Abstract
Adenosine is an endogenous molecule that regulates many physiological processes via the activation of four specific G-protein-coupled ADORA receptors. Extracellular adenosine may originate either from the hydrolysis of released ATP by the ectonucleotidases or from cellular exit via the equilibrative nucleoside transporters (SLC29A). Adenosine extracellular concentration is also regulated by its successive hydrolysis into uric acid by membrane-bound enzymes or by cell influx via the concentrative nucleoside transporters (SLC28A). All of these members constitute the adenosine signaling pathway and regulate adenosine functions. Although the roles of this pathway are quite well understood in adults, little is known regarding its functions during vertebrate embryogenesis. We have used Xenopus laevis as a model system to provide a comparative expression map of the different members of this pathway during vertebrate development. We report the characterization of the different enzymes, receptors, and nucleoside transporters in both X. laevis and X. tropicalis, and we demonstrate by phylogenetic analyses the high level of conservation of these members between amphibians and mammals. A thorough expression analysis of these members during development and in the adult frog reveals that each member displays distinct specific expression patterns. These data suggest potentially different developmental roles for these proteins and therefore for extracellular adenosine. In addition, we show that adenosine levels during amphibian embryogenesis are very low, confirming that they must be tightly controlled for normal development.
Electronic supplementary material
The online version of this article (doi:10.1007/s11302-014-9431-6) contains supplementary material, which is available to authorized users.
Keywords: Adenosine metabolism, Adenosine receptor, Embryogenesis, Extracellular adenosine, Nucleotide transporter, Xenopus
Introduction
Extracellular adenosine, an endogenous purine nucleoside, is one of the key mediators of the purinergic signaling pathway [1]. Its inhibitor effect on neurotransmitter release was discovered four decades ago [2]. This molecule plays multiple roles in adult physiology and pathophysiology and regulates various processes through the activation of four specific P1 receptors named ADORA (A) 1, 2A, 2B, and 3 [1, 3–6]. These receptors are members of the rhodopsin-like family of G-protein-coupled receptors. ADORA1 and ADORA3 are negatively coupled to adenylate cyclase and their activation inhibits cAMP production, whereas ADORA2A and ADORA2B are positively coupled to adenylate cyclase and elevates cAMP.
Different actors of the purinergic signaling pathway regulate adenosine concentration in the extracellular space. Ectonucleotidases on the cell surface allow the production of adenosine from the sequential degradation of extracellular ATP in ADP and AMP [7]. Four major families exist and work in concert or consecutively. Ecto-nucleoside triphosphate diphosphohydrolase family (NTPDases) includes nine mammalian proteins which can hydrolyze extracellular nucleoside 5’-triphosphate (NTP) and 5’-diphosphate (NDP) in nucleoside monophosphate (NMP) with specific substrate specificity. Seven ecto-nucleotide pyrophosphates/phosphodiesterases (ENPPs) were found in mammals, but only three (ENPP1, 2, and 3) can hydrolyze nucleotides and generate NMP or NDP from NTP, and NMP from NDP. ENPP2 displays a wider catalytic capacity by being able to hydrolyze AMP to adenosine. However, a recent study demonstrated that ENPP4 could generate ADP and ATP from diadenosine polyphosphates [8]. The functional role of each enzyme is very variable and depends on the specific nucleotide or non-nucleotide substrate [9]. Alkaline phosphatases (APs) reveal hydrolysis properties with broad substrate specificity [7]. Several paralogs are expressed in vertebrate and their number varies between species. In human, four APs have been characterized and named according to their predominant distribution in adult tissues. ALPL or TNAP (tissue-non-specific AP) is ubiquitous but highly expressed in the liver, bone and kidney, while ALPP or PLAP (placental AP), ALPP2 or GCAP (germ cell AP) and ALPI or IAP (intestinal AP) show a more restricted tissue distribution. APs can sequentially dephosphorylate extracellular ATP, ADP, and AMP to adenosine and are the only ectonucleotidases with this property. The major function of NT5E (ecto-5’-nucleotidase) or CD73 is the hydrolysis of extracellular AMP in adenosine like other ribonucleotidases and deoxyribonucleotidases [7]. Recently, it was demonstrated an ectonucleotidase function to prostatic acid phosphatases (PAP), producing adenosine from extracellular AMP, a minor extent ADP, but not ATP [10]. Four other isoenzymes have been identified: erythrocytic, macrophagic, lysosomal and testicular acid phosphatases [11], but no ectonucleotidase activity have been demonstrated to date. Degradation enzymes hydrolyze successively adenosine in inosine, hypoxanthine, xanthine and finally uric acid that is excreted by the kidney. Adenosine deaminase (ADA) catalyzes the irreversible deamination of adenosine and 2’-deoxyadenosine to inosine and 2’-deoxyinosine, respectively. The (deoxy)inosine is then metabolized into hypoxanthine by the purine nucleoside phosphorylase (PNP) enzyme [12]. These two enzymes display a similar subcellular localization and are expressed in the cytosol but can also be found on the cell surface. Adenosine kinase (ADK) is a cytosolic enzyme which catalyzes the phosphorylation of adenosine to AMP + ADP, using ATP [13].
With adenosine being a hydrophilic molecule, specific nucleotide transporters facilitate its movement across the cell membranes and are indispensable for its recycling into the cell [14, 15]. Equilibrative nucleoside transporters (SLC29A or ENTs) mediate the transport of adenosine and other nucleosides in both directions, according to the nucleoside gradient concentration. Four SLC29A were distinguished: SLC29A1, 2, and 3 (ENT1, 2, and 3) are selective for purine and pyrimidine nucleosides and SLC29A4 (ENT4) is specifically selective for adenosine. Concentrative nucleoside transporters (SLC28A or CNTs) mediate nucleoside uni-directional transport, coupled with sodium ion gradient. Three different transporters have been described: SLC28A1 and 3 (CNT1 and CNT3) are selective for pyrimidine nucleosides and for adenosine or purines, respectively, and SLC28A2 (CNT2) is selective for purine nucleosides and uridine.
The roles of extracellular adenosine in adult are quite well known, especially in the modulation of the immune and nervous systems, although other organ physiology, such as the kidney and heart, is also regulated by this purine. Adenosine regulates both innate and adaptive immune system cell functions [5]. Indeed, the accumulation of extracellular adenosine drives a strong inhibition of T cell proliferation and immunodeficiency, as observed in the severe combined immunodeficiency disease (SCID) due to adenosine deaminase deficiency [16]. In the nervous system, adenosine has a pervasive and generally inhibitory effect on neuronal activity and modulates neuronal functions and normal behavior mainly through ADORA1 and ADORA2A receptor activation [17]. Its actions generally antagonize those induced by ATP, as in the sleep/wake cycle. Aberrant adenosine signaling has been implicated in several neuropathological conditions ranging from degenerative neuronal diseases to psychiatric disorders and also in non-neuronal pathologies such as cancer [18–22]. However, neuroprotective action of adenosine has been demonstrated in neuroinflammation, epileptic seizures, ischemia via the activation of the ADORA1 receptor, and also in neurodegenerative diseases via the inhibition of ADORA2A receptor [17, 21–23]. Therefore, adenosine receptors are now therapeutic targets with several ligands being in clinical trials or used already in medicine [20, 21]. Furthermore, recent evidences have placed the purinergic signaling pathway as key regulator of stem cell proliferation and differentiation and may open door to novel cell therapy and tissue repair applications [6].
Recent evidence has implicated adenosine into the proliferation of neural progenitors and their differentiation into neurons or oligodendrocytes [3, 24]. However, despite the generation of knockout mice for the ADORA receptors or the enzymes involved in adenosine metabolism, their functions during embryonic development have been poorly studied. Nevertheless, modification of adenosine signaling can have powerful effects on development. Indeed, Ada−/− mice die prenatally with profound disturbances in purine metabolism affecting not only the developing immune system but also the renal, neural, skeletal, and pulmonary systems [16]. Moreover, constitutive overexpression of Adora1 in mice caused the development of cardiac dilatation and death within 6 to 12 weeks [25]. Muscle-tissue-specific overexpression of Adora3 in mice is lethal before E8.5 [26]. Furthermore, an excess of exogenous adenosine blocks mouse and starfish development [27, 28]. These data suggest that the adenosine levels must be tightly regulated during embryogenesis. Nevertheless, no comprehensive expression profile for the adenosine signaling members during vertebrate embryogenesis is available, and the implication of adenosine during embryogenesis remains still largely unknown.
Xenopus laevis provides a powerful model organism for the establishment of the roles of a signaling pathway during vertebrate embryogenesis. X. laevis embryo has many advantages, namely the large size, its abundance, and rapid external development. Moreover, this model was used for the first demonstration of the roles of the purinergic signaling pathway during embryogenesis [29]. We have conducted here a comprehensive study of the embryonic expression profile for each actor of the adenosine pathway in the amphibian X. laevis, thus realizing the obligatory step before any in vivo functional analysis. Although we have previously described the embryonic expression profiles of Xenopus entpdase [30] and enpp [31] gene families, the expression of the other purinergic actors remained poorly documented [32–34].
Here, we report the cloning and characterization of thirteen enzymes involved in the metabolism of adenosine, together with nine adora receptors and seven nucleoside transporters in X. laevis. We also present and compare their temporal and spatial expression patterns in frog embryos and their distribution in adult tissues. The identification of these actors in Xenopus tropicalis allowed us to provide an in-depth phylogenic analysis. Furthermore, adenosine concentration was measured during X. laevis embryogenesis. This work is the first study that describes and compares the complete embryonic expression pattern of the actors of the adenosine pathway during the development of a vertebrate model organism.
Materials and methods
Ethic statement
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the European Community. The protocol was approved by the “Comité d’éthique en experimentation de Bordeaux” Nu33011005-A.
Bioinformatics
Sequences were identified on the NCBI and Xenbase databases [35]. Basic Local Alignment Search Tool (BLAST) searches were performed on the NBCI Nucleotide and on the Xenbase X. laevis 7.1 Scaffolds genome databases [36]. Conceptual translation of complementary DNA (cDNA) was performed on the ExPASy internet site using the program Translate Tool (web.expasy.org/translate/). Accession numbers of all sequences used in this study are given in Supplementary Table S1 and in figure legends.
Alignments were performed using the program CLUSTAL W2 [37]. A phylogenetic tree was created on the Phylogeny.fr platform, using the tree builder program “PhyML” using maximum likelihood [38] and the Drosophila melanogaster sequences as root sequences. Synteny maps were generated by comparison of X. tropicalis, Danio rerio, Gallus gallus, Mus musculus, and Homo sapiens genomics context using the NCBI Entrez Gene of each gene.
Embryos culture and dissection
Embryos were obtained by in vitro fertilization of eggs collected in 1× Marc’s Modified Ringer’s (MMR) saline solution, from a hormonally stimulated X. laevis female by adding crushed testis isolated from a killed male. Fertilized eggs were dejellied in 3 % L-cysteine hydrochloride, pH 7.8 (Sigma-Aldrich), and washed several times with 0.1× MMR. Embryos were then cultured to the required stage in 0.1× MMR in presence of 10 μg/mL of gentamycin sulfate. The embryos were staged according to Nieuwkoop and Faber [39]. Dissections of anesthetized embryos were performed in 0.1× MMR using forceps and an eyebrow hair knife.
RT-PCR
RNA extraction from whole or dissected embryos and adult tissues, and cDNA synthesis were performed as described by Massé et al. [31]. For each gene, specific primers were designed on two different exons, in order to discriminate genomic from cDNA amplification (Supplementary Table S2). After optimization of the PCR conditions using a gradient PCR machine (Biorad), RT-PCR products were verified by sequencing (Beckman Coulter Genomics Company). For each experiment, the quantity of input cDNA was determined by equalization of the samples with a constant gene, either odc (ornithine decarboxylase) or ef1α (elongation factor 1α). Linearity of the signal was controlled by carrying out PCR reactions on doubling dilutions of cDNA, illustrated by the triangle in Figs. 4, 5, and 6. Negative controls (−RNA, −RT, and −cDNA) were also performed.
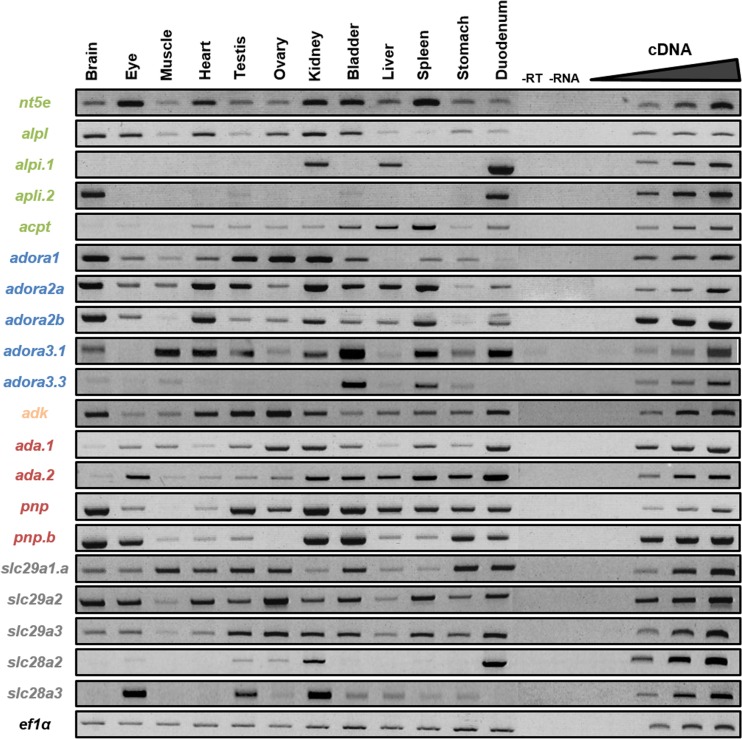
Fig. 4.
Spatial expression profiles of adenosine pathway actors in adult frog. The expression profile of each gene in adult tissues was determined by RT-PCR. Negative controls (−RT, −RNA) were performed. The linearity was performed with doubling dilutions of cDNA from the following tissues: adora 1, 2A, and 2B and slc29a2 with the brain; ada.2 with the eye; NT5E with the heart; slc29a1 and slc28a3 with the testis; pnp with the ovary; alpl, alpi.1, pnp.a, and slc28a2 with the kidney; adora3.1 and 3.3 with the bladder; acpp with the liver; ada.1 and slc29a3 with the spleen; and alpi2 and adk with the duodenum. Ef1α was used as a loading control. Adenosine production enzymes are in green, adenosine receptors are in blue, adenosine degradation enzymes are in orange (intracellular) and red (extracellular), and nucleoside transporters are in gray
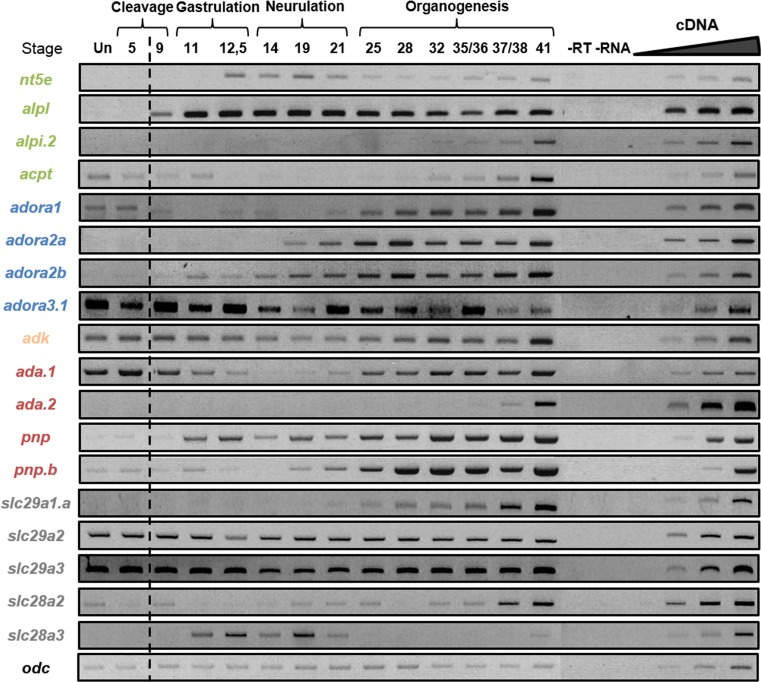
Fig. 5.
Temporal expression profiles of adenosine pathway actors during embryogenesis. The expression profile of each gene was determined by RT-PCR from cDNA of unfertilized egg (Un) and whole embryo at different stages covering the different phases of X. laevis embryogenesis. Negative controls (−RT, −RNA) were performed. Mid-blastula transition is indicated by the dotted line. The linearity was performed with doubling dilutions of cDNA from stages 37–38, except acpt and adk with stage unfertilized; slc28a3 with stage 11; nt5e with stage 14; adora3.1 and slc29a3 with stage 21; slc29a1 and slc29a2 with stage 32; and alpi.2, ada.2, and slc28a2 with stage 41. Odc was used as a loading control. Adenosine production enzymes are in green, adenosine receptors are in blue, adenosine degradation enzymes are in orange (intracellular) and red (extracellular), and nucleosides transporters are in gray
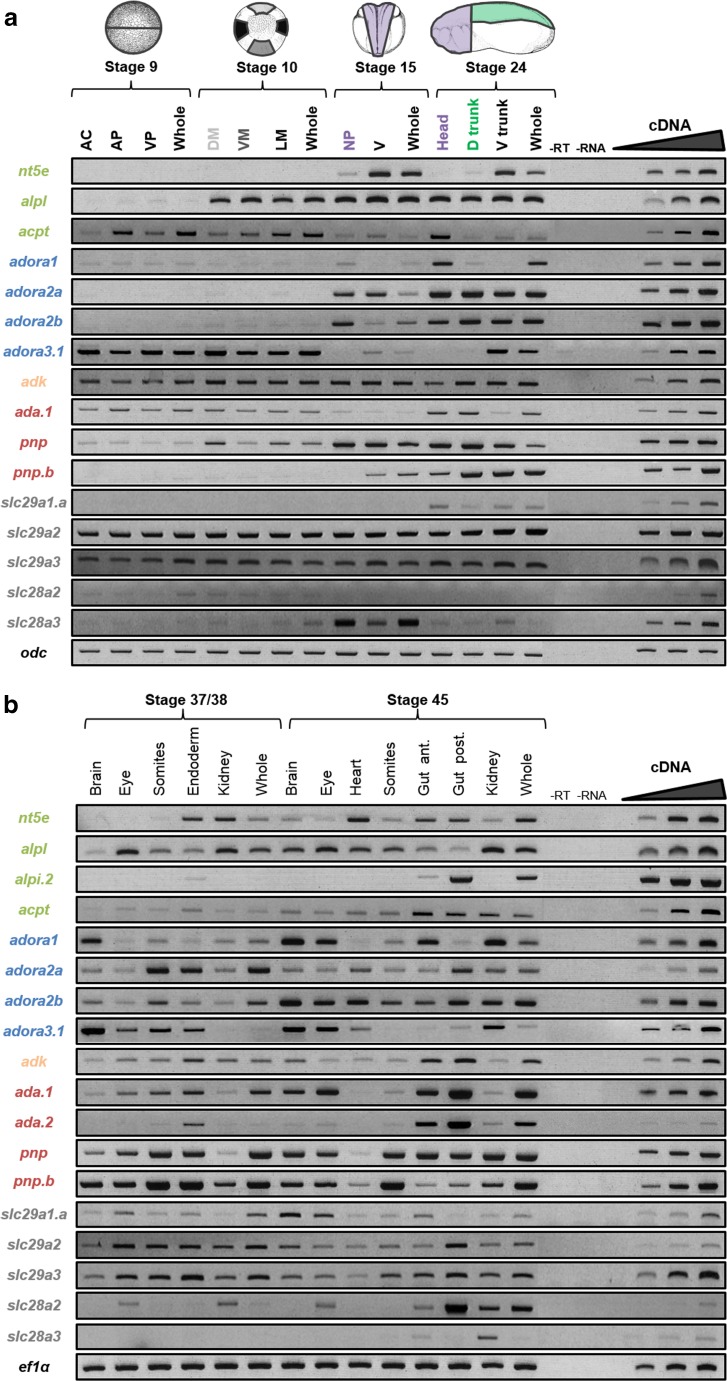
Fig. 6.
Spatio-temporal expression profiles of adenosine pathway actors in dissected embryos. The expression profile of each gene was determined by RT-PCR from cDNA of a dissected tissues of a blastula (stage 9), gastrula (stage 10), neurula (stage 15), and tadpole (stage 24) as illustrated on the schematic embryos, and b dissected tissues of tadpoles (stages 37/38 and 45). Amplification from cDNAs of whole embryos was used as a control. Negative controls (−RT, −RNA) were also performed. Linearity was performed with doubling dilutions of cDNA from the following tissues: (panel a) enpp2B, ADORA3.1, ADA.1, ADK, and SLC28A2 with whole embryo st9; ALPL and ACPT with whole embryo st10; SLC28A3 whole embryo st15; ADORA1 and PNP with head st24; ADORA2B with somites st24; nt5e and ADORA2A with rest st24; and PNP.a, SLC29A1, 2, and 3 with whole embryo st24. (Panel b) SLC29A2 with brain st37; PNP.a with somites st37; nt5e with endoderm st37; ALPL and ADORA2A with kidney st37; ADK and SLC28A2 with whole embryo st37; ADORA1, 2, 3.1 and ADA.1 with brain st45; ADA.2 with eye st45; PNP with somites st45; enpp2B, ACPT and SLC29A1 with anterior gut st45; ALPI.2 and SLC29A3 with posterior gut st45; and SLC28A3 with whole embryo st45. Odc and ef1α were used as loading controls. AC animal cap, AP animal pole, DM dorsal mesoderm, D Trunk dorsal trunk, Gut Ant. anterior gut, Gut Post. posterior gut, LM lateral mesoderm, NP neural plate, VM ventral mesoderm, VP vegetative pole, V Trunk ventral trunk. Color code of adenosine pathway actors: adenosine production enzymes in green, adenosine receptors in blue, adenosine degradation enzymes in orange (intracellular) and red (extracellular), and nucleosides transporters in gray
HPLC
Cellular extracts from embryos were prepared by an ethanol extraction method adapted from Loret et al.[40]. In brief, whole embryos were dropped into 5 ml ethanol/10 mM Hepes, pH 7.2 (4:1 v/v), and incubated at 80 °C for 3 min. Samples were evaporated using a rotavapor apparatus. The residue was resuspended in 500 μL of water and insoluble particles were eliminated by centrifugation and the supernatant was ultra-filtrated on a Nanosep10K® cartridge (Pall). Metabolites were separated on a carbopacPA1 column, linked to an ICS3000 chromatography workstation (Dionex) using a sodium adetate gradient (from 50 to 800 mM) in 50 mM NaOH as described [41]. ATP, ADP, AMP, adenosine, inosine, and hypoxanthine peaks were identified by their retention time as well as by co-injection with standards and/or their UV spectrum signature.
Results
Isolation and sequence characterization of adenosine pathway genes and proteins
Table 1 summarizes the official symbol provided by HGNC together with their synonyms of the different actors, enzymes, receptors, and transporters involved in adenosine signaling pathway. We were able to identify the amphibian orthologs for all the members of this pathway except for the SLC281 and SLC29A4 transporters and the ALPP and ALPPL2 phosphatases (Table 1). The amphibian members involved in adenosine signaling pathways were cloned using different strategies (Supplementary Table S3). The amphibian enpp2A, enpp2B, and pnp sequences have been already published [31, 32]. The majority of the X. laevis and X. tropicalis sequences were available on the Xenbase or NCBI websites (accession numbers given in Supplementary Table S1). However, the remaining sequences were identified by BLASTN on the X. laevis genome version 7.1 databases using the X. tropicalis orthologous sequences as query sequences. Moreover, two X. laevis ada (ada.a and ada.2) and three adora3 (adora.1–3) genes respectively located on the scaffold 14557 and 137317 were identified. These genes are also present in X. tropicalis genome and certainly have arisen from genome duplication. They were numbered according to their positions within their scaffolds. Furthermore, this genomic sequence search allowed us to identify a second gene encoding similar adora1, adora2, ada.1, pnp, slc29a1 and slc29a3 proteins, respectively. These duplicated genes are located on separate scaffolds and must be homeologs, arisen from the X. laevis genome polyploidization event [42]. According to the nomenclature gene guidelines, we tagged these homeologs with .a and .b, with the suffix .a being allocated to the published or identified on Xenbase. The different protein sequences were deduced from conceptual translation of the nucleotide sequences.
Table 1.
Adenosine signaling pathway members studied. The table summarizes the official symbols provided by HGNC (HUGO Gene Nomenclature Committee) of the genes as used on the NCBI website and by the Gene Nomenclature Guidelines from Xenbase website
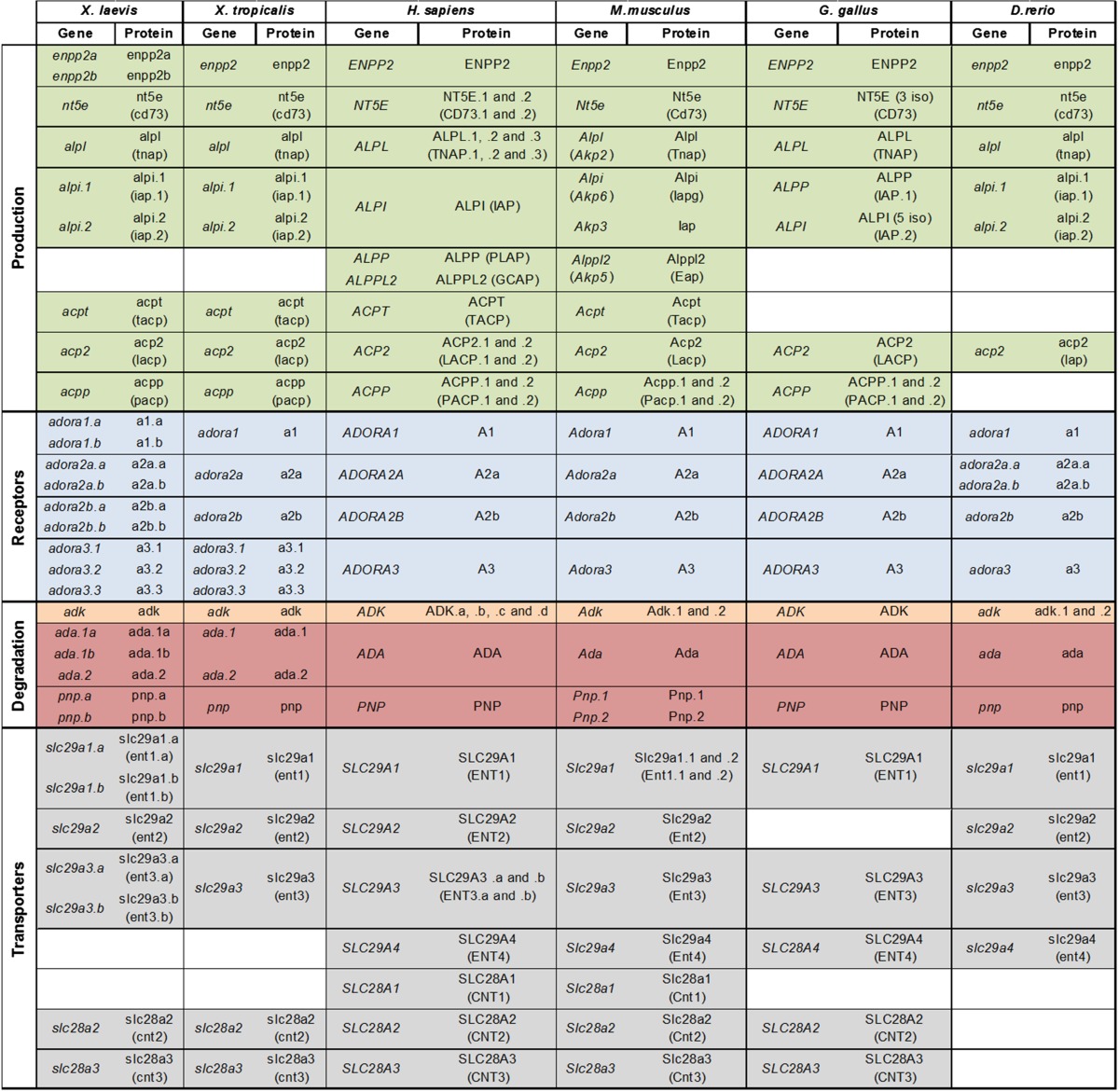
Synonyms are also indicated in brackets. D.rerio = Danio rerio, G.gallus = Gallus gallus, H.sapiens = Homo sapiens, M.musculus = Mus musculus, X.laevis = Xenopus laevis, X.tropicalis = Xenopus tropicalis
Each X. laevis and X. tropicalis sequence was further verified as true ortholog of mammalian genes by sequence alignments, synteny and phylogenetic analyses. We chose to only represent in the following section these analyses for the multigenic families.
Synteny and phylogenic analysis of adenosine pathway actors
Adenosine production
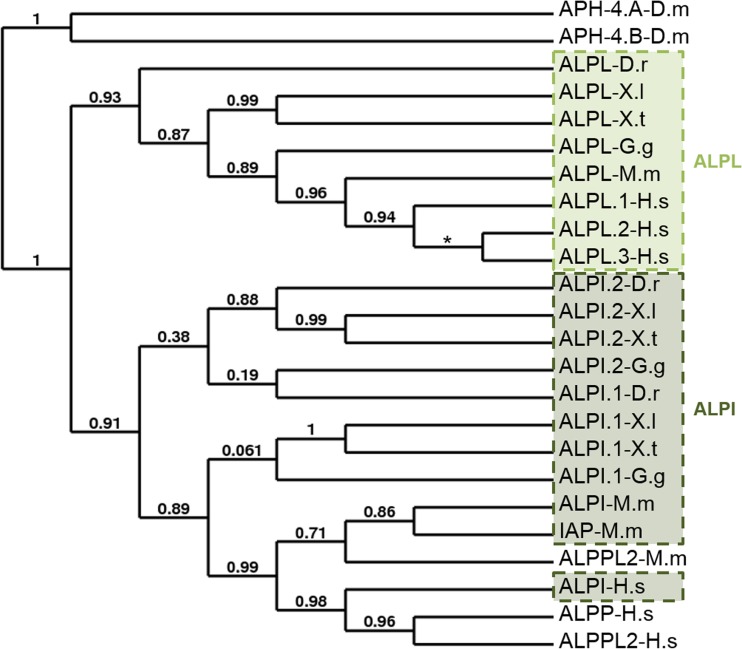
As we have previously described the phylogenetic analyses of the entpdases and enpp enzyme families [30, 31], we conducted a synteny and phylogenetic analysis for the alkaline and acid phosphatase multigenic families and used the Drosophila melanogaster enzymes as root for the phylogenetic tree (Figs. 1, S1, S2 and Table 2 and Supplementary Table S4). Since X. laevis genome assembly is not finished, synteny analyses were conducted with the X. tropicalis genome. As shown in Fig. 1, there is a clear separation between the tissue-non-specific (ALPL) and the tissue specific (ALPI, ALPP, and ALPPL2) alkaline phosphatases. In contrast to the ALPL proteins, the other alkaline phosphatases seem to be less conserved during evolution. Two ALPI genes have been identified in frog, chick, and zebrafish (ALPI.1 and ALPI.2). Although five ALPI.2 transcripts have been reported in chick, we decided to use only the isoform 5 for our phylogenetic analysis because of its higher level of identity with the human protein. There is a clear separation between the three mammalian enzymes and the non-mammalian ones, which can be correlated with the restricted mammalian expression of the mouse ALPPL2 and human ALPPL and ALPPL2 enzymes. However, the human and mouse ALPI genes are more related to their paralog genes than to their orthologs genes. Regarding the non-mammalian enzymes, the ALPI.1 proteins are more related to their mammalian orthologs. For example, X. laevis and X. tropicalis alpi.1 share more than 60 % of identity with the human protein, respectively, whereas the percentage of identity between Xenopus and human ALPI.2 sequence is less than 55 % (Table 2). Synteny analysis confirmed that Xenopus identified intestinal alkaline phosphatase genes are the true orthologs of the human, mouse, and chick ALPI genes (Supplementary Fig. S1). However, the orientation of these genes is different in mouse. Interestingly, the other alkaline phosphatase genes, human ALPPL2 and ALPP and mouse Alppl2, are located on the same chromosomes than the ALPI genes.
Fig. 1.
Relatedness of alkaline phosphatase (AP) proteins. A phylogenetic tree of vertebrate AP proteins was constructed on the Phylogeny.fr platform. A cluster algorithm was used to build the tree. Likelihood ratio is indicated at each node of the tree. The star represents a trifurcation (likelihood ratio null). The GenBank accession numbers of AP proteins are given in Supplementary Table S1 and as follows: APH-4.A: NP_524601; APH-4.B: NP_733413; ALPL-D.r: NP_957301; ALPL-G.g: NP_990691; ALPL-M.m: NP_031457; ALPL.1-H.s: NP_000469; ALPL.2-H.s: NP_001120973; ALPL.3-H.s: NP_001170991; ALPI.1-D.r: NP_001014375; ALPI.2-D.r: NP_001020359; ALPI.1-G.g: XP_003641809; ALPI.2-G.g: XP_422743 (isoform 5); ALPI-M.m: NP_001074551; IAP-M.m: NP_031458.2; ALPI-H.s: NP_001622; ALPPL2-M.m: NP_031459; ALPP-H.s: NP_001623; and ALPPL2-H.s: NP_112603. ALPPL2-M.m, embryonic alkaline phosphatase; ALPPL2-H.s, germ cell alkaline phosphatase; ALPI and IAP-M.m, intestinal alkaline phosphatase; ALPI-M.m, intestinal alkaline phosphatase global; ALPP, placental alkaline phosphatase; ALPL, tissue-non-specific alkaline phosphatase. D.m, Drosophila melanogaster; D.r, Danio rerio; G.g, Gallus gallus; H.s, Homo sapiens; M.m, Mus musculus; X.l, Xenopus laevis; X.t, Xenopus tropicalis
Table 2.
Identity conservation between the Xenopus tissue specific alkaline phosphatase proteins and their human orthologs
| X.l | X.t | M.m | H.s | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ALPI.1 | ALPI.2 | ALPI.1 | ALPI.2 | IAP | ALPI | ALPPL2 | ALPI | ALPP | ALPPL2 | ||
| X.l | ALPI.1 | 100 | |||||||||
| ALPI.2 | 56.8 | 100 | |||||||||
| X.t | ALPI.1 | 88.1 | 58.6 | 100 | |||||||
| ALPI.2 | 57.6 | 89.0 | 59.4 | 100 | |||||||
| M.m | IAP | 60.4 | 56.8 | 61.4 | 55.5 | 100 | |||||
| ALPI | 62.6 | 56.7 | 61.1 | 57.3 | 78.7 | 100 | |||||
| ALPPL2 | 58.4 | 55.5 | 58.0 | 55.0 | 74.1 | 76.4 | 100 | ||||
| H.s | ALPI | 63.3 | 53.8 | 61.9 | 54.9 | 77.5 | 78.2 | 76.1 | 100 | ||
| ALPP | 59.3 | 51.5 | 58.2 | 49.4 | 72.9 | 73.8 | 74.7 | 85.5 | 100 | ||
| ALPPL2 | 58.1 | 51.0 | 57.1 | 47.6 | 74.3 | 75.0 | 74.5 | 86.9 | 96.8 | 100 | |
The percentage of amino acid identity between proteins was determined by pairwise alignment using Clustal W2 software on the EMBL-EBI Internet site. The accession numbers of the different adenosine member clones are given in Supplementary Table S1 and in the legend of Fig. 1
H.s Homo sapiens, M.m Mus Musculus, X.l Xenopus laevis, X.t Xenopus tropicalis
The phylogenetic analysis of the acid phosphatases demonstrates the separation between the prostatic (ACPP), testicular (ACPT), and lysosomal (ACP2) acid phosphatase groups (Supplementary Fig. S2). Each of these proteins is more related to its orthologs than to the other acid phosphatases in the same species. Our phylogenetic study revealed that the Xenopus acpt genes, identified under the acpp gene symbol on Xenbase and NCBI websites, are indeed encoding the amphibian orthologs of human ACPT enzyme. Sequence analysis demonstrates that the ACP2 protein is the most conserved enzyme during vertebrate evolution. Xenopus and human proteins share more than 55 % of identity, whereas the Xenopus and human ACPT or ACPP share less than 50 % of identity (Supplementary Table S4).
Adenosine receptors
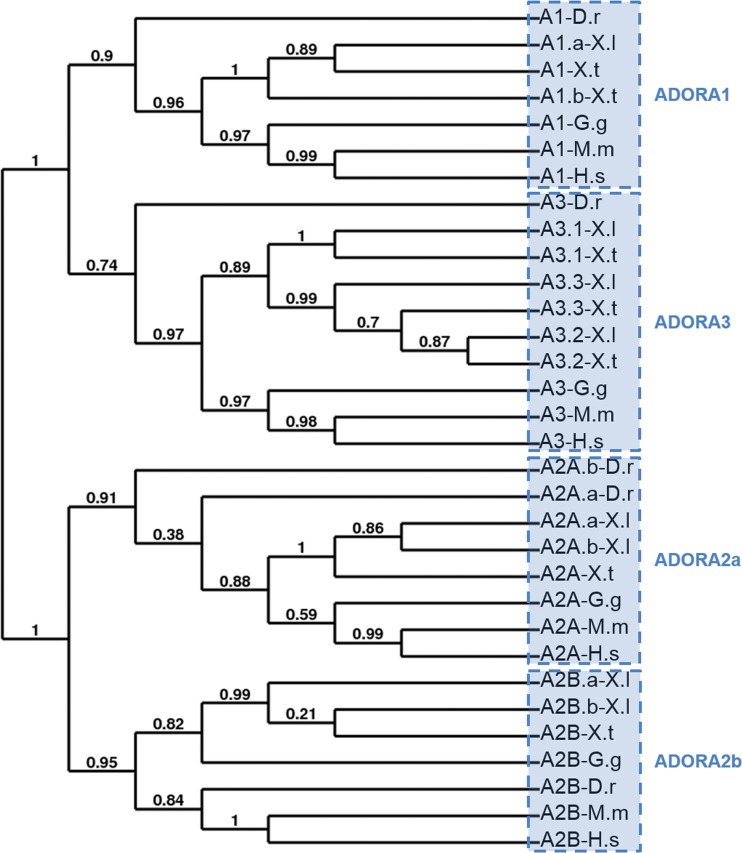
We have identified nine putative ADORA receptors in X. laevis and six in X. tropicalis. As expected, adora1, adora2a and adora2b homeologs are highly similar, with 94 to 97 % of identity between them (Table 3). The duplicated adora3 receptors are less conserved, sharing only 51.1 to 85.4 % of identity. The phylogenetic analysis of these adora receptors in Xenopus and other vertebrates and invertebrates was carried out using the full sequence of each protein. This analysis demonstrates the clear separation between adora1-adora3 and adora2a-adora2b receptors receptors (Fig. 2a). Each member is more related to its orthologous than to the other family members in the same species. Analysis of the amino acid conservation between Xenopus proteins and their mammalian orthologs showed a higher level of identity among species for a specific ADORA receptor (Table 3). For example, X. laevis adora1.a shares 96.3 and 68.4 % of identity with X. tropicalis and human ADORA1 protein, and only 44.9 to 51.8 % of identity with the three other X. laevis adenosine receptors. ADORA1 protein is the most conserved receptor during vertebrate evolution with X. laevis and human proteins sharing 68.4 % of identity (Table 3). The three adora3 proteins are the least conserved receptors, showing less than 55 % with the human ADORA3 protein. Among these three receptors, the adora3.3 is the most conserved, sharing 93.2 and 54.7 % of identity with its orthologous X. tropicalis and human sequence.
Table 3.
Identity conservation between the Xenopus ADORA (A) proteins and their human orthologs
| A1 | A2a | A2b | A3 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .a | .b | X.t | H.s | .a | .b | X.t | H.s | .a | .b | X.t | H.s | .1 | .2 | .3 | H.s | ||||||
| X.l | X.l | X.l | X.l | X.t | X.l | X.t | X.l | X.t | |||||||||||||
| A1 | .a | X.l | 100 | ||||||||||||||||||
| .b | 96.6 | 100 | |||||||||||||||||||
| X.t | 96.3 | 97.2 | 100 | ||||||||||||||||||
| H.s | 68.4 | 69.2 | 69.0 | 100 | |||||||||||||||||
| A2a | .a | X.l | 45.1 | 45.2 | 45.7 | 47.9 | 100 | ||||||||||||||
| .b | 45.4 | 45.5 | 46.0 | 48.5 | 94.3 | 100 | |||||||||||||||
| X.t | 45.7 | 46.5 | 47.9 | 47.9 | 94.4 | 92.8 | 100 | ||||||||||||||
| H.s | 44.2 | 44.6 | 45.1 | 48.2 | 60.2 | 61.2 | 60.2 | 100 | |||||||||||||
| A2b | .a | X.l | 46.9 | 47.4 | 46.9 | 49.4 | 60.7 | 58.3 | 59.5 | 58.6 | 100 | ||||||||||
| .b | 46.6 | 46.8 | 46.6 | 48.8 | 61.1 | 58.5 | 59.5 | 57.6 | 97.0 | 100 | |||||||||||
| X.t | 46.0 | 46.5 | 46.0 | 48.8 | 60.8 | 57.9 | 59.4 | 57.6 | 94.9 | 94.7 | 100 | ||||||||||
| H.s | 42.6 | 42.8 | 42.9 | 44.6 | 55.7 | 56.0 | 56.6 | 57.8 | 63.0 | 62.1 | 61.1 | 100 | |||||||||
| A3 | .1 | X.l | 44.9 | 44.9 | 45.2 | 46.5 | 35.4 | 36.3 | 36.9 | 38.1 | 38.2 | 38.2 | 38.2 | 35.7 | 100 | ||||||
| X.t | 44.6 | 44.9 | 45.5 | 48.0 | 37.9 | 38.2 | 38.8 | 39.4 | 41.2 | 41.5 | 41.5 | 37.9 | 86.5 | 100 | |||||||
| .2 | X.l | 46.6 | 47.1 | 46.0 | 44.3 | 37.1 | 37.1 | 37.4 | 39.2 | 38.3 | 38.0 | 38.0 | 34.9 | 51.1 | 51.7 | 100 | |||||
| X.t | 49.2 | 49.2 | 48.9 | 46.3 | 40.8 | 41.5 | 40.8 | 44.4 | 43.1 | 43.1 | 42.4 | 38.9 | 52.1 | 52.4 | 81.7 | 100 | |||||
| .3 | X.l | 51.8 | 51.7 | 51.2 | 47.9 | 41.1 | 41.5 | 41.8 | 42.4 | 42.7 | 42.1 | 41.5 | 38.1 | 53.5 | 53.2 | 85.4 | 82.3 | 100 | |||
| X.t | 50.0 | 50.2 | 50.3 | 48.0 | 40.3 | 40.7 | 40.7 | 43.4 | 42.2 | 42.2 | 41.6 | 39.1 | 52.9 | 53.2 | 85.6 | 84.9 | 93.3 | 100 | |||
| H.s | 47.8 | 48.1 | 48.1 | 48.1 | 39.6 | 39.3 | 41.2 | 41.2 | 40.6 | 40.9 | 40.2 | 37.7 | 44.3 | 46.9 | 51.9 | 53.7 | 54.7 | 53.5 | 100 | ||
The percentage of amino acid identity between proteins was determined by pairwise alignment using Clustal W2 software on the EMBL-EBI Internet site. The accession numbers of the different adenosine member clones are given in Supplementary Table S1 and in the legend of Fig. 2
H.s Homo sapiens, X.l Xenopus laevis, X.t Xenopus tropicalis
Fig. 2.
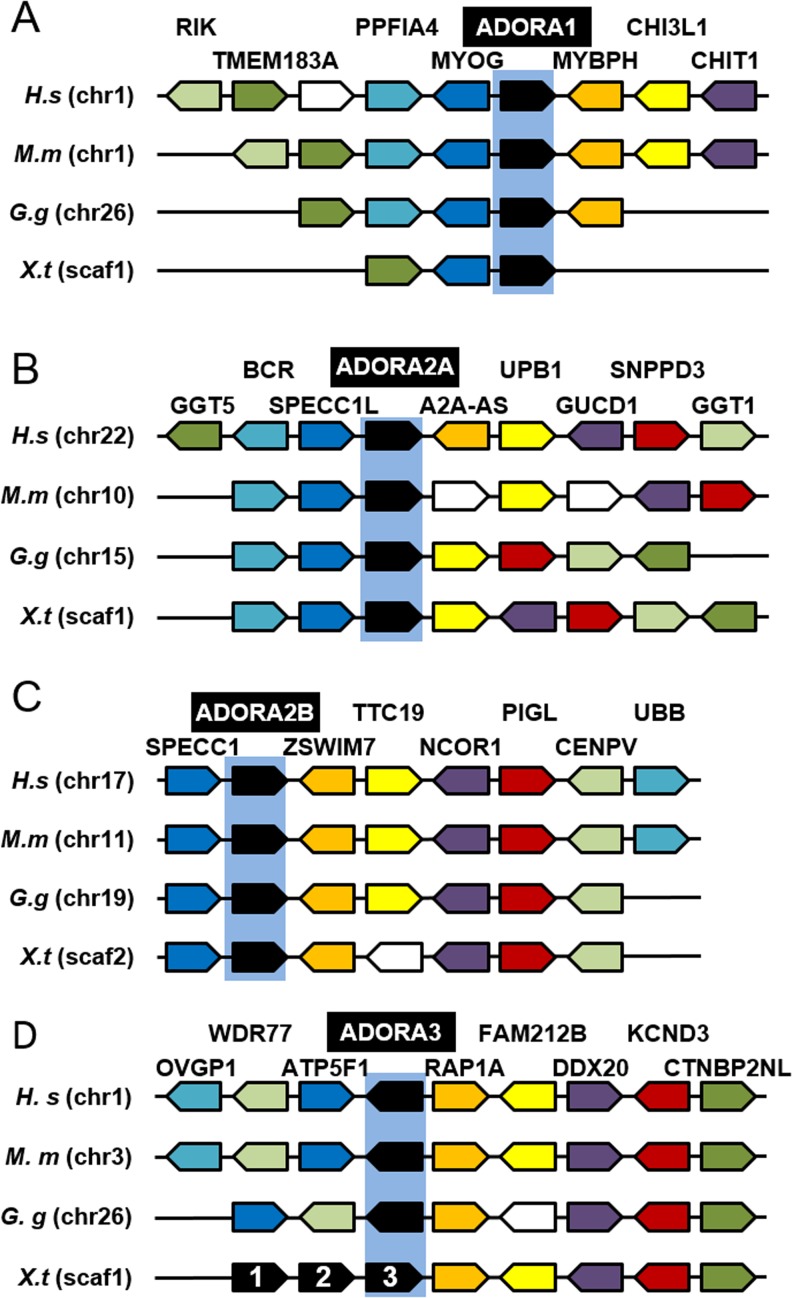
Conservative evolution of ADORA receptor genes.a Phylogenetic tree of vertebrate ADORA (A) protein. A phylogenetic tree was constructed on the Phylogeny.fr platform. A cluster algorithm was used to build the tree. Likelihood ratio is indicated at each node of the tree. The GenBank accession numbers of ADORA proteins are given in Supplementary Table S1 and as follows: A1-D.r: NP_001122056; A1-G.g: NP_989647; A1-M.m: NP_001008533; A1-H.s: NP_001041695; A2a.a-D.r: NP_001034904; A2a.b-D.r: NP_001035125; A2a-G.g: XP_425280; A2a-M.m: NP_033760; A2a-H.s: NP_001041695; A2b-D.r: NP_001034902; A2b-G.g: NP_990418; A2b-M.m: NP_031439; A2a-H.s: NP_000667; A3-D.r: XP_700086; A3-G.g: NP_989482; A3-M.m: NP_033761; A3-H.s: NP_000668. b Synteny map for the ADORA receptor genes in vertebrate genomes. The map was generated by comparison of X. tropicalis, G.gallus, M.musculus, and H.sapiens chromosome regions containing ADORA1 (A), ADORA2A (B), ADORA2B (C) and ADORA3 (D) genes. The NCBI GeneID of ADORA (A) genes is given in Supplementary Table S1 and as follows: A1-G.g: 374212; A1-M.m: 11539; A1-H.s: 134; A2a-G.g: 427705; A2a-M.m: 11540; A2a-H.s: 135; A2b-G.g: 395971; A2b-M.m: 11541; A2b-H.s: 136; A3-G.g: 373956; A3-M.m: 11542; A3-H.s: 140. Each conserved gene is color-coded; the unconserved genes are indicated by white boxes. The orientation of the genes on the different chromosome (chr) or scaffold (scaf) is indicated. D.r Danio rerio; G.g: Gallus gallus; H.s, Homo sapiens; M.m, Mus musculus; X.l, Xenopus laevis; X.t, Xenopus tropicalis
We confirmed the conservation of ADORA genes in the vertebrate lineage at the genomic level (Fig. 2b). The synteny is conserved for each ADORA gene, confirming that the identified amphibian sequences are true orthologs of the human adenosine receptors. However, the adora3 gene has been duplicated in amphibian species and the orientation of these genes is different in the X. tropicalis genome.
Adenosine degradation
We identified two genes, named ada.1 and ada.2, coding for ada enzymes in X. tropicalis and three in X. laevis, called ada1.a, ada1.b, and ada.2. Synteny analysis confirmed the conservation of these genes during vertebrate evolution and a duplication event in the amphibian lineage (Supplementary Fig. S3a). Phylogenetic analysis demonstrated that each Xenopus ada gene is more related to its amphibian orthologs than to its paralog (Supplementary Fig. S3b). Indeed, Xenopus ada.1 proteins share around 95 % of identity but less than 70 % of identity with amphibian ada.2 (Supplementary Table S5). Moreover, the phylogenetic tree shows a clear separation between the non-mammalian and the mammalian ADA proteins (Supplementary Fig. S3b). Xenopus ada.1 proteins are more related than Xenopus ada.2 proteins to the human enzyme, with a percentage of identity of 69 versus 57.8 % (Supplementary Table S5).
We identified one pnp gene in X. tropicalis and the two homologs pnp.a and pnp.b in X. laevis. Our genomic sequence analysis allowed us to complete the published pnp.a 3’ end sequence [32]. Moreover, we identified a putative exon, encoding 34 extra amino acids in the pnp.a N-terminal region, which are not conserved in any other PNP sequences. We decided to carry out our study without this extra sequence. The two X. laevis pnp.a and pnp.b homeologs encode highly similar proteins, sharing 90 % of identity and with only 31 amino acid differences along their coding region (Supplementary Fig. S4 and Supplementary Table S6). Sequence analysis showed that there are 43 amino acid differences (substitution and deletion) between X. laevis pnp.a and X. tropicalis pnp but only 32 differences between X. laevis pnp.b and X. tropicalis pnp proteins (Supplementary Fig. S4a). As expected, the percentage of identity beween X. laevis pnp.b and X. tropicalis pnp is higher than the one between X. laevis pnp.a and X. tropicalis pnp (Supplementary Table S6). However, the percentages of identity between X. laevis pnp.a or pnp.b and human PNP are very similar. The phylogenetic tree shows clearly that the two pnp X. laevis sequences are derived from one X. tropicalis sequence, which has diverged from the other vertebrate sequences (Supplementary Fig. S4b).
Adenosine transporters
All members of the SCL28 and SCL29 gene families, excepted for SLC28A1 and SLC29A4, were cloned in X. laevis and X. tropicalis. We have identified homeologs genes encoding the X. laevis slc29a1 and slc29a3 transporters, which we named slc29a1.a and slc29a1.b, and slc29a3.a and slc29a3.b. The two slc29a1 proteins are 91.8 % identical, but sequence analysis suggests that slc29a1.b is more related to its X. tropicalis and human orthologs than slc29a1.a (Supplementary Table S7). The X. laevis slc29a3.a and slc29a3.b transporters are less conserved, sharing less than 90 % of identity.
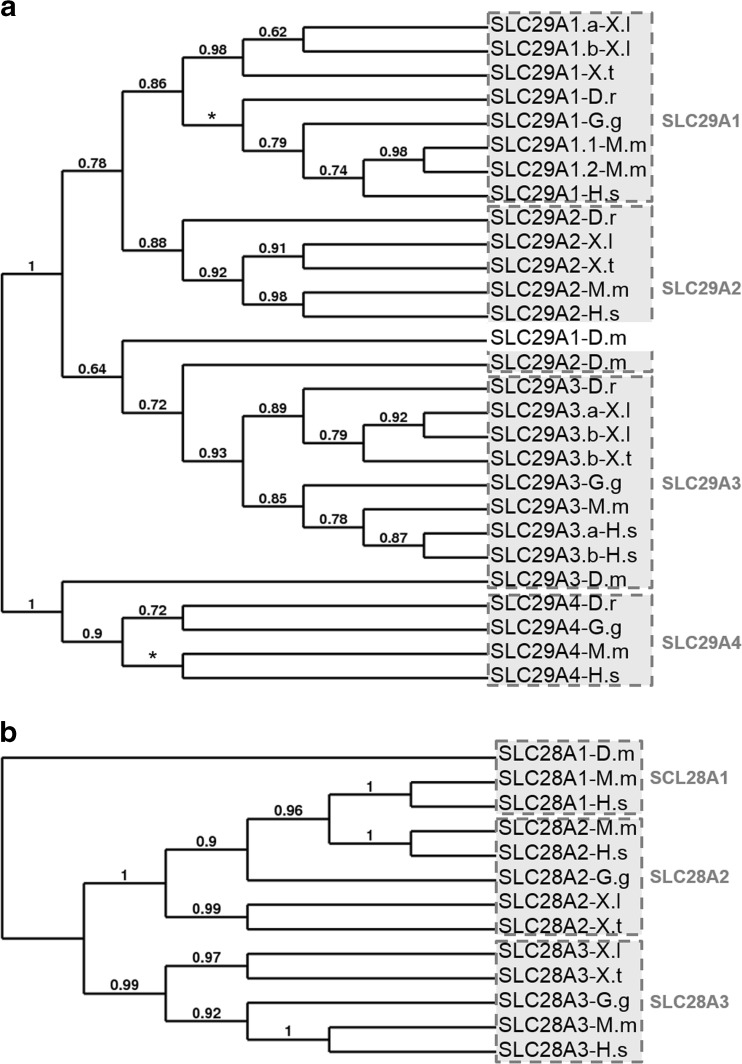
The phylogenetic trees illustrate clearly the relatedness of each transporter family (Fig. 3). Each member of these gene families is more related to its orthologs than to the other family members in the same species. Although four branches can be distinguished in the SLC29A family, there are clearly two major subgroups, with SLC29A1, SLC29A2, and SLC29A3 belonging to the first group and SLC29A4 forming the second group (Fig. 3a). The phylogenetic analysis shows the three Drosophila melanogaster SLC29A proteins in this tree, but our analysis suggest that these sequences were certainly wrongly annotated. The existence of two major subgroups in the SLC28A family can also be proposed, with SLC28A1 and 2 forming the first group and SLC28A3 the second group (Fig. 3b). These results are supported by the amino acid conservation studies (Supplementary Tables S7 and S8). Synteny analyses confirmed the vertebrate conservation of these SLC28A and SLC29A gene families (Supplementary Figs. S5 and S6).
Fig. 3.
Conservative evolution of the two nucleoside transporter family genes. The phylogenetic trees of vertebrate equilibrative nucleoside transporter (SLC29A) (a) and concentrative nucleoside transporter (SLC28A) (b) were constructed on the Phylogeny.fr platform. A cluster algorithm was used to build the tree. Likelihood ratio is indicated at each node of the tree. The star represents a trifurcation (likelihood ratio null). The GenBank accession numbers of nucleoside transporter proteins are given in Supplementary Table S1 and as follows: SLC29A1-D.m: NP_722628 (isoform A); SLC29A1-D.r: NP_001025348; SLC29A1-G.g: XP_419491 (isoform 6); SLC29A1.1-M.m: NP_001186042; SLC29A1.2-M.m: NP_001186044; SLC29A1-H.s: NP_001071642 (isoform 4); SLC29A2-D.m: NP_609049; SLC29A2-D.r: NP_001012519; SLC29A2-M.m: NP_031880; SLC29A2-H.s: NP_001523; SLC29A3-D.m: NP_648608 (isoform A); SLC29A3-D.r: XP_005156582; SLC29A3-G.g: XP_421594; SLC29A3-M.m: NP_076085; SLC29A3.a-H.s: NP_060814; SLC29A3.b-H.s: NP_001167569; SLC29A4-D.r: NP_001074041; SLC29A4-G.g: XP_004945212 (isoform 4); SLC29A4-M.m: NP_666369; SLC29A4-H.s: NP_001035751; SLC28A1-D.m: NP_610447; SLC28A1-M.m: NP_001004184; SLC28A1-H.s: NP_004204; SLC28A2-G.g: XP_004943665 (isoform 6); SLC28A2-M.m: NP_766568; SLC28A2-H.s: NP_004203.2; SLC28A3-G.g: XP_425033 (isoform 6); SLC28A3-M.m: NP_071712; SLC28A3-H.s: NP_071410. D.r, Danio rerio; G.g, Gallus gallus; H.s, Homo sapiens; M.m, Mus musculus; X.l, Xenopus laevis; X.t, Xenopus tropicalis
Spatial expression profiles of adenosine pathway genes in the adult frog
The expression of the different genes in the adult frog was analyzed by RT-PCR (Fig. 4). As the X. laevis genome sequencing is not complete and because of the high level of sequence identity between duplicate genes, we were not able to amplify all the identified members listed in the Table 1. However, we tried at least to amplify each couple of homeologs and the duplicate genes, except for adora3.2. To distinguish the expression profile of the pnp homeologs, we managed to amplify both pnp genes and specifically pnp.b gene and considered that any expression profile differences might be due to pnp.a expression.
All of the adenosine signaling pathway members are expressed in the adult frog and display different expression patterns. The genes coding for enzymes able to generate adenosine appeared to be expressed in a wide range of organs in the adult frog, except for the intestinal alkaline phosphatase alpi.1 and alpi2 genes that are highly expressed in the duodenum. However, these genes are also expressed at a high level in the kidney and liver for alpi.1 and in the brain for alpi.2.
The adora genes, except for adora3.3, are ubiquitously expressed and their transcripts can be detected in every tested organ, except for the liver for adora1 and in the eye for adora3.1. Adora3.3 displays a more restricted expression pattern as its expression is only detected at a high level in the bladder and duodenum and at a very low level in the brain, muscle, and stomach.
The genes encoding enzymes involved in adenosine degradation display a wide expression pattern and are expressed at a high level in the kidney. The two ada genes display a similar expression profile as their transcripts can be detected in all tested tissues. However, the level of expression for each of these genes is higher in some organs with ada.2 expression higher in the eye, bladder, liver, and stomach and ada.1 expression higher in the ovary. Both pnp genes are highly expressed in most tissues, except for the muscle, heart, and eye. Their high level expression in the testis, ovary, liver, and spleen is certainly due to pnp.a expression, as pnp.b expression is faintly detected in these tissues.
The three members of the slc29a family display a wide expression profile, being expressed in every organ analyzed. In contrast, the slc28a transporter transcripts can only be detected in a few organs. Slc28a2 is highly expressed in the kidney and duodenum while slc28a3 expression is mainly detected in the eyes, testis, and kidney.
Temporal expression profiles of adenosine pathway genes in the X. laevis embryos
The temporal expression profile of the genes was assessed by RT-PCR using whole unfertilized eggs and embryos from stage 5 to stage 41 in order to cover all embryonic phases of X. laevis development (Fig. 5).
The genes encoding enzymes generating adenosine are not expressed maternally, except for acpt, which is expressed at low level in unfertilized eggs and in stage 5 embryo. Nt5e expression starts to be detected during neurulation and the expression level decreases during organogenesis, from stage 21 to 35/36 before a re-increase from stage 37/38. The zygotic alkaline phosphate alpl is expressed from stage 9 and remains constant until stage 41. No expression at any embryonic stage tested could be detected for alpi.1 whereas the zygotic alpi.2 expression is only and weakly detected from stage 32. The zygotic expression profile of acpt is similar to that of alpi.2, one where transcripts are only detected during late organogenesis phase.
The adora receptors encoding genes display distinct temporal expression profile in Xenopus embryos. Adora2a and Adora2b are not expressed maternally and their zygotic expression is first detected during neurulation and gastrulation, respectively. Their level of expression increases slowly until stage 41. Adora1 and adora3.1 are both expressed maternally. The zygotic expression of adora1 is weakly detected during neurulation, with an increase of expression from stage 25 to stage 41 while adora3.1 is expressed at all stages of development. No expression of adora3.3 could be detected at any stage suggesting that adora3.3 is dispensable for embryogenesis.
In contrast to the genes encoding adenosine-generating enzymes, several genes coding for enzymes metabolizing adenosine are maternally expressed. Indeed, expression of adk, ada.1, and pnp genes is detected in unfertilized eggs and during early cleavage phases. Adk expression is detected at all tested stages with a dynamic expression pattern. After a decrease of maternal expression from unfertilized egg to the end of neurulation, adk expression level increases slowly during organogenesis until stage 41. The two ada genes display also different expression profiles during X. laevis embryogenesis. Ada.1 maternal expression is detected until the end of gastrulation while zygotic expression is first detected at the end of neurulation. On the contrary, ada.2 expression is only detected at the end of embryonic development. Pnp gene expression is detected at all stages tested with maternal contribution being low whereas the zygotic expression level increases until the end of organogenesis. Zygotic pnp.b expression is only weakly detected during gastrulation and neurulation phases but is upregulated at stage 19 and increases until stage 41. Therefore, the pnp transcripts amplified during gastrulation and neurulation phases might correspond to pnp.a expression.
The equilibrative and concentrative nucleoside transporter family members display different expression profiles during X. laevis embryogenesis. The slc29a genes can be divided into two groups according to their expression patterns. The first group includes slc29a1.a which is not expressed maternally and whose zygotic expression is only detected during organogenesis. The second group is composed of slc29a2 and slc29a3, which are expressed at a similar level at all stages tested. The slc28a genes display distinct expression profile where slc28a2 is weakly maternally expressed and then its zygotic expression level increases from the end of neurulation until stage 37/38, whereas slc28a3 is only expressed during gastrulation and neurulation phases.
Spatial expression profiles of adenosine pathway genes during development of X. laevis
The spatial expression of the genes was assessed by RT-PCR on a series of X. laevis embryo dissections as indicated in Fig. 6. The presumptive nervous system was dissected from the rest of the embryo at stage 9 (animal cap) and stage 15 (neural plate). At stage 10, the different mesodermal tissues (dorsal, lateral and ventral) were dissected. At stage 24, the brain and anterior structures (head) and spinal cord with somitic tissues (dorsal trunk) were separated from the ventral region of the embryo (ventral trunk) (Fig. 6a). At later stages, selected organs were isolated from stage 37/38 and stage 45 embryos (Fig. 6b).
Among the genes encoding enzymes involved in adenosine production, nt5e displays the most restricted expression profile in the early phases of embryogenesis, with no expression or at low level in neural and somitic tissues (Fig. 6a). At late organogenesis stages, nt5e transcripts are detected in mesodermal derived tissues such as the heart and kidney and also endodermal derived tissues (Fig. 6b). Alpl is ubiquitously expressed from the end of cleavage (stage 10) until the beginning of organogenesis (stage 24) (Fig. 6a). At stage 37/38, alpl expression remains detected in all dissected regions tested, but at a higher level in the eye and kidney (Fig. 6b). At the end of embryogenesis, alpl transcripts are detected in all tissues analyzed although at a weaker level in the posterior gut. As expected, the intestinal alkaline phosphatase alpi.2 is highly and specifically expressed in the posterior gut at stage 45 (Fig. 6b) but with a weaker expression in the ventral region of stage 37/38 embryo and in stage 45 anterior gut. The acid phosphate acpt displays a ubiquitous expression profile except at stage 24, with a high expression in the head (Fig. 6).
Based on their expression profiles during the early phases of embryogenesis, the adora receptors can be divided into three groups (Fig. 6a). Adora1 gene constitutes the first group and is ubiquitously and weakly expressed in blastula and gastrula stage embryo, and then, its expression becomes restricted to the nervous system from neurula stages. The second group is composed of adora2a and adora2b, which are expressed in all dissected regions of stages 15 and 24 embryo. Adora3 belongs to the third group and is ubiquitously detected in stages 9 and 10, but in stage 15 embryo, it becomes the only adenosine receptor not expressed in neural tissues. At later stages, adora genes display a ubiquitous expression profile and are all expressed in the brain and eye (Fig. 6b).
Genes encoding enzymes involved in adenosine catabolism, except for ada.2, are ubiquitously expressed in X. laevis embryo, although their expression level may vary between dissected regions (Fig. 6). Ada.2 is only expressed at late stages with a highly level detected in the endodermal tissues (Fig. 6b).
The slc29a genes are expressed ubiquitously in embryos, although slc29a1.a expression seems more restricted to the nervous system (Fig. 6). Slc28a genes display a more stage and tissue specific expression profile. Slc28a3 transcripts are highly detected in the neural plate in neurula stage embryo and in stage 45 kidney. Slc28a2 transcripts are found in renal tissues and in dissected eyes at late organogenesis phases.
Concentration of adenosine compared toother purines during Xenopus development
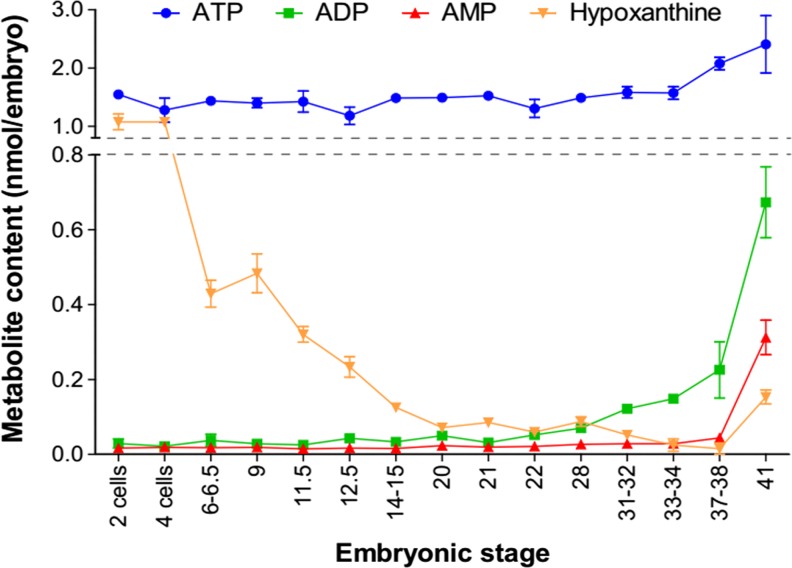
The embryonic content of ATP and its derivatives was measured by liquid chromatography from two-cell stage to stage 41 in order to cover all X. laevis embryonic development (Fig. 7). The ATP content remains constant at around 1.3 nmol/embryo until stage 33/34, before an increase during late phases of organogenesis to reach 2.4 nmol/embryo. ADP and AMP contents remain low during early phases of development but increase from stage 37/38 (>0.04 nmol/embryo) to reach 0.7 and 0.3 nmol/embryo, respectively, at stage 41. In two-cell-stage embryo, hypoxanthine and ATP contents are similar (around 1 nmol/embryo) before a dramatic drop of hypoxanthine from four-cell-stage embryos to reach its lower value of 0.01 nmol/embryo in stage 37/38 embryo, before a slight re-increase. Adenosine and inosine contents are undetectable during Xenopus development, with a value inferior to the detection value limit of the technique (<1 pmol/embryo for these two metabolites).
Fig. 7.
Evolution of purine content in embryo during development. Total embryo extracts were prepared from indicated stages, and metabolites were separated and quantified by liquid chromatography. Each point corresponds to the mean of 3 to 8 extractions done on independent batches of embryos (10 embryos per extract) and from two independent females. Error bars indicate variation to the mean
Discussion
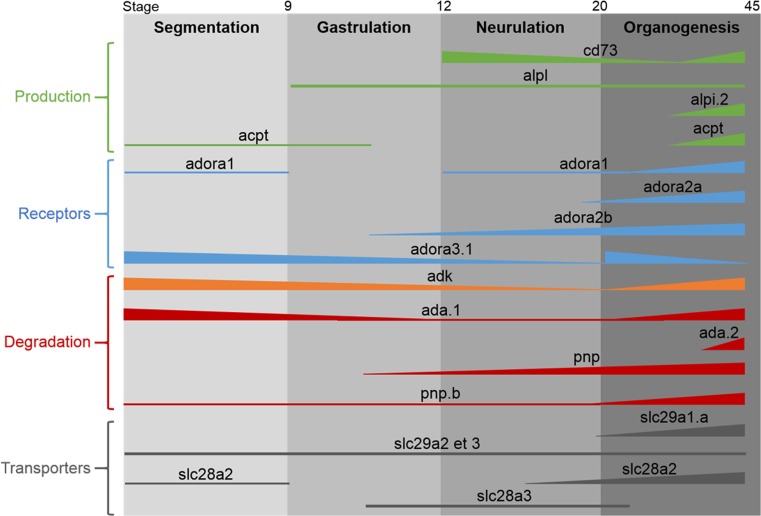
In this paper, we report the identification and expression in X. laevis embryo and adult of thirteen enzymes involved in the metabolism of adenosine, of nine adora receptors, and of the seven nucleoside transporters. Although the sequences for the majority of these members were available on Xenbase, none of their embryonic expression profile had been characterized yet, except for pnp and adora2a [32, 33]. The expression profiles for some of the genes analyzed were achieved through a comparative transcriptome study [43]. However, our data not only confirmed their results, available on Xenbase website, but is also more complete including later developmental stages. Although we have not analyzed the expression profiles of all the identified members of the adenosine signaling pathway, we were able through our in-depth analysis to provide the first comparative and comprehensive expression map (Fig. 8).
Fig. 8.
Comparative map of the expression profiles of the members forming the adenosine signaling pathway during X. laevis development. The full lines represent the expression for each gene detected in the study whereas the dotted lines represents the expression for which the level may be too low for being detected by our RT-PCR. The thickness of the lines indicates the expression level of each gene
Evolutionary conserved members of the adenosine signaling pathway are present in Xenopus
Our study reveals that all the identified members regulating this pathway (see Table 1) are present in X. laevis and X. tropicalis except for four actors. As expected, no amphibian orthologous were found for human placental and germ cell alkaline phosphatases, the two mammalian specific enzymes. Our BLAST genomic search failed to identify amphibian sequences encoding the transporters SLC28A1 and SLC29A4. However, our study revealed the presence of slc29a1 and slc29a3 homeologs that may compensate for the loss of slc29a4 gene. Bioinformatic analyses indicate that all amphibian slc29a, as their mammal orthologs, display the characteristic 11 transmembrane domains (TM) of these transporters. Moreover, the residues important for mammalian slc29a functions are conserved in the amphibian sequences, such as G79 (numbered on the human SLC29A1 sequence), which is involved in transport activity [44]. The residue 33 is a determinant of overall functional difference between SLC29A1 and SLC29A2 [45]. In human SLC29A2, this residue, an isoleucine, is a functional determinant for purine nucleoside transport and is conserved in both X. laevis and X. tropicalis sequences. In the three amphibian slc29a1 sequences, this residue is a methionine as in human SLC29A1. Interestingly, this residue, which is also an isoleucine in human SLC29A3 sequence, is not conserved in X. laevis and X. tropicalis slc29a3 proteins. No duplicated genes were identified for slc28a2 and slc28a3. The residues 319 and 353 (numbered in human SLC28A1) are involved in pyrimidine and purine selectivity [46], and the presence of glycine and serine respectively at these positions in all Xenopus slc28a sequences suggests that these transporters may be purine selective. Thus, this may compensate for the absence of slc28a1 which, although being pyrimidine selective, can transport adenosine in a high-affinity, low-capacity manner [46].
Surprisingly, nine adora receptors are present in X. laevis, with three homeologs and three duplicated adora3 genes. Bioinformatic analysis indicated that all of the receptors display the characteristic 7 TM domains of these receptors, each 21 to 28 amino acids long [47]. As for their mammalian orthologs, the percentages of identity between the different Xenopus adora proteins are weak, ranging from 45 to 50 %. However, the residues in the TM essential for ligand recognition, such as A66, T91, M180, W247, N254, I274, and H278 (numbered on the human ADORA1 sequence), are conserved in all Xenopus adora receptors [20].
Ada enzyme is another member of the adenosine pathway that has been duplicated in Xenopus. Moreover, the ada.1 homologs have been conserved, making to three the number of ada proteins in X. laevis, suggesting the importance of this enzyme. Several residues have been characterized to be essential for adenosine deaminase activity [48–50]. The majority of these residues, especially G184, E217, and D295 (numbered on human ADA gene), are conserved in all Xenopus ada sequences [50]. Surprisingly, D296, which is required for proper binding of the substrate in the active site, is not conserved in ada.2 proteins. This difference may suggest different catalytic properties for the amphibian ada.1 and ada.2 enzymes.
Our study reveals that each member of the amphibian adenosine signaling pathway is more related to its orthologs than to other family members in the same species. This suggests that any future functional test performed in X. laevis will be relevant to a conserved function in other vertebrates.
Several duplicate members in Xenopus
Among the 29 members of the adenosine signaling pathway identified in this study, 7 are duplicate genes also found in X. tropicalis, arisen from a duplication event that has taken place before X. laevis and X. tropicalis divergence 50 million years ago, and 7 are homeologs which arose from the duplication of the two parent species 40 million years ago [51]. Although we may have not identified all the homeolog sequences, our work demonstrates that 39 % of the duplicate genes of this pathway have been conserved and encode proteins sharing 91 to 97 % of amino acid identity. This is in agreement with previous bioinformatics analyses [42]. However, Morin et al. demonstrate that one of the gene ontology categories that have lost the tetraploidization-derived paralog is nucleoside metabolism [52]. Indeed, only two enzymes have kept their duplicate gene whereas three out of the four adora receptors and two out of the three slc29a transporters are present with their homeologs.
These duplicated genes may be functional, and because they display different expression pattern, they might have acquired sub-functionalization. Although we could not analyze the expression profiles of all of them because of the lack of specific sequences, we show that pnp.a and pnp.b have distinct expression profiles, suggesting that these 2 proteins may fulfill different parts of their original ancestral function. Three adora3, two alpi, and two ada duplicates were identified in both Xenopus species. Two intestinal alkaline phosphatases, Alpi.1 and Alpi.2, are also present in mouse, chick, and zebrafish. In mice, these two genes have been shown to have distinct expression profiles and regulation suggesting different biological functions [53]. We show here that these two genes have distinct expression profiles during X. laevis development. In adult, they are highly expressed in adult X. laevis intestine but each alpi gene is also expressed in the kidney or brain. This second domain of expression, specific to each Xenopus duplicate gene, has been identified for human ALPI via ESTs sequences, indicating sub-functionalization (Unigene Hs39009). To our knowledge, the ADORA3 and ADA genes have not been duplicated in other vertebrate species and no information is available regarding a physiological relevance for their duplication in Xenopus. However, we show that these genes are differentially expressed in amphibian adult and embryos, suggestive of sub-functionalization.
Specific functions for the different members in Xenopus
The majority of the former descriptive studies on adenosine signaling pathway actors have been carried out in adults. Our work shows that the majority of these genes are expressed ubiquitously in the adult frog, although their expression level may vary between tissues. Similar expression profiles have been shown for their mammalian ortholog genes, such as SLC29A, PNP, ADA, and ADK [44, 54, 55]. However, less is known regarding the expression profile of these actors during vertebrate development. Our present work shows that these members can be divided into five groups based on their embryonic temporal expression (Fig. 8). (1) The first group includes genes such alpi.1, alpi.2, adora3.3, and ada.2 whose expression pattern precludes any roles during Xenopus development. No ESTs encoding the enzymes ALPI or the receptor Adora3 have been identified during mouse development (Mm. 58068 and Mm.235024), and only ESTs from organogenesis phase have been found for the enzyme Ada (Unigene Mm.388), suggesting that these proteins are not or not much involved in mammal embryogenesis. (2) The second group includes genes such as adk, slc29a2, and slc29a3 whose maternal and zygotic expression is constant. These genes are also expressed in all dissected tissues analyzed, suggesting that they may have housekeeping functions in Xenopus embryos. Not much information is available regarding their embryonic expression profiles in mammals, but analyses of EST databases and mouse transcriptome revealed that Adk is strongly expressed during mouse development whereas the two transporters seem to be weakly or not expressed ([56]; Unigene Mm.188734, Mm.4930, Mm.284462). (3) The third group gathers genes such as adora2a and adora2b receptors, slc29a1 transporter, and alkaline phosphatase alpl which are not expressed maternally but whose zygotic expression level increases and reaches a maximum level at tadpole stages. These genes are also widely expressed in dissected embryos, although their expression level varies between tissues. The pnp.a gene might be part of this group. (4) The fourth group contains genes such as acpt, pnp.b, adora1, ada1.a, and slc28a2, which are maternally expressed and then zygotically expressed from late neurulation or organogenesis. These genes display a more tissue-specific expression in both embryo and adult, suggesting a specific function. Mouse Slc28a2 high expression has been reported in gut during organogenesis phase [56], suggesting that it might be implicated in gut formation in vertebrates. Interestingly, Xenopus acpt is not highly expressed in frog testis, as its mouse orthologs (Unigene: Mm.367353), ruling out a possible implication in testis physiology as it has been proposed for the human gene [11]. (5) The last group is composed of genes such as nt5e and slc28a3 whose zygotic expression level peaks during neurulation before a decrease during development. The transporter slc28a3 is highly expressed in the neural plate, suggesting a role in neural tissues formation. However, in mouse, Slc28a3 expression has only been detected in morula and blastocyst stages (Mm.18188). Surprisingly, nt5e is not expressed in the developing nervous system but mainly in derivated mesoderm tissues. Similar expression has also been found in mouse, chick, and human embryos [56–58].
Implication of adenosine during Xenopus development
Our measurement of ATP content in the embryo ends up with a value of 1.5 nmol/embryo at two-cell-stage embryo. This is in agreement with other data that measured an ATP content between 0.74 and 1.03 nmol/oocyte [59]. Despite the presence in the embryo of the different ectonucleotidases, entpdases, enpps, and alkaline phosphatases, the content of ATP and other nucleotides remains constant during embryogenesis until stage 37/38. This is quite expected, as a balanced supply of purine nucleotides is needed in growing tissues such as during embryogenesis. The rapid decline of hypoxanthine from 1 nmol/embryo (two-cell stage) to 0.015 nmol/embryo (stage 37/38) suggests that this metabolite is the major source of purines during X. laevis development and purine nucleotides are therefore made available for cells via the salvage pathway. Indeed, high level of expression of hypoxanthine phosphoribosyltransferase 1 has been detected by RTqPCR during Xenopus development [43]. Moreover, it has been shown that an increase in HPRT activity is related to developmental changes [60]. As salvage of the purine ring is six times more efficient in terms of ATP equivalents than de novo purine synthesis, our data suggests that the salvage pathway compensates for the nucleotide requirement to the embryo for growth and development [61].
Our study suggests that the adenosine signaling pathway may regulate the physiology of all Xenopus adult organs, in a similar way to mammals [3, 5, 62–64]. In particular, the kidney, intestine, and brain appear to be major sites of expression in the adult for the pathway members, including the enzymes involved in generating or hydrolyzing adenosine, adora receptors and nucleotide receptors. This observation may also account for tadpole’s organs. Interestingly, the ectonucleotidase nt5e is highly expressed in developing renal tissues and may be therefore involved in pronephric formation. This would not be unexpected as renal dysfunctions have been observed in Nt5e null mice [65, 66]. Our data demonstrate that the different actors of the adenosine signaling pathway are present during Xenopus development, suggesting that this pathway is functional during vertebrate embryogenesis. This pathway might be particularly involved in the nervous system formation, as several members of this pathway, such as adora1, 2a, and 2b receptors, are expressed in developing neural tissues. However, our work raises the question of the origin of the extracellular adenosine in this tissue as nt5e, the major enzyme producing extracellular adenosine from AMP, is not expressed there [7]. In the adult mouse dorsal spinal cord, the Alpl, Acpt, and Nt5e are the main AMP ectonucleotidases to generate extracellular adenosine [67]. As the alpl and acpt phosphatases are both expressed in the neural plate, we could speculate that they may act redundantly to generate extracellular adenosine. However, we have previously shown that enpp2b is expressed in neural tissues, placing this enzyme as the potential major source of extracellular adenosine [31]. We have shown that the embryonic expression profiles of the different actors are dynamic and therefore the extracellular/intracellular adenosine content must be tightly regulated temporally and spatially. At late stages of organogenesis, all the different types of actors, e.g., adenosine anabolic and catabolic enzymes, receptors, and nucleotides, are expressed, suggesting that the adenosine signaling pathway is fully on. However, during segmentation, our data suggest that this pathway may be off due to the presence of the catabolic enzymes adk and ada and the slc29a transporters. Moreover, the same observation can be made in the ovary, suggesting that any adenosine molecule can be metabolized into either hypoxanthine or AMP/ADP/ATP, as it has been previously shown in Xenopus oocytes [68]. Therefore, during early stages of development, the adenosine content must be low. Indeed, we observed that adenosine content is lower than 0.001 nmol/embryo,which is a very low level compared to the adenylic nucleotide AMP (0.015 to 0.310 nmol/embryo), ADP (0.02 to 0.68 nmol/embryo), and ATP (1.1 to 2.4 nmol/embryo). Therefore, during vertebrate development, a sophisticated regulatory system is probably in place in order to maintain this low level of adenosine. Any disruption of this system would have severe consequences for development. Indeed, addition of exogenous adenosine blocks early development, and increase of adenosine signaling is lethal (reviewed in [69]), these two observations being in good agreement with a regulatory system keeping adenosine at a low level all along the embryonic development.
We have previously published the expression profiles of the entpdase and enpp gene members during Xenopus embryos and demonstrated the power of this model to establish previously unknown functions for the purinergic signaling pathway [29–31]. This study brings novel insights into the implications of the purinergic signaling pathway during vertebrate development. Functional analyses to decipher the embryonic roles of extracellular adenosine are in progress in the laboratory.
Electronic supplementary material
(PDF 1165 kb)
Acknowledgments
This work was supported by funding from the University of Bordeaux, the CNRS, and the “Association Française contre les Myopathies.” AT was supported by a PhD fellowship from the French Ministry for Research and Education. The funders had no role in study design, data analysis, or manuscript writing. We thank Lionel Parra Iglesias for taking care of the Xenopus colony.
References
- 1.Burnstock G. Purine and pyrimidine receptors. Cell Mol Life Sci. 2007;64(12):1471–1483. doi: 10.1007/s00018-007-6497-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vizi ES, Knoll J. The inhibitory effect of adenosine and related nucleotides on the release of acetylcholine. Neuroscience. 1976;1(5):391–398. doi: 10.1016/0306-4522(76)90132-9. [DOI] [PubMed] [Google Scholar]
- 3.Burnstock G, Krugel U, Abbracchio MP, Illes P. Purinergic signalling: from normal behaviour to pathological brain function. Prog Neurobiol. 2011;95(2):229–274. doi: 10.1016/j.pneurobio.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 4.Castrop H. Mediators of tubuloglomerular feedback regulation of glomerular filtration: ATP and adenosine. Acta Physiol (Oxf) 2007;189(1):3–14. doi: 10.1111/j.1748-1716.2006.01610.x. [DOI] [PubMed] [Google Scholar]
- 5.Di Virgilio F. Purines, purinergic receptors, and cancer. Cancer Res. 2012;72(21):5441–5447. doi: 10.1158/0008-5472.CAN-12-1600. [DOI] [PubMed] [Google Scholar]
- 6.Glaser T, Cappellari AR, Pillat MM, Iser IC, Wink MR, Battastini AM, Ulrich H. Perspectives of purinergic signaling in stem cell differentiation and tissue regeneration. Purinergic Signal. 2012;8(3):523–537. doi: 10.1007/s11302-011-9282-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zimmermann H, Zebisch M, Strater N. Cellular function and molecular structure of ecto-nucleotidases. Purinergic Signal. 2012;8(3):437–502. doi: 10.1007/s11302-012-9309-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Albright RA, Chang WC, Robert D, Ornstein DL, Cao W, Liu L, Redick ME, Young JI, De La Cruz EM, Braddock DT. NPP4 is a procoagulant enzyme on the surface of vascular endothelium. Blood. 2012;120(22):4432–4440. doi: 10.1182/blood-2012-04-425215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stefan C, Jansen S, Bollen M. Modulation of purinergic signaling by NPP-type ectophosphodiesterases. Purinergic Signal. 2006;2(2):361–370. doi: 10.1007/s11302-005-5303-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zylka MJ, Sowa NA, Taylor-Blake B, Twomey MA, Herrala A, Voikar V, Vihko P. Prostatic acid phosphatase is an ectonucleotidase and suppresses pain by generating adenosine. Neuron. 2008;60(1):111–122. doi: 10.1016/j.neuron.2008.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yousef GM, Diamandis M, Jung K, Diamandis EP. Molecular cloning of a novel human acid phosphatase gene (ACPT) that is highly expressed in the testis. Genomics. 2001;74(3):385–395. doi: 10.1006/geno.2001.6556. [DOI] [PubMed] [Google Scholar]
- 12.Yegutkin GG. Nucleotide- and nucleoside-converting ectoenzymes: important modulators of purinergic signalling cascade. Biochim Biophys Acta. 2008;1783(5):673–694. doi: 10.1016/j.bbamcr.2008.01.024. [DOI] [PubMed] [Google Scholar]
- 13.Park J, Gupta RS. Adenosine kinase and ribokinase—the RK family of proteins. Cell Mol Life Sci. 2008;65(18):2875–2896. doi: 10.1007/s00018-008-8123-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Podgorska M, Kocbuch K, Pawelczyk T. Recent advances in studies on biochemical and structural properties of equilibrative and concentrative nucleoside transporters. Acta Biochim Pol. 2005;52(4):749–758. [PubMed] [Google Scholar]
- 15.Young JD, Yao SY, Baldwin JM, Cass CE, Baldwin SA. The human concentrative and equilibrative nucleoside transporter families, SLC28 and SLC29. Mol Aspects Med. 2013;34(2–3):529–547. doi: 10.1016/j.mam.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 16.Blackburn MR, Kellems RE. Adenosine deaminase deficiency: metabolic basis of immune deficiency and pulmonary inflammation. Adv Immunol. 2005;86:1–41. doi: 10.1016/S0065-2776(04)86001-2. [DOI] [PubMed] [Google Scholar]
- 17.Dunwiddie TV, Masino SA. The role and regulation of adenosine in the central nervous system. Annu Rev Neurosci. 2001;24:31–55. doi: 10.1146/annurev.neuro.24.1.31. [DOI] [PubMed] [Google Scholar]
- 18.Porkka-Heiskanen T, Alanko L, Kalinchuk A, Stenberg D. Adenosine and sleep. Sleep Med Rev. 2002;6(4):321–332. doi: 10.1053/smrv.2001.0201. [DOI] [PubMed] [Google Scholar]
- 19.Antonioli L, Pacher P, Vizi ES, Hasko G. CD39 and CD73 in immunity and inflammation. Trends Mol Med. 2013;19(6):355–367. doi: 10.1016/j.molmed.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacobson KA, Balasubramanian R, Deflorian F, Gao ZG. G protein-coupled adenosine (P1) and P2Y receptors: ligand design and receptor interactions. Purinergic Signal. 2012;8(3):419–436. doi: 10.1007/s11302-012-9294-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen JF, Eltzschig HK, Fredholm BB. Adenosine receptors as drug targets—what are the challenges? Nat Rev Drug Discov. 2013;12(4):265–286. doi: 10.1038/nrd3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sperlagh B, Vizi ES. The role of extracellular adenosine in chemical neurotransmission in the hippocampus and Basal Ganglia: pharmacological and clinical aspects. Curr Top Med Chem. 2011;11(8):1034–1046. doi: 10.2174/156802611795347564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yun J, Li J, Zuo Z. Transferred inter-cell ischemic preconditioning-induced neuroprotection may be mediated by adenosine A1 receptors. Brain Res Bull. 2014;103:66–71. doi: 10.1016/j.brainresbull.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kermer V, Ritter M, Albuquerque B, Leib C, Stanke M, Zimmermann H. Knockdown of tissue nonspecific alkaline phosphatase impairs neural stem cell proliferation and differentiation. Neurosci Lett. 2010;485(3):208–211. doi: 10.1016/j.neulet.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 25.Funakoshi H, Chan TO, Good JC, Libonati JR, Piuhola J, Chen X, MacDonnell SM, Lee LL, Herrmann DE, Zhang J, Martini J, Palmer TM, Sanbe A, Robbins J, Houser SR, Koch WJ, Feldman AM. Regulated overexpression of the A1-adenosine receptor in mice results in adverse but reversible changes in cardiac morphology and function. Circulation. 2006;114(21):2240–2250. doi: 10.1161/CIRCULATIONAHA.106.620211. [DOI] [PubMed] [Google Scholar]
- 26.Zhao Z, Yaar R, Ladd D, Cataldo LM, Ravid K. Overexpression of A3 adenosine receptors in smooth, cardiac, and skeletal muscle is lethal to embryos. Microvasc Res. 2002;63(1):61–69. doi: 10.1006/mvre.2001.2366. [DOI] [PubMed] [Google Scholar]
- 27.Nureddin A, Epsaro E, Kiessling AA. Purines inhibit the development of mouse embryos in vitro. J Reprod Fertil. 1990;90(2):455–464. doi: 10.1530/jrf.0.0900455. [DOI] [PubMed] [Google Scholar]
- 28.Tsuchimori N, Miyashiro S, Shibai H, Ikegami S. Adenosine induces dormancy in starfish blastulae. Development. 1988;103(2):345–351. doi: 10.1242/dev.103.2.345. [DOI] [PubMed] [Google Scholar]
- 29.Masse K, Bhamra S, Eason R, Dale N, Jones EA. Purine-mediated signalling triggers eye development. Nature. 2007;449(7165):1058–1062. doi: 10.1038/nature06189. [DOI] [PubMed] [Google Scholar]
- 30.Masse K, Eason R, Bhamra S, Dale N, Jones EA. Comparative genomic and expression analysis of the conserved NTPDase gene family in Xenopus. Genomics. 2006;87(3):366–381. doi: 10.1016/j.ygeno.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 31.Masse K, Bhamra S, Allsop G, Dale N, Jones EA. Ectophosphodiesterase/nucleotide phosphohydrolase (Enpp) nucleotidases: cloning, conservation and developmental restriction. Int J Dev Biol. 2010;54(1):181–193. doi: 10.1387/ijdb.092879km. [DOI] [PubMed] [Google Scholar]
- 32.Bourdelas A, Li HY, Carron C, Shi DL. Dynamic expression pattern of distinct genes in the presomitic and somitic mesoderm during Xenopus development. Int J Dev Biol. 2009;53(7):1075–1079. doi: 10.1387/ijdb.072474ab. [DOI] [PubMed] [Google Scholar]
- 33.Iijima R, Kunieda T, Yamaguchi S, Kamigaki H, Fujii-Taira I, Sekimizu K, Kubo T, Natori S, Homma KJ. The extracellular adenosine deaminase growth factor, ADGF/CECR1, plays a role in Xenopus embryogenesis via the adenosine/P1 receptor. J Biol Chem. 2008;283(4):2255–2264. doi: 10.1074/jbc.M709279200. [DOI] [PubMed] [Google Scholar]
- 34.Brown P, Dale N. Adenosine A1 receptors modulate high voltage-activated Ca2+ currents and motor pattern generation in the xenopus embryo. J Physiol. 2000;525(Pt 3):655–667. doi: 10.1111/j.1469-7793.2000.00655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bowes JB, Snyder KA, Segerdell E, Gibb R, Jarabek C, Noumen E, Pollet N, Vize PD. Xenbase: a Xenopus biology and genomics resource. Nucleic Acids Res. 2008;36(Database issue):D761–D767. doi: 10.1093/nar/gkm826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McGinnis S, Madden TL. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004;32(Web Server issue):W20–W25. doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36(Web Server issue):W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nieuwkoop PD, Faber J. Normal table of Xenopus laevis (Daudin) New York: Garland Publishing Inc; 1994. [Google Scholar]
- 40.Loret MO, Pedersen L, Francois J. Revised procedures for yeast metabolites extraction: application to a glucose pulse to carbon-limited yeast cultures, which reveals a transient activation of the purine salvage pathway. Yeast. 2007;24(1):47–60. doi: 10.1002/yea.1435. [DOI] [PubMed] [Google Scholar]
- 41.Hurlimann HC, Laloo B, Simon-Kayser B, Saint-Marc C, Coulpier F, Lemoine S, Daignan-Fornier B, Pinson B. Physiological and toxic effects of purine intermediate 5-amino-4-imidazolecarboxamide ribonucleotide (AICAR) in yeast. J Biol Chem. 2011;286(35):30994–31002. doi: 10.1074/jbc.M111.262659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hellsten U, Khokha MK, Grammer TC, Harland RM, Richardson P, Rokhsar DS. Accelerated gene evolution and subfunctionalization in the pseudotetraploid frog Xenopus laevis. BMC Biol. 2007;5:31. doi: 10.1186/1741-7007-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yanai I, Peshkin L, Jorgensen P, Kirschner MW. Mapping gene expression in two Xenopus species: evolutionary constraints and developmental flexibility. Dev Cell. 2011;20(4):483–496. doi: 10.1016/j.devcel.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baldwin SA, Beal PR, Yao SY, King AE, Cass CE, Young JD. The equilibrative nucleoside transporter family, SLC29. Pflugers Arch. 2004;447(5):735–743. doi: 10.1007/s00424-003-1103-2. [DOI] [PubMed] [Google Scholar]
- 45.Visser F, Zhang J, Raborn RT, Baldwin SA, Young JD, Cass CE. Residue 33 of human equilibrative nucleoside transporter 2 is a functionally important component of both the dipyridamole and nucleoside binding sites. Mol Pharmacol. 2005;67(4):1291–1298. doi: 10.1124/mol.104.005884. [DOI] [PubMed] [Google Scholar]
- 46.Gray JH, Owen RP, Giacomini KM. The concentrative nucleoside transporter family, SLC28. Pflugers Arch. 2004;447(5):728–734. doi: 10.1007/s00424-003-1107-y. [DOI] [PubMed] [Google Scholar]
- 47.Ralevic V, Burnstock G. Receptors for purines and pyrimidines. Pharmacol Rev. 1998;50(3):413–492. [PubMed] [Google Scholar]
- 48.Wilson DK, Rudolph FB, Quiocho FA. Atomic structure of adenosine deaminase complexed with a transition-state analog: understanding catalysis and immunodeficiency mutations. Science. 1991;252(5010):1278–1284. doi: 10.1126/science.1925539. [DOI] [PubMed] [Google Scholar]
- 49.Mohamedali KA, Kurz LC, Rudolph FB. Site-directed mutagenesis of active site glutamate-217 in mouse adenosine deaminase. Biochemistry. 1996;35(5):1672–1680. doi: 10.1021/bi9514119. [DOI] [PubMed] [Google Scholar]
- 50.Sideraki V, Mohamedali KA, Wilson DK, Chang Z, Kellems RE, Quiocho FA, Rudolph FB. Probing the functional role of two conserved active site aspartates in mouse adenosine deaminase. Biochemistry. 1996;35(24):7862–7872. doi: 10.1021/bi952920d. [DOI] [PubMed] [Google Scholar]
- 51.Schmitt SM, Gull M, Brandli AW. Engineering Xenopus embryos for phenotypic drug discovery screening. Adv Drug Deliv Rev. 2014;69–70:225–246. doi: 10.1016/j.addr.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 52.Morin RD, Chang E, Petrescu A, Liao N, Griffith M, Chow W, Kirkpatrick R, Butterfield YS, Young AC, Stott J, Barber S, Babakaiff R, Dickson MC, Matsuo C, Wong D, Yang GS, Smailus DE, Wetherby KD, Kwong PN, Grimwood J, Brinkley CP, 3rd, Brown-John M, Reddix-Dugue ND, Mayo M, Schmutz J, Beland J, Park M, Gibson S, Olson T, Bouffard GG, Tsai M, Featherstone R, Chand S, Siddiqui AS, Jang W, Lee E, Klein SL, Blakesley RW, Zeeberg BR, Narasimhan S, Weinstein JN, Pennacchio CP, Myers RM, Green ED, Wagner L, Gerhard DS, Marra MA, Jones SJ, Holt RA. Sequencing and analysis of 10,967 full-length cDNA clones from Xenopus laevis and Xenopus tropicalis reveals post-tetraploidization transcriptome remodeling. Genome Res. 2006;16(6):796–803. doi: 10.1101/gr.4871006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Narisawa S, Hoylaerts MF, Doctor KS, Fukuda MN, Alpers DH, Millan JL. A novel phosphatase upregulated in Akp3 knockout mice. Am J Physiol Gastrointest Liver Physiol. 2007;293(5):G1068–G1077. doi: 10.1152/ajpgi.00073.2007. [DOI] [PubMed] [Google Scholar]
- 54.Boison D. Adenosine kinase: exploitation for therapeutic gain. Pharmacol Rev. 2013;65(3):906–943. doi: 10.1124/pr.112.006361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moriwaki Y, Yamamoto T, Higashino K. Enzymes involved in purine metabolism—a review of histochemical localization and functional implications. Histol Histopathol. 1999;14(4):1321–1340. doi: 10.14670/HH-14.1321. [DOI] [PubMed] [Google Scholar]
- 56.Diez-Roux G, Banfi S, Sultan M, Geffers L, Anand S, Rozado D, Magen A, Canidio E, Pagani M, Peluso I, Lin-Marq N, Koch M, Bilio M, Cantiello I, Verde R, De Masi C, Bianchi SA, Cicchini J, Perroud E, Mehmeti S, Dagand E, Schrinner S, Nurnberger A, Schmidt K, Metz K, Zwingmann C, Brieske N, Springer C, Hernandez AM, Herzog S, Grabbe F, Sieverding C, Fischer B, Schrader K, Brockmeyer M, Dettmer S, Helbig C, Alunni V, Battaini MA, Mura C, Henrichsen CN, Garcia-Lopez R, Echevarria D, Puelles E, Garcia-Calero E, Kruse S, Uhr M, Kauck C, Feng G, Milyaev N, Ong CK, Kumar L, Lam M, Semple CA, Gyenesei A, Mundlos S, Radelof U, Lehrach H, Sarmientos P, Reymond A, Davidson DR, Dolle P, Antonarakis SE, Yaspo ML, Martinez S, Baldock RA, Eichele G, Ballabio A. A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol. 2011;9(1):e1000582. doi: 10.1371/journal.pbio.1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gonzales C, Ullrich ND, Gerber S, Berthonneche C, Niggli E, Pedrazzini T. Isolation of cardiovascular precursor cells from the human fetal heart. Tissue Eng Part A. 2012;18(1–2):198–207. doi: 10.1089/ten.tea.2011.0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robin E, Sabourin J, Marcillac F, Raddatz E (2013) Involvement of CD73, equilibrative nucleoside transporters and inosine in rhythm and conduction disturbances mediated by adenosine A1 and A2A receptors in the developing heart. J Mol Cell Cardiol [DOI] [PubMed]
- 59.Woodland HR, Pestell RQ. Determination of the nucleoside triphosphate contents of eggs and oocytes of Xenopus laevis. Biochem J. 1972;127(3):597–605. doi: 10.1042/bj1270597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Adams A, Harkness RA. Adenosine deaminase activity in thymus and other human tissues. Clin Exp Immunol. 1976;26(3):647–649. [PMC free article] [PubMed] [Google Scholar]
- 61.Linder N, Lundin J, Isola J, Lundin M, Raivio KO, Joensuu H. Down-regulated xanthine oxidoreductase is a feature of aggressive breast cancer. Clin Cancer Res. 2005;11(12):4372–4381. doi: 10.1158/1078-0432.CCR-04-2280. [DOI] [PubMed] [Google Scholar]
- 62.Abbracchio MP, Burnstock G. Purinergic signalling: pathophysiological roles. Jpn J Pharmacol. 1998;78(2):113–145. doi: 10.1254/jjp.78.113. [DOI] [PubMed] [Google Scholar]
- 63.Burnstock G, Evans LC, Bailey MA. Purinergic signalling in the kidney in health and disease. Purinergic Signal. 2014;10(1):71–101. doi: 10.1007/s11302-013-9400-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Burnstock G, Vaughn B, Robson SC (2013) Purinergic signalling in the liver in health and disease. Purinergic Signal [DOI] [PMC free article] [PubMed]
- 65.Castrop H, Huang Y, Hashimoto S, Mizel D, Hansen P, Theilig F, Bachmann S, Deng C, Briggs J, Schnermann J. Impairment of tubuloglomerular feedback regulation of GFR in ecto-5’-nucleotidase/CD73-deficient mice. J Clin Invest. 2004;114(5):634–642. doi: 10.1172/JCI21851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huang DY, Vallon V, Zimmermann H, Koszalka P, Schrader J, Osswald H. Ecto-5’-nucleotidase (cd73)-dependent and -independent generation of adenosine participates in the mediation of tubuloglomerular feedback in vivo. Am J Physiol Renal Physiol. 2006;291(2):F282–F288. doi: 10.1152/ajprenal.00113.2005. [DOI] [PubMed] [Google Scholar]
- 67.Street SE, Kramer NJ, Walsh PL, Taylor-Blake B, Yadav MC, King IF, Vihko P, Wightman RM, Millan JL, Zylka MJ. Tissue-nonspecific alkaline phosphatase acts redundantly with PAP and NT5E to generate adenosine in the dorsal spinal cord. J Neurosci. 2013;33(27):11314–11322. doi: 10.1523/JNEUROSCI.0133-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Riegelhaupt PM, Cassera MB, Frohlich RF, Hazleton KZ, Hefter JJ, Schramm VL, Akabas MH. Transport of purines and purine salvage pathway inhibitors by the Plasmodium falciparum equilibrative nucleoside transporter PfENT1. Mol Biochem Parasitol. 2010;169(1):40–49. doi: 10.1016/j.molbiopara.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Masse K, Dale N. Purines as potential morphogens during embryonic development. Purinergic Signal. 2012;8(3):503–521. doi: 10.1007/s11302-012-9290-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 1165 kb)