, Chang JW, Chen CY, Chen JT, Wang YC. Alteration of DNA methyltransferases contributes to 5′CpG methylation and poor prognosis in lung cancer. Lung Cancer. 2007 Feb;55(2):205–13. doi: 10.1016/j.lungcan.2006.10.022. [DOI] [PubMed] [Google Scholar]
- 52.Vallböhmer D, Brabender J, Yang D, Schneider PM, Metzger R, Danenberg KD, et al. DNA methyltransferases messenger RNA expression and aberrant methylation of CpG islands in non-small-cell lung cancer: association and prognostic value. Clin Lung Cancer. 2006 Jul;8(1):39–44. doi: 10.3816/CLC.2006.n.031. [DOI] [PubMed] [Google Scholar]
- 53.Hakuma N, Kinoshita I, Shimizu Y, Yamazaki K, Yoshida K, Nishimura M, et al. E1AF/PEA3 activates the Rho/Rho-associated kinase pathway to increase the malignancy potential of non-small-cell lung cancer cells. Cancer Res. 2005 Dec;65(23):10776–82. doi: 10.1158/0008-5472.CAN-05-0060. [DOI] [PubMed] [Google Scholar]
- 54.Hiroumi H, Dosaka-Akita H, Yoshida K, Shindoh M, Ohbuchi T, Fujinaga K, et al. Expression of E1AF/PEA3, an Ets-related transcription factor in human non-small-cell lung cancers: its relevance in cell motility and invasion. Int J Cancer. 2001 Sep;93(6):786–91. doi: 10.1002/ijc.1410. [DOI] [PubMed] [Google Scholar]
- 55.Ratovitski EA. LKB1/PEA3/ΔNp63 pathway regulates PTGS-2 (COX-2) transcription in lung cancer cells upon cigarette smoke exposure. Oxid Med Cell Longev 2010. 2010 Sep-Oct;3(5):317–24. doi: 10.4161/oxim.3.5.13108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Upadhyay S, Liu C, Chatterjee A, Hoque MO, Kim MS, Engles J, et al. LKB1/STK11 suppresses cyclooxygenase-2 induction and cellular invasion through PEA3 in lung cancer. Cancer Res. 2006 Aug;66(16):7870–9. doi: 10.1158/0008-5472.CAN-05-2902. [DOI] [PubMed] [Google Scholar]
- 57.Rehan VK, Torday JS, Peleg S, Gennaro L, Vouros P, Padbury J, et al. 1Alpha,25-dihydroxy-3-epi-vitamin D3, a natural metabolite of 1alpha,25-dihydroxy vitamin D3: production and biological activity studies in pulmonary alveolar type II cells. Mol Genet Metab. 2002 May;76(1):46–56. doi: 10.1016/s1096-7192(02)00022-7. [DOI] [PubMed] [Google Scholar]
- 58.Li S, Huang X, Zhang D, Huang Q, Pei G, Wang L, et al. Requirement of PEA3 for Transcriptional Activation of FAK Gene in Tumor Metastasis. PLoS One. 2013;8(11):e79336. doi: 10.1371/journal.pone.0079336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim SH, Chen G, King AN, Jeon CK, Christensen PJ, Zhao L, et al. Characterization of vitamin D receptor (VDR) in lung adenocarcinoma. Lung Cancer. 2012 Aug;77(2):265–71. doi: 10.1016/j.lungcan.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Phokela SS, Peleg S, Moya FR, Alcorn JL. Regulation of human pulmonary surfactant protein gene expression by 1alpha,25-dihydroxyvitamin D3. Am J Physiol Lung Cell Mol Physiol. 2005 Oct;289(4):L617–26. doi: 10.1152/ajplung.00129.2004. [DOI] [PubMed] [Google Scholar]
- 61.Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci U S A. 2002 Mar;99(6):3740–5. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of CpG sites in the promoter regions (5000bp) of SFTPA2 (upper panel, NCBI Reference Sequence: NG_013046.1) and SFTPA1 (lower panel, NCBI Reference Sequence NG_021189.1) sequences using the online software CpG Island Searcher (http://cpgislands.usc.edu/) (61). The sequence of interest for the present study is indicated between arrows, and contains a region with dense CpG sites, shown in blue (CpG island 1).