Abstract
The expression of mepA, encoding the Staphylococcus aureus MepA multidrug efflux protein, is repressed by the MarR homologue MepR. Repression occurs through binding of two MepR dimers to an operator with two homologous and closely approximated pseudopalindromic binding sites (site 1 [S1] and site 2 [S2]). MepR binding is impeded in the presence of pentamidine, a MepA substrate. The effects of various mepA operator mutations on MepR binding were determined using electrophoretic mobility shift assays and isothermal titration calorimetry, and an in vivo confirmation of the effects observed was established for a fully palindromic operator mutant. Altering the S1-S2 spacing by 1 to 4 bp severely impaired S2 binding, likely due to a physical collision between adjacent MepR dimers. Extension of the spacing to 9 bp eliminated the S1 binding-mediated DNA allostery required for efficient S2 binding, consistent with positive cooperative binding of MepR dimers. Binding of a single dimer to S1 was maintained when S2 was disrupted, whereas disruption of S1 eliminated any significant binding to S2, also consistent with positive cooperativity. Palindromization of binding sites, especially S2, enhanced MepR affinity for the mepA operator and reduced MepA substrate-mediated MepR induction. As a result, the on-off equilibrium between MepR and its binding sites was shifted toward the on state, resulting in less free MepR being available for interaction with inducing ligand. The selective pressure(s) under which mepA expression is advantageous likely contributed to the accumulation of mutations in the mepA operator, resulting in the current sequence from which MepR is readily induced by MepA substrates.
INTRODUCTION
Efflux of antimicrobial agents and biocides is an important bacterial resistance mechanism (1). The ability of selected efflux proteins to recognize multiple structurally diverse substrates amplifies the problem, resulting in a multidrug resistance (MDR) phenotype. Several MDR-conferring efflux proteins, here referred to as MDR-EPs, have been studied in Staphylococcus aureus, including NorA-B-C, MdeA, and QacA (2). All of these proteins are members of the major facilitator superfamily and are secondary transporters that are dependent on the proton motive force for substrate efflux. More recently, a novel multidrug and toxic compound extrusion (MATE)-family protein named MepA was identified (3, 4). This protein also is an MDR-EP and has select fluoroquinolones, biocides, and tigecycline as substrates.
Expression of mepA is regulated by MepR, a winged helix-turn-helix repressor belonging to the MarR family, that is encoded by a sequence immediately upstream of mepA (3). The mepR and mepA genes each have their own promoter elements and operator sequences to which MepR binds (5). The interactions of MepR with operator sequences upstream of mepR and mepA differ considerably, in that the protein binds as a single dimer to the operator of mepR but as a pair of dimers to that of mepA (6). Binding of MepR to the mepA operator is readily prevented by the presence of MepA substrates, whereas this effect is greatly attenuated at the mepR operator (5). It has been shown for other MarR-family proteins that ligand binding results in a conformational change, likely locking the structure in a conformation that is unable to interact with cognate DNA (7). The DNA-binding domains of ligand-free (apo)MepR are highly flexible and often are too widely separated to interact with consecutive major grooves of target DNA (6). However, upon specific DNA binding these winged helix-turn-helix motifs move to positions that allow efficient, high-affinity interaction (6, 8). On the basis of this structural plasticity, it is presumed that ligand-bound MepR is in a form incompatible with DNA docking, but confirmation of this hypothesis awaits the structural determination of complexes of MepR bound to germane MepA substrates.
The mepA operator consists of two pseudopalindromic inverted repeats (IRs), which incorporate both the −35 and −10 promoter elements (Fig. 1A). Spacing between the mepA IRs, here referred to as site 1 and site 2 (S1 and S2, respectively) consists of a single T·A base pair. Recent structural data for the highly analogous but single IR in the mepR operator identified a signature subsequence (GTTAG) with which specific residues of the MepR DNA-binding helices interact via van der Waals contacts, and isothermal titration calorimetry (ITC) experiments confirmed the importance of this signature sequence to efficient MepR binding (8). Positive cooperative binding of MepR dimers to each signature motif in the mepA operator may be the basis for the high-affinity binding of MepR to this site and also may play a role in easier substrate induction from it. Cooperativity may be achieved by protein-protein interactions or, more likely, by DNA allosteric effects, such as those observed upon the binding of a pair of QacR dimers to the qacA operator (9, 10).
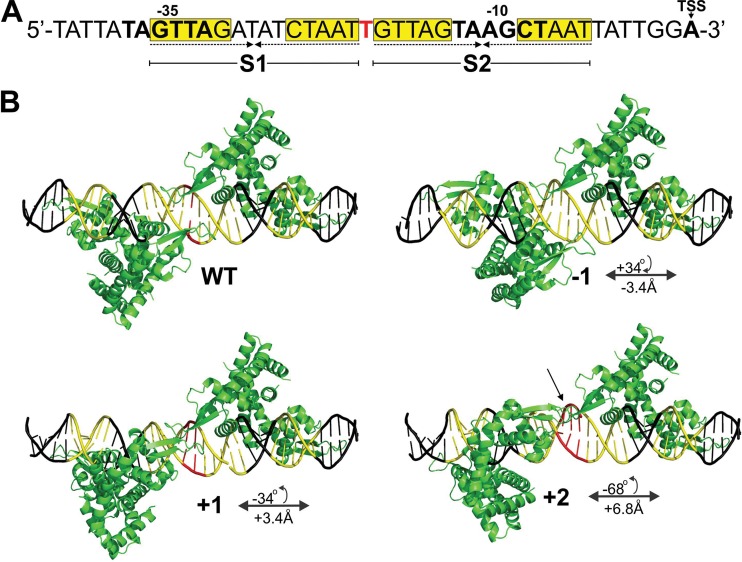
FIG 1.
Model of the mepA operator based on the known MepR-mepR operator structure. (A) Sequence of the wild-type mepA operator. For clarity, only the sequence of the positive strand is shown. Inverted repeats (IRs; dashed arrows) and MepR signature sequences (yellow boxes) making up S1 and S2 are indicated. The −35 and −10 motifs and the transcription start site (TSS) are identified in boldface, and the single-base-pair inter-IR spacer is identified in red. This sequence was modified as described in Materials and Methods. (B) Structural effects of changing the length of the inter-IR spacer region. The GTTAG signature sequence regions are indicated in yellow, and the inter-IR base pairs are indicated in red. WT, wild-type operator; −1, +1, and + 2, number of base pairs by which the inter-IR spacer was altered. The change in the overall operator length and position of the dimer bound to S1 relative to that bound to S2 is indicated. The clockwise or counterclockwise rotation of the S1 dimer as a result of each mutation is indicated by positive or negative numbers of degrees, respectively. The potential collision of the MepR dimers is especially evident for the +2 construct (arrow).
Previous work employing florescence polarization and ITC has revealed that S1 is the primary MepR binding site and S2 is the secondary MepR binding site, consistent with a positive cooperative mode of target interaction (6, 8). Data from our laboratory employing clinical mepA-overexpressing strains also support S1 as the primary site (11, 12). In those studies, one strain each with a mutation in the signature subsequence of S1 (GTTAG → GTTAA) or S2 (GTTAG → GTTGG) (where the mutations are underlined) were identified. MepR binding to these altered operators was assessed using electrophoretic mobility shift assays (EMSAs), and the efficiency of the target shift obtained using the S1 mutant was reduced by 60%, whereas that obtained using the S2 mutant was unaffected (unpublished). These data are consistent with the findings of ITC experiments examining the same S1 subsequence mutation, which revealed a 200-fold reduction in MepR binding affinity (8).
In this report, we describe the effects of various mepA operator mutations on MepR binding and induction by pentamidine, a MepA substrate. The data reveal that the sequences of S1 and S2 as well as the spacing between them profoundly affect the affinity of MepR for the mepA operator as well as the ease of its induction by substrate. They also support the presence of a positive cooperative mechanism of interaction of MepR dimers with this operator.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and reagents.
The strains used in the construction of various S. aureus derivatives are provided in Table 1. The plasmids employed in the construction of these strains, as well as the plasmid used to produce large quantities of the mepA promoter and examine the in vivo functional effect(s) of the fully palindromic (FP) mepA operator mutant, are also provided. Details on strain and plasmid construction are provided subsequently. Escherichia coli One-Shot TOP10 cells and the pCR2.1-TOPO plasmid were used for site-directed mutagenesis of the mepA operator (Life Technologies, Grand Island, NY). Large quantities of MepR with a C-terminal hexahistidine tag were produced using E. coli BL21 Star(DE3) cells and the pET101/D-TOPO plasmid as described previously (Life Technologies) (5). Hexahistidine-tagged MepR is referred to here simply as MepR. E. coli strains were grown in LB broth with appropriate antibiotic selection. Unless otherwise noted, all media and reagents were the highest grade available and were obtained from Sigma Chemical Co. (St. Louis, MO) or BD Biosciences (Sparks, MD).
TABLE 1.
S. aureus strains and plasmids used
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| S. aureus strains | ||
| NCTC 8325-4 | Wild-type strain cured of known prophages; rsbU mutant | 23 |
| SA-K2979 | NCTC 8325-4 mepA::lacZ | This study |
| SA-K6208 | NCTC 8325-4 mepRAB knockout; Spcr | This study |
| SA-K6331 | SA-K6208/pK1199 | This study |
| SA-K6365 | SA-K6208/pK1206 | This study |
| Plasmids | ||
| pIC156 | Bacillus subtilis vector used for chromosomal integration; Spcr | 24 |
| pTStetK | Temperature-sensitive vector used for allele replacement; Tcr | 14 |
| pALC2073 | S. aureus-E. coli shuttle vector containing a tetracycline-inducible promoter controlling expression of cloned genes; Cmr | 15 |
| pK481 | TOPO Blunt II vector containing 1.2 kb of mepRAB flanking DNA and internal SalI/SspI sites | This study |
| pK837 | pTStetK containing mepRAB 5′ and 3′ flanking DNA interrupted by the Spcr cassette | This study |
| pK918 | pCR2.1-TOPO vector containing the mepA operator (244 bp) | This study |
| pK1199 | pALC2073-mepRA::lacZ; carries wild-type mepA promoter | This study |
| pK1206 | pK1199 with fully palindromic S1 and S2 mepA operator | This study |
Spcr, Tcr, and Cmr, spectinomycin, tetracycline, and chloramphenicol resistance, respectively.
Genetic procedures.
The nucleotide sequences of all constructs were confirmed, using an automated dideoxy chain termination method, by the Applied Genomics Technology Center, Wayne State University (13). A 244-bp PCR product encompassing the mepA operator and surrounding sequence was cloned into pCR2.1-TOPO as described previously, producing pK918 (5). A QuikChange Lightning site-directed mutagenesis kit was used to introduce desired changes into the mepA operator within pK918 following the directions provided by the manufacturer (Agilent Technologies, Santa Clara, CA).
Following sequence confirmation, a 90-bp subsequence including all of the mepA operator sequence as well as flanking sequences was amplified from pK918. This amplicon corresponds to positions 329277 to 329366 of the S. aureus NCTC 8325 genome (GenBank accession number NC_007795 [http://www.ncbi.nlm.nih.gov/nuccore/88193823]). The primers used also added 12 bp to the 5′ end of each product, consisting of CCGGCCCCGGGC and CGGCGCGGGCGG for the forward and reverse primers, respectively. This was done to reduce the potential for intramolecular annealing within PCR products having highly palindromic sequences as well as the possibility of residual single-stranded products confounding subsequent experiments. This 114-bp product was used for EMSAs.
PCR was used to generate amplicons consisting of ∼600 bp of DNA flanking the mepRAB locus of S. aureus NCTC 8325-4. Primers used to produce amplicon 1 incorporated BamHI and sequential SalI/SspI sites at the 5′ and 3′ ends, respectively. Primers used to create amplicon 2 incorporated the same restriction endonuclease sites but in the reverse orientation, namely, SalI/SspI and BamHI sites at the 5′ and 3′ ends, respectively. These products were joined at their overlapping SalI/SspI sites by PCR-based overlap extension, producing a 1.2-kb product with flanking BamHI sites and centrally located SalI/SspI sites. This product was cloned into the BamHI site of the TOPO Blunt II vector, producing pK481 (Life Technologies).
The spectinomycin resistance (Spcr) cassette of pIC156 was amplified by PCR, incorporating the flanking SalI and SspI sites that the plasmid possesses. These enzymes were used to open pK481, followed by insertion of the Spcr cassette. BamHI was used to excise the resultant 2.5-kb fragment consisting of up- and downstream mepRAB flanking DNA separated by the Spcr cassette, which then was cloned into the temperature-sensitive allele replacement vector pTStetK, creating pK837. Allele replacement was carried out as previously described, resulting in S. aureus NCTC 8325-4 with the mepRAB locus deleted and replaced by the Spcr cassette (SA-K6208) (14).
A transcriptional fusion between mepA and lacZ was constructed using pAZ106 as described previously, followed by transduction into S. aureus NCTC 8325-4 to produce SA-K2979 (3). The mepRA::lacZ construct from SA-K2979 was amplified by PCR and cloned into pALC2073, producing pK1199 and placing the expression of mepRA::lacZ under the control of a tetracycline-inducible promoter (15). The wild-type (WT) mepA operator within pK1199 was altered to produce a fully palindromic sequence using site-directed mutagenesis, producing pK1206. The resultant sequence is provided in Fig. 4. Both pK1199 and pK1206 were expressed in the SA-K6208 (mepRAB knockout, Spcr) background (SA-K6331 and SA-K6365, respectively). The efficiency of mepA expression from these plasmids was quantitated using three replicates of a β-galactosidase assay as previously described, modified only by a reduction in the duration of the experiments from 10 to 8 h (14). These data are expressed as 4-methylumbelliferyl-β-d-galactopyranoside (MUG) units, where 1 MUG unit equals 1 pmol of MUG cleaved per min per optical density at 600 nm unit. Expression data for SA-K6331 and SA-K6365 were compared using the t-test function embedded within SigmaPlot (version 12.5) software (Systat Software, Inc., San Jose, CA).
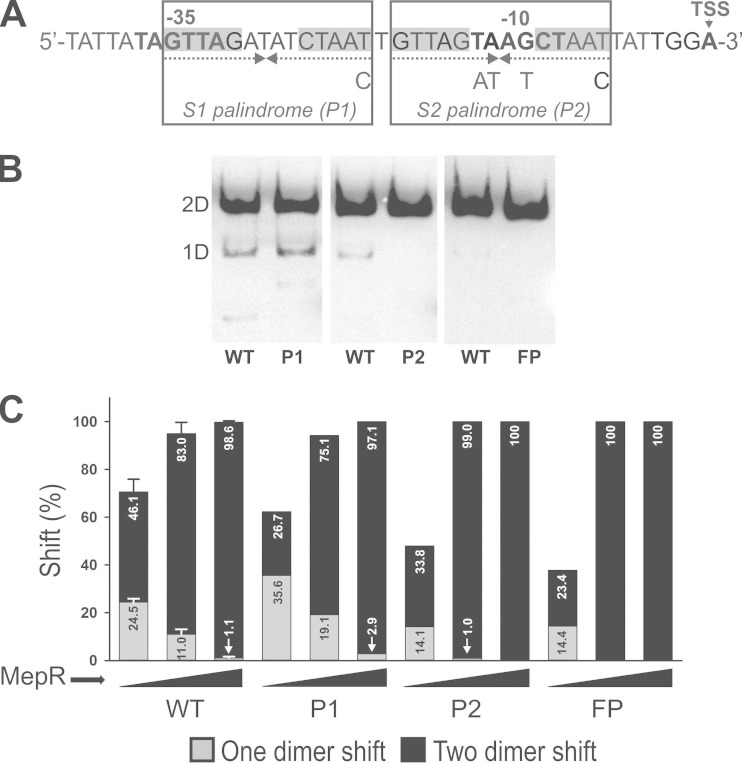
FIG 4.
Binding of MepR to mepA operator variants with binding sites converted to perfect palindromes. (A) WT mepA operator with the positions of the mutations introduced into S1, S2, or both indicated (producing P1, P2, and a fully palindromic [FP] operator, respectively; the sequence of the positive strand is shown). The operator site annotation is as described in the legend to Fig. 1. (B) Representative EMSAs for P1, P2, and FP operator sequences (12.5 ng input MepR). 1D and 2D, one- and two-dimer binding, respectively. (C) Efficiency of MepR-mediated target shift (in percent) for WT and each operator derivative with 3.1, 12.5, and 50 ng of input MepR. Data represent the means from two independent experiments for all but the WT, as described in the legend to Fig. 1.
The effect of substrate exposure on mepA expression was assessed using the same approach, except that tetracycline induction of mepR expression was not done and seven replicates were performed. In this case, the normal leakiness of the tetracycline-inducible promoter of the pALC2073 backbone was used so as to not produce so much MepR that the effect of substrate relating to a very low inducer/protein concentration ratio would be obscured (16). Data were collected in the absence and presence of 0.4 μg/ml pentamidine, which was 1/32× MIC of both SA-K6331 and SA-K6365. These expression data were first analyzed using the fourth-spread method to identify any outliers, and none were found (17). Changes in expression were determined by subtracting the number of MUG units in the absence of pentamidine from that in its presence and then comparing these data using the t test.
EMSA procedures.
MepR was purified using a Probond purification system (Life Technologies). EMSAs employing MepR and mepA operator derivatives were performed essentially as described previously, but some experiments were modified by use of a range of input MepR amounts (3.1 to 50 or 100 ng) to assess binding affinity and elimination of salmon sperm DNA as a nonspecific competitor (5). The effect of a concentration range of pentamidine (25 to 100 μM), a MepA substrate previously shown to induce MepR, on the affinity of a 50-ng input amount of MepR for mepA operator derivatives also was evaluated (5). Each EMSA included the WT mepA operator as a control, and modified operator sequences were evaluated twice. Representative images of the results of experiments performed on a single day are provided in Fig. 2 to 4 and 6. For clarity, the sequence data provided in these figures are for the positive strand only. Unless otherwise noted, all bar graphs providing EMSA data represent the means of two independent replicates. For experiments other than pentamidine induction, the WT sequence was evaluated a sufficient number of times to allow calculations of means and standard errors.
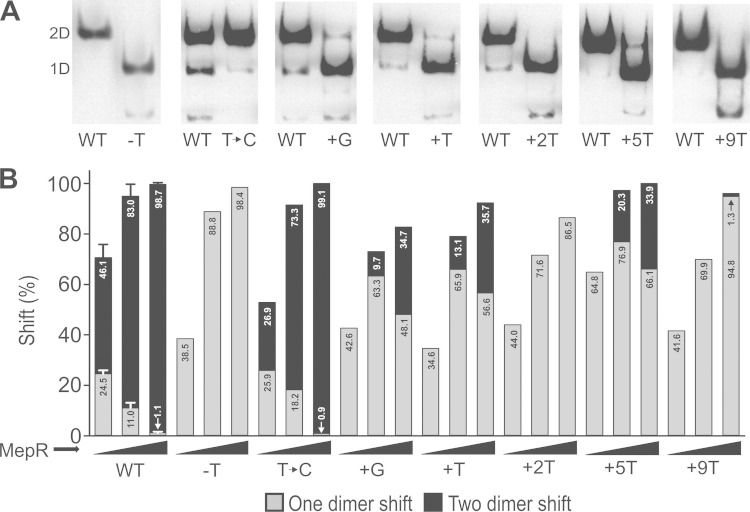
FIG 2.
Binding of MepR to mepA operator variants with changes introduced in the inter-IR spacer region. (A) Representative results of EMSAs illustrating MepR binding (12.5 ng input protein) to WT and modified mepA operators. 1D and 2D, one- and two-dimer binding to target DNA, respectively. The operator mutations introduced are indicated. (B) Efficiency of the shift (in percent) observed for the WT and each operator derivative with 3.1, 12.5, and 50 ng input MepR. Data represent the means from two independent experiments for all but the WT, for which multiple replicates were done. Standard errors for WT binding are indicated.
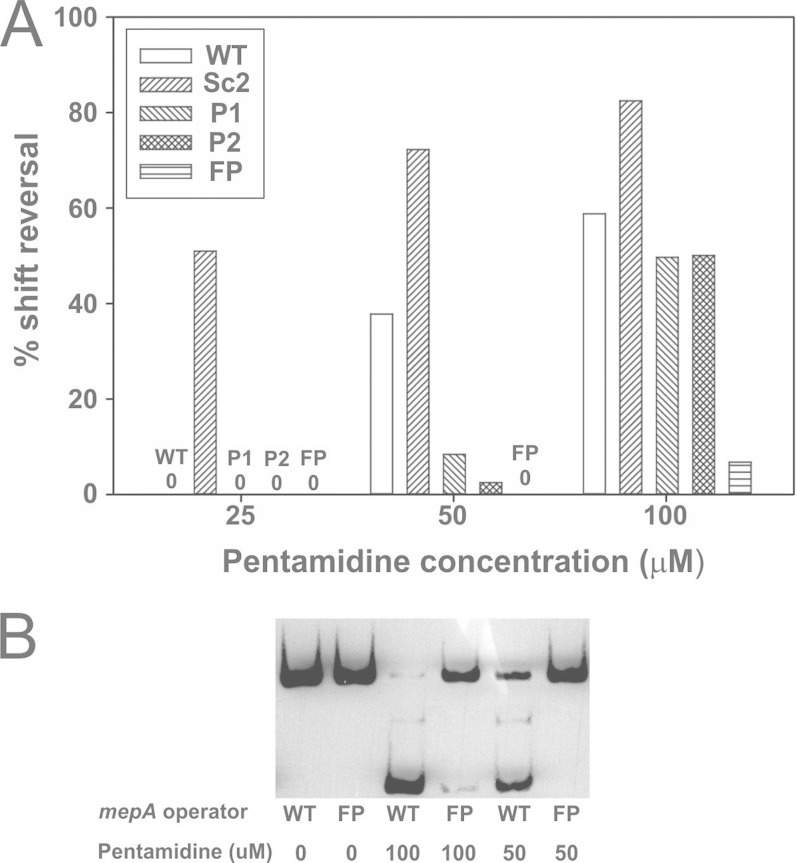
FIG 6.
Effect of palindromicity of binding sites on MepR induction by pentamidine. Data represent the means from two independent experiments. (A) Proportion of target DNA returning to the position observed in the absence of MepR, with data presented as the percentage of shift reversal. WT, wild type; Sc2, scrambled S2 site; P1 and P2, palindromization of S1 and S2, respectively; FP, fully palindromized mepA operator sequence. The absence of reversal of the MepR-mediated shift for a particular construct at each pentamidine concentration is indicated by the numeral 0. (B) Representative results of EMSAs employing pentamidine concentrations of 50 and 100 μM and 50 ng of input MepR, illustrating the profoundly reduced shift reversal for the FP operator variant.
ITC.
Isothermal titration calorimetry (ITC) experiments were performed exactly as described previously using a Microcal VP-ITC calorimeter (GE Healthcare Bio-Sciences, Pittsburgh, PA) (8). DNA was purchased commercially (Integrated DNA Technologies, Coralville, IA). DNA was annealed by heating at 95°C for 5 min, followed by slow cooling to room temperature. The buffer used in the ITC experiments was 20 mM Tris HCl (pH 7.5), 150 mM NaCl, and 5 mM MgCl2.
Bioinformatics.
The CLC Main Workbench (version 7.5) program (CLC bio, Boston MA) was used for analyses of DNA sequences. Quantification of the efficiency of the band shift in EMSAs was done using the Phoretix 1D Pro (version 11.8) program (Nonlinear Dynamics, Newcastle upon Tyne, United Kingdom), and ITC data were analyzed using the Origin (version 7.0) program (OriginLab Corporation, Northampton, MA). Solution of the structure of the MepR-mepR operator complex allowed a model of the mepR-mepA operator to be built (Fig. 1B), which was done using the PyMOL molecular graphics system (version 1.3; Schrödinger, LLC) (8). Structural data for the apo- and DNA-bound forms of MepR were obtained from the RCSB Protein Data Bank (www.rcsb.org; PDB accession numbers 3ECO and 4L9J, respectively).
RESULTS AND DISCUSSION
Importance of a single-base-pair spacer between MepR binding sites.
As noted previously and illustrated in Fig. 1, the mepA operator has profound symmetry that includes two IR pairs separated by a single base pair. These IRs include the MepR GTTAG signature sequence and its complementary mate, and for S1 it also includes a 4-bp palindromic intersignature sequence spacer (ATAT). S2 is less palindromic due to degeneracy in this intersignature sequence spacer (TAAG). This configuration is relatively unique among MarR-family operator sites. Similar to MepR, the MarR-family repressors MexR of Pseudomonas aeruginosa and MarR of E. coli bind as a pair of dimers to their respective operators (18, 19). However, the structures of both operators differ from the structure of mepA by longer inter-IR spacing (3 and 13 bp, respectively).
Using the structure of the MepR-mepR operator complex as a foundation (8), the structure of the mepA operator is hypothesized to take a B-form DNA conformation, whereby the average rise per nucleotide is approximately 3.4 Å and the helical twist of each base pair is approximately 34°. In silico modeling of the MepR-mepA operator complex revealed that addition or removal of 1 bp or more between the S1 and S2 sites would result in the translation and rotation of the dimer bound to S1 relative to the dimer bound to S2. Hence, removal of the inter-IR T·A pair would result in the 3.4-Å closer approach of the S1- and S2-bound dimers as well as a relative 34° clockwise rotation between them. Conversely, the addition of a single base pair would provide the opposite effect. Regardless, changes in the position of the S1-bound dimer relative to that of the dimer bound to S2 are predicted to result in steric clashes between dimers primarily involving the wing motifs, which impede binding of a dimer to S2 in the presence of a dimer already bound to the higher-affinity S1 site. Even the addition of 2 bp to the spacer separating S1 and S2 results in a steric clash between the wings of the dimers, which would be unable to bind minor grooves effectively if they were repositioned to alleviate these unfavorable protein-protein interactions. These effects are illustrated in Fig. 1.
EMSA data for operator mutants in which the inter-IR distance was altered are presented in Fig. 2. These data reveal that the nature of the single-base-pair spacer was not important, as substitution of a C·G pair for the native T·A pair did not alter the binding efficiency significantly (compare the data for the WT and the T → C substitution). Increasing the spacing by 1 bp, regardless of its identity (+G or +T; positive-strand base), markedly reduced the binding of two dimers, while it increased the proportion of one-dimer binding. Increasing the spacer by 2 bp (+2T) or deletion of the S1-S2 spacer altogether (−T) eliminated two-dimer binding completely. These data provide general support for our collision hypothesis, especially for the addition of 2 bp. However, these results do not rule out completely the possibility that changing the inter-IR spacer interferes with the occurrence of S1 binding-mediated allosteric changes to operator DNA that may be required for efficient S2 binding.
The MepR-mepA operator model predicts that a point will be reached at which further extension of the inter-IR spacing will eliminate the possibility of collision between dimer pairs. This was tested by addition of 4 and 5 T·A bp to increase the size of the spacer as well as the construction of two different +9-bp mutants, including a mutant to which 9 T residues were added (+9T mutant) and a mutant to which the mixed-sequence spacer GAACTTGAT was added (positive-strand bases; complementary bases are inferred). The mixed-sequence spacer was examined to control for the possibility of potential changes in DNA topology resulting from A tracts, which can introduce sequence-dependent structural effects that could confound the results (20). Two-dimer binding remained absent in the +4T mutant (not shown) but was partially recovered in the +5T mutant, providing support for our collision hypothesis and for the conclusion that protein-protein interactions are not required for MepR binding to S2. Two-dimer binding was much less efficient for the +9T mutant than the +5T mutant, with similar results being observed for the +9-bp mixed-sequence spacer mutant. The latter data eliminate the possibility of A-tract-mediated changes to the DNA conformation and suggest that when the spacer length is increased beyond a critical threshold, the necessary S1 binding-mediated positive allostery utilized for efficient S2 binding is severely compromised.
Binding to S1 is a prerequisite for binding to S2 in the context of the native mepA operator.
Scrambling of the S1 sequence (Sc1) completely eliminated two-dimer binding (Fig. 3, Sc1). Inefficient one-dimer binding was maintained at large amounts of input MepR (100 ng) but disappeared at smaller amounts. These data confirmed previous ITC data, where dissociation constant (Kd) values of 42 and 3,800 nM were observed for the intact mepA operator and the presence of a scrambled S1, respectively (8). In contrast, highly efficient one-dimer binding was maintained when S2 was scrambled, even with small amounts of input MepR (12.5 ng) (Fig. 3, Sc2). Intriguingly, ITC studies employing a 26-bp fragment encompassing the WT S2 sequence alone revealed that MepR binds with an affinity identical to that observed for the intact mepA operator (Kd = 40 nM for 26-mer mepA S2; Fig. 3C). The addition of 3 bp from the WT to the 5′ end (toward S1) resulted in a dramatic 14-fold loss of binding affinity, as reflected in the observed Kd (Kd = 570 nM for 29-mer mepA S2). The addition of an additional 3 bp from the WT to this end of the sequence, introducing all but 3 bp of the intact S1, resulted in a dramatic reduction in binding affinity (Kd = 1,900 nM for 32-mer mepA S2). In all three ITC experiments, the stoichiometry of binding was one MepR dimer per double-stranded DNA site (molar ratio, 2), consistent with the EMSA data generated using targets with a single, intact binding site that consistently demonstrated one-dimer binding.
FIG 3.
Binding of MepR to mepA operator variants with scrambled binding sites. (A) WT mepA operator (positive strand) with the positions of the mutations introduced into S1 or S2 (producing Sc1 and Sc2, respectively) indicated. The operator annotation is as described in the legend to Fig. 1. (B) Representative results of EMSAs employing 100 and 12.5 ng of input MepR for Sc1 and Sc2, respectively. 1D and 2D, one- and two-dimer binding, respectively. (C) Binding thermograms and the associated isotherms of MepR and different lengths of S2. These ITC experiments revealed that for the WT mepA operator, a complete S1 sequence must be present and occupied by a MepR dimer to effect physiologically relevant binding of MepR to S2. MepR signature sequences or portions thereof are highlighted by gray boxes below the binding isotherms.
These EMSA and ITC data, as well as those for the +9-bp spacer mutant discussed above, reinforce each other and establish the critical need for S1 to be present and occupied by a MepR dimer to effect the physiologically relevant binding of MepR to S2. They are also completely consistent with earlier work in which various deletions of the mepA operator were examined and which indicated that S1 is the primary MepR binding site (6). They thus provide strong support for the notion that MepR binding to the mepA operator exhibits positive cooperativity (8).
Palindromic binding sites and substrate induction.
The sequence of the mepA operator reveals that the IR forming S1 has only one mismatch, whereas that of S2 has four (Fig. 4). Conversion of S1 to a perfect palindrome (P1) modestly augmented the binding of one dimer, presumably to S1, by 1.6- to 1.8-fold at a small amount of input MepR (at 3.1 and 12.5 ng of input MepR, 35.6 and 19.1%, respectively, for P1 and 24.5 and 11.0%, respectively, for the WT) (Fig. 4, gray bars). However, it did not appreciably affect one- or two-dimer binding at 50 ng of input MepR. These data are concordant with the fact that the WT S1 sequence is already a nearly perfect palindrome, with the single change present in P1 providing minimal additional benefit. In sharp contrast, palindromization of S2 (producing P2) had a greater effect, in that it clearly increased the proportion of two-dimer binding relative to that of one-dimer binding and the efficiency of the complete shift, best seen at 12.5 ng input MepR. At this amount of input MepR, the ratios of two- to one-dimer binding were 7.5 (83/11) and 99 (99/1) for the WT and P2, respectively. A reasonable explanation for these observations is that the more extensive changes necessary to convert S2 to a perfect palindrome contribute to more efficient recruitment of the second MepR dimer when S1 is already occupied. The mechanism of this effect is related in greater part to the better binding affinity of MepR.
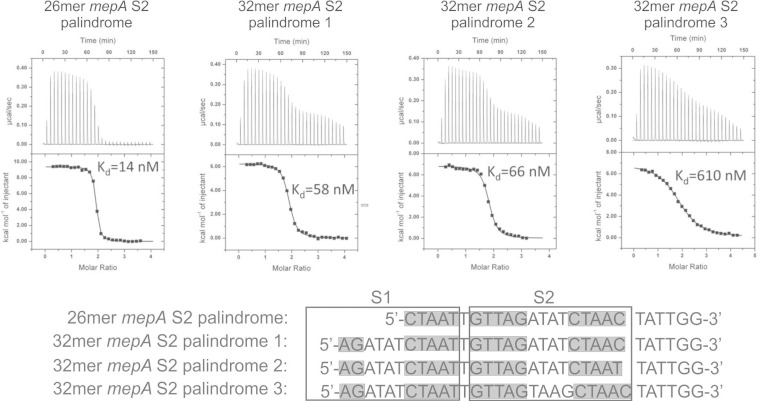
Indeed, ITC studies using a full-length 46-bp mepA operator in which S1 was scrambled and S2 was fully palindromized to contain the S1-like ATAT sequence linking the GTTAG/CTAAT signature sequences revealed a Kd of 195 nM and a stoichiometry of 2 (data not shown). This is an improvement of more than 19-fold compared to the previously reported Kd of 3,800 nM for MepR binding to the mepA operator with a scrambled S1 and a WT pseudopalindromic S2 sequence (8). Figure 5 provides additional ITC data on MepR binding to fully palindromized S2 sites in isolation. These data support the critical importance of palindromicity, whereby a fully palindromic 26-bp S2 binds a single MepR dimer with a Kd of 14 nM, a 3-fold increase in affinity compared to that observed for the same target with a WT S2 sequence (Kd = 40 nM) (described above and illustrated in Fig. 3). Increasing the length of the DNA target to 32 bp by adding back a partial S1 site resulted in a Kd of 58 nM (32-mer mepA palindrome 1), a result which poorly mirrored that observed when assaying the WT 32-bp S2 site (Kd = 1,900 nM) (Fig. 3, 32-mer mepA S2). Thus, compared to the Kd values for the respective 26-bp sequences, the 32-bp palindromized mepA S2 showed an increase in Kd of only 4-fold (14 versus 58 nM), whereas the WT 32-mer mepA S2 showed a 47-fold increase in Kd (40 versus 1,900 nM). Further modification of the 32-bp palindromized S2 sequence revealed that the key to the increased binding affinity achieved by palindromization lay in the central ATAT linker. Changing the 3′-most C to T, found in the WT S2 sequence, did not change the binding affinity (Kd = 58 and 66 nM for 32-mer mepA palindromes 1 and 2, respectively). However, changing the central ATAT back to the WT TAAG resulted in an 11-fold reduction in affinity (Kd = 58 and 610 nM for 32-mer mepA palindromes 1 and 3, respectively). Although this particular palindromic sequence of the central linker might play some role in affinity, it would be through indirect readout. MepR in complex with the mepR operator and almost certainly in complex with the mepA operator does not directly contact any of these minor groove bases, which lie in the region between the DNA recognition helices of each subunit that creates the functional MepR dimer (8, 21, 22). Further, we cannot completely rule out the possibility that palindromization of S2 results in an alteration in S1 binding-mediated allosteric effects to better receive the second dimer at S2 or a combination of these events. The EMSA data revealed that full palindromization of both S1 and S2 did not improve the results observed when only S2 was made palindromic (compare the results for P2 and FP in Fig. 4), consistent with the mechanistic hypotheses just advanced.
FIG 5.
Binding thermograms and the associated isotherms of MepR bound to a variety of palindromes of S2. The palindromicity of S2 has a profound effect on the MepR binding affinity for isolated S2. MepR signature sequences (or portions thereof) are highlighted by gray boxes below the binding isotherms.
Palindromization of binding sites also affected the ability of pentamidine, a MepA ligand and known MepR inducer, to interfere with MepR binding (Fig. 6) (5). The input quantity of MepR used (50 ng) shifted both WT and palindromized sequences completely. The differences were most evident at 50 μM pentamidine, where, compared to the results obtained with the WT sequence, palindromization of S1, S2, or S1 and S2 substantially reduced the effect of the substrate on MepR binding (Fig. 6, P1, P2, and FP, respectively). The reduced substrate effect was maintained for the fully palindromized operator even at a high pentamidine concentration (100 μM). The most plausible explanation for these data is the increased stability of the MepR-DNA complex with palindromized binding sites, especially for the FP sequence. The increased stability will shift the normal on-off equilibrium between MepR and its target DNA more toward on, resulting in less free MepR available to interact with ligands. For P2 and the FP sequence, the data without and with inducer are fully compatible with this hypothesis. Both constructs had improved two-dimer binding at lower input amounts of MepR (12.5 ng) compared with that for the WT (Fig. 4), and both were less responsive to pentamidine (Fig. 6). However, for P1 an apparent inconsistency was observed, in that in the absence of inducer there were minimal differences from the results for the WT, whereas in the presence of 50 μM pentamidine, a reduced effect on MepR binding was observed. The reason(s) for this discrepancy is not immediately apparent but may be related more to the number of EMSA replicates performed (two) than to a real biological phenomenon.
We also observed that binding to S1 alone, which occurred when the S2 sequence was scrambled, was more easily prevented by pentamidine at all concentrations tested (Fig. 6, Sc2). This observation is consistent with a reduced affinity of MepR bound only to S1, leading to more available free MepR. While the sequence of a scrambled S2 differed from that which we employed, previously performed ITC experiments revealed a modest reduction in MepR affinity for isolated S1 binding (Kd, 42 versus 71 nM for the WT and S2 scrambled operator, respectively), providing general support for our hypothesis (8).
Transcriptional efficiency and effect of pentamidine exposure on the FP mepA operator derivative.
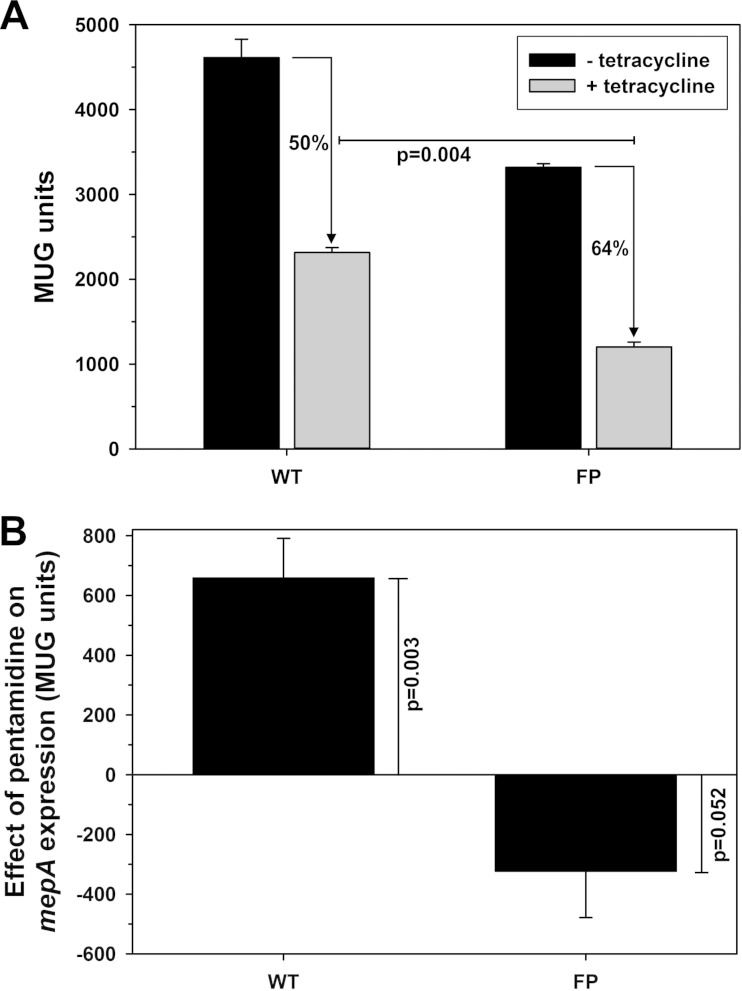
β-Galactosidase assays revealed that MepR repression of mepA expression was greater for the FP than the WT operator derivatives (50 versus 64%; P = 0.004) (Fig. 7). Thus, the findings of both in vitro (EMSA) and in vivo analyses were consistent with the conclusion that the affinity of MepR for the mepA operator is enhanced when the operator is fully palindromic. The 28% higher level of expression of mepA from the WT operator without the induction of mepR expression is also consistent with the increased MepR affinity for the FP operator derivative. As mentioned previously, it is known that the tetracycline-responsive promoter of pALC2073 is leaky, allowing some mepR transcription to occur in the absence of tetracycline induction (16). The low level of available MepR in the uninduced state will have a greater effect on mepA transcription from the FP operator than that from the WT operator as a result of its greater affinity there, resulting in lower expression in the uninduced state. We do concede, however, that reduced transcriptional efficiency as a result of the mutations introduced into the mepA operator to produce the FP derivative may contribute, as some of the base pair changes altered the −10 hexamer (TAAGCT → ATATCT, where the alterations are underlined) (Fig. 4). Even if this was a contributing factor, it does not change the fact that the greater effect of MepR in the induced state is consistent with a greater MepR affinity for the FP operator sequence.
FIG 7.
mepA expression analyses employing the WT and FP mepA operator sequences. (A) mepA expression in the absence (− tetracycline) and presence (+ tetracycline) of induction of mepR transcription, expressed as the mean number of MUG units ± SE. The MepR effect on mepA expression was greater for the FP than the WT operator derivative (50 versus 64% reduction, P = 0.004). (B) Effect of pentamidine (0.4 μg/ml; 1/32× MIC) on mepA expression from the WT and FP operator derivatives, expressed as the mean increase or decrease in the number of MUG units ± SE. Pentamidine significantly increased mepA expression from the WT but not the FP operator derivative.
Exposure to 0.4 μg/ml pentamidine (1/32× MIC) resulted in a significant increase in mepA expression in the presence of the WT operator (P = 0.003), whereas a modest, but not significant, reduction was observed for the FP operator derivative (Fig. 7). These data are consistent with the EMSA data obtained with pentamidine, which showed a reversal of the MepR-WT operator interaction and resistance to this effect for the FP derivative (Fig. 6). The reason for the small reduction in mepA expression observed for the FP derivative is not apparent, but our data clearly reveal that substrate-mediated induction of MepR does not occur.
Model of the MepR-mepA operator interaction.
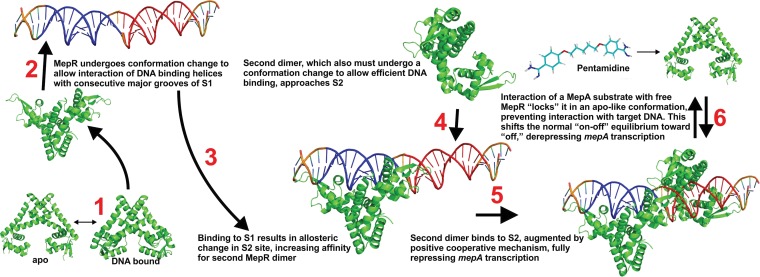
Figure 8 provides a model of the interaction between MepR and its mepA operator target as well as between MepR and the inducer. Confirmation of this model, especially for the occurrence of the allosteric changes in DNA structure associated with the binding of MepR to S1 that recruit a second dimer to S2 and the effect of ligand binding on the conformation of MepR, awaits solution of MepR-mepA operator and MepR-ligand structures.
FIG 8.
Model of the interaction of MepR with the mepA operator. MepR must first undergo a conformational change to allow high-affinity interaction with cognate DNA (step 1). This conformation would be stabilized by target sequences only. The MepR dimer then binds with a high affinity to S1 (blue; step 2). Binding to S1 induces an allosteric change to downstream DNA (step 3), augmenting the recruitment of a second MepR dimer to S2 (red; steps 4 and 5). The interaction of MepA substrates such as pentamidine with MepR locks the dimer into a low-affinity DNA-binding conformation. This shifts the normal on-off equilibrium between MepR and target DNA toward the off state, resulting in derepression of mepA transcription (step 6).
Concluding remarks.
The current WT mepA operator sequence may have evolved through the accumulation of favorable mutations at times of exposure of S. aureus to MepA substrates. The WT sequence is completely conserved across all sequenced S. aureus genomes, indicating that it represents the most favorable balance between MepR binding affinity, the induction of MepR in the presence of MepA substrates, and the maintenance of functional −10 and −35 promoter elements. The positive cooperativity of MepR binding to the mepA operator is supported by our data, confirming those generated previously by deletion analyses and ITC, and likely occurs as a result of the S1-MepR binding event mediating a conformational alteration of the downstream double helix such that subsequent S2 binding by MepR is augmented. Direct interdimer interactions are not necessary for two-dimer binding and, on the basis of our data, do not contribute directly to the cooperativity observed. Our current data also uncover the fact that palindromicity is a critical determinant in the regulation of the mepA operator, and perhaps the sequence degeneracy present in S2 is necessary for a proper and timely response to MepA substrates. The structure of the mepA operator is unique among those with which MarR proteins interact, and the data presented herein provide further insight into the biological behavior of this family of regulatory proteins.
ACKNOWLEDGMENT
This work was supported by VA Biomedical Laboratory Research & Development grant 1IO1BX000465.
REFERENCES
- 1.Li XZ, Nikaido H. 2009. Efflux-mediated drug resistance in bacteria: an update. Drugs 69:1555–1623. doi: 10.2165/11317030-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hassan KA, Skurray RA, Brown MH. 2007. Active export proteins mediating drug resistance in staphylococci. J Mol Microbiol Biotechnol 12:180–196. doi: 10.1159/000099640. [DOI] [PubMed] [Google Scholar]
- 3.Kaatz GW, McAleese F, Seo SM. 2005. Multidrug resistance in Staphylococcus aureus due to overexpression of a novel multidrug and toxin extrusion (MATE) transport protein. Antimicrob Agents Chemother 49:1857–1864. doi: 10.1128/AAC.49.5.1857-1864.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McAleese F, Petersen P, Ruzin A, Dunman PM, Murphy E, Projan SJ, Bradford PA. 2005. A novel MATE family efflux pump contributes to the reduced susceptibility of laboratory-derived Staphylococcus aureus mutants to tigecycline. Antimicrob Agents Chemother 49:1865–1871. doi: 10.1128/AAC.49.5.1865-1871.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaatz GW, DeMarco CE, Seo SM. 2006. MepR, a repressor of the Staphylococcus aureus MATE family multidrug efflux pump MepA, is a substrate-responsive regulatory protein. Antimicrob Agents Chemother 50:1276–1281. doi: 10.1128/AAC.50.4.1276-1281.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumaraswami M, Schuman JT, Seo SM, Kaatz GW, Brennan RG. 2009. Structural and biochemical characterization of MepR, a multidrug binding transcription regulator of the Staphylococcus aureus multidrug efflux pump MepA. Nucleic Acids Res 37:1211–1224. doi: 10.1093/nar/gkn1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perera IC, Grove A. 2010. Molecular mechanisms of ligand-mediated attenuation of DNA binding by MarR family transcriptional regulators. J Mol Cell Biol 2:243–254. doi: 10.1093/jmcb/mjq021. [DOI] [PubMed] [Google Scholar]
- 8.Birukou I, Seo SM, Schindler BD, Kaatz GW, Brennan RG. 2014. Structural mechanism of transcription regulation of the Staphylococcus aureus multidrug efflux operon mepRA by the MarR family repressor MepR. Nucleic Acids Res 42:2774–2788. doi: 10.1093/nar/gkt1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schumacher MA, Miller MC, Grkovic S, Brown MH, Skurray RA, Brennan RG. 2002. Structural basis for cooperative DNA binding by two dimers of the multidrug-binding protein QacR. EMBO J 21:1210–1218. doi: 10.1093/emboj/21.5.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim S, Brostromer E, Xing D, Jin J, Chong S, Ge H, Wang S, Gu C, Yang L, Gao YQ, Su XD, Sun Y, Xie XS. 2013. Probing allostery through DNA. Science 339:816–819. doi: 10.1126/science.1229223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeMarco CE, Cushing LA, Frempong-Manso E, Seo SM, Jaravaza TA, Kaatz GW. 2007. Efflux-related resistance to norfloxacin, dyes, and biocides in bloodstream isolates of Staphylococcus aureus. Antimicrob Agents Chemother 51:3235–3239. doi: 10.1128/AAC.00430-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kosmidis C, Schindler BD, Jacinto PL, Patel D, Bains K, Seo SM, Kaatz GW. 2012. Expression of multidrug resistance efflux pump genes in clinical and environmental isolates of Staphylococcus aureus. Int J Antimicrob Agents 40:204–209. doi: 10.1016/j.ijantimicag.2012.04.014. [DOI] [PubMed] [Google Scholar]
- 13.Sanger F, Nicklen S, Coulson AR. 1977. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A 74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaatz GW, Thyagarajan RV, Seo SM. 2005. Effect of promoter region mutations and mgrA overexpression on transcription of norA, which encodes a Staphylococcus aureus multidrug efflux transporter. Antimicrob Agents Chemother 49:161–169. doi: 10.1128/AAC.49.1.161-169.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bateman BT, Donegan NP, Jarry TM, Palma M, Cheung AL. 2001. Evaluation of a tetracycline-inducible promoter in Staphylococcus aureus in vitro and in vivo and its application in demonstrating the role of sigB in microcolony formation. Infect Immun 69:7851–7857. doi: 10.1128/IAI.69.12.7851-7857.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corrigan RM, Foster TJ. 2009. An improved tetracycline-inducible expression vector for Staphylococcus aureus. Plasmid 61:126–129. doi: 10.1016/j.plasmid.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 17.Devore J. 2012. Probability and statistics for engineering and the sciences, 8th ed Brooks/Cole, Boston, MA. [Google Scholar]
- 18.Evans K, Adewoye L, Poole K. 2001. MexR repressor of the mexAB-oprM multidrug efflux operon of Pseudomonas aeruginosa: identification of MexR binding sites in the mexA-mexR intergenic region. J Bacteriol 183:807–812. doi: 10.1128/JB.183.3.807-812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin RG, Rosner JL. 1995. Binding of purified multiple antibiotic-resistance repressor protein (MarR) to mar operator sequences. Proc Natl Acad Sci U S A 92:5456–5460. doi: 10.1073/pnas.92.12.5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haran TE, Mohanty U. 2009. The unique structure of A-tracts and intrinsic DNA bending. Q Rev Biophys 42:41–81. doi: 10.1017/S0033583509004752. [DOI] [PubMed] [Google Scholar]
- 21.Rohs R, Jin X, West SM, Joshi R, Honig B, Mann RS. 2010. Origins of specificity in protein-DNA recognition. Annu Rev Biochem 79:233–269. doi: 10.1146/annurev-biochem-060408-091030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rohs R, West SM, Sosinsky A, Liu P, Mann RS, Honig B. 2009. The role of DNA shape in protein-DNA recognition. Nature 461:1248–1253. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kreiswirth BN, Lofdahl S, Betley MJ, O'Reilly M, Schlievert PM, Bergdoll MS, Novick RP. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 24.Steinmetz M, Richter R. 1994. Plasmids designed to alter the antibiotic resistance expressed by insertion mutations in Bacillus subtilis, through in vivo recombination. Gene 142:79–83. doi: 10.1016/0378-1119(94)90358-1. [DOI] [PubMed] [Google Scholar]