Abstract
Penicillium digitatum is the most destructive postharvest pathogen of citrus fruits, causing fruit decay and economic loss. Additionally, control of the disease is further complicated by the emergence of drug-resistant strains due to the extensive use of triazole antifungal drugs. In this work, an orthologus gene encoding a putative sterol regulatory element-binding protein (SREBP) was identified in the genome of P. digitatum and named sreA. The putative SreA protein contains a conserved domain of unknown function (DUF2014) at its carboxyl terminus and a helix-loop-helix (HLH) leucine zipper DNA binding domain at its amino terminus, domains that are functionally associated with SREBP transcription factors. The deletion of sreA (ΔsreA) in a prochloraz-resistant strain (PdHS-F6) by Agrobacterium tumefaciens-mediated transformation led to increased susceptibility to prochloraz and a significantly lower EC50 value compared with the HS-F6 wild-type or complementation strain (COsreA). A virulence assay showed that the ΔsreA strain was defective in virulence towards citrus fruits, while the complementation of sreA could restore the virulence to a large extent. Further analysis by quantitative real-time PCR demonstrated that prochloraz-induced expression of cyp51A and cyp51B in PdHS-F6 was completely abolished in the ΔsreA strain. These results demonstrate that sreA is a critical transcription factor gene required for prochloraz resistance and full virulence in P. digitatum and is involved in the regulation of cyp51 expression.
Introduction
Fungal infection is one of the three main diseases of crops, other than bacteria and viruses that can result in reductions in agricultural output [1]. Green mold caused by the ascomycete fungi P. digitatum is the most destructive disease of citrus fruit, responsible for up to 90% of total crop losses during postharvest packing, storage, transportation, and marketing [2]. Control of P. digitatum is critical to solving this worldwide problem; however, the emergence of drug-resistant strains due to excessive use of demethylation inhibitor (DMI) fungicides has resulted in less efficient control of this disease [3–5]. Under this circumstance, an understanding of the potential molecular mechanisms involved in DMI resistance is of great significant because it will provide a basis for the designing of novel antifungal chemicals with greater efficacy.
Fungal resistance to azole reagents has been attributed variously to genetic mutations in its target erg11 (cyp51), and/or the upregulation of efflux pump genes such as MDR1, CDR1, and CDR2 [6]. Filamentous fungi, particularly Ascomycetes, often possess two or more CYP51 paralogous: in Aspergillus fumigatus (two), A. nidulans (two), A. flavus (three), Magnaporthe oryzae (two) and species of Fusarium, including F. verticillioides, F. oxysporum f. sp. lycopersici and F. graminearum (three) [7]. Three sterol 14α-demethylase (CYP51) genes were found in P. digitatum, and evidence on the transcriptional regulation of these target genes has emerged to explain the drug-resistant mechanisms of P. digitatum [8]. Hamamoto et al. [9] reported that duplication of a 126-bp DNA element in the cyp51 promoter region led to the increasing resistance of P. digitatum strains to the antifungal drug imazalil. Another case of imazalil-resistance is associated with up-regulated CYP51 expression caused by the insertion of a 199-bp miniature inverted-repeat transposable element (MITE) in the promoter region [10]. In addition to the overexpression of the cyp51, transporter genes from the ATP-binding cassette (ABC) transporter family and the major facilitator super family (MFS) have also been associated with fungicide resistance in P. digitatum. As reported, seven ABC proteins induced by imazalil in P. digitatum contributed to DMI fungicide efflux, and PdMFS1, a typical MFS member, is involved in imazalil-resistance and pathogenicity of P. digitatum [11–14].
The drug resistance mechanisms of fungi may rely on transcription factors acting on effector genes that have been characterized in a number of clinical species [15]. CaUpc2 is a well-characterized transcription factor in Candida albicans that is associated with drug resistance and sterol metabolism. CaUpc2 is required for induction of the erg2 and erg11 ergosterol biosynthesis genes. CaUpc2 deletion strains exhibit reduced ergosterol levels and no induced expression of cyp51 orthologs, which may explain the increased susceptibilities of these strains [16–17]. It was also reported that gain-of-function mutations in CaUpc2 could contribute to azole resistance [18–19]. However, orthologs of upc2 do not appear to exist in P. digitatum, thus it is possible that other transcription factors in P. digitatum serve similar functions as Upc2 in C. albicans.
Sterol regulatory element-binding proteins (SREBPs) contain a basic helix-loop-helix domain with a specific tyrosine residue and function as membrane-bound transcription factors required for virulence, resistance to antifungal drugs, and hypoxia responses in fungi [20]. Sre1, an SREBP transcription factor first characterized in the fission yeast Schizosaccharomyces pombe, is required for adaptation to hypoxia and anaerobic conditions [21]. Sre1, an SrbA ortholog identified in A. fumigates, a well investigated opportunistic pathogenic mold, is not only required for hypoxia response, cell polarity, and full virulence, but also regulates resistance to the azole antifungal drugs [22–24]. A null mutant of SrbA was unable to grow under hypoxia and displayed increased susceptibility to the azole antifungal drugs, demonstrating that SrbA mediate triazole susceptibility through the direct regulation of erg11A expression [24]. Although Upc2 is not an ortholog of SREBPs, these two classes of transcription factors have analogous functions, similar localization and activation patterns, and are proposed to be an example of convergent evolution in the fungal kingdom [24]. Based on these reports, we deduced that P. digitatum might also have a SREBP-like transcript factor involved in antifungal drug responses.
Prochloraz is a type of triazole fungicide that is widely used in Europe, Australia, Asia and South America for gardening and agriculture [25]. However, little is known about prochloraz resistance mechanisms of in P. digitatum. In this study, we report the identification and characterization of an ortholog of Aspergillus SrbA, SreA, in P. digitatum. By constructing an sreA-disrupted strain and a complemented strain, we analyzed the effects of SreA on full virulence, prochloraz (PRC) resistance and ergosterol biosynthetic genes. Our results provide further insight into the molecular mechanisms of fungicide resistance in P. digitatum.
Materials and Methods
Strains and media
The P. digitatum strain HS-F6 previously isolated by our research group [26] was used in this study. All mutant strains were generated from PdHS-F6 through A. tumefaciens-mediated transformation. Conidial suspensions of wild-type HS-F6 and the mutant strains were stored in 20% glycerol solution at -80°C. PdHS-F6 is highly resistant to triazole drug prochloraz with an EC50 value of 7.896 mg/l. P. digitatum strains were cultured on potato dextrose agar (PDA) medium (extract of 200 g potato boiled water, 20 g dextrose, and 15 g agar per liter) at 25°C. The mycelium used for DNA and RNA extraction was obtained by inoculating 20 μl of a conidial suspension (106 spores ml-1) into 100 ml liquid potato dextrose medium (PDA without agar) and growing on a rotary shaker (160 rpm) at 25°C for three days. The A. tumefaciens EHA105 strain, which was generously provided by Dr. Daohong Jiang (Huazhong Agricultural University, China), was grown in YEP medium [26], minimal medium (MM) (K2HPO4 2 g/l, KH2PO4 1.45 g/l, MgSO4·7H2O 0.6 g/l. NaCl 0.3 g/l, CaCl2·2H2O 0.01 g/l, glucose 2 g/l, FeSO4 0.001 g/l, ZnSO4·7H2O 0.005 g/l, CuSO4·5H2O 0.005 g/l, H3BO3 0.005 g/l, MnSO4·H2O 0.005 g/l, Na2MoO4·2H2O 0.005 g/l, NH4NO3 0.5 g/l) and induction medium (IM) (MM salts with 40 mM 2-[N-morpholino] ethanesulfonic acid (MES) pH 5.3, 10 mM glucose, 0.5% (v/v) glycerol) supplemented with 10 μg/ml kanamycin and 60 μg/ml rifampicin at 28°C.
Cloning and sequencing of sreA from P. digitatum
Based on the DNA sequence of A. fumigatus SrbA (GenBank accession no.XM_744169), we identified an SREBP protein-encoding gene Pc20g05880 (GenBank accession no.XM_002563071) in P. chrysogenum. Given that the genome of P. digitatum and P. chrysogenum share high similarity [27], two pairs of specific primers sreA-a, sreA-b, sreA-c and sreA-d (Table 1) were designed according to the relatively conserved sequences of A. fumigatus sreA and P. chrysogenum Pc20g05880 after sequence alignment with ClustalW. Two approximately 1000-bp DNA fragments were amplified from genomic DNA of P. digitatum by PCR, and then cloned into pMD18-T vector (TaKaRa Biotech. Co., Dalian, China) for sequencing. Then an approximately 2000-bp DNA fragment of P. digitatum sreA was obtained after sequence-assembling. The 5’ flanking DNA sequence of sreA was amplified by genome walking using the Genome Walking Kit (TaKaRa Biotech. Co., Dalian, China) with specific primers sreA-e, sreA-f and sreA-g (Table 1). The 3’ flanking unknown DNA sequence of sreA was amplified using SMATer RACE 5’/3’ Kit (TaKaRa Biotech. Co., Dalian, China) with specific primers sreA-h and sreA-i. The DNA sequence of sreA has been deposited in GenBank under accession number KJ939329.
Table 1. Primers used in this study.
| Name | Sequence(5’-3’) | Purpose |
|---|---|---|
| sreA-a | ATTTGAACTACAAAGACTTCTC | PCR primers used to amplify the DNA fragment of sreA. |
| sreA-b | TACCACTCTCGGAAGAACCTATG | PCR primers used to amplify the DNA fragment of sreA. |
| sreA-c | GCCGGTCTGATGGTTCTTGAAGG | PCR primers used to amplify the DNA fragment of sreA. |
| sreA-d | TCCAATGAGAGAAAGCTGGACTGG | PCR primers used to amplify the DNA fragment of sreA. |
| sreA-e | ACACGAGGCCTAAGTTTTGTTGCTG | PCR primers used to amplify the 5’ unknown DNA sequence of sreA. |
| sreA-f | ATCAAGCCGACCGGAAGATTAGGC | PCR primers used to amplify the 5’ unknown DNA sequence of sreA. |
| sreA-g | TTGATCTCCTTCCGAGAACATGGGC | PCR primers used to amplify the 5’ unknown DNA sequence of sreA. |
| sreA-h | ATACTGGGCCCGAAATGCCTACACC | PCR primers used to amplify the 3’ unknown DNA sequence of sreA. |
| sreA-i | TAAGAAATACAGGACCCGTCGACGC | PCR primers used to amplify the 3’ unknown DNA sequence of sreA. |
| sreA-1 | CCCTCGAGATGTCTGGCCCCAATATGGAG | PCR primers used to amplify 5’ fragments of sreA. |
| sreA-2 | GGACTAGTCAGCAAAGACTCCATGGTTTGC | PCR primers used to amplify 5’ fragments of sreA. |
| sreA-3 | CGAGCTCTTTCGAAGCAAGCGAGAAGG | PCR primers used to amplify 3’ fragments of sreA. |
| sreA-4 | GGGGTACCTCAGGCAGGGACATTTTGCA | PCR primers used to amplify 3’ fragments of sreA. |
| sreA-F | ATGGATGTCTGGCCCCAATATGGAG | PCR primers used to amplify the ORF of sreA gene. |
| sreA-R | TCAGGCAGGGACATTTTGCAC | PCR primers used to amplify the ORF of sreA gene. |
| sreA-F1 | GGACTAGTGGCAATAGTGGAGACTA GCAC | PCR primers used to amplify sreA, including its promoter and terminator. |
| sreA-R1 | GGACTAGTCTGATAACATTCCATTTCCC | PCR primers used to amplify sreA, including its promoter and terminator. |
| cyp51A-F | CACTGGATTCCTTTCATTGGG | PCR primers used to amplify cyp51A gene by quantitative real-time PCR. |
| cyp51A-R | TCCGAAGACGGGGGTTGTAA | PCR primers used to amplify cyp51A gene by quantitative real-time PCR. |
| cyp51B-F | GAGTTCATCCTCAATGGCAAGC | PCR primers used to amplify cyp51B gene by quantitative real-time PCR. |
| cyp51B-R | CTTAGAGTTGGGGCAATCGTAGAC | PCR primers used to amplify cyp51B gene by quantitative real-time PCR. |
| cyp51C-F | TGTTCAAGCAGCCATTCAAGC | PCR primers used to amplify cyp51C gene by quantitative real-time PCR. |
| cyp51C-R | CAAGTTGGGTCCGACGAAATA | PCR primers used to amplify cyp51C gene by quantitative real-time PCR. |
| β-actin-F | TGTCACCAACTGGGACGATA | PCR primers used to amplify β-actin gene by quantitative real-time PCR. |
| β-actin-R | GAGCTTCGGTCAAGAGGATG | PCR primers used to amplify β-actin gene by quantitative real-time PCR. |
Note: The underlined sequences represent different restriction enzyme sites.
DNA and protein analysis
The DNA sequence of sreA was analyzed using NCBI BLAST and BioEdit software. A protein analysis was performed using the NCBI BLAST and Interpro (http://www.ebi.ac.uk/interpro/) programs. TMHMM software (version 2.0) was used to predict the transmembrane domains of SreA.
Construction of an sreA disruption plasmid
The plasmid pTFCM containing the PtrpC promoter and TtrpC terminator from A. nidulans and carrying the hph gene which confers resistance to hygromycin B as selective marker was generously provided by Dr. Daohong Jiang (Huazhong Agricultural University, China). The sreA disruption plasmid was constructed by inserting the up-stream and down-stream flanking sequences of sreA into the pTFCM vector [28]. To this end, a 1-kb DNA fragment of the sreA 5’ coding sequence was amplified from P. digitatum genomic DNA using primers sreA-1 and sreA-2 (Table 1) and cloned into pMD18-T vector. After sequencing, the fragment containing sreA 5’ coding sequence was digested by SpeI and XhoI and sub-cloned into pTFCM between SpeI and XhoI sites to generate the pTFCM-L plasmid. Next, another 1-kb fragment containing the sreA 3’ coding sequence was amplified and cloned into the pMD18-T vector for sequencing using primers sreA-3 and sreA-4 (Table 1). After digestion by restriction enzymes KpnI and SacI, the fragment containing sreA 3’ coding sequence was inserted between the KpnI and SacI sites of pTFCM-L to generate the sreA disruption plasmid pTFCM-L-R (Fig. 1A).
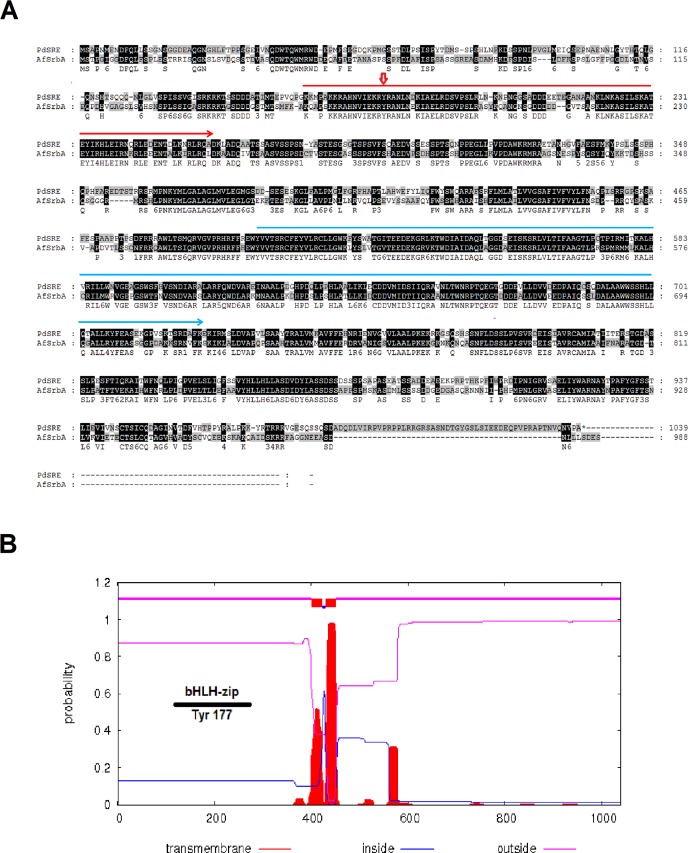
Fig 1. Sequence analysis of sreA from PdHS-F6.
A: Homology between the amino acid sequences of SreA from P. digitatum and SrbA in A. fumigatus. The sequence under the red line indicates basic helix-loop-helix-leucine zipper (bHLH-zip) domain located at the N-terminus of the protein. The sequence under the blue line indicates the DUF2014 domain specific to the ER membrane-bound transcription factor Sterol Regulatory Element Binding Proteins (SREBPs). The arrow indicates the amino acid position of the highly conserved tyrosine in the bHLH zipper domain (bHLH-zip). B: Transmembrane domain prediction plots of SreA.
Construction of sreA complementation plasmid
The sreA complementation plasmid was obtained by a PCR strategy (Fig. 1A). A DNA fragment containing the sreA open reading frame as well as its promoter and terminator (4385bp) was amplified using genomic DNA of P. digitatum as a template and sreA-F1/sreA-R1 as primers (Table 1). The PCR conditions were as follows: 3 min at 94°C followed by 30 cycles each of 30 s at 94°C, 1 min at 60°C, and 2 min at 72°C. A GeneAmp 9700 thermal cycler (Perkin-Elmer [PE] Applied Biosystems, Foster City, Calif., USA) was used with LA Taq polymerase (TaKaRa Biotech. Co., Dalian, China). The amplified fragment was digested by SpeI and cloned into plasmid pTFCM-neo to obtain the complementation plasmid pTFCM-neo-sreA. The pTFCM-neo plasmid was constructed by replacing the hph cassette between two XbaI sites in pTFCM with a neo cassette that confers resistance to the antibiotic G418.
Transformation of P. digitatum
Prior to the transformation, the recombinant plasmids pTFCM-L-R and pTFCM-neo-sreA were transformed into A. tumefaciens strain EHA105 by a heat-shock method [29]. Next, A. tumefaciens-mediated transformation was performed to obtain sreA disruption (ΔsreA) and complementation (COsreA) strains [28, 30]. First, A. tumefaciens strains harboring the disruption plasmids were recovered on YEP plates at 28°C for two days. A single colony containing pTFCM-L-R plasmids was selected and cultured in MM medium before transfer into IM medium cointaining 200 μM acetosyringone; the cell density was adjusted to OD 600 = 0.15. After incubating at 28°C with shaking at 180 rpm for 6 h, equal volumes of A. tumefaciens culture and P. digitatum conidial suspensions (106 spores ml-1) were mixed and transferred to a lens paper on an IM agar plate for cultivation at 25°C for 3 days. Finally, the lens paper was transferred onto the PDA medium containing 50 μg/ml hygromycin B and 50 μg/ml cefoxitin to select for sreA disruption strains (ΔsreA). To obtain sreA complementation strains (COsreA), one selected ΔsreA strain was transformed with the complementation plasmid pTFCM-neo-sreA according to the protocol of ΔsreA strain construction, with the modification that G418 (200 μg/ml) was used to select transformants instead of hygromycin B. The ΔsreA and COsreA transformants were confirmed by PCR using primers sreA-F and sreA-R (Table 1) and a Southern blot analysis.
Southern blot analysis
Genomic DNA was extracted from the HS-F6 wild-type, ΔsreA, and COsreA strains using Biospin Fungus DNA Extraction Kit (BioFlux, Tokyo, Japan). Approximately 30 μg genomic DNA was digested with HindIII at 37°C for 12 hours, electrophoresed on a 1% agarose gel and transferred to a positively charged nylon membrane. A 1003-bp digoxigenin-labeled probe specific to the 5’ region of sreA was generated using the DNA Probe Labeling Kit (TIANDZ, Beijing, China) by PCR with primers sreA-1 and sreA-2 (Table 1) and hybridized with the genomic DNA blot. DIG Random Labeling and Detection Kit II (BOSTER, Wuhan, China) was used for color detection following the manufacturer’s protocol.
Assays of vegetative growth and prochloraz EC50
To compare the growth of HS-F6 wild-type, ΔsreA, and COsreA strains, a 0.8-mm mycelial plug was obtained from a PDA plate with 50 μl of the corresponding conidial suspension (1×106 spores ml-1) coated on the surface and then cultured on a new PDA plate. After four days’ cultivation at 25°C, the diameters of different colonies were measured.
EC50 values of prochloraz (1-{N-propyl-N-[2-(2,4,6-trichlorophenoxy)ethyl]carbamoyl}; PRC) for the different strains were measured according to [31] with modifications. Briefly, 50 μl of a conidial conidia suspension (106 spores ml-1) of the HS-F6 wild-type and mutant strains was coated onto a PDA plate respectively and cultivated at 25°C for 24 h. Mycelial plugs (approximately 0.8 mm diameter) were then obtained from the plate using a punch and placed on the center of PDA plates containing different concentrations of prochloraz. After incubation at 25°C for 6 days, the diameters of the colonies were measured. Three replicates were used for each experiment. The average of the colony diameters in each independent test was used for EC50 calculation by software SPSS 10.0.
Virulence assay
Mature citrus fruits (Citrus sinensis) were purchased from a fruit market in Hongshan district, Wuhan. The fruits were washed with distilled water and dried at room temperature before inoculation. Virulence assays for the HS-F6 and mutant strains were performed directly on citrus fruits. Firstly, a 2-mm deep hole was made on the pericarp using a 1-ml pipett tip. Then 3 μl of a conidial suspension (106 spores ml-1) of the HS-F6 wild-type or mutants was injected into the hole. After incubation at 25°C for three days, the diameters of the disease spots formed were measured and compared.
RNA extraction and quantitative real-time PCR (qRT-PCR)
qRT-PCR was used to analyze cyp51 gene expression. Before RNA extraction, 20 μl of a conidial suspension (106 spores ml-1) of P. digitatum HS-F6 and ΔsreA strains was cultured in PDB medium at 25°C for 72 h. In the prochloraz-treatment experiment, 7 μg/ml prochloraz (about the concentration of EC50) was added to the PDB medium with shaking for an extra 6 h after cultured at 25°C for 48 h. The mycelia were then filtered and washed several times using double distilled water. Total RNA was extracted using RNAiso Plus (TaKaRa Biotech. Co., Dalian, China) according to the manufacturer’s protocol. All RNA samples were treated with DNase I (TaKaRa Biotech. Co., Dalian, China). First-strand cDNA was prepared using All-in-one First strand cDNA Synthesis Kit (Genecopoeia, Guangzhou, China) following the manufacturer’s protocol. qRT-PCR was performed using a BIO-RAD CFX96 q-PCR system with SYBR Green I fluorescent dye detection. The mRNA abundance was normalized using the housekeeping gene β-actin, and the relative expression levels were calculated using the 2-ΔΔCt method [32]. The primers used to amplify cyp51A/B/C and β-actin genes in qRT-PCR are listed in Table 1.
Statistical analysis
All results presented with statistical significance were analyzed with an unpaired two-tailed Student’s t test and two-way ANOVA. P<0.05 was considered significant.
Results
Cloning and sequence analysis of sreA from PdHS-F6
A 3120-bp DNA fragment of sreA ORF was amplified from PdHS-F6 genomic DNA using primers sreA-F and sreA-R (Table 1), sequenced, and then analyzed by NCBI BLAST. The sreA ORF is predicted to encode a protein of 1040 amino acids that is a putative HLH transcription factor and an ortholog of A. fumigatus transcription factor SrbA. The nucleotide homology between sreA and srbA is 63%; protein homology is 61% (Fig. 1). The nucleotide homology between sreA and P. chrysogenum Pc20g05880 is 89%.
Two highly conserved domains in SreA were identified. A basic helix-loop-helix-leucine zipper (bHLH-zip) domain located in the N-terminus of the protein contains a unique tyrosine residue (at site 177) that distinguishes SREBPs from other bHLH transcriptional factors [33]; a domain DUF2014 of unknown function is located in the C-terminus of the protein. This domain is found at the C-terminus of a family of ER membrane bound transcription factors called sterol regulatory element binding proteins (SREBP). SreA is predicted to contain two transmembrane domains, suggesting that this protein is a membrane-bound transcriptional factor with a topology similar to vertebrate SREBPs.
Construction of sreA-disruption and -complementation strains
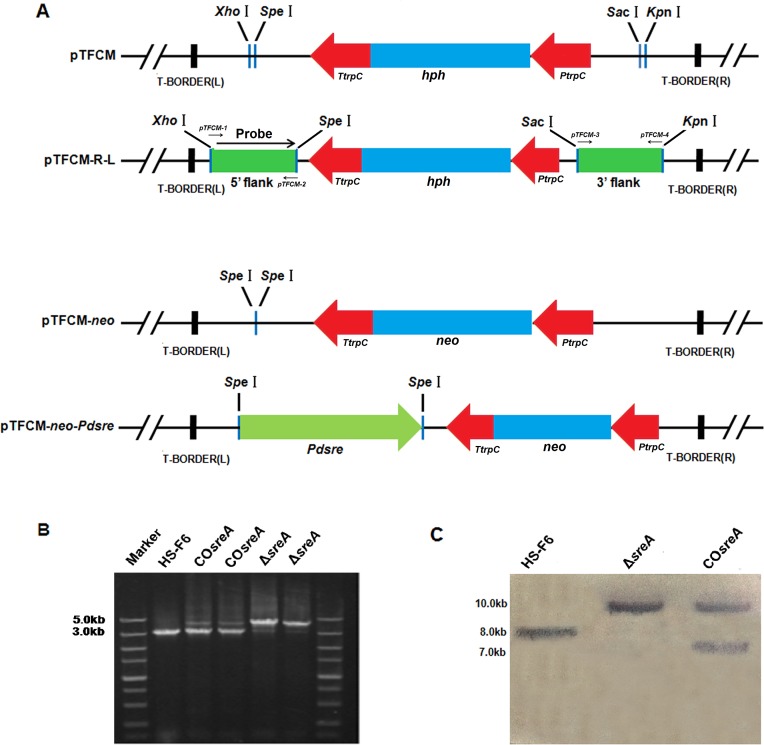
To analyze the function of SreA in P. digitatum, sreA disruption strains (ΔsreA) of HS-F6 (PdHS-F6m) were constructed (Fig. 2A). Twenty putative mutants were selected on PDA medium supplemented with 50 μg/ml hygromycin B and screened by PCR using primers sreA-1 and sreA-4 (Table 1). The 3.0-kb fragment of sreA in HS-F6 was replaced by a 4.0-kb fragment (the sreA gene disrupted with the hph gene) in the recombinant strains (ΔsreA) (as shown in Fig. 2B). Southern blot analysis further confirmed that a single copy was integrated. Only one strain was chosen and used for further study (Fig. 2C).
Fig 2. Construction and analysis of sreA strains.
A: Construction of pTFCM-R-L and pTFCM-neo-sreA plasmids. B: PCR analysis of ΔsreA, and COsreA transformants using primers sreA-F and sreA-R (Table 1). C: Southern blot analysis of P. digitatum wild-type PdHS-F6 and ΔsreA, COsreA strains using a probe specific to the 5’ region of Pdsre. 30μg genomic DNA was digested with HindIII and detected using a probe specific to the 5’ region of sreA gene.
Twenty complementation strains (COsreA) were obtained using G418 as selective marker. The insertion of sreA in the COsreA strains was confirmed by PCR using primers sreA-1 and sreA-4. The occurrence of a 3.0-kb band (sreA) and a 4.0-kb band (sreA disrupted with the hph) indicated the successful integration of the sreA gene into the genome of the ΔsreA strains (Fig. 2B). Southern blotting confirmed that the ΔsreA and COsreA strains were successfully constructed and that a single copy of sreA was integrated into the genome of the ΔsreA strain to generate the COsreA strain (Fig. 2C).
Deletion of sreA renders P. digitatum more susceptible to triazole drug prochloraz
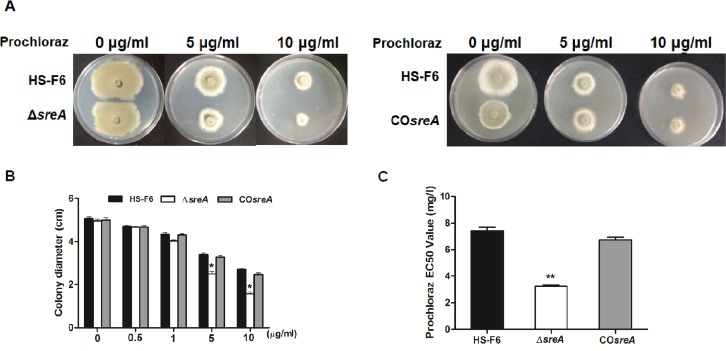
As stated above, PdHS-F6 is highly-resistant to triazole drug prochloraz and has an EC50 value of 7.896 mg/l, which is more than 800 times higher than prochloraz-susceptible strains. As shown in Fig. 3, PdHS-F6 and ΔsreA strains showed similar growth on the PDA plate without prochloraz, the diameters of colonies were not significantly different. However, the diameters of colonies of ΔsreA strains on PDA plates supplemented with prochloraz (5 μg/ml, 10 μg/ml) were smaller than those of HS-F6. After being cultured on PDA plates supplemented with different concentration of prochloraz (0, 0.5, 1, 5, 10 μg/ml) for 6 days, the diameters of the colonies were measured (Fig. 3B), and the EC50 value was calculated. The average EC50 value of prochloraz for the ΔsreA strain was 3.2 mg/l, which was less than half of the value of HS-F6 (Fig. 3C). However, complementation of sreA (PdHS-F6c) restored the EC50 value of the ΔsreA strain (average EC50 value of prochloraz for the COsreA strain was 6.7 mg/l). These results demonstrated that the deletion of sreA renders P. digitatum more susceptible to triazole drug prochloraz, suggesting that sreA plays an important role in the resistance of P. digitatum to triazole drugs.
Fig 3. The sensitivity of P. digitatum wild-type (PdHS-F6), ΔsreA, and COsreA strains to prochloraz.
A: Growth assay of P. digitatum HS-F6 wild-type, ΔsreA and COsreA strains on PDA plates with or without prochloraz (concentrations: 0, 5 and 10μg/ml). All strains were cultured at 25°C for three days. B: Bars represent the average diameter plus standard errors of colonies grown on PDA plates supplemented with different concentrations of prochloraz (concentrations: 0, 0.5, 1, 5,10μg/ml). C: Comparison of prochloraz EC50 values of the PdHS-F6 wild-type, ΔPdsre, and COPdsre strains. Each bar represents the EC50 value plus standard error of three measurements. (*P<0.05; **P<0.01)
Gene sreA is required for full virulence in P. digitatum
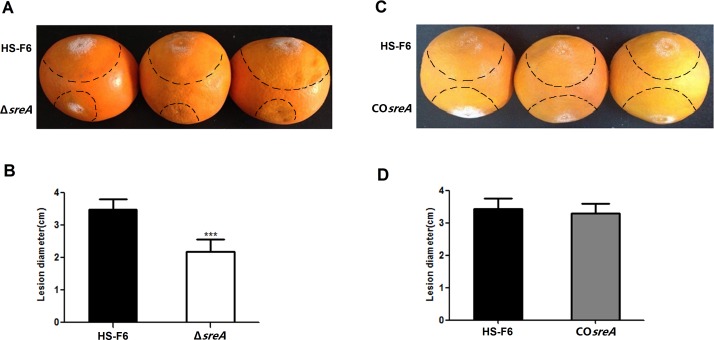
To determine whether sreA plays a role in the virulence of P. digitatum toward citrus fruits, a virulence assay was performed directly on these fruits. The assay results demonstrated that the symptoms in the fruits incubated with conidial suspension of the ΔsreA strains developed more slowly than in the fruits incubated with the wild-type conidial suspension. The mean diameter of the macerated lesions of the fruits incubated with the ΔsreA conidial suspension was approximately 1.90 cm at 3 days post inoculation, whereas the mean macerated lesion diameter of the fruits incubated with the HS-F6 conidial suspension was about 3.4 cm (Fig. 4A, B). The virulence assay results revealed that the deletion of sreA rendered the ΔsreA strain less virulent compared with the wild-type HS-F6. A further experiment was performed to confirm this result. The virulence assay demonstrated that average diameter of the macerated lesions induced by COsreA was comparable to that of wild type HS-F6 (3.2 cm) (Fig. 4C, D). These results indicated that SreA is required for full virulence in P. digitatum.
Fig 4. Virulence assay for the P. digitatum HS-F6, the ΔsreA strian, and the COsreA strain.
A: A 3μL aliquot of a conidial suspension (106 spores ml1) of the HS-F6 or ΔsreA strain was injected into citrus fruits and incubated at 25°C for three days. B: Bars represent the mean diameter plus standard errors of 20 disease spots. (***P<0.001); C: 3μl conidial suspension (106 spores ml-1) of HS-F6 or COsreA strains was injected into the citrus fruits and incubated at 25°C for three days; D: Bars represent the mean diameter plus standard errors of 20 disease spots.
Transcriptional abundance of the cyp51 genes is significantly decreased in the ΔsreA strain
Because SrbA regulates the expression of erg11A gene in A. fumigatus [21], we hypothesized that its ortholog, SreA, plays a similar role in P. digitatum. To test this hypothesis, the mRNA abundance of the cyp51 genes in PdHS-F6 wild-type and the ΔsreA strain was analyzed using qRT-PCR (Fig. 5). The results showed that transcriptional abundance of the three cyp51 genes were all decreased in the ΔsreA strain, especially with regard to cyp51A. The normalized expression values of cyp51A, cyp51B, and cyp51C in the ΔsreA strain were 0.02, 0.29 and 0.47, respectively.
Fig 5. Differential cyp51A, B, and C transcriptional abundance for P. digitatum wild-type (HS-F6) and ΔsreA strains.
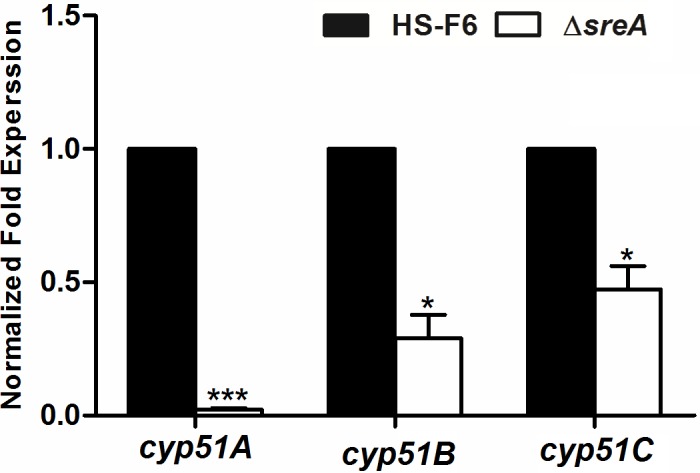
RNA was isolated from three-day-old mycelium of HS-F6 and ΔsreA strains for qRT-PCR analysis. The mRNA abundance was normalized by the housekeeping gene β-actin. The relative expression levels were calculated using the 2-ΔΔCt method. Three biological replicates were performed. (*P<0.05; ***P<0.001)
SreA is required for transcriptional response to prochloraz in P. digitatum
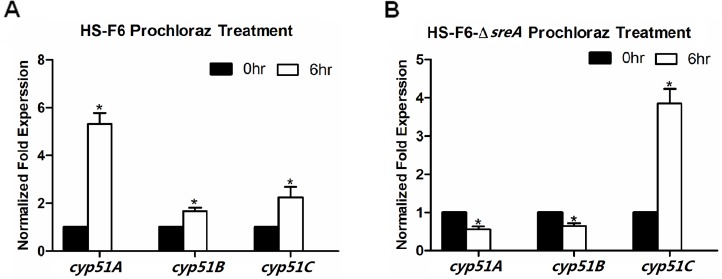
To investigate the role of sreA in the transcriptional response of P. digitatum to prochloraz, a prochloraz induction experiment was performed with the ΔsreA and HS-F6 strains. Total RNA was isolated from the PdHS-F6 and ΔsreA strains with or without prochloraz treatment and used for qRT-PCR. In the case of wild-type HS-F6, the expression levels of the cyp51 genes were all increased after 6 h of prochloraz treatment (Fig. 6A); the normalized expression values in the cyp51A, cyp51B and cyp51C expression levels after prochloraz treatment were 5.3, 1.8, and 2.2, respectively. However, the induction of cyp51A and cyp51B by prochloraz was abolished in the ΔsreA strain. As shown in Fig. 6B, the normalized expression values for cyp51A and cyp51B mRNA abundances were 0.56 and 0.65, respectively. Notably, the increasing fold-change in cyp51C mRNA abundance after prochloraz treatment was higher in the ΔsreA strain than in the wild-type strain HS-F6 (Fig. 6A, B), which might be due to regulation by other transcription factors that maintained cyp51C transcript level in response to prochloraz.
Fig 6. Transcriptional abundance of cyp51 genes in HS-F6 wild-type and Δsre strains after prochloraz treatment.
The wild-type and Δsre strain were treated with or not with prochloraz (7μg/ml) and shaking at 25°C for 6 hours. Total RNA was isolated for qRT-PCR as described in Materials and methods. A: Bars represent the relative expression levels of CYP51 genes in HS-F6. B: Bars represent the relative expression levels of CYP51 genes in the ΔsreA strain; mRNA abundance was normalized by the housekeeping gene β-actin. The relative expression levels were calculated using the 2-ΔΔCt method. Three biological replicates were performed. (*P<0.05)
Discussion
A number of transcription factors associated with sterol metabolism and drug resistance have been well characterized in different clinical fungi. Upc2, a typical zinc cluster protein identified in S. cerevisiae and C. albicans, was the first documented transcription factor highly correlated with fungal drug resistance [34–37]. Deletion of CaUpc2 rendered C. albicans increased susceptibility to a range of common antifungal drugs used in clinical therapy, and this increased susceptibility is observed for drugs targeting the ergosterol biosynthesis pathway [16]. A previous report also provided direct evidence for Upc2 in transcriptional regulation of C. albicans erg genes involved in ergosterol biosynthesis, including erg7, erg11 and erg25 [38]. The erg11 overexpression in the Upc2 gain-of-function mutants led to strain resistance to azole fungicides [39]. SREBPs, functionally conserved in fungal kingdom, have been revealed to regulate sterol synthesis in fission yeast S. pombe as well as a number of fungal pathogens particularly grown under hypoxia conditions [20, 40]. Sre1 undergoes sterol-dependent proteolytic activation and regulates genes required for maintaining cellular sterol homeostasis. Sre1 is also required for many other fungi to regulate sterol biosynthesis, and SREBP is required for mammalian cells to regulate cholesterol and fatty acid anabolism [41–42]. SrbA from the SREBP super family is associated with hypoxia, cell polarity, full virulence, and ergosterol homeostasis in A. fumigatus. It is notable that SrbA plays a critical role for triazole drug interactions in A. fumigates, which may have clinical importance [23–24]. In most eukaryotes, including the majority of fungi, expression of sterol biosynthesis genes is regulated by SREBPs. However, in yeasts such as S. cerevisiae and C. albicans, sterol synthesis is regulated by Upc2 instead [43]. Even though Upc2 functions similarly to SrbA, they are totally different from each other in terms of the protein structure: Upc2 is a zinc finger transcription factor with a typical Gal4-type zinc finger, while SREBPs contain a bHLH domain and a characteristic tyrosine residue.
In the present study, we characterized an SREBP transcriptional factor, SreA, in P. digitatum as an ortholog of SrbA that is associated with triazole susceptibility, full virulence, and regulation of cyp51 genes [24]. Our results demonstrated that SreA in P. digitatum may play a role similar to SrbA. The sreA deletion strains of P. digitatum (ΔsreA) were generated using A. tumefaciens-mediated genetic transformation based on the prochloraz-resistant P. digitatum strain PdHS-F6. Prochloraz is a triazole fungicide that is widely used in Europe, Australia, Asia and South America within gardening and agriculture [25]. However, with the extensive and excessive use of prochloraz, drug-resistant fungi have appeared. As reported in 2013, 78 strains of P. digitatum were isolated from citrus fruits collected in Hubei Province and 25 isolates were identified to be prochloraz-resistant, with a proportion as high as 32% [26]. For reducing the increasing economic loss due to fungicide resistance, understanding the molecular mechanisms of resistance is of practical significance.
Our results in P. digitatum demonstrated that the deletion of sreA increases susceptibility of PdHS-F6 to prochloraz. As shown in Fig. 3, the prochloraz EC50 value of the ΔsreA strain was significantly lower than HS-F6. The growth assay on PDA plates revealed that the ΔsreA strain grew as well as PdHS-F6 without prochloraz. However, the growth rate of ΔsreA strain on PDA plates supplemented with prochloraz (5 μg/ml, 10 μg/ml) was much slower than that of the HS-F6 (Fig. 3A, B), revealing that drug response was altered in the ΔsreA strain. CYP51 proteins are the targets of triazole fungicides, and the susceptibility of fungi to triazole drugs is closely related with the mutations in or the expression level of cyp51 genes [44]. To confirm the role of SreA in the regulation of cyp51 gene expression, the transcriptional abundance of cyp51 genes in HS-F6 and ΔsreA strain were analyzed by qRT-PCR. The results revealed that the expression level of the three cyp51 genes was decreased in the ΔsreA strain, particularly cyp51A, which almost could not be detected (Fig. 6). Our study thus demonstrated that SreA is an important regulator of cyp51 genes: the deletion of sreA reduced the expression levels of cyp51 genes, which are the targets of trizole fungicide prochloraz, rendering the strain more susceptible to the drug. However, SreA is not the only transcription factor regulating the expression of cyp51 genes. In the ΔsreA strain, other regulators may control the expression of cyp51 genes in response to the drug and to some extent may compensate the loss of sreA. In support of this notion, the EC50 value of the ΔsreA strain was not as low as the prochloraz-susceptible strain, and the ΔsreA strain could still grow on PDA plates supplemented with prochloraz at a relatively high concentration; nevertheless, the growth rate was much slower than that of HS-F6.
To determine whether sreA is associated with pathogenicity of P. digitatum, virulence assays were performed directly on citrus fruits using the HS-F6 wild-type and mutant strains. The results demonstrated that the ΔsreA strain was less virulent than the wild type. The symptoms in fruits incubated with a ΔsreA conidial suspension developed more slowly and the mean diameter of the macerated lesions was almost half of those produced by the HS-F6 wild-type strain (Fig. 4). As expected, the COsreA strain displayed symptoms similar to the wild type, indicating that the complementation of sreA restored the virulence of ΔsreA strain. Therefore, we can conclude that SreA is required for full virulence in P. digitatum.
Fungi have evolved sophisticated mechanisms to cope with environmental stresses, including antifungal drugs, and respond to triazole drugs by altering the expression levels of effector genes. DMI-resistant isolates were initially thought to exhibit fitness penalties that would preclude DMI resistance and becoming wide-spread in nature, as reported in the studies of several important fungal pathogens of different crops [45]. In the past few years, DMI-resistant strains have also been identified in human pathogenic fungi. For example, erg25 transcription is induced in response to itraconazole treatment in C. albicans [44]. Besides, the triazole-induced overexpression of cyp51 genes in fungi has been regarded as an important fungal self-protective mechanism towards a range of fungicides [24]. In this study, we compared the cyp51 mRNA expression in PdHS-F6 and ΔsreA strains before and after prochloraz treatments. In the HS-F6 wild-type strain, the expression level of cyp51 genes were all increased after prochloraz induction for 6 h (Fig. 6A). However, the increase in cyp51 expression levels, except cyp51C, was abolished in the ΔsreA strain. The further increased mRNA abundance of cyp51C in response to prochloraz-treatment in the ΔsreA strain might be attributed to the regulating by other transcription factors in response to the drug. However, the relatively decreased expression level of cyp51C in the ΔsreA strain demonstrated that sreA could regulate the expression of cyp51C to some extent. These results indicated that SreA is an important regulator of cyp51 genes. The deletion of sreA renders P. digitatum unable to cope with prochloraz properly, making it more susceptible to the antifungal drug.
With the widespread and constant use of triazloe antifungal drugs, resistance has been selected in many fungal species. Most of this resistance is due to the mutation and overexpression of target cyp51 genes or the mutation of their regulatory genes [5–6, 18–19]. Besides, the overexpression of transporter-encoding genes also contributed to fungi resistance [46]. The documented alterations in transcription factors, particularly in their DNA binding sites, could affect drug resistance in the pathogenic yeast C. albicans, tumorigenesis in hosts, and for resistance of the malarial parasite Plasmodium vivax [18, 47–51]. Therefore, identifying new target genes and the designing of new drugs is of great significance to control the spread of resistant fungi. Previously, we reported the cloning, expression, and characterization of cyp51 from P. digitatum and Ustilago maydis [52–55]. The structural characteristics of the interaction between heterologous CYP51 and commercial azoles were also analyzed by binding assays. A series of new 2-azolyl-3, 4-dihydroquinazolines 6 was synthesized by the direct cyclization of imidazole or 1, 2, 4-triazole with carbodiimides 4, and the preliminary bioassay results demonstrated that these compounds exhibited well to significant fungicidal activity against P. digitatum [54]. The screening of new DMI fungicides based on optimized expression has been carried out for the first time in Ustilago maydis [55]. Recently, a cell-based high-throughput screen has identified small-molecule inhibitors of the Upc2-dependent induction of sterol gene expression in response to azole drug treatment. The compounds were growth-inhibitory and could attenuate antifungal-induced sterol gene expression in vivo [56]. Thus, as a regulator of cyp51, SreA, with an essential function in P. digitatum fungicide resistance, represents a promising research direction to uncover a new fungus-specific antifungal drug targets.
Acknowledgments
The authors sincerely thank Dr. Christopher D. Rock (Department of Biological Sciences, Texas Tech University, TX, USA) for the suggestions and critical reading on the manuscript.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was supported by the National Natural Science Foundations of China (NO. 31371893, 31071653, 31101595). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Oerke EC, Dehne HW, Schonbeck F, Weber A (1994) Crop Protection and Crop Production. Amsterdam: Elsevier; [Google Scholar]
- 2. Eckert J, Sievert J, Ratnayake M (1994) Reduction of imazalil effectiveness against citrus green mold in California packinghouses by resistant biotypes of Penicillium digitatum . Plant Dis 78: 971–974. [Google Scholar]
- 3. Sanchez-Torres P, Tuset JJ (2011) Molecular insights into fungicide resistance in sensitive and resistant Penicillium digitatum strains infecting citrus. Posthavest Biol Tec 59: 159–165. [Google Scholar]
- 4. Smilanick JL, Mansour MF, Mlikota GF, Goodwine WR (2006) The effectiveness of pyrimethanil to inhibit germination of Penicillium digitatum and to control citrus green mold after harvest. Posthavest Biol Tec 42: 75–85. [Google Scholar]
- 5. Tim JH, Derek WH (1997) Molecular mechanisms of azole resistance in fungi. FEMS Microbiol Lett 149: 141–149. [DOI] [PubMed] [Google Scholar]
- 6. White TC, Marr KA, Bowden RA (1998) Bowden, Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin Microbiol Rev 11: 382–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fan JR, Martin U, Josie EP, Helen CB, Steven L, et al. (2013) Characterization of the sterol 14a-demethylases of Fusarium graminearum identifies a novel genus-specific CYP51 function. New Phytol 198: 821–835 10.1111/nph.12193 [DOI] [PubMed] [Google Scholar]
- 8. Sun X, Wang J, Feng D, Ma Z, Li H, et al. (2011) PdCYP51B, a new putative sterol 14α-demethylase gene of Penicillium digitatum involved in resistance to imazalil and other fungicides inhibiting ergosterol synthesis. Applied Microbiol Biot 9: 1107–1119. [DOI] [PubMed] [Google Scholar]
- 9. Hamamoto H, Hasegawa K, Nakaune R, Lee YJ, Makizumi Y, et al. (2000) Tandem repeat of a transcriptional enhancer upstream of the sterol 14alpha-demethylase gene (CYP51) in Penicillium digitatum . Appl Environ Microbiol 66: 3421–3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ghosoph JM, Schmidt LS, Margosan DA, Smilanick JL (2007) Imazalil resistance linked to a unique insertion sequence in the PdCYP51 promoter region of Penicillium digitatum . Posthavest Biol Tec 44: 9–18. [Google Scholar]
- 11. Wolfger H, Mamnun YM, Kuchler K (2001) Fungal ABC proteins: pleiotropic drug resistance, stress response and cellular detoxification. Res. Microbiol 152: 375–389. [DOI] [PubMed] [Google Scholar]
- 12. Hamamoto H, Nawata O, Hasegawa K, Nakaune R, Lee YJ, et al. (2001) The Role of the ABC transporter gene PMR1 in demethylationinhibitor resistance in Penicillium digitatum . Pestic Biochem Phys 70: 19–26. [Google Scholar]
- 13. Sun X, Ruan R, Lin L, Zhu C, Zhang T, et al. (2013) Genomewild investigation into DNA elements and ABC transporters involved in imazalil resistance in Penicillium digitatum . FEMS Microbiol Lett 348: 11–18. 10.1111/1574-6968.12235 [DOI] [PubMed] [Google Scholar]
- 14. Wang JY, Sun XP, Lin LY, Zhang TY, Ma ZH, et al. (2012) PdMfs1, a major facilitator superfamily transporter from Penicillium digitatum, is partially involved in the imazalil-resistance and pathogenicity. AFR J Microbiol Res 6: 96–105 [Google Scholar]
- 15. Sanglard D, Coste A, Ferrari S (2009) Antifungal drug resistance mechanisms in fungal pathogens from the perspective of transcriptional gene regulation. FEMS Yeast Res 9: 1029–1050. 10.1111/j.1567-1364.2009.00578.x [DOI] [PubMed] [Google Scholar]
- 16. Silver PM, Oliver BG, White TC (2004) Role of Candida albicans transcription factor Upc2p in drug resistance and sterol metabolism. Eukaryot Cell 3:1391–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. White TC, Silver PM (2005) Regulation of sterol metabolism in Candida albicans by the Upc2 gene. Biochem Soc Trans 33: 1215–1218. [DOI] [PubMed] [Google Scholar]
- 18. Dunkel N, Liu TT, Barker KS, Homayouni R, Morschhäuser J, et al. (2008) A gain-of-function mutation in the transcription factor Upc2p causes upregulation of ergosterol biosynthesis genes and increased fluconazole resistance in a clinical Candida albicans isolate. Eukaryot Cell 7: 1180–1190. 10.1128/EC.00103-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hoot SJ, Smith AR, Brown RP, White TC (2011) An A643V amino acid substitution in Upc2p contributes to azole resistance in well-characterized clinical isolates of Candida albicans . Antimicrob Agents Ch 55: 940–942. 10.1128/AAC.00995-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bien CM, Espenshade PJ. (2010) Sterol regulatory element binding proteins in fungi: hypoxic transcription factors linked to pathogenesis. Eukaryot Cell 9: 352–359. 10.1128/EC.00358-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hughes AL, Todd BL, Espenshade PJ (2005) SREBP pathway responds to sterols and functions as an oxygen sensor in fission yeast. Cell 120: 831–842. [DOI] [PubMed] [Google Scholar]
- 22. Blatzer M, Barker BM, Willger SD, Beckmann N, Blosser SJ, et al. (2011) SREBP coordinates iron and ergosterol homeostasis to mediate triazole drug and hypoxia responses in the human fungal pathogen Aspergillus fumigatus . PLoS Genet 7: e1002374 10.1371/journal.pgen.1002374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Willger SD, Puttikamonkul S, Kim KH, Burritt JB, Grahl N, et al. (2008) A sterol-regulatory element binding protein is required for cell polarity, hypoxia adaptation, azole drug resistance, and virulence in Aspergillus fumigatus . PLoS Pathog 4: e1000200 10.1371/journal.ppat.1000200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Blosser SJ, Cramer RA (2012) SREBP-dependent triazole susceptibility in Aspergillus fumigatus is mediated through direct transcriptional regulation of erg11A (cyp51A). Antimicrob Agents Ch 56: 248–257. 10.1128/AAC.05027-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vinggaard AM, Hass U, Dalgaard M, Andersen HR, Bonefeld-Jørgensen E, et al. (2006) Prochloraz: an imidazole fungicide with multiple mechanisms of action. Int J Androl 29: 186–192. [DOI] [PubMed] [Google Scholar]
- 26. Wang JL, Yu JH, Liu J, Yuan YZ, Li N, et al. (2014) Novel Mutations in CYP51B from Penicillium digitatum Involved in Prochloraz Resistance. J Microbiol 52: 762–770 10.1007/s12275-014-4112-2 [DOI] [PubMed] [Google Scholar]
- 27. Marcet-Houben M, Ballester AR, de la Fuente B, Harries E, Marcos JF, et al. (2012) Genome sequence of the necrotrophic fungus Penicillium digitatum, the main postharvest pathogen of citrus. BMC GENOMICS 13:646 10.1186/1471-2164-13-646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang JY, Li HY (2008) Agrobacterium tumefaciens-mediated genetic transformation of the phytopathogenic fungus Penicillium digitatum . J Zhejiang Univ Sci 9: 823–828. 10.1631/jzus.B0860006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bowyer P (2001) DNA-mediated transformation of fungi. Molecular and Cellular Biology of Filamentous Fungi Oxford: 33–46.
- 30. Malonek S, Meinhardt F (2001) Agrobacterium tumefaciens-mediated genetic transformation of the phytopathogenic ascomycete Calonectria morganii. Curr Genet 40: 152–155. [DOI] [PubMed] [Google Scholar]
- 31. Holmes GJ, Eckert JW (1999) Sensitivity of Penicillium digitatum and Penicillium italicum to Postharvest Citrus Fungicides in California. Phytopathology 89: 716–721. 10.1094/PHYTO.1999.89.9.716 [DOI] [PubMed] [Google Scholar]
- 32. Livak KJ, Schmittgen TD (2001) Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2 (-Delta Delta C (T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 33. Parraga A, Bellsolell L, Ferre-D’Amare AR, Burley SK (1998) Co-crystal structure of sterol regulatory element binding protein 1a at 2.3 Å resolution. Structure 6: 661–672. [DOI] [PubMed] [Google Scholar]
- 34. Marie C, Leyde S, White TC (2008) Cytoplasmic localization of sterol transcription factors Upc2p and Ecm22p in S. cerevisiae . Fungal Genet.Biol 45: 1430–1438. 10.1016/j.fgb.2008.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hoot SJ, Smith AR, Brown RP, White TC (2011) An A643V amino acid substitution in Upc2p contributes to azole resistance in well-characterized clinical isolates of Candida albicans . Antimicrob Agents Ch 55: 940–942. 10.1128/AAC.00995-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vasicek EM, Berkow EL, Flowers SA, Barker KS, Rogers PD (2014) UPC2 is Universally Essential for Azole Antifungal Resistance in Candida albicans . Eukaryot Cell 10.1128/EC.00221-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lohberger A, Coste AT, Sanglard D (2014) Distinct roles of Candida albicans drug resistance transcription factors TAC1, MRR1, and UPC2 in virulence. Eukaryot Cell 13: 127–142. 10.1128/EC.00245-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. MacPherson S, Akache B, Weber S, De Deken X, Raymond M (2005) Candida albicans zinc cluster protein Upc2p confers resistance to antifungal drugs and is an activator of ergosterol biosynthetic genes. Antimicrob Agents Ch 49: 1745–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Flowers SA, Barker KS, Berkow EL,Toner G, Chadwick SG, et al. (2012) Gain-of-function mutations in UPC2 are a frequent cause of ERG11 upregulation in azole-resistant clinical isolates of Candida albicans . Eukaryot Cell 11: 1289–1299. 10.1128/EC.00215-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hughes AL,Todd BL,Espenshade PJ (2005) SREBP pathway responds to sterols and functions as an oxygen sensor in fission yeast. Cell 120:831–842. [DOI] [PubMed] [Google Scholar]
- 41. Horton JD, Goldstein JL, Brown MS (2002) SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest 109: 1125–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Todd BL, Stewart EV, Burg JS, Hughes AL, Espenshade PJ (2006) Sterol regulatory element binding protein is a principal regulator of anaerobic gene expression in fission yeast. Mol Cell Biol 26:2817–2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Maguire SL, Wang C, Holland LM, Brunel F, Neuvéglise C, et al. (2014) Zinc Finger Transcription Factors Displaced SREBP Proteins as the Major Sterol Regulators during Saccharomycotina Evolution. PLoS Genet 10: e1004076 10.1371/journal.pgen.1004076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Luo CX, Schnabel G (2008) The cytochrome p450 lanosterol 14 alpha-demethylase gene is a demethylation inhibitor fungicide resistance determinant in Monilinia fructicola field isolates from Georgia. Appl Environ Microb 74: 359–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Flaherty JE, Payne GA (1997) Overexpression of aflR leads to upregulation of pathway gene transcription and increased aflatoxin production in Aspergillus flavus. Appl Environ Microb 63: 3995–4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. De Backer MD, Ilyina T, Ma XJ, Vandoninck S, Luyten WH, et al. (2001). Genomic profiling of the response of Candida albicans to itraconazole treatment using a DNA microarray. Antimicrob Agents Ch 45: 1660–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dharia NV, Bright AT, Westenberger SJ, Barnes SW, Batalov S, et al. (2010). Whole-genome sequencing and microarray analysis of ex vivo Plasmodium vivax reveal selective pressure on putative drug resistance genes. Proc. Natl. Acad. Sci. 107: 20045–20050. 10.1073/pnas.1003776107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dunkel N, Blaß J, Rogers PD, Morschhäuser J (2008) Mutations in the multi‐drug resistance regulator MRR1, followed by loss of heterozygosity, are the main cause of MDR1 overexpression in fluconazole-resistant Candida albicans strains. Molecular microbiology 69: 827–840. 10.1111/j.1365-2958.2008.06309.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mogavero S, Tavanti A, Senesi S, Rogers PD, Morschhäuser J (2011) Differential requirement of the transcription factor Mcm1 for activation of the Candida albicans multidrug efflux pump MDR1 by its regulators Mrr1 and Cap1. Antimicrob Agents Ch 55: 2061–2066. 10.1128/AAC.01467-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schubert S, Barker KS, Znaidi S, Schneider S, Dierolf F, et al. (2011) Regulation of efflux pump expression and drug resistance by the transcription factors Mrr1, Upc2, and Cap1 in Candida albicans . Antimicrob Agents Ch 55: 2212–2223. 10.1128/AAC.01343-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Znaidi S, De Deken X, Weber S, Rigby T, Nantel A, et al. (2007) The zinc cluster transcription factor Tac1p regulates PDR16 expression in Candida albicans . Molecular microbiology 66: 440–452. [DOI] [PubMed] [Google Scholar]
- 52. Zhao L, Liu DL, Zhang QY, Zhang S, Wan J, et al. (2007) Expression and homology modeling of sterol14α-demethylase from Penicillium digitatum . FEMS Microbiol Lett 277: 37–43 [DOI] [PubMed] [Google Scholar]
- 53. Zhang JH, Zhao L, Zhang J, Han R, Li SX, et al. (2010) Optimised expression and spectral analysis of the target enzyme CYP51 from Penicillium digitatum with possible new DMI fungicides. Pest Manag. Sci. 66: 1344−1350. 10.1002/ps.2021 [DOI] [PubMed] [Google Scholar]
- 54. Li WJ, Li Q, Liu DL, Ding MW (2013) Synthesis, Fungicidal Activity, and Sterol 14α-Demethylase Binding Interaction of 2-Azolyl-3,4-dihydroquinazolines on Penicillium digitatum . J. Agric. Food Chem. 61: 1419−1426. 10.1021/jf305355u [DOI] [PubMed] [Google Scholar]
- 55. Han R, Zhang JH, Li SX, Cao SF, Geng H, et al. Homology Modeling and Screening of New 14r-Demethylase Inhibitor (DMI) Fungicides Based on Optimized Expression of CYP51 from Ustilago maydis in Escherichia coli . J. Agric. Food Chem. 58: 12810–12816. 10.1021/jf103243m [DOI] [PubMed] [Google Scholar]
- 56. Gallo-Ebert C, Donigan M, Stroke IL, Swanson RN, Manners MT, et al. (2014). Novel Antifungal Drug Discovery Based on Targeting Pathways Regulating the Fungus-Conserved Upc2 Transcription Factor. Antimicrob Agents Ch 58: 258–266 10.1128/AAC.01677-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.