Abstract
Human pluripotent stem cell derived models that accurately recapitulate neural development in vitro and allow for the generation of specific neuronal subtypes are of major interest to the stem cell and biomedical community. Notch signaling, particularly through the Notch effector HES5, is a major pathway critical for the onset and maintenance of neural progenitor cells (NPCs) in the embryonic and adult nervous system1-3. This can be exploited to isolate distinct populations of human embryonic stem (ES) cell derived NPCs4. Here, we report the transcriptional and epigenomic analysis of six consecutive stages derived from a HES5-GFP reporter ES cell line5 differentiated along the neural trajectory aimed at modeling key cell fate decisions including specification, expansion and patterning during the ontogeny of cortical neural stem and progenitor cells. In order to dissect the regulatory mechanisms that orchestrate the stage-specific differentiation process, we developed a computational framework to infer key regulators of each cell state transition based on the progressive remodeling of the epigenetic landscape and then validated these through a pooled shRNA screen. We were also able to refine our previous observations on epigenetic priming at transcription factor binding sites and show here that they are mediated by combinations of core and stage- specific factors. Taken together, we demonstrate the utility of our system and outline a general framework, not limited to the context of the neural lineage, to dissect regulatory circuits of differentiation.
We utilized the human ES cell line WA9 (or H9) expressing GFP under the HES5 promoter5 to isolate defined neural progenitor populations of neuroepithelial (NE), early radial glial (ERG), mid radial glial (MRG) and late radial glial (LRG) cells based on their Notch activation state4, as well as long term neural progenitors (LNP) based on their EGFR expression (Fig. 1a, Extended Data Fig. 1a). We took these defined stages to create strand-specific RNA-Seq data, chromatin immunoprecipitation followed by sequencing (ChIP-Seq) maps for H3K4me1, H3K4me3, H3K27ac, and H3K27me3 as well as DNA methylation (DNAme) data by whole genome bisulfite sequencing (WGBS) for the first four stages and reduced representation bisulfite sequencing (RRBS) for the last two (LRG and LNP) stages (Fig. 1a, Supplementary Table 1).
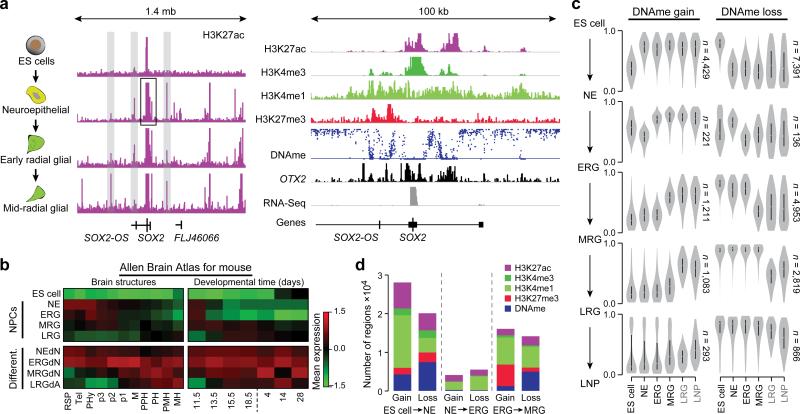
Figure 1. Consecutive stages of ES cell derived neural progenitors are characterized by distinct epigenetic states.
a. Left: Schematic of the cell system. Middle: Normalized read-count level for H3K27ac over a 1.4 mega base (mb) region around the SOX2 locus (chr3:180,854,252-182,259,543). ChIP-Seq read counts were normalized to 1 million reads and scaled to the same level (1.5) for all tracks shown. Right: Additional tracks for H3K4me3, H3K4me1 and H3K27me3 as well as DNAme (scale 0-100%), OTX2 and expression covering a 100 kilo base (kb) sub-region (chr3:181,389,523-181,490,148) of this locus. Histone and RNA-Seq data were normalized to 1 million reads and are shown on distinct scales.
b. Maximum gene set activity levels shown as z-scores for genes expressed in defined brain structures (left) and developmental time points (right) based on the mouse Allen Brain Atlas. Gene set activity was defined as average expression level of all member genes followed by z-score computation across all nine cell types.
Abbreviations: Rostral secondary prosencephalone (RSP), Telencephalon (Tel), peduncular (caudal) hypothalamus (PHy), Hypothalamus (p3), prethalamus (p2), pre-tectum (p1), midbrain (M), prepontine hindbrain (PPH), pontine hindbrain (PH), pontomedullary hindbrain (PMH), medullary hindbrain (MH); and embryonic (E)11.5, E13.5, E15.5, E18.5 as well postnatal P4, P14 and P28.
c. Distribution of DNAme levels for differentially methylated regions (delta meth≥0.2, p≤0.01) across state transitions, For instance, distributions for regions gaining methylation in the transition from ES cell to NE (top left) at all stages of differentiation. Distinct methylation level trace plots are shown for regions gaining methylation (left) during the specific transitions (indicated on the side) and loss of methylation (right). Black labeled samples are based on WGBS data and grey color samples (LRG and LNP) were profiled by RRBS.
d. Barplot of the frequency and associated mark of epigenetic changes for all cell state transitions broken up into gain and loss for consecutive differentiation stages.
Global transcriptional analysis of the undifferentiated ES cells and the first four NPC stages identified 3,396 differentially expressed genes (Extended Data Fig. 1b, c, Supplementary Table 2). Pluripotency associated genes such as OCT4 and NANOG are, as expected, rapidly downregulated, and pan-neural genes are induced early and maintained throughout (Extended Data Fig. 1c). Using data from the mouse Allen Brain Atlas as an in vivo reference for genes expressed in different brain compartments and developmental stages, we observe a consecutive shift of expression signatures along our NPC differentiation trajectory (Fig. 1b). NE through LRG transcripts suggest anterior neural fates, while the MRG and LRG stages show in addition some posterior identities (Fig. 1b, left). Accordingly, differentiated progeny derived from these populations express deep cortical layer neuronal markers (NEdN and ERGdN) such as FEZF2 and BCL11B and superficial layer neuronal markers (MRGdN) such as SATB2 (Extended Data Fig. 1d). Progression from early (NE) to late (LRG) stages was also accompanied by a transition from predominantly neurogenic to mainly gliogenic potential, although LRG cells can still generate neurons (Extended Data Fig. 1d). This progressive change in NPC identity aligns well with the in vivo order developmental events4.
In line with these observations, our WGBS data show changes in DNAme that can be separated into two overall patterns: the first is characterized by widespread loss and retention of the resulting hypomethylated state throughout subsequent differentiation stages (Fig. 1c, top right). This pattern coincides with major cell fate decisions such as commitment from ES cells to the neural fate and the transition from ERG to MRG, the latter demarcating both peak of neurogenesis and onset of gliogenic potential (Fig. 1c, right middle). The second pattern is defined by a stage-specific loss with subsequent gain at the next stage as observed during the transition from NE to ERG and also from MRG to LRG (Fig. 1c, right). Conversely, regions gaining DNAme during transition from one stage to another frequently reside in a hypomethylated state in all preceding stages, indicating the possible silencing of stem cell or pan-neural gene regulatory elements (Fig. 1c, left). At the histone modification level we also observe the most widespread changes during the initial neural induction (Fig. 1d), although it is worth noting that the biggest gain of the repressive mark H3K27me3 occurs at the MRG stage.
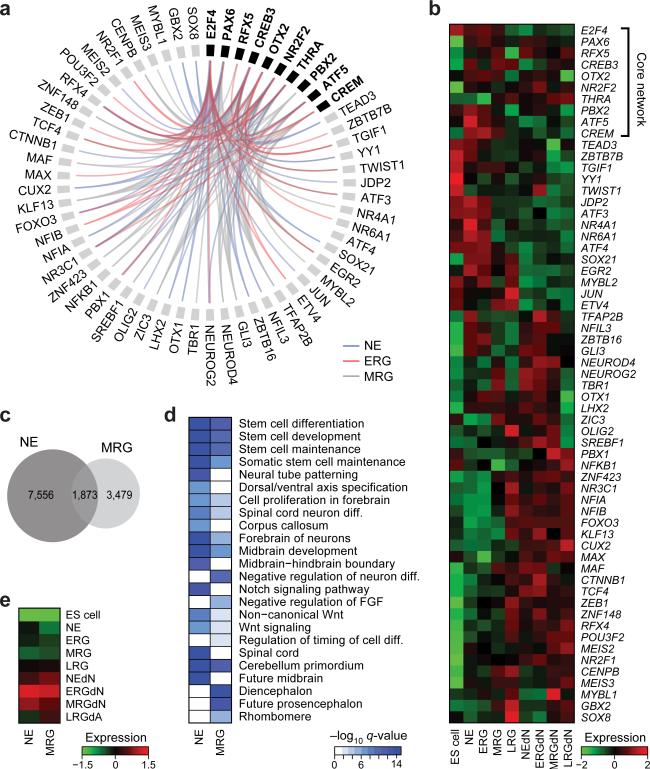
These coordinated epigenetic changes are likely the result of differential transcription factor (TF) activity6-8. We therefore developed a computational method to attribute the genome wide changes in histone modifications and DNAme at regions termed footprints (FPs) to particular TFs and quantified this remodeling potential (TERA: Transcription factor Epigenetic Remodeling Activity; (Fig. 2a, Extended Data Fig. 2a, b and Online Methods). Interestingly, TF FPs in our NPC model were highly enriched for single nucleotide polymorphisms previously reported to be implicated in Alzheimer's disease (p≤0.001, Extended Data Fig. 2c) and bipolar disorders (p≤0.001) by genome wide association studies, suggesting the possibility to utilize this differentiation system to study the genetic component of complex diseases in vitro9,10. Next, in order to identify potential key regulators of onset, maintenance and transition through distinct NPC populations, we ranked all motifs and their associated TFs based on their TERA scores between consecutive time points (Supplementary Table 3). We then retrieved the highest scoring 40 TFs for each cell state transition (Fig. 2b). This analysis confirmed many well known key regulators of in vivo neural development and forebrain specification that are induced at the NE stage such as PAX6, OTX2, FOXG1 (Refs 11-13) as well as various SOX proteins14. Interestingly, we also find differential activity of distinct downstream components of signaling pathways such as a decrease of SMAD4 activity at the NE stage, consistent with inhibition of TGFb signaling that promotes neural induction15. Another example is POU3F2 known to be involved in sub ventricular zone expansion and superficial layer neuronal specification, and TCF12, which ishighly expressed in germinal zones during brain development16 (Fig. 2b, Supplementary Table 3).
Figure 2. Distinct transcription factor modules are associated with stage specific epigenetic transitions.
a. Illustration of epigenomic footprinting across the PAX6 locus (chr11:31,780,014-31,842,503) for dips in H3K27ac regions (right). Black boxes highlight footprints (FP) determined for H3K27ac peaks that harbor various putative transcription factor (TF) binding sites based on motif matching.
b. The 40 top ranked TFs predicted to be activated during the cell state transition indicated on the bottom. Color-coding represents normalized TF epigenetic remodeling scores, averaging over all TERAs based on H3K4me3, H3K4me1, H3K27ac and DNAme. In addition, predictions were filtered for factors expressed at least at the stage of predicted induction.
To obtain a higher-level overview of the processes and roles associated with the distinct putative regulators, we decomposed the H3K27ac data into seven distinct modules, each corresponding to a unique epigenetic dynamic, genomic region and upstream regulator set (Extended Data Fig. 2d, top). Gene set enrichment analysis17 on the genomic regions associated with each of the distinct modules revealed that the module activated upon neural induction and sustained throughout the MRG stage is strongly associated with stem cell maintenance and differentiation related processes as well as Notch signaling (Extended Data Fig. 2d,e; module 2). Further analysis of upstream regulators of this module revealed a strong association with PAX6 and FOXG1, suggesting a role for these factors in the general establishment and maintenance of the telencephalic cortical identity of the NPC states (Extended Data Fig. 2e).
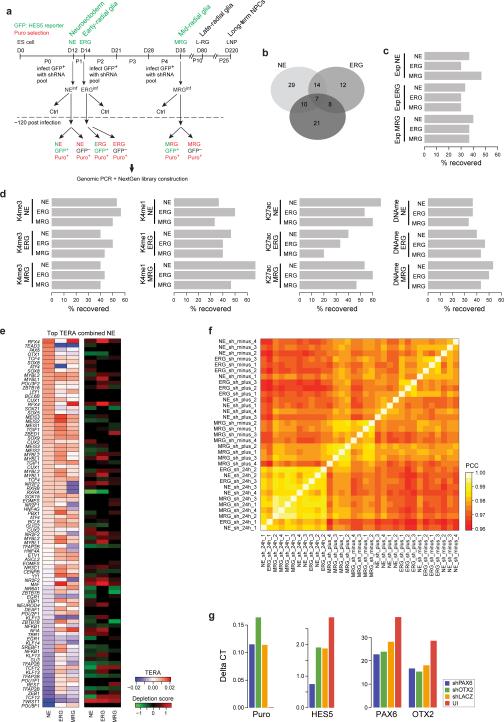
To explore the relevance of predicted factors for each cellular state, we carried out a pooled shRNA screen against 244 TFs and epigenetic modifiers selected based on our RNA-Seq data (Fig. 3a, Extended Data Fig. 3a, Supplementary Table 4). In total, we recovered 110 factors with a significant (Fig. 3b, q-value≤0.05, mean empirical FDR=0.045 see Online Methods) negative impact on the number of HES5+ cells in at least one differentiation stage (Supplementary Table 4), with high overlap between the distinct stages (Fig. 3c, Extended Data Fig. 3b). Despite the expected high false negative rate18 our screen consistently validated more than 50% of the predicted TFs with a known motif for the top 20 motifs found at each stage (Fig. 3d, Extended Data Fig. 3c-d), while an expression based identification yielded only ~30% recovery (Extended Data Fig. 3c). Among the top factors recovered from the predictions at the early stage (NE and ERG) are the RFX proteins including RFX4, which has been implicated in cortical and brain development19,20, FOXG1, as well as NR2F2, whose paralog NR2F1 has been shown to serve as an intrinsic factor for early regionalization of the neocortex21,22. Gene set enrichment analysis of putative genomic targets of NR2F2 (see Online Methods) in the NE cells further expands this role suggesting involvement in telencephalon, diencephalon and posterior hindbrain development (Supplementary Table 5). At the MRG stage, we recover genes involved in extensive neurogenesis but also in commencing early gliogenesis such as NFIA and NFIB, which are involved in both repressing the neuronal progenitor state through Notch signaling concomitantly with activating glial fates23, as well as REST – a major pleotropic epigenetic regulator of neural cell fate decisions24.
Figure 3. A pooled shRNA screen recovers predicted regulators of in vitro NPC differentiation.
a. Simplified schematic of the pooled shRNA screen (see Extended Data Fig. 3 for more details).
b. Depletion scores for all genes that are significantly reduced (q-value≤ 0.05 for at least 2 different shRNAs per gene) in at least one stage for FACS purified HES5+ cells 6 days after knockdown compared to FACS sorted HES5- obtained from the same infection or compared to cells collected 24h after infection (see Extended Data Fig. 3a). Depletion score indicates the extent to which shRNAs targeting a particular gene were lost during the knockdown period relative to the control, indicating potential relevance of a particular gene for HES5+ maintenance, NPC state progression and proliferation or cell survival. Higher depletion scores (red) indicate stronger reduction in shRNA presence; scores were capped at 1.5 and computed based on at least three technical replicates per condition.
c. Overlap of genes detected to be significantly depleted in the HES5+ population relative to at least one of the control conditions.
d. Performance of combined regulator predictions based on TERA ranking averaged over H3K4me3, H3K4me1, H3K27ac and DNAme. Performance is measured as percentage of the top 20 predicted activating or repressing motifs for each stage mapping to TFs included in the shRNA library.
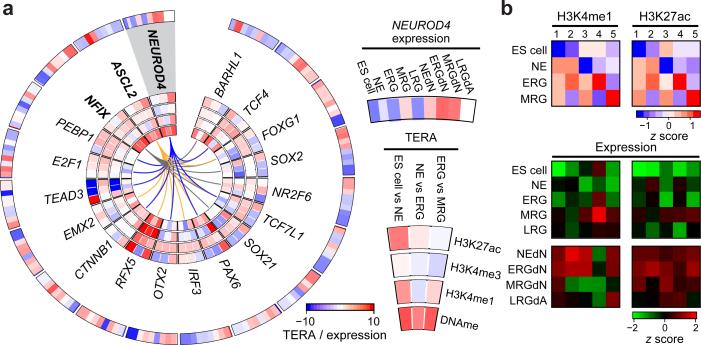
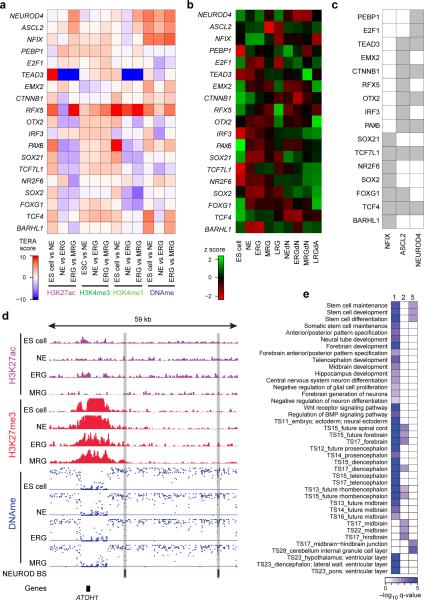
Next, we selected a set of 22 core factors with evidence to be functional at all stages as assessed by RNA-Seq and the shRNA screening results (Extended Data Fig. 4a, Online Methods). In order to determine whether the subset of core factors with a DNA binding motif available (10/22) exerts the same function at each stage, we performed a co-binding analysis based on the predicted binding sites of 523 TFs in dynamically regulated H3K27ac footprints. This analysis uncovered highly stage-specific relationships that were also supported by the observed knockdown effect at each stage (Fig. 4a, Extended Data Fig. 4b). Interestingly, most of the identified co-binding partners are either expressed in a more stage-specific fashion or are only activated in more mature neuronal or glial cell types (Fig. 4b). To further validate some of these findings, we focused on OTX2 due to its high expression in all NPC populations (Fig. 4b). OTX2 was enriched at more targets in NE of which around 35% overlapped with MRG bound sites (Fig. 4c, Extended Data Fig. 4c). The shared target set is highly enriched for genes involved in stem cell maintenance and differentiation as well as various pro-neural gene sets known to act during advanced stages of forebrain and midbrain progenitor cell maturation (Fig. 4d, Extended Data Fig. 4d). This binding pattern combined with the observation that the OTX2 target gene set reaches peak transcriptional activity in the NEdN and ERGdN populations implies a role for OTX2 in the preparation of pro-neural genes expressed at later stages (Fig. 4d, e). These findings further suggest a model where a core set of TFs helps sustain NPC identity throughout the differentiation time course and at the same time participates in the progression and modulation of NPC differentiation potential through cooperation with stage-specific regulators.
Figure 4. A set of core TFs dynamically associates with stage-specific factors to modulate NPC identity and differentiation potential.
a. Predicted top 10 significant (p≤0.01, odds ratio≥1.5) co-binding relationships in dynamically regulated H3K27ac footprints for a set of 10 TFs (bold) essential for HES5+ cells at each stage. Stage-specific predicted co-binding relationships are indicated in blue (NE), red (ERG) and grey (MRG). All predicted relations are supported by a knockdown effect of each gene at the relevant stage.
b. Gene expression patterns shown as z-scores for the core network TFs as well as all predicted co-binding partners across ES cells, all NPCs and more mature cellular states.
c. Venn diagram showing the overlap of OTX2 binding sites determined by ChIP-Seq in early NE and MRG cells.
d. Gene set enrichment analysis results for OTX2 binding sites in early NE and MRG cells.
e. Median expression patterns for ES cells, all NPCs and more mature cell populations shown as z-scores for putative downstream target genes of OTX2 binding sites.
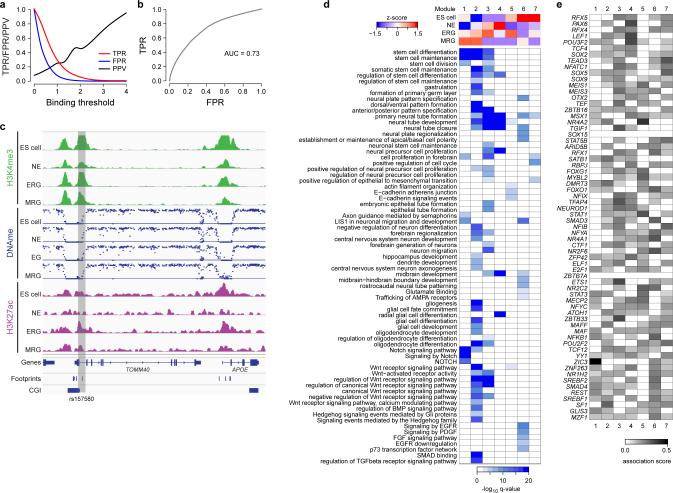
To gain a better understanding of how factors that are active at distinct NPC stages contribute to their corresponding neuronal and glial cell propensities, we took advantage of the fact that many TFBSs exhibit a gain of H3K27ac or H3K4me1 and loss of DNAme at the early NPC stages prior to increased expression of their associated genes in more differentiated cell types (hence referred to as epigenetic priming) (Fig. 5a, Extended Data Fig. 5a-c). For instance, we identified three pro-neural factors that show evidence of priming, are induced only at a later stage, and possess TFBS that are also significantly (p≤0.05 permutation test) associated with other genes differentially expressed at a later stage (Fig. 5a, bold genes). Because these pro-neural genes are not expressed at the early NPC stages but at more mature cell types or later NPC stages derived from these early NPCs, the identification of such priming events highlights that the epigenetic state is useful for predicting key regulators and their downstream targets. In order to pinpoint TFs potentially involved in facilitating these priming events at the respective NPC stages, we determined significant co-binding relationships between the subset of pro-neural genes and other TFs that are concurrently expressed (Fig. 5a).
Figure 5. Binding of core and stage-specific NPC TFs is associated with epigenetic priming of pro-neural genes.
a. Characterization of TFs associated with motifs gaining H3K4me1 or losing DNAme at the NE stage prior to their expression at a later or more differentiated cell state as determined by high TERA scores (bold), termed priming. In addition, significant (p≤0.01, odds ratio≥1.5) co-binding relationships with factors expressed at the NE are indicated by colored lines. For each TF (from outer to inner circles, see example below for NEUROD4) heatmaps indicating the relative expression level as z-score in all cell types as well as normalized TERA scores for H3K27ac, H3K4me3, H3K4me1 and DNAme.
b. Heatmaps depicting the H3K4me1 (left) and H3K27ac (right) enrichment level for predicted NEUROD binding sites at each NPC stage for 5 distinct dynamic patterns. Here, none of the NEUROD family proteins is expressed (<2.5 FPKM). Bottom: Heatmap showing the z-scores of the median gene expression levels for predicted NEUROD downstream target genes for each of the 5 dynamic patterns in the more mature neuron and astrocyte-like populations.
To specifically investigate the hypothesis that a part of the pro-neural binding site landscape is epigenetically primed at the NPC stages, we focused on predicted NEUROD binding sites within H3K27ac footprints and defined five patterns of H3K27ac and H3K4me1 enrichments across these sites (Fig. 5b). We found that genes associated with predicted NEUROD binding sites in regions gaining H3K27ac or H3K4me1 enrichment at distinct stages of NPC progression are up-regulated in more mature populations derived from the respective NPC stage (Fig. 5b and Extended Data Fig. 5d). Consistent with the idea of a comprehensive preparation of the epigenetic landscape during lineage specification, NEUROD binding sites associated with NPC related genes that retain high levels of H3K27ac and H3K4me1 throughout the time course, are associated with various anterior and posterior cortical structures as well as early and late developmental time points (Extended Data Fig. 5e).
These results support a model where selected TFs at the NPC stage remodel the binding site repertoire for pro-neural factors by preparing the epigenetic landscape at their respective targets. First the general lineage landscape is established upon commitment to the neural fate, followed by the stage-specific modulation of primed pro-neural binding sites. This in turn restricts their binding space as a mechanism to ensure proper neuronal and glial differentiation capacity. In addition to these mechanistic insights, we provide a general analysis strategy to interpret differences in epigenetic landscapes based on cell fate regulatory TFs. This strategy can be readily applied to other datasets including the extensive collection of the NIH Roadmap Epigenomics Project (Supplementary Table 3).
Online Methods
Culturing undifferentiated human ES cells
HES5::eGFP BAC transgenic human ES cells (H9; WA-09; Wicell) expressing GFP under the HES5 promoter were cultured on mitotically inactivated mouse embryonic fibroblasts (MEFs) (Globalstem). Undifferentiated ES cells were maintained as described previously25 in medium containing DMEM/F12, 20% KSR, 1mM Glutamine, 1% Penicillin/Streptomycin, non essential amino acids and beta-mercaptoethanol. Undifferentiated ES cells were purified with pluripotency markers Alexa 647-conjugated Tra-1-60 and PE-conjugated SSEA-3 (BD Pharmingen).
Neural induction and long-term propagation of NPCs
Neural differentiation of ES cells was performed as described in refs4,15,25. Briefly, neuroepithelial cells were generated either by monolayer induction – with dissociated ES cells plated on Matrigel (BD biosciences), or by co-culture on MS5 stromal cells. In both cases neural fate was directed by dual SMAD inhibition protocol15. NE cells and Neural rosettes were harvested mechanically during all stages of differentiation and replated on culture dishes pre-coated with 15 μg/mL polyornithine (Sigma), 1 μg/mL Laminin (BD Biosciences) and 1ug/ml Fibronectin (BD Biosciences) (Po/Lam/FN) in N2 medium composed of DMEM/F12 and N2 supplement (Invitrogen). N2 supplement contained Insulin, Apo-transferin, Sodium Selenite, Putrecine and Progesterone. This medium was supplemented with SHH (30 ng/mL), FGF8 (100 ng/mL) and BDNF (20 ng/mL) (all from R&D Systems) to induce and maintain early anterior regionalization of the neural plate. These factors were gradually replaced by FGF2 (20 ng/mL) and EGF (20 ng/mL) in the following two weeks of differentiation in order to maintain a proliferative (FGF and EGF responsive) NPC state. NPCs from all stages were collected at indicated days and FACS purified for HES5::GFP (NE to LRG) or EGFR for LNPs to purify for the highest NPC state for each stage. NE cells were collected at day 12 of differentiation, ERG were collected at day 14, mid neurogenesis radial glial (ERG) cells were collected at day 35, late gliogenic radial glial (LRG) cells were collected at day 80, and long term NPCs (LNP) were collected at day 220. At each stage cells were either split for the next passage or subjected to FACS purification for HES5::GFP as described. All replating was performed on Po/Lam/FN coated dishes. For generating mature differentiated populations, HES5+ sorted NPCs were seeded at high density and subjected to mitogen withdrawal differentiation medium for 17 days which included N2 supplemented with Ascorbic Acid (AA)/BDNF (neuronal; NEdN, ERGdN, MRGdN) or 5% Fetal Bovine Serum (FBS) (Invitrogen) (glial) (LRGdA). For additional details, see Edri et al.4.
Chromatin Immunoprecipitation followed by sequencing (ChIP Seq)
For the histone ChIP experiments, we used similar approaches to ref26. Specifically, around 160.000 cells were crosslinked in 1% formaldehyde for 10 min at 37°C, followed by quenching with 125mM glycine for 5 min at 37°C, washed with PBS containing protease inhibitor (Roche, 04693159001) and flash frozen in liquid nitrogen. To lyse the cells, we used 1% SDS, 10mM EDTA and 50mM Tris-HCl pH 8.1 complemented with protease inhibitor. The chromatin was then fragmented with a Branson Sonifier (model S-450D) at 4°C, calibrated to a size range of 200 and 800bp. Chromatin was mixed with antibody and incubated at 4°C overnight. Protein-A and Protein-G Dynabeads were added to chromatin/antibody mix (Invitrogen, 100-02D and100-07D, respectively) and incubated for 1-2 hours at 4°C. Samples were washed 6 times with RIPA buffer (10mM Tris-HCl pH 8.0, 1mM EDTA pH 8.0, 14mM NaCl, 1% TritonX-100, 0.1% SDS, 0.1% DOC), twice with RIPA buffer containing 500mM NaCl, twice with LiCl buffer (10 mM TE, 250mM LiCl, 0.5% NP-40, 0.5% DOC), twice with TE (10Mm Tris-HCl pH 8.0, 1mM EDTA), and then eluted in elution buffer (10mM Tris-Cl pH 8.0, 5mM EDTA, 300mM NaCl, 0.1% SDS; pH 8.0) at 65°C. Eluate was treated with RNaseA (Roche, 11119915001) and Proteinase K (NEB, P8102S) overnight at 65°C.
For the OTX2 ChIP cells were collected and crosslinked in 1% formaldehyde for 15 minutes on ice, quenched with 125mM glycine for 5 minutes at room temperature and pelleted. Nuclei were then isolated and chromatin was digested at 37°C with MNase enzyme until the majority of the DNA was between 50 and 800bp. Specifically, 25U and 35U of MNase enzyme were used to digest NE cells and RNS/RG cells, respectively. The chromatin was then incubated with the antibodies over night at 4°C and co-immunoprecipitation of antibody-protein complexes was performed with Protein A or G beads for 1-2 hours at 4°C.
All antibody catalog and lot numbers are listed next to the dataset for which they were used in Supplementary Table 1.
ChIP-Seq library preparation and sequencing
To extract DNA and create the Illumina libraries we used Solid-Phase Reversible Immobilization (SPRI) beads. The SPRI beads were added to the samples, mixed 15 times, incubated for 2 minutes at room temperature. Supernatant was extracted from the beads on a magnet (4 minutes). 70% ethanol was used to wash the beads and then dried for another 4 minutes. 40μl EB buffer (10 mM Tris-HCl pH 8.0) was used to elute the DNA. The next steps of Illumina library construction include end-repair, addition of A-base, ligation of barcoded adaptors and PCR enrichment. To minimize the loss of ChIP material throughout this procedure, we used a general SPRI cleanup procedure after each reaction step reusing the same beads. PEG buffer (20% PEG and 2.5 M NaCl) was used to rebind chIP material to SPRI following each reaction, and washing and extraction occured as stated above. The enzymatic reactions were carried as follows: 1. DNA end-repair: Epicenter End-IT Repair kit incubated at room temperature for 45 min. 2. A-base addition: Klenow (3’->5’ exonuclease; New England Biolabs) incubated at 37°C for 30 min. 3. Adaptor ligation: DNA ligase (New England Biolabs) and indexed oligo adaptors and incubated 25C for 15 min, followed by 0.7X SPRI/reaction to remove non-ligated adaptors. 4. PCR enrichment: PCR mastermix (primer set, dNTP mix, Pfu Ultra Buffer (Agilent), Pfu Ultra-II Fusion (Agilent), water), for 20 cycles. The PCR amplified libraries we cleaned up using 0.7X SPRI/reaction (size selection mode) to remove excessive primers. Roughly 5 picomoles of DNA library was then applied to each lane of the flow cell and sequenced on Illumina HiSeq 2000 sequencers according to standard Illumina protocols.
For the OTX2 ChIP, DNA libraries were constructed using standard Illumina protocols for blunt-ending, polyA extension, and ligation. MyOne Silane beads (Life Technologies 37002D), were used to purify DNA fragments following each step of the library preparation. Adapter ligation was performed overnight at 16C. Ligated DNA was then PCR amplified and gel size selected for fragments between 150 and 700bp. Samples were sequenced using Illumina HiSeq at a target sequencing depth of 20 million uniquely aligned reads.
Strand Specific RNA-Sequencing Library Construction
RNA was extracted using the miRNeasy kit (Qiagen, 217004). Poly(A) RNA was isolated using Oligo d (T25) beads (NEB, E7490L). The Poly(A) fraction was then fragmented (Invitrogen, AM8740). Fragments smaller than 200 bps were eliminated (Zymo, R1016) and the remaining fraction was treated with FastAP Thermosensitive Alkaline Phosphatase (Thermo Scientific, EF0652) and T4 Polynucleotide Kinase (NEB, M0201L). RNA was then ligated to a RNA adaptor as reporter previously27 using T4 RNA Ligase 1 (NEB, M0204L), which was then used to facilitate cDNA synthesis using Affinity Script Multiple Temperature Reverse Transcriptase (Agilent, 600105). More specifically, we used the following adaptors reported in ref27: RNA sequencing - RiL-19 3’ RNA adaptor: /Phosphate/rArGrArUrCrGrGrArArGrArGrCrGrUrCrGrUrG/ddC RNA sequencing - AR17 RT primer: ACACGACGCTCTTCCGA RNA sequencing - 3Tr3 5’ DNA adaptor: /Phosphate/AGATCGGAAGAGCACACGTCTG/ddC RNA sequencing - PCR enrichment: AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTC CGATCTCAAGCAGAAGACGGCATACGAGATNNNNNNNNGTGACTGGAGTTC AGACGTGTGCTCTTCCGATCT.
RNA was then degraded and the cDNA was ligated to a DNA adaptor using T4 RNA Ligase 1 as described previously27. Final library amplification was completed using NEB Next High Fidelity 2X PCT Master Mix (M054L). To clean up the final PCR and removed adapter dimers, two subsequent 1X and .8X SPRI reactions were completed to prepare the final library for sequencing.
Pooled shRNA screen
We selected 244 transcription factors and epigenetic modifiers that were differentially or continuously highly expressed during our in vitro differentiation time course in an otherwise unbiased fashion (Supplementary Table 4). In addition, we included GFP, RFP, lacZ and luciferase as internal controls. We then obtained a sub-pool of the human 45K shRNA pool28 distributed by the Broad Institute Genomic Perturbations Platform and the RNAi Consortium (TRC) against these genes. For each gene, 5 distinct shRNAs were included as well as 5 scrambled and 3 empty control vectors, amounting to a total of 1230+8 shRNAs. The plasmid for shRNA expression under the control of the constitutive U6 shRNA promoter was the lentiviral vector pLKO.1. shRNA pool production and infection conditions were performed as previously described28. Subsequently, we performed calibration experiments to determine to optimal combination of MOI and Puromyocin concentration to ensure efficient selection. We identified MOI 0.4 and 1ug/ml of Puromycin as optimal parameters for all stages. We then infected 26 million cells at each stage of NE, ERG and MRG to ensure sufficient shRNA integration events to recover the complexity of the shRNA library. 24h post infection and prior to full expression but after integration of the lentivirus into the genome we collected 3 million cells to determine our baseline shRNA library representation. Subsequently, we subjected the cells to 5 days of Puromycin selection and then FACS sorted the resulting populations into HES5+ and HES5- compartments. Next, we assessed the representation of the shRNA library in each of the 9 populations by retrieving all shRNA integration events from genomic DNA isolated from each sample using PCR followed by next generation sequencing as previously described29. More specifically, we performed two rounds of PCR using the following primers for the primary PCR: Primary R :CTTTAGTTTGTATGTCTGTTGCTATTAT Primary F: AATGGACTATCATATGCTTACCGTAAC For the second, nested PCR we used: Nested F: GGCTTTATATATCTTGTGGAAAGGA Nested R: GGATGAATACTGCCATTTGTCTC.
Next, we performed standard Illumina sequencing library construction as outlined above for 4 technical replicates for NE and MRG and 3 technical replicates for ERG, each comprising HES5+, HES5- and 24h control, amounting to a total of 33 libraries. We then sequenced these amplicon libraries on a HiSeq2500 with a PhiX spike in of 25%.
Individual shRNA validation for OTX2 and PAX6
RNA was extracted using miRNeasy kit (Qiagen) followed by Maxima reverse transcription reaction kit (Fermentas). 1ng of cDNA was subjected to qPCR using our homemade designed primers and the ABsolute QPCR SYBR Green ROX Mix (ABgene) on a ViiA-7 cycler (ABI). Threshold cycle values were determined in triplicates and presented as average compared to HPRT. Fold changes were calculated using the 2−ΔCT method.
WGBS and RRBS library production
WGBS libraries were generated as previously described in ref 8. RRBS was carried out using the multiplexed, gel free protocol described in ref 30.
Data processing
For RNA-Seq data processing, reads were trimmed to 80, 60 or 30bp depending on their per-base quality distribution in order to achieve maximum alignment rates. Reads were mapped to the human genome (hg19) using TopHat v2.0 (Ref 31) (http://tophat.cbcb.umd.edu) employing the unfiltered gencode.v19.annotation.gtf annotation as the transcriptome reference. TopHat was run with default parameters except for the coverage search being turned off. Transcript expression was estimated with Cuffdiff 2 (Ref 32). The workflow used to analyze the data is described in detail in Trapnell et al. (2012) (alternate protocol B)33.
WGBS libraries were aligned using BSMap 2.7 (Ref 34) to the hg19/GRCh37 reference assembly. Subsequently, CpG methylation calls were made using custom software as previously described7, excluding duplicate, low-quality reads as well as reads with more than 10% mismatches. Only CpGs with more than 5x coverage were considered for further analysis.
ChIP-Seq data were aligned to the hg19/GRCh37 reference genome using MAQ35 version 0.7.1 with default parameter settings or Bowtie 2 version 2.05 (Ref 36). Reads were filtered for duplicates and extended by 200 bp at the end of the read. Visualization of read count data was performed by converting raw bam files to .tdf files using IGV tools37 and normalizing to 1 million reads. Fragment length extended, duplicate and quality-filtered reads were used for subsequent analysis.
shRNA screen data analysis
For the screen data analysis, we followed the protocol outlined by Dai et al.38 employing the R package limma39. First, we extracted and counted the number of times each shRNA was observed in each library using the shRNA sequence as barcode and the R function processHairpinReads(). Next, we normalized the shRNA counts to the total number of reads observed harboring a shRNA to counts per million (cpm) and retained only those shRNAs with more than 0.5 cpm in more than 2 samples. After further QC showing excellent reproducibility (Extended Data Fig. 3f), we performed differential shRNA count analysis between the HES5+ and 24h control and the HES5+ and HES5- populations for each stage. To that end we first estimate the dispersion for each condition and then fit a negative binomial generalized linear model using the R package edgeR. We then conduct a likelihood ratio test for each contrast and only retain those shRNAs as differentially enriched at a FDR≤0.05. To determine genes with significant positive or negative impact on HES5+ maintenance or cell survival, we determined all genes that were targeted by at least two independent shRNAs which showed a significant effect (FDR≤0.05) in the same direction. We then computed a mean effect score in order to rank genes by computing the weighted mean of the log fold change between the two conditions weighted by the log cpm across all significant shRNAs and targeting a particular gene with an effect in the same direction. If an equal number of shRNAs showed a significant effect in positive or negative direction, we classified the gene as not significantly affected. Otherwise we chose the effect direction based on the majority of the shRNAs. We then combined the results from the HES5+ to 24h control and HES5- comparison into one by taking the maximum mean effect score observed in either comparison. The resulting mean effect scores are then used for visualization and analysis purposes in main text and figures and are reported in Supplementary Table 3. In addition, we also calculated an empirical FDR by determining the fraction of shRNAs with a statistical significant effect based on the generalized linear model but were not expressed based on the RNA-Seq data for the condition where the significant effect was observed.
For the TERA validation analysis, we ranked all motifs according to their TERA scores at each stage. Next, we filtered out motifs that were not associated with at least one TF that was covered in our screen design. We then determined the fraction of top 20 motifs (by absolute TERA values) that were linked to TFs which showed a significant effect in the corresponding stage specific shRNA screen. We report this number as the percentage of motifs recovered. Only motif-knockdown results that have a straightforward interpretation were considered as hits. These include: 1. positive TERA score and positive depletion score (gene is involved HES5+ maintenance, progression or cell survival); 2. negative TERA score and negative depletion score (impedes HES5+ maintenance, progression or apoptosis); 3. negative TERA score and positive depletion score (gene is involved HES5+ maintenance, progression or cell survival but most likely acts as a repressor by causing H3K27ac or H3K4me3/1 loss). For the comparison with the expression based analysis, we ranked all significantly differentially expressed genes by their absolute fold change and determined the fraction of top 20 TFs observed among the differentially enriched shRNAs in the screen.
Differential expression analysis
Differential expression analysis was carried out using Cuffidff 2 (ref 32) and genes differentially expressed at a FDR ≤ 0.1 for each comparison and a minimal expression level of 1 FPKM in at least one of the conditions were considered. Clustering analysis was performed using the csCluster() function in the cummeRbund40 package version 2.6.1 (http://compbio.mit.edu/cummeRbund/) with the Jensen-Shannon distance as metric. The number of clusters for the NPC set (ESC, NE, ERG, MRG, LRG) and the differentiated populations (NEdN, ERGdN, MRGdN, LRGdA) was determined as the number of clusters between 10 and 20 with the minimum average silhouette width across all clusters. Subsequently, a pseudocount of 1 was added to all FPKM counts followed by a log2 transformation. The resulting values were used for all further expression analysis.
ChIP-Seq data analysis and normalization
For H3K27ac and H3K4me3 histone marks, the Irreproducible Discovery Rate (IDR) framework41 with a cutoff of 0.1 in combination with the MACS242 peak caller version 2.1 was used to identify peaks taking advantage of both replicates for each condition. For MACS2 peak calling, we used an initial p-value cutoff of 0.01 and the corresponding whole cell extract (WCE) control library as background. All IDR peak sets can be obtained from GEO under GSE62193.
For the broad histone marks H3K27me3 and H3K4me1, we first determined all 1kb tiles of the human genome (hg19) that were significantly enriched over background in at least one of the replicates. To that end we used a Poisson model43 with the WCE as background to model the fragment count distribution in each genomic To that end we defined a nominal p-value for enrichment within a given region i in sample k harboring rik ChIP fragments compared to the WCE control sample l with ril ChIP fragments as P(C≥ rik) where43:
and eil = ril / λl, λk = (region size) × (total number of ChIP fragments in sample k)/(corrected genome size), λl = (region size) × (total number of ChIP fragments in sample l)/(corrected genome size). In order to account for regions with no/minimal WCE read counts due to sampling, we chose eil = max(eil,1). Resulting p-values were adjusted for multiple testing using the Benjamini-Hochberg44 correction and the q-value R package45. Only regions significant at a q-value ≤ 0.05 and with an enrichment level over background ≥ 1.5 were considered to be enriched.
For differential enrichment analysis of histone marks between consecutive conditions, we used the R-package diffBind46. To normalize read counts, we used the effective library size, counting only reads in peak regions (either the IDR peaks for H3K27ac, H3K4me3 or the enriched 1kb tiles for H3K27me3 or H3K4me1). The differential analysis was then conducted using the DBA_DESEQ2 method, taking full advantage of both replicates per condition with the bTagwise parameter set to true. Only regions differentially between consecutive conditions at a p-value of 0.05 were reported.
In addition, we created a union peak set for each mark separately by joining overlapping peaks/enriched regions in preparation for the transcription factor epigenetic remodeling activity (TERA) analysis. For H3K4me1, we computed the enrichment over the union of all H3K27ac regions since we wanted to focus on well more sharply defined promoter and putative enhancer regions for this mark. For H3K27ac, we focused on distal regions only (≥1kb of nearest TSS) since we were specifically interested in putative enhancer regions for this mark. For H3K4me3, we used the union of all H3K4me3 IDR based peaks regardless of distance, accounting for most promoters and CpG islands. We then determined the enrichment level for all regions in the union set in each replicate across all marks separately. Region enrichment was computed as follows: First, the number of tag counts in each region was determined and normalized to reads per kilobase per million reads (RPKM) sequenced using the full library size of non-duplicate reads. Next, RPKM read counts were divided by the mean RPKM counts across all WCE libraries. Subsequently, the resulting enrichment levels were log2 transformed after adding a pseudo enrichment of 1. Finally, the resulting enrichment values were quantile normalized across the entire dataset for each mark separately. The resulting values were then average across replicates to obtain a region x condition normalized enrichment matrix. The resulting matrix was used as input for the TERA analysis. We tested several ChIP normalization strategies by assessing between replicate correlation and between condition discriminative power on a large dataset of 70 REMC H3K27ac samples and identified this strategy as best performing one.
Footprinting detection
To determine small regions depleted of histone modifications but surrounded by regions of much greater enrichment, termed footprints, we extended an approach used for the analysis of DNAse I HS data47. Our footprints identification algorithm consisted of three main phases: In the first phase, we identify peaks using the IDR framework (see previous section) for H3K27ac and H3K4me3 and use these as baseline regions in which footprints could be detected. In the second phase, we identified footprints located within/around peak regions in the following manner:
For each peak, extend by 400 bp from apex in either direction
Split entire resulting region into bins of size 20 bp
Compute number of RPKM counts for a central sliding window across the entire region (shifting by increments of one bin) for different window sizes ranging from two bins to ten bins in increments of one.
For each position of the central window and for each window size, compute the following three quantities: Cij – RPKM count for central window at current position i and window size j, Rij – RPKM count for a 200 bp stretch directly to the right of the central window and Lij – RPKM count for a 200 bp stretch directly to the left of the central window.
- For each resulting position i and window size j compute the depletion score:
With the footprint size normalization factor f = s / b, with s the size of the central window and b the size of the border regions. Identify non-overlapping, non-adjacent footprint candidates starting from small to larger central window sizes and recording footprint candidate iff eij > 0 & eij < 1 & Lij > Cij & Rij > Cij , followed by removing all other potential footprints (central window+borders) of larger size overlapping the current candidate.
Finally, all resulting candidate footprints with a footprinting score eij ≤0.9 were reported.
The latter procedure was carried out for H3K27ac and H3K4me3 independently for each sample. Subsequently, we merged all footprints from individual samples into consensus footprints set for each epigenetic mark separately, collapsing overlapping footprints by taking the union of all regions with non-zero overlap.
DMR detection
DMR detection was carried out as previously described with slight modifications8. Pairwise comparisons of consecutive samples (hESC, NE, ERG, MRG, LRG, LNP) were carried out on a single CpG level using a beta-binomial model and the beta difference distribution requiring a maximum q-value below 0.05 and an absolute methylation difference greater than 0.1. q-values were computed based on beta-binomial model p-values using Benjamini-Hochberg44 method. Only CpGs covered by at least 5 reads in either sample were considered. Subsequently, differentially methylated CpGs within 500 bp were merged into discrete regions. Differential CpGs without neighbors were embedded into a 100 bp region surrounding each CpG. Next, differential methylation analysis was repeated on the region level using a random effects model. Only regions significant at q-value below 0.01, an absolute methylation difference above 0.2 and harboring at least 2 differentially methylated CpGs were considered differentially methylated and used for subsequent analysis. For the DNA methylation analysis in the context of the TERA framework, we restricted our analysis to DMRs consistently covered across all conditions, including those only assessed by RRBS. This left us with 7,929 regions.
Gene set enrichment analysis
Gene set enrichment analysis for genomic regions was carried out using the GREAT toolbox17 and only categories with q-values ≤ 0.05 for both the hypergeometric and the binomial test as well as a minimal region enrichment level greater than 2 were considered, following the GREAT recommendations. Due to the large number of enriched gene sets, a selected subset of the results is shown in the different figures. In addition, we utilized the Allen Brain atlas48 to determine enrichment for distinct brain structures and developmental time points. To that end we derived gene sets from the Brain atlas data in the following fashion:
We obtained in situ hybridization counts for the developing mouse brain at 7 distinct fetal time points and 11 different brain substructures through direct correspondence with alleninstitute.org. Specifically, we investigated the following structures and time points: Rostral secondary prosencephalone (RSP), Telencephalon (Tel), peduncular (caudal) hypothalamus (PHy), Hypothalamus (p3), pre-thalamus (p2), pre-tectum (p1), midbrain (M), prepontine hindbrain (PPH), pontine hindbrain (PH), pontomedullary hindbrain (PMH), medullary hindbrain (MH); and embryonic (E)11.5, E13.5, E15.5, E18.5 as well postnatal P4, P14 and P28. In total, we had 14,585 measurements for 2,105 different genes across these different regions and time points. In order to define sets of genes characteristic for each combination of time point and structure, we computed the z-scores as well as the maximum observed variation for each gene across the entire matrix of structure and developmental time point combinations. Only genes that exhibited a maximum observed variation (maximum activity – minimum activity) ≥ 1 were considered for gene set definition. Next, we mapped all mouse genes to their human orthologs using the biomaRt database. Finally, we defined gene sets for each region-time point combination using genes that exhibited a z-score ≥ 2 in that particular combination. Since the Allen brain atlas gene sets are defined for each developmental time point and regional identity, we next simplified the visualization by focusing either exclusively on structures or developmental time points. Therefore, we determined the gene set with the maximum gene set activity at each differentiation stage across all gene sets associated with distinct developmental time points for each structure separately. Similarly, we determined the gene set with maximum activity for each developmental time point now taking the maximum across all structures at each stage. The gene set activity was determined as the mean log2 transformed expression level of all gene set members in for each condition.
Motif library construction and mapping to transcription factors
We combined the position weight matrices from Transfac professional database49 (2011) with the PWM collection reported in Jolma et al.50, only retaining motifs annotated for homo sapiens or mouse. To eliminate redundant motifs, we determined pairwise motif similarities for all resulting 1,886 PWMs using the TOMTOM51 program which is part of the MEME52 suite with default parameters. Next, we compiled a pseudo-distance matrix based on the resulting pairwise motif similarities. As a proxy for motif similarity, we used the log10 transformed TOMTOM q-value which was capped at 10. To convert the resulting motif similarities into a distance matrix, we inverted the scale by subtracting the transformed q-values from 10. We then used the resulting matrix to perform hierarchical clustering with Euclidean distance and Ward's method. Finally, we employed the cutree() function with a threshold of 7 to partition the resulting clustering dendrogram into discrete clusters of motifs. For each cluster, we then determined the motif with the highest complexity based on the relative entropy compared to a genome background model with the following base frequencies: A=0.2725, C=0.189, G= 0.189,T= 0.2728. Only motifs with a relative entropy greater or equal than 8 were retained for subsequent analysis. After identification of the candidate with the highest complexity for each motif cluster, we assigned all genes mapping to any motif in each corresponding cluster to the cluster representative motif. This lead to a final motif list of 557 motifs. In order to obtain a more quantitative association of each motif with its linked genes, we computed the ETFA scores across 70 REMC H3K27ac or H3K4me3 cell types and correlated the results with RNA-Seq expression data across 40 cell types. This analysis gave rise to a correlation matrix containing the pearson correlation coefficient of each motif with its linked genes. This matrix was used in combination with the plain gene mapping reported in primary motif sources. For Fig. 2b, we uniquely map each motif to a corresponding linked gene by computing an association score as the product of the absolute pearson correlation coefficient and the average gene expression level of the corresponding gene. We then chose the gene with the highest association score. For motifs without an entry in the H3K27ac correlation matrix (due to the inability to determine suitable GEV parameters on the REMC dataset), we chose the gene with the highest gene expression level. In Fig. 2b, only genes expressed with at least 10 FKPM in the respective condition are considered. We then report the top 35 genes for each condition, where TERA scores of motifs mapping the same gene were averaged.
In Fig. 4 and 5, we incorporate the results of the shRNA screen to uniquely map motifs apply the aforementioned mapping strategy only on the genes identified as hits. If it does not map to any gene hit by the screen, we use the standard assignment strategy outlined above.
Identification of putative transcription factor binding sites
In order to determine putative binding sites in a given genomic region, we used a biophysical model of transcription factor affinities to DNA53,54 to determine putative binding to our footprint sets. This biophysical model requires the training of generalized extreme value (GEV) distributions of binding affinities based on a PWM matrix for each transcription factor and each set of genomic regions in order to generate a suitable background model. In order to take the distinct properties of footprints determined from different epigenetic marks, we determined the GEV parameters for footprints arising from H3K27ac, H3K4me3 and DNAme using the framework outlined by Manke et al.53,54. The resulting three binding matrices were then filtered for minimal significant binding affinity at p-values below 0.05. All other entries with higher p-values were set to one. Next, we took the negative log10 of the entire matrix as a quantitative measure of binding affinity in subsequent analysis.
Inference of transcription factor activities based on epigenetic data
In order to infer transcription factor epigenetic remodeling activities (TERA), we first computed epigenetic transcription factor activities (ETFA) from our epigenetic data. To that end, we first focused on motif activity analysis and associated each motif in a second step with its corresponding transcription factor. For each epigenetic mark, we used the normalized epigenetic enrichment scores as well as DMRs with a minimal DNA methylation difference of at least 0.2 and covered consistently in all datasets. For the DNA methylation data, we inverted the scale to obtain de-methylation scores (1=fully de-methylated, 0=fully methylated) since usually the de-methylated states coincides with gene regulatory element activity. To determine the unobserved activity of a transcription factor binding motif, we took advantage of recent developments in the microarray field55,56 and adapted this approach to epigenetic data. To that end we modeled the enrichment level yit of a particular epigenetic mark at genomic region i and time point t as a linear function the unknown transcription factor activities. Considering p predictor variables (epigenetic motif/transcription factor activities -ETFA) and k time points we describe the unknown TFA X as a p × k matrix. Incorporating all regions n meeting the above listed criteria, we employ the linear model:
With the observed matrix of epigenetic enrichment scores Y (n × k), a constant offset matrix A (n × k), the connectivity matrix B (n × p), describing the filtered binding affinities for all transcription factor motifs to all regions and an error term matrix E. Subsequently, we followed the approach outlined by Boulesteix and Strimmer55 and applied partial least square (PLS) regression and specifically the SIMPLs algorithm57 to determine the unknown transcription factor motif activities. The idea in PLS is to employ a linear dimensionality reduction
where the p predictors in X are mapped onto c ≤ rank(X) ≤ min(p,n) latent components T (n × c matrix) and to compute the weight matrix R not only based on the data matrix B but explicitly taking into account the response matrix Y. The latter strategy maximizes predictive power even for a small number of latent components.
In order to determine the number of latent components for each epigenetic mark and genomic context, we performed cross validation by randomly partitioning the dataset 20 times into 2/3 training and 1/3 test set. We then chose the number of components such that it minimized the prediction error. The corresponding analysis methodology was implemented in the statistical programming language R adapting the implementation provided by Boulesteix and Strimmer55. To assess the significance of the resulting ETFA scores, we perform a permutation test by randomly permuting the epigenetic enrichment scores for each gene regulatory element and recomputed the ETFA values on the permuted values. This process is repeated 100 times. Positive ETFA scores are considered to be insignificant and set to 0 if a greater ETFA score is observed more than once on the randomly permuted set and vice versa for negative ETFA scores.
Finally, we determined the TERA scores by computing the differential ETFA scores between consecutive conditions. These scores were determined by subtracting ETFA scores of consecutive time points from each other. Subsequently, the significance of this difference using a permutation test by randomly permuting the epigenetic enrichment scores across all regions, recomputing the ETFA scores for each conditions and assessing the TERA score between consecutive conditions for each motif. Positive TERA scores are considered to be insignificant and set to 0 if a greater TERA score is observed more than once on the randomly permuted set and vice versa for negative TERA scores.
Co-binding analysis
Co-binding relationships were evaluated using an empirical approach with the entire set of footprints for each epigenetic mark as background. For a given factor i, we determined the footprints set Fi relevant for the current comparison (e.g. changing their epigenetic state in particular cell state transition) that were predicted to harbor a TFBS based on the binding model outlined above. Next, we computed the frequency of motif co-occurrence sFij across Fi for all other motifs j in our database. To generate a proper null distribution, we randomly sampled K = 100 size standardized footprint sets Gk of cardinality |Fi| from the entire footprint collection for the epigenetic mark under study and computed the same test statistic sGkij on these sets. Finally, we determined an empirical p-value and odds ratio based on these quantities by counting the number of instances for which sGkij ≥ sFij :
Only co-binding relationships significant at p-value ≤ 0.01 were retained.
Validation analysis on ENCODE data
To validate the outlined strategy in silico we took advantage of publically available transcription factor ChIP-Seq data in four cell lines from the ENCODE58 project as well as H3K27ac and RNA-Seq data for 70 cell types from the REMC project. We downloaded H3K27ac data as well as processed transcription factor binding data from the ENCODE project for the cell line K562 since abundant transcription factor binding data based on ChIP-Seq was available. In addition, this dataset has been successfully used in several studies to benchmark TF binding predictions59,60. We then applied our TERA-pipeline to the H3K27ac datasets and computed the TF-binding affinities for a set of 557 distinct motifs. With these datasets at hand, we computed the true positive rate (TPR), the false positive rate (FPR) and the positive predictive values (PPV) for all transcription factors that could be matched to at least one motif with available binding affinities (46/117). In the event that one factor matched multiple motifs, we chose the motif with the highest AUC.
GWAS analysis
The GWAS analysis was conducted using 11,027 GWAS SNPs from the GWAS catalog (August 2013). For each footprint set, we sampled K=100 randomly selected, H3K27ac footprints determined across 57 epigenome roadmap datasets processed in the same fashion as our neural dataset. Next, we determined the overlap with GWAS SNPs for control and neural H3K27ac footprint sets. Subsequently, we computed an empirical p-value for each trait/disease i in the catalog by determining the number of trait associated SNPs sCij overlapping with each control region set Cj and the number overlapping with the corresponding footprint set si according to
Determination of core network
The core network was defined as those transcription factors that were differentially expressed during neural induction from ES cell to NE and not differentially expressed between consecutive stages of NE, ERG and MRG. We did not consider the LRG stage. Furthermore, we required that each factor was expressed at least 10 FPKM or more in NE, ERG and MRG and that it's mean normalized, maximum difference in expression levels between any of the stages did not exceed one standard deviation computed across the entire dataset of 9 cell types.. In addition, we also considered genes that were not differentially expressed between any consecutive stages including the ESC stage but fulfilled all other criteria. This identification procedure gave rise to the candidate list of core factors. We then intersected this list with the results of our shRNA screen and retained only those factors that were significantly depleted in the HES5+ population relative to the respective HES5- or control population in at least two stages. Since the literature supported a role for PAX6 and OTX2 for which our shRNAs showed no effect due to the pooled setup or absent knockdown (Fig. 3f), we included these genes as well. Finally, we merged this list will all TFs that were depleted in our shRNA screen at all 3 stages in the HES5+ population relative to the controls and were expressed at least at 10 FPKM or more in NE, ERG and MRG. This algorithm yielded a list of 22 transcription factors or epigenetic modifiers (Fig. 4a). We then carried out co-binding analysis in H3K27ac footprints dynamically regulated at each stage in order to obtain putative stage specific co-binding relationships. To determine significant co-binding events, we used the permutation procedure outlined above and retained all co-binding partners with an odds-ratio ≥ 1.5 that were significant at p≤0.01 that were also identified as a significant hit in the shRNA screen at the particular stage under investigation.
Transcription factor binding site priming analysis
To determine transcription factors associated with transcription factor binding site priming prior to factor activation, we determined all transcription factors at each stage that were significantly up-regulated at the consecutive NPC time point or induced in the corresponding more differentiated cell type (q-value≤0.1) and showed an increase in H3K4me1 or DNAme derived TERA activity at the current stage under investigation. In addition, we required that the corresponding motif did not map to any TF that was expressed more than 2 FPKM at the current stage under investigation. From this list, we picked the pro-neural genes NEUROD4, ASCL2 andNFIX for further investigation due to their literature support for their pro-neural functions. Finally, we required that the potential downstream target genes were significantly enriched for differentially regulated genes at the next NPC stage or in the corresponding more differentiated cell types. To that end, we determined all putative transcription factor binding sites for a particular factor in dynamically regulated H3K27ac or H3K4me1 footprints at the stage of potential priming. We then associated each of these putative binding sites with the nearest TSS and determined the number of differentially expressed genes for each factor. To assess significance, we randomly drew 100 sets of equally sized H3K27ac footprints with no motif of the factor under investigation and determined the number of differentially expressed genes for the subsequent stages. Only factors that exhibited more differentially expressed genes compared to the control sets in more than 99 % of the cases were retained.
Next, we performed co-binding analysis in H3K27ac peaks differentially regulated between the ES cell and NE stage as outlined above and display the top 10 co-binding relationships per factor with an odds-ratio ≥ 1.5 that were significant at an permutation test based p≤0.01 in Fig. 5a.
Extended Data
Extended Data Fig. 1 related to Fig. 1. Isolation and characterization of ES cell derived neural progenitor cells.
a. Schematic of our differentiation model including the specific days of sample collection. Human ES cells were differentiated into neuroepithelial (NE) cells using dual inhibition of TGFb and BMP followed by the transition to neural base media. Subsequently, sonic hedgehog and FGF8, are used to transition to the early radial glial stage (ERG). For the rest of the differentiation experiment the cells were constantly maintained in FGF2 and EGF2 neural base media to reach the mid radial glia (MRG) stage after 35 days, the late radial glia (LRG) stage after 80 and the long term neural progenitor (LNP) stage after about 200 days of in vitro culture. Cell type names indicated in red were profiled for gene expression, histone modifications as well as DNAme by WGBS, while names shown in grey for gene expression only and names in black for DNAme by RRBS only.
b. Hierarchical clustering for all RNA-Seq datasets collapsing replicates using the Jensen-Shannon divergence as metric.
c. Gene expression patterns shown as z-scores for all differentially expressed genes (q-value≤ 0.1) across ES cells and four neural precursor differentiation stages for genes expressed with at ≥ 2 FPKM in at least one stage (n=20,306). Genes were grouped into 18 clusters based on minimal average silhouette width using PAM clustering and Jensen-Shannon divergence based metric. Pie charts below indicate fraction of up (red) and down-regulated (green) genes during each transition.
e. Gene expression patterns shown as z-scores for all significantly differentially expressed genes (q-value≤ 0.1) across four more mature cell populations obtained through differentiation of NE, ERG or MRG cells to neuronal like cells (NE/ERG/MRGdN) and astrocyte like cells (LRGdA) derived from the LRG stage. Genes were grouped into 12 clusters based on minimal average silhouette width using PAM clustering and Jensen-Shannon divergence based metric.
Extended Data Fig. 2 related to Fig. 2. Epigenetic dynamics and TF footprints.
a. Median TPR (red), FPR (blue) and PPV (black) for n=46 TFs with matching motif for H3K27ac footprints (n=27,292) in K562 cells as a function of confidence in predicted binding (-log10 p-value). True positives were defined as predicted binding events overlapping with peaks determined by ChIP-Seq and false positives accordingly. The entire set of positives was defined as all TF ChIP-Seq peaks for a particular factor that overlapped with any H3K27ac footprint.
b. ROC curve of the median TPR/FPR values from a.
c. Epigenetic dynamics across the APOE locus (chr19:45,391kb – 45,414kb) for ES cells and three stages of the NPCs. H3K4me3 read counts normalized to 1 million reads are shown on a scale of 0 to 2 (green). DNAme levels for single CpGs are indicated as blue dots on a scale of 0 to 100% of methylation (y-axis). H3K27ac read counts normalized to 1 million reads are shown on a scale of 0 to 1 (purple). For reference footprints (FP) and CpG islands (CGIs) are indicated as blue boxes (bottom). Shaded gray box indicates the position of the putative enhancer element overlapping with the Alzheimer related SNP rs157580.
d. Top: Decomposition of H3K27ac dynamics into 7 distinct modules based on PLS regression. Colors indicate median epigenetic enrichment level of gene regulatory elements assigned to each module for each cellular state for H3K27ac. Bottom: Gene set enrichment analysis results for gene regulatory elements associated with each module.
e. Connectivity matrix showing the association strength of each of the factors listed in Fig. 2b with each of the 7 modules identified by the partial least square (PLS) regression.
Extended Data Fig. 3 related to Fig. 3. Functional validation using a pooled shRNA screen.
a. Detailed outline of the pooled shRNA screen. Each stage (NE, ERG and MRG) was infected with an optimized virus titer aiming for an average of one shRNA integration per cell. Immediately after infection, cells were subjected to puromycin (puro) selection and bulk population material was collected 24h after infection and prior to efficient shRNA knockdown. Five days after infection and selection, cells were FACS sorted for HES5-GFP and both GFP+ and GFP- were collected for analysis. Subsequently, genomic DNA was extracted and all integrated shRNAs were amplified by PCR for each population separately. The resulting material was then used to construct libraries for next generation sequencing to count the number of shRNA integrations for each shRNA in each cell population.
b. Overlap of genes identified to facilitate HES5+ cell maintenance, progression or proliferation determined by genes with at least two shRNAs significantly (q≤0.05) overrepresented in the HES5+ population with respect to the 24h or HES5- control.
c. Regulator predictions based on differential gene expression. Performance is measured as percentage of the top 20 differentially expressed factors for each stage linked to the TF included in the shRNA library.
d. Regulator predictions based on TERA ranking for H3K4me3, H3K4me1, H3K27ac or DNAme. Performance is measured as percentage of the top 20 predicted activating or repressive motifs for each stage mapping to a TF included in the shRNA library.
e. Detailed heatmap showing the top 20 predicted motifs and corresponding TFs differentially active between the ES cell and NE stage based on the combined TERA scores for H3K27ac, H3K4me3, H3K4me1 and DNAme. In addition, knockdown results as depletion scores (green-red heatmap) obtained at each stage are shown on the right.
f. Heatmap showing the pairwise pearson-correlation coefficient (PCC) of the log2 read-count normalized shRNA libraries across all conditions and replicates.
g. Individual validation for shRNAs against OTX2 and PAX6 at the NE stage, which showed no effect in our pooled screening approach at any stage. Shown are qPCR levels for OTX2 or PAX6, HES5 and Puromycin relative to HPRT. Each gene was measured in an independent knockdown experiment for a pool of the 5 shRNAs against PAX6 (blue), OTX2 (green), lacZ (orange) as well as the uninfected control (red).
Extended Data Fig. 4 related to Fig. 4. Co-binding analysis.
a. Gene expression levels reported as z-scores for core network TFs and epigenetic modifiers with and without a known DNA binding motif.
b. Illustration of predicted significant co-binding relationships (p≤0.01, odds ratio≥1.5) of core factors (rows) with more stage-specific or proneuronal/glial factors (columns). Color-coding indicates whether binding is stage-specific or occurs at multiple stages.
c. Overlap of predicted binding sites in dynamic putative enhancer regions based on H3K27ac for OTX2 in NE and ERG.
d. Gene set enrichment analysis results for predicted OTX2 binding sites in dynamic putative enhancer regions at the NE and MRG stage.
Extended Data Fig. 5 related to Fig. 5. Epigenetic priming.
a. TERA scores for H3K27ac, H3K4me3, H3K4me1 and DNAme for TFs showing evidence of priming (top bold) and TFs predicted to significantly co-occur in these primed binding sites.
b. Gene expression levels shown as z-scores for primed and co-binding TFs from panel a.
c. Detailed predicted co-binding relationship (p≤0.01, odds ratio≥1.5) of primed TFs (columns) with significantly associated co-binding factors (rows).
d. Illustration of a potential priming event and the associated predicted target gene at the ATOH1 locus (chr4:94,740-94,800). For each stage, H3K27ac, H3K27me3 and DNAme patterns are shown along with predicted NEUROD binding sites (black boxes) in putative gene regulatory elements marked by a loss of DNAme (highlighted by the grey bars).
e. Gene set enrichment analysis results for predicted NEUROD binding sites split up by dynamic patterns defined in Fig. 5b top. Binding sites in patterns 3 and 4 showed no significant enrichment.
Supplementary Material
Acknowledgments
We would like to thank all members of the Meissner and Elkabetz laboratories; we also thank Fontina Kelley and other members of the Broad Sequencing Platform, John Doench and members of the Genome Perturbation Platform at the Broad Institute, Dan-Avi Landau for critical reading of the manuscript, as well as to Irena Shur and Orit Sagi-Assif at Tel Aviv University for their extensive FACS operation. We also thank Leslie Gaffney for graphical support. This work was funded by the NIH Common Fund (U01ES017155), NHGRI (HG006911), NIGMS (P01GM099117), the New York Stem Cell Foundation, the Israel Science Foundation (ISF1126/10) and a Marie Curie International Reintegration Grant (IRG277151). A.M. is a New York Stem Cell Foundation Robertson Investigator.
Footnotes
Author contributions:
The study was designed by M.J.Z, Y.E. and A.M. R.E. Y.Y and Y.E. developed the NPC system, performed consecutive cell isolation, propagation and differentiation and conducted the shRNA screen. M.J.Z performed the analysis and designed shRNA screen. W.M and J.R. helped with RNA-Seq data processing and analysis. J.D. performed TF ChIP-Seq experiments. R.P. and C.G. performed RNA-Seq library construction. R.P and D.C. performed shRNA library construction. T.S.M. provided experimental advice. R.I. J.X. and A.G. conducted histone ChIP-Seq experiments. H.G. performed WGBS and RRBS library construction, A.G. and A.M. supervised the DNA methylation profiling, C.E. and B.E.B. provided experimental input and advice for the ChIP-Seq experiments. A.T. provided the TF-ChIP Seq protocol. O.K. assisted in design of analytical methods. M.J.Z, Y.E. and A.M. interpreted the data and wrote the manuscript.
All data are available through GEO (GSE62193), the NIH Roadmap (http://www.roadmapepigenomics.org/data) and NCBI Epigenomics portal (http://www.ncbi.nlm.nih.gov/epigenomics). The authors declare no competing financial interests.
References
- 1.Imayoshi I, Sakamoto M, Yamaguchi M, Mori K, Kageyama R. Essential roles of Notch signaling in maintenance of neural stem cells in developing and adult brains. J Neurosci. 2010;30:3489–3498. doi: 10.1523/JNEUROSCI.4987-09.2010. doi:10.1523/JNEUROSCI.4987-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shimojo H, Ohtsuka T, Kageyama R. Dynamic expression of notch signaling genes in neural stem/progenitor cells. Frontiers in neuroscience. 2011;5:78. doi: 10.3389/fnins.2011.00078. doi:10.3389/fnins.2011.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carlen M, et al. Forebrain ependymal cells are Notch-dependent and generate neuroblasts and astrocytes after stroke. Nature neuroscience. 2009;12:259–267. doi: 10.1038/nn.2268. doi:10.1038/nn.2268. [DOI] [PubMed] [Google Scholar]
- 4.Edri R, et al. Analyzing human neural stem cell ontogeny by consecutive isolation of Notch active neural progenitors. Under Review (Nature Comms.) 2014 doi: 10.1038/ncomms7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Placantonakis DG, et al. BAC transgenesis in human embryonic stem cells as a novel tool to define the human neural lineage. Stem Cells. 2009;27:521–532. doi: 10.1634/stemcells.2008-0884. doi:10.1634/stemcells.2008-0884. [DOI] [PubMed] [Google Scholar]
- 6.Voss TC, Hager GL. Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat Rev Genet. 2014;15:69–81. doi: 10.1038/nrg3623. doi:10.1038/nrg3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ziller MJ, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500:477–481. doi: 10.1038/nature12433. doi:10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gifford CA, et al. Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. Cell. 2013;153:1149–1163. doi: 10.1016/j.cell.2013.04.037. doi:10.1016/j.cell.2013.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maurano MT, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. doi:10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Claussnitzer M, et al. Leveraging cross-species transcription factor binding site patterns: from diabetes risk loci to disease mechanisms. Cell. 2014;156:343–358. doi: 10.1016/j.cell.2013.10.058. doi:10.1016/j.cell.2013.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gotz M, Stoykova A, Gruss P. Pax6 controls radial glia differentiation in the cerebral cortex. Neuron. 1998;21:1031–1044. doi: 10.1016/s0896-6273(00)80621-2. [DOI] [PubMed] [Google Scholar]
- 12.Hanashima C, Li SC, Shen L, Lai E, Fishell G. Foxg1 suppresses early cortical cell fate. Science. 2004;303:56–59. doi: 10.1126/science.1090674. doi:10.1126/science.1090674. [DOI] [PubMed] [Google Scholar]
- 13.Martinez-Barbera JP, et al. Regionalisation of anterior neuroectoderm and its competence in responding to forebrain and midbrain inducing activities depend on mutual antagonism between OTX2 and GBX2. Development. 2001;128:4789–4800. doi: 10.1242/dev.128.23.4789. [DOI] [PubMed] [Google Scholar]
- 14.Pevny LH, Sockanathan S, Placzek M, Lovell-Badge R. A role for SOX1 in neural determination. Development. 1998;125:1967–1978. doi: 10.1242/dev.125.10.1967. [DOI] [PubMed] [Google Scholar]
- 15.Chambers SM, et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. doi:10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uittenbogaard M, Chiaramello A. Expression of the bHLH transcription factor Tcf12 (ME1) gene is linked to the expansion of precursor cell populations during neurogenesis. Brain Res Gene Expr Patterns. 2002;1:115–121. doi: 10.1016/s1567-133x(01)00022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McLean CY, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. doi:10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sims D, et al. High-throughput RNA interference screening using pooled shRNA libraries and next generation sequencing. Genome Biol. 2011;12:R104. doi: 10.1186/gb-2011-12-10-r104. doi:10.1186/gb-2011-12-10-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blackshear PJ, et al. Graded phenotypic response to partial and complete deficiency of a brain-specific transcript variant of the winged helix transcription factor RFX4. Development. 2003;130:4539–4552. doi: 10.1242/dev.00661. doi:10.1242/dev.00661. [DOI] [PubMed] [Google Scholar]
- 20.Zarbalis K, et al. A focused and efficient genetic screening strategy in the mouse: identification of mutations that disrupt cortical development. PLoS Biol. 2004;2:E219. doi: 10.1371/journal.pbio.0020219. doi:10.1371/journal.pbio.0020219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou C, Tsai SY, Tsai MJ. COUP-TFI: an intrinsic factor for early regionalization of the neocortex. Genes Dev. 2001;15:2054–2059. doi: 10.1101/gad.913601. doi:10.1101/gad.913601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faedo A, et al. COUP-TFI coordinates cortical patterning, neurogenesis, and laminar fate and modulates MAPK/ERK, AKT, and beta-catenin signaling. Cereb Cortex. 2008;18:2117–2131. doi: 10.1093/cercor/bhm238. doi:10.1093/cercor/bhm238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piper M, et al. NFIA controls telencephalic progenitor cell differentiation through repression of the Notch effector Hes1. J Neurosci. 2010;30:9127–9139. doi: 10.1523/JNEUROSCI.6167-09.2010. doi:10.1523/JNEUROSCI.6167-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qureshi IA, Gokhan S, Mehler MF. REST and CoREST are transcriptional and epigenetic regulators of seminal neural fate decisions. Cell Cycle. 2010;9:4477–4486. doi: 10.4161/cc.9.22.13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elkabetz Y, et al. Human ES cell-derived neural rosettes reveal a functionally distinct early neural stem cell stage. Genes Dev. 2008;22:152–165. doi: 10.1101/gad.1616208. doi:10.1101/gad.1616208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garber M, et al. A high-throughput chromatin immunoprecipitation approach reveals principles of dynamic gene regulation in mammals. Molecular cell. 2012;47:810–822. doi: 10.1016/j.molcel.2012.07.030. doi:10.1016/j.molcel.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Engreitz JM, et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341:1237973. doi: 10.1126/science.1237973. doi:10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo B, et al. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci U S A. 2008;105:20380–20385. doi: 10.1073/pnas.0810485105. doi:10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strezoska Z, et al. Optimized PCR conditions and increased shRNA fold representation improve reproducibility of pooled shRNA screens. PLoS One. 2012;7:e42341. doi: 10.1371/journal.pone.0042341. doi:10.1371/journal.pone.0042341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boyle P, et al. Gel-free multiplexed reduced representation bisulfite sequencing for large-scale DNA methylation profiling. Genome Biol. 2012;13:R92. doi: 10.1186/gb-2012-13-10-r92. doi:10.1186/gb-2012-13-10-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. doi:10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trapnell C, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nature Biotechnology. 2013;31:46. doi: 10.1038/nbt.2450. doi:Doi 10.1038/Nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature protocols. 2012;7:562–578. doi: 10.1038/nprot.2012.016. doi:10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xi Y, Li W. BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics. 2009;10:232. doi: 10.1186/1471-2105-10-232. doi:10.1186/1471-2105-10-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. doi:gr.078212.108 [pii] 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. doi:10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2012 doi: 10.1093/bib/bbs017. doi:10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dai Z, et al. shRNA-seq data analysis with edgeR. F1000Res. 2014;3:95. doi: 10.12688/f1000research.4204. doi:10.12688/f1000research.3928.110.12688/f1000research.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smyth GK. In: Bioinformatics and computational biology solutions using R and Bioconductor Statistics for biology and health. Gentleman Robert., editor. Springer Science+Business Media; 2005. [Google Scholar]
- 40.Goff L, Trapnell C, Kelley D. cummeRbund: Analysis, exploration, manipulation, and visualization of Cufflinks high-throughput sequencing data. 2012 < http://compbio.mit.edu/cummeRbund/>.
- 41.Li QH, Brown JB, Huang HY, Bickel PJ. Measuring Reproducibility of High-Throughput Experiments. Annals of Applied Statistics. 2011;5:1752–1779. doi:Doi 10.1214/11-Aoas466. [Google Scholar]
- 42.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. doi:10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mikkelsen TS, et al. Comparative epigenomic analysis of murine and human adipogenesis. Cell. 2010;143:156–169. doi: 10.1016/j.cell.2010.09.006. doi:10.1016/j.cell.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met. 1995;57:289–300. [Google Scholar]
- 45.Alan Dabney J. D. S. a. w. a. f. G. R. W. Q-value estimation for false discovery rate control. R package version 1.34.0 [Google Scholar]
- 46.Ross-Innes CS, et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481:389–393. doi: 10.1038/nature10730. doi:10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Neph S, et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012;489:83–90. doi: 10.1038/nature11212. doi:10.1038/nature11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thompson CL, et al. A high-resolution spatiotemporal atlas of gene expression of the developing mouse brain. Neuron. 2014;83:309–323. doi: 10.1016/j.neuron.2014.05.033. doi:10.1016/j.neuron.2014.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fogel GB, et al. A statistical analysis of the TRANSFAC database. Bio Systems. 2005;81:137–154. doi: 10.1016/j.biosystems.2005.03.003. doi:10.1016/j.biosystems.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 50.Jolma A, et al. DNA-binding specificities of human transcription factors. Cell. 2013;152:327–339. doi: 10.1016/j.cell.2012.12.009. doi:10.1016/j.cell.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 51.Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS. Quantifying similarity between motifs. Genome Biol. 2007;8:R24. doi: 10.1186/gb-2007-8-2-r24. doi:10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–373. doi: 10.1093/nar/gkl198. doi:34/suppl_2/W369 [pii]10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Manke T, Roider HG, Vingron M. Statistical modeling of transcription factor binding affinities predicts regulatory interactions. PLoS Comput Biol. 2008;4:e1000039. doi: 10.1371/journal.pcbi.1000039. doi:10.1371/journal.pcbi.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Manke T, Heinig M, Vingron M. Quantifying the effect of sequence variation on regulatory interactions. Human mutation. 2010;31:477–483. doi: 10.1002/humu.21209. doi:10.1002/humu.21209. [DOI] [PubMed] [Google Scholar]
- 55.Boulesteix AL, Strimmer K. Predicting transcription factor activities from combined analysis of microarray and ChIP data: a partial least squares approach. Theoretical biology & medical modelling. 2005;2:23. doi: 10.1186/1742-4682-2-23. doi:10.1186/1742-4682-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boulesteix AL, Strimmer K. Partial least squares: a versatile tool for the analysis of high-dimensional genomic data. Brief Bioinform. 2007;8:32–44. doi: 10.1093/bib/bbl016. doi:10.1093/bib/bbl016. [DOI] [PubMed] [Google Scholar]
- 57.Dejong S. Simpls - an Alternative Approach to Partial Least-Squares Regression. Chemometr Intell Lab. 1993;18:251–263. doi:Doi 10.1016/0169-7439(93)85002-X. [Google Scholar]
- 58.Bernstein BE, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. doi:10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sherwood RI, et al. Discovery of directional and nondirectional pioneer transcription factors by modeling DNase profile magnitude and shape. Nat Biotechnol. 2014;32:171–178. doi: 10.1038/nbt.2798. doi:10.1038/nbt.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thurman RE, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. doi:10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.