Abstract
Background: Prospective studies consistently link low magnesium intake to higher type 2 diabetes (T2D) risk.
Objective: We examined the association of common genetic variants [single nucleotide polymorphisms (SNPs)] in genes related to magnesium homeostasis with T2D risk and potential interactions with magnesium intake.
Methods: Using the Women's Health Initiative-SNP Health Association Resource (WHI-SHARe) study, we identified 17 magnesium-related ion channel genes (583 SNPs) and examined their associations with T2D risk in 7287 African-American (AA; n = 1949 T2D cases) and 3285 Hispanic-American (HA; n = 611 T2D cases) postmenopausal women. We performed both single- and multiple-locus haplotype analyses.
Results: Among AA women, carriers of each additional copy of SNP rs6584273 in cyclin mediator 1 (CNNM1) had 16% lower T2D risk [OR: 0.84; false discovery rate (FDR)-adjusted P = 0.02]. Among HA women, several variants were significantly associated with T2D risk, including rs10861279 in solute carrier family 41 (anion exchanger), member 2 (SLC41A2) (OR: 0.54; FDR-adjusted P = 0.04), rs7174119 in nonimprinted in Prader-Willi/Angelman syndrome 1 (NIPA1) (OR: 1.27; FDR-adjusted P = 0.04), and 2 SNPs in mitochondrial RNA splicing 2 (MRS2) (rs7738943: OR = 1.55, FDR-adjusted P = 0.01; rs1056285: OR = 1.48, FDR-adjusted P = 0.02). Even with the most conservative Bonferroni adjustment, two 2-SNP-haplotypes in SLC41A2 and MRS2 region were significantly associated with T2D risk (rs12582312-rs10861279: P = 0.0006; rs1056285-rs7738943: P = 0.002). Among women with magnesium intake in the lowest 30% (AA: ≤0.164 g/d; HA: ≤0.185 g/d), 4 SNP signals were strengthened [rs11590362 in claudin 19 (CLDN19), rs823154 in SLC41A1, rs5929706 and rs5930817 in membra; HA: ≥0.313 g/d), rs6584273 in CNNM1 (OR: 0.71; FDR-adjusted P = 0.04) and rs1800467 in potassium inwardly rectifying channel, subfamily J, member 11 (KCNJ11) (OR: 2.50; FDR-adjusted P = 0.01) were significantly associated with T2D risk.
Conclusions: Our findings suggest important associations between genetic variations in magnesium-related ion channel genes and T2D risk in AA and HA women that vary by amount of magnesium intake.
Keywords: genes, ion channel, magnesium intake, diabetes, women
Introduction
Magnesium is an essential mineral found in whole grains, leafy green vegetables, legumes, and nuts. Large prospective studies of men and women have associated greater magnesium intake with lower risk of type 2 diabetes (T2D)15 and metabolic abnormalities (1, 2). Absorption of magnesium, a positively charged cation (Mg2+), relies on ion channels, including the transient receptor potential (TRP) family of cation channels. In patients with hypomagnesemia and secondary hypocalcemia, several mutations in TRP cation channel, subfamily M, member 6 (TRPM6) were identified, indicating the importance of genes in the TRP family (TRPM6 and TRPM7) in the regulation of magnesium homeostasis. In particular, 2 variants of the TRPM6 coding region were recently identified as T2D susceptibility loci in Caucasian women with low magnesium intake (<250 mg), further highlighting a potential nutrient-gene interaction in affecting T2D risk. However, this magnesium-gene interaction was not consistently observed nor examined (2, 3) in other racial/ethnic groups.
Moreover, pancreatic β-cell ATP-sensitive K+ (KATP) channels play a central role in glucose-induced insulin secretion (4). Common variants in potassium inwardly rectifying channel, subfamily J, member 11 (KCNJ11) and ATP-binding cassette, sub-family C, member 8 (ABCC8) genes that encode the major KATP channel subunits SUR1 and Kir6.2, respectively, were identified as T2D susceptibility loci (5). KATP channel activity is regulated by the intracellular balance of magnesium-ATP and magnesium-ADP and depends on the presence of Mg2+ ions (6). Low magnesium intake and imbalance in intracellular Mg2+ ion concentrations may interact with KATP ion channel variants in affecting T2D risk; however, no studies to date have examined these interactions.
According to their activity at the protein level (e.g., magnesium and other divalent cation transport, reabsorption, mitochondrial magnesium homeostasis, regulating magnesium-dependent enzymes, and handling renal sodium and magnesium), at least 13 additional genes (in the families of claudin, solute carrier family, cyclin, nonimprinted in Prader-Willi/Angelman syndrome, zinc finger DHHC-type, mitochondrial RNA splicing, membrane magnesium transporter, and FXYD domain-containing ion transport regulator) may play a role in magnesium homeostasis (Supplemental Table 1).
We therefore hypothesized that genetic variations in ion channels related to magnesium homeostasis and glucose metabolism are associated with an increased risk of T2D. Because sufficient magnesium intake may partially compensate for magnesium deficiency caused by genetic mutations (2), we further hypothesized that any potential effect on T2D risk because of magnesium-related genetic variants may be more evident among individuals with low magnesium intake. To test these hypotheses, we conducted a genetic association study of T2D risk in 7287 African American (AA) and 3285 Hispanic American (HA) postmenopausal women aged 50–79 y from the National Women’s Health Initiative SNP Health Association Resource (WHI-SHARe). With the use of a whole-genome scan, we identified 583 common single nucleotide polymorphisms (SNPs) in 17 candidate genes of ion channels, including TRPM6, TRPM7, ABCC8, KCNJ11, claudin 16 (CLDN16), CLDN19, solute carrier family 41 (anion exchanger), member 1 (SLC41A1), SLC41A2, solute carrier family 12 (anion exchanger), member 3 (SLC12A3), cyclin mediator 1 (CNNM1), CNNM2, nonimprinted in Prader-Willi/Angelman syndrome 1 (NIPA1), NIPA2, zinc finger, DHHC-type containing 17 (ZDHHC17), mitochondrial RNA splicing 2 (MRS2), membrane magnesium transporter 1 (MMGT1), and FXYD domain-containing ion transport regulator 2 (FXYD2), and investigated their associations with T2D risk.
Methods
Study population.
The WHI is a long-term national health study that focuses on strategies to prevent cardiovascular disease, cancer, diabetes, and fractures in postmenopausal women. Details on the design and recruitment of the WHI are published elsewhere (7). The WHI-SHARe includes 7287 AA and 3285 HA participants aged 50–79 y who participated in the WHI clinical trials or observational study and for whom genome-wide association study genotyping was conducted and who provided supplemental consent that allowed for data sharing. Prevalent T2D cases were identified from self-report of diabetes treatment. Incident T2D was identified as first-time use of hypoglycemic medication (insulin or oral hypoglycemic agents) or hospitalization for previously unreported T2D (7). The procedures of this study were in accordance with the ethical standards of Brown University on human experimentation, and approval was obtained from the institutional review board on human subjects.
Data collection.
Standardized questionnaires, including information on age, ethnicity, education, income, occupation, medical and family history, smoking status, alcohol use, recreational physical activity, and medication and supplement use, were administered at the baseline visit. Dietary magnesium intake was assessed with a semiquantitative FFQ that estimated average dietary intake for the past 3 months. Total magnesium represented the sum of magnesium intake from both dietary and supplemental sources.
Fasting blood specimens were collected from all participants at baseline according to a standardized protocol. Participants were instructed to fast for 12 h before collection, take all regular medications except for diabetes medication, take no aspirin or nonsteroidal anti-inflammatory drugs for 48 h before the visit except for those medications taken regularly, refrain from smoking for 1 h before the visit, and perform no vigorous physical activity for 12 h before the visit.
Genotyping.
DNA extraction and genotyping were conducted at the Fred Hutchinson Cancer Research Center in Seattle. The Affymetrix Genome-wide Human SNP Array 6.0 was used to genotype >1.8 million markers of genetic variation. To test the hypotheses in this study, we identified a total of 583 SNPs that include 68 SNPs on the TRPM6 gene, 36 on TRPM7, 65 on ABCC8, 12 on KNCJ11, 40 on CLDN16, 5 on CLDN19, 19 on SLC41A1, 62 on SLC41A2, 29 on SLC12A3, 48 on CNNM1, 36 on CNNM2, 23 on NIPA1, 18 on NIPA2, 71 on ZDHHC17, 23 on MRS2, 14 on MMGT1, and 14 on FXYD2.
Statistical analyses.
Baseline demographic and lifestyle characteristics were examined by T2D case-control status with the use of χ2 or t tests. Deviations from Hardy-Weinberg equilibrium were assessed with a χ2 goodness-of-fit test in PLINK (8). Relatedness was determined with the method-of-moments approach with an identity-by-descent model (8). Confirmatory analysis (9) was also performed with a pairwise kinship coefficient estimator. On the basis of these coefficients, pairs of parent-offspring (22 pairs and 2 trios), monozygotic twins (5 pairs), and siblings (192 pairs and 5 trios) were identified. The ones with the largest call rate of each pair of relatives were included in the subsequent analysis; 234 individuals with lower call rates of each pair of relatives (parent-offspring pairs, monozygotic twins, and siblings) were excluded on the basis of the relatedness analysis. To correct for population stratification because of admixture within AA and HA populations, we conducted principal component analyses (10) of global ancestry and included 3 principal components in all multivariable-adjusted models.
We used logistic regression to calculate ORs and 95% CIs for single-locus (SNP) associations with T2D risk under an additive genetic model. We also used dominant model for assessing single-SNP association with T2D risk. All multivariable models were adjusted for age, geographic region, and 3 principal components of global ancestry. To account for potential false positives because of multiple comparisons in this study, we calculated the false discovery rate (FDR) by incorporating all P values from multiple tests performed for the association of SNPs in each gene and T2D risk. FDR is defined as the proportion of false positives among all significant results and is estimated by setting some rejection region so that on average FDR < the level of significance (α = 0.05); another commonly used and more conservative multiple comparison adjustment method, Bonferroni’s correction, sets the significance cutoff at α/n, where n is the number of hypotheses to test. The FDR statistics were obtained for each P value, and the FDR statistics with adjusted P ≤ 0.05 were considered significant (11). All models were run separately in AA and HA women. Stratified analyses were performed to examine whether the genetic associations with T2D were modified by magnesium intake by using the magnesium intake in the lowest 30% (≤0.164 g/d for AA women and ≤0.185 g/d for HA women) and highest 70% (≥0.291 g/d for AA women and ≥0.313 g/d for HA women) categories as cutoffs. Sliding window haplotype-based analyses (12) (window width = 2) were used to examine the joint contribution of 2-SNP windows on T2D risk. For each window, a Wald test based on t distribution was used. The test performed N-1 haplotype-specific tests for N haplotypes (of each vs. all others), depending on the number of SNPs in each gene. All statistical analyses were conducted with PLINK (8) and R open access software version 2.13.0 (13).
Results
Baseline characteristics of the study population are shown in Table 1. Compared with controls, cases had a higher BMI, lower current alcohol intake, a lower level of physical activity, and higher percentage of family history of T2D (P < 0.05). Total magnesium intake was lower among cases than among controls (mean intake = 0.248 g/d for AA women and 0.267 g/d for HA women), and AA women had lower magnesium intake than HA women.
TABLE 1.
Baseline characteristics of AA and HA women in the WHI-SHARe cohort by diabetes status1
| AA women |
HA women |
|||||
| Cases (n = 1949) | Controls (n = 6161) | P2 | Cases (n = 611) | Controls (n = 2855) | P2 | |
| Total magnesium intake,3 g/d | 0.243 ± 0.1514 | 0.254 ± 0.162 | 0.005 | 0.255 ± 0.154 | 0.276 ± 0.169 | 0.003 |
| Total energy intake, kcal/d | 1490 ± 962 | 1490 ± 881 | 0.96 | 1550 ± 964 | 1550 ± 932 | 0.99 |
| Age, y | 62.4 ± 6.80 | 61.4 ± 7.10 | <0.001 | 60.4 ± 6.70 | 60.2 ± 6.70 | 0.52 |
| BMI, kg/m2 | 33.1 ± 6.50 | 30.4 ± 6.20 | <0.001 | 31.5 ± 5.80 | 28.3 ± 5.40 | <0.001 |
| Smoking, % | 0.23 | 0.49 | ||||
| Never | 46.7 | 48.8 | 60.8 | 63.1 | ||
| Past | 41.6 | 39.5 | 31.6 | 30.2 | ||
| Current | 11.7 | 11.7 | 7.68 | 6.68 | ||
| Alcohol consumption, % | <0.001 | <0.001 | ||||
| Never | 18.0 | 15.5 | 25.1 | 17.2 | ||
| Past | 43.0 | 30.2 | 32.6 | 19.1 | ||
| Current | 39.1 | 54.3 | 42.3 | 63.7 | ||
| Physical activity, METs/wk | 7.97 ± 11.2 | 10.2 ± 13.1 | <0.001 | 8.52 ± 12.4 | 11.2 ± 14.0 | <0.001 |
| Family history of diabetes, % | 63.0 | 41.2 | <0.001 | 62.4 | 38.5 | <0.001 |
n = 11,576. AA, African American; HA, Hispanic American; MET, metabolic equivalent; SHARe, SNP (single nucleotide polymorphism) Health Association Resource; WHI, Women’s Health Initiative.
Determined by χ2 or t tests.
Includes dietary and supplemental magnesium.
Means ± SDs (all such values).
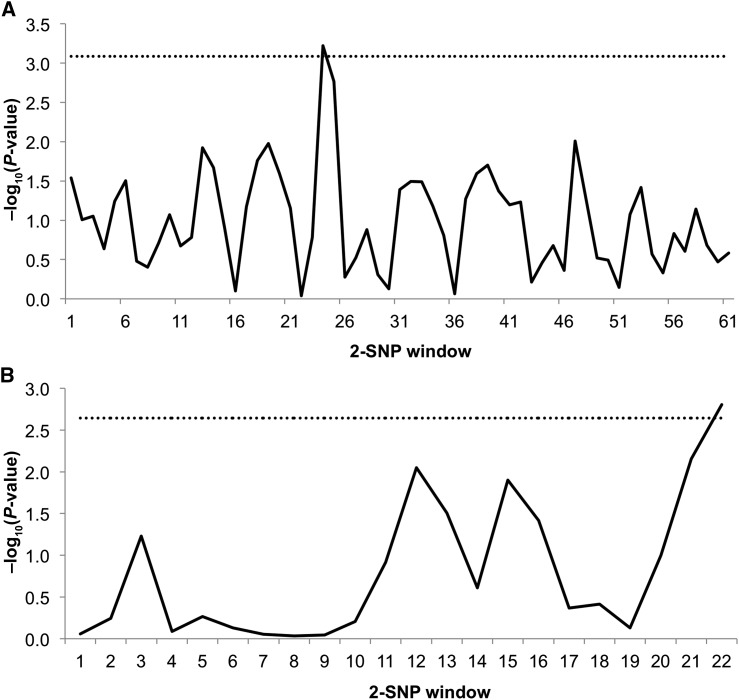
After adjustment for age, region, principal components of global ancestry, and multiple comparisons with the FDR, carriers of each additional copy of a variant allele of the SNP rs6584273 (in the CNNM1 gene) had 16% lower T2D risk in AA women. Among HA women, 4 variants were significantly associated with T2D risk; these include rs10861279 in SLC41A2, rs7174119 in NIPA1, and 2 SNPs in MRS2 gene (rs7738943 and rs1056285) (Tables 2, 3). Results were similar with the use of a dominant genetic model (Supplemental Tables 2, 3). Sliding window analyses revealed two 2-SNP sub-haplotypes (SLC41A2 rs12582312-rs10861279: P = 0.0006; MRS2 rs1056285-rs7738943: P = 0.002) significantly associated with T2D risk after the most conservative Bonferroni adjustment [threshold for SLC41A2 = −log10(0.05/61) = 3.09; threshold for MRS2 = −log10(0.05/22) = 2.64] (Figure 1A, B).
TABLE 2.
Top 10 associations (ranked by nominal P values among AA women) between ion channel-related genes and diabetes risk in the WHI-SHARe cohort1
| Minor allele frequency (AA/HA) |
AA women (n = 7287) |
HA women (n = 3285) |
||||||
| Locus | SNP | Major/minor allele | Cases | Controls | OR (95% CI) | P (FDR-adjusted P) | OR (95% CI) | P |
| CNNM1 | rs6584273 | G/A | 0.20/0.04 | 0.22/0.05 | 0.84 (0.76, 0.92) | 0.0005 (0.02)* | 1.05 (0.76, 1.44) | 0.76 |
| TRPM6 | rs577787 | T/C | 0.30/0.09 | 0.27/0.08 | 1.15 (1.10, 1.25) | 0.002 (0.09) | 1.14 (0.91, 1.44) | 0.26 |
| ABCC8 | rs7947462 | G/A | 0.07/0.23 | 0.09/0.24 | 0.79 (0.68, 0.92) | 0.002 (0.14) | 0.92 (0.79, 1.08) | 0.29 |
| TRPM6 | rs546862 | T/C | 0.30/0.09 | 0.27/0.08 | 1.14 (1.00, 1.25) | 0.003 (0.09) | 1.17 (0.93, 1.47) | 0.19 |
| CLDN16 | rs4438632 | G/A | 0.04/0.25 | 0.03/0.22 | 1.38 (1.10, 1.70) | 0.003 (0.11) | 1.06 (0.90, 1.24) | 0.48 |
| TRPM6 | rs7046143 | T/C | 0.30/0.24 | 0.27/0.22 | 1.13 (1.00, 1.24) | 0.004 (0.10) | 1.05 (0.90, 1.23) | 0.53 |
| TRPM7 | rs2011064 | C/G | 0.19/0.46 | 0.17/0.45 | 1.16 (1.00, 1.28) | 0.005 (0.09) | 1.02 (0.89, 1.16) | 0.80 |
| CLDN16 | rs1593313 | T/C | 0.06/0.32 | 0.05/0.29 | 1.28 (1.10, 1.53) | 0.006 (0.12) | 1.01 (0.87, 1.18) | 0.87 |
| TRPM7 | rs3109894 | C/T | 0.30/0.47 | 0.28/0.46 | 1.13 (1.00, 1.23) | 0.007 (0.09) | 1.00 (0.88, 1.15) | 0.95 |
| TRPM7 | rs1395297 | T/C | 0.30/0.47 | 0.28/0.46 | 1.12 (1.00, 1.22) | 0.008 (0.09) | 0.99 (0.87, 1.14) | 0.95 |
n = 10,572. ORs and 95% CIs were computed by using additive model with multivariate logistic regression models and were adjusted for age, region, and 3 principal components of global ancestry. *FDR-adjusted P ≤ 0.05. AA, African American; FDA, false discovery rate; HA, Hispanic American; SHARe, SNP Health Association Resource; SNP, single nucleotide polymorphism; WHI, Women’s Health Initiative.
TABLE 3.
Top 10 associations (ranked by nominal P values among HA women) between ion channel-related genes and diabetes risk in the WHI-SHARe cohort1
| Minor allele frequency (AA/HA) |
AA women (n = 7287) |
HA women (n = 3285) |
||||||
| Locus | SNP | Major/minor allele | Cases | Controls | OR (95% CI) | P | OR (95% CI) | P (FDR-adjusted P) |
| SLC41A2 | rs10861279 | T/G | 0.08/0.03 | 0.07/0.07 | 1.11 (0.96, 1.28) | 0.17 | 0.54 (0.38, 0.77) | 0.0006 (0.04)* |
| MRS2 | rs7738943 | G/C | 0.13/0.08 | 0.12/0.06 | 1.02 (0.90, 1.14) | 0.79 | 1.55 (1.20, 2.00) | 0.0006 (0.01)* |
| NIPA1 | rs7174119 | G/A | 0.22/0.30 | 0.22/0.27 | 1.00 (0.91, 1.10) | 1.00 | 1.27 (1.10, 1.47) | 0.002 (0.04)* |
| MRS2 | rs1056285 | G/A | 0.14/0.08 | 0.14/0.06 | 1.02 (0.91, 1.14) | 0.74 | 1.48 (1.20, 1.90) | 0.002 (0.02)* |
| FXYD2 | rs479991 | T/C | 0.27/0.19 | 0.28/0.23 | 0.99 (0.91, 1.08) | 0.84 | 0.79(0.67, 0.93) | 0.005 (0.07) |
| NIPA1 | rs6606830 | C/T | 0.21/0.29 | 0.22/0.27 | 1.00 (0.91, 1.10) | 0.94 | 1.23 (1.10, 1.43) | 0.01 (0.07) |
| SLC41A2 | rs11112191 | T/C | 0.27/0.17 | 0.27/0.20 | 1.00 (0.91, 1.09) | 0.96 | 0.78 (0.66, 0.93) | 0.01 (0.15) |
| KCNJ11 | rs1800467 | G/C | 0.01/0.06 | 0.009/0.04 | 1.01 (0.65, 1.55) | 0.98 | 1.49 (1.10, 2.01) | 0.01 (0.11) |
| SLC12A3 | rs8061631 | C/T | 0.38/0.22 | 0.37/0.25 | 1.02 (0.94, 1.11) | 0.62 | 0.81 (0.69, 0.95) | 0.01 (0.32) |
| SLC41A2 | rs11112211 | G/C | 0.09/0.05 | 0.08/0.07 | 1.05 (0.92, 1.21) | 0.47 | 0.67 (0.49, 0.92) | 0.01 (0.15) |
n = 10,572. ORs and 95% CIs were computed by using additive model with multivariate logistic regression models and were adjusted for age, region, and 3 principal components of global ancestry. *FDR-adjusted P ≤ 0.05. AA, African American; FDA, false discovery rate; HA, Hispanic American; SHARe, SNP Health Association Resource; SNP, single nucleotide polymorphism WHI, Women’s Health Initiative.
FIGURE 1.
(A) Sliding window analysis for SLC41A2 haplotype-T2D associations among 3285 Hispanic American WHI women. Window size is 2 SNPs. Sliding window haplotype-based analyses (12) (window width = 2) were used to examine the joint contribution of 2 SNP windows on T2D risk. For each window, a Wald test on the basis of t distribution was used. The test performed N-1 haplotype-specific tests for N haplotypes (of each vs. all others), depending on the number of SNPs in each gene. The dashed line represents a −log10P of 3.08 (corresponding to P = 0.001), which was used as the global significance threshold by Bonferroni’s correction for 61 window frames. (B) Sliding window analysis for MRS2 haplotype-T2D associations among 3285 Hispanic American WHI women. Window size is 2 SNPs. Sliding window haplotype-based analyses (12) (window width = 2) were used to examine the joint contribution of 2 SNP windows on T2D risk. For each window, a Wald test on the basis of t distribution was used. The test performed N-1 haplotype-specific tests for N haplotypes (of each vs. all others), depending on the number of SNPs in each gene. The dashed line represents a −log10P of 2.64 (corresponding to P = 0.002), which was used as the global significance threshold by Bonferroni’s correction for 22 window frames. MRS2, mitochondrial RNA splicing 2 gene; SLC41A2, solute carrier family 41 (anion exchanger), member 2 gene; SNP, single nucleotide polymorphism; T2D, type 2 diabetes; WHI, Women’s Health Initiative.
Among women with total magnesium intake in the lowest 30% (≤0.164 g/d for AA women; ≤0.185 g/d for HA women) and after FDR adjustment, 4 SNPs signals were significantly strengthened among HA women (CLDN19 rs11590362, SLC41A1 rs823154, MMGT1 rs5929706 and MMGT1 rs5930817) (Table 4). TRPM6 coding region variants, IIe1393Val and Lys1584Glu, previously identified as susceptibility loci for T2D among women with low magnesium intake (2), were not statistically significant among women with low magnesium intake in single-SNP or haplotype sliding window analyses. Among women with magnesium intake in the highest 70% of magnesium intake (≥0.291 g/d for AA women; ≥0.313 g/d for HA women) and after FDR adjustment, CNNM1 rs6584273 was significantly associated with T2D risk among AA women with magnesium intake ≥291 mg/d. A missense mutation in KCNJ11 (rs1800467) was also significantly associated with T2D risk among HA women with magnesium intake ≥0.313 g/d. Results were similar with the use of a dominant genetic model (Supplemental Table 4).
TABLE 4.
Significant associations (FDR q ≤ 0.05) between ion channel-related genes and diabetes risk stratified by total magnesium intake in the WHI-SHARe cohort1
| AA women |
HA women |
||||||||
| Low intake (≤0.164 g/d; n = 2154) |
High intake (≥0.291 g/d; n = 2120) |
Low intake (≤0.185 g/d; n = 1015) |
High intake (≥0.313 g/d; n = 954) |
||||||
| Locus | SNP | OR (95% CI) | P | OR (95% CI) | P (FDR-adjusted P) | OR (95% CI) | P (FDR q) | OR (95% CI) | P (FDR-adjusted P) |
| KCNJ11 | rs1800467 | 0.97 (0.46, 2.04) | 0.93 | 1.13 (0.55, 2.32) | 0.74 | 1.11 (0.66, 1.86) | 0.70 | 2.50 (1.50, 4.28) | <0.001 (0.01)* |
| CLDN19 | rs11590362 | 1.24 (0.79, 1.93) | 0.34 | 1.23 (0.80, 1.88) | 0.34 | 2.01 (1.20, 3.37) | 0.01 (0.04)** | 1.38 (0.79, 2.41) | 0.26 |
| SLC41A1 | rs823154 | 1.06 (0.9, 1.26) | 0.47 | 1.02 (0.85, 1.23) | 0.84 | 1.46 (1.10, 1.86) | 0.002 (0.04)** | 1.01 (0.75, 1.37) | 0.93 |
| CNNM1 | rs6584273 | 0.88 (0.74, 1.06) | 0.18 | 0.71 (0.59, 0.87) | <0.001 (0.04)** | 0.93 (0.52, 1.66) | 0.80 | 0.88 (0.46, 1.71) | 0.71 |
| NIPA2 | rs7170784 | 1.05 (0.88, 1.26) | 0.58 | 0.94 (0.77, 1.13) | 0.50 | 1.07 (0.86, 1.34) | 0.55 | 0.62 (0.47, 0.80) | <0.001 (0.01)** |
| NIPA2 | rs8028189 | 1.05 (0.88, 1.26) | 0.60 | 0.93 (0.77, 1.13) | 0.46 | 1.08 (0.87, 1.35) | 0.48 | 0.65 (0.50, 0.84) | 0.001 (0.01)* |
| MMGT1 | rs5929706 | 0.89 (0.60, 1.32) | 0.57 | 0.73 (0.47, 1.14) | 0.17 | 4.66 (1.50, 14.3) | 0.01 (0.05)** | 1.42 (0.29, 6.90) | 0.67 |
| MMGT1 | rs5930817 | 1.09 (0.83, 1.43) | 0.54 | 0.90 (0.66, 1.23) | 0.52 | 3.10 (1.40, 6.85) | 0.01 (0.05)** | 0.88 (0.19, 4.03) | 0.87 |
ORs and 95% CIs were computed by using additive model with multivariate logistic regression models and were adjusted for age, region, and 3 principal components of global ancestry. *FDR-adjusted P ≤ 0.01; **FDR-adjusted P ≤ 0.05. AA, African American; FDA, false discovery rate; HA, Hispanic American; SHARe, SNP Health Association Resource; SNP, single nucleotide polymorphism WHI, Women’s Health Initiative.
Discussion
In this large prospective study of postmenopausal women, we observed genetic associations between magnesium-related ion channel genes and risk of T2D that were modified by total magnesium intake and ethnicity.
The significant signal in the KCNJ11 gene (rs1800467) identified in our study is consistent with a previous report that identified KCNJ11 as an important susceptibility locus for T2D (14, 15). Mutations in KCNJ11 cause familial persistent hyperinsulinemic hypoglycemia in infancy and permanent neonatal diabetes (16–18). In addition, the E23K single nucleotide variant (K23, rs5219) in the KCNJ11 gene is also associated with diabetes (19, 20) and impaired insulin action and secretion (21–24). The rs1800467 polymorphism is a missense mutation (C-G substitution) located on chromosome 11 (codon 3), which appears to result in an L/V substitution that is not predicted to be deleterious (25). In particular, this SNP was significantly associated with T2D risk with high magnesium intake (≥313 mg/d) among HA women. Although there are as yet no functional links about this particular SNP, rs1800467 is in tight linkage disequilibrium (D’ = 0.913) with rs5125, a known type 1 diabetes risk variant. Previously, rs5215 was linked to T2D risk among 924 diabetic cases and 2938 population controls in the Wellcome Trust Case Control Consortium study (28). Previous work has also associated genetic variants and micro-RNA involved in β cells with T2D development. A haplotype consisting of SNPs within KCNJ11 and ABCC8 genes (rs5210, rs5115, and rs5219), involved in pancreatic β-cell development and function, was reported to be significantly associated with T2D among 2025 North Indian subjects of Indo-European ancestry (27). A β-cell commotion of the micro-RNA network, which allowed for normal β-cell development, led to impairment of insulin secretion, impaired glucose homeostasis, and development of diabetes in mice (28).
Several rare mutations in TRPM6 lead to autosomal recessive hypomagnesemia with secondary hypocalcemia, a severe magnesium-wasting disorder caused by impaired intestinal magnesium reabsorption through TRPM6 channels. Oral magnesium intake can compensate for severe magnesium deficiency caused by this disorder through the non–TRPM6-mediated paracellular uptake pathway. Experimental evidence suggests this compensation may be because of dual pathways of intestinal magnesium absorption; a passive paracellular route active when intestinal magnesium concentrations are high and an active transport system when magnesium intake is low. In our sample of AA and HA women, however, we did not observe TRPM6 SNPs to pose significant increased T2D risk among those with low magnesium intake as previously reported in white women (2). In contrast, SNPs in CNNM1, SLC41A2, and MRS2, genes that code for proteins that function in divalent cation and magnesium homeostasis, were significantly associated with T2D risk among AA and HA women. Genetic variants in SLC41A2 and MRS2 lead to changes in corresponding proteins that control Mg2+ influx. Previous studies have shown that increased Mg2+ efflux through dysregulation of the vascular exchange of Na+ and Mg2+ and decreased Mg2+ influx because of defective vascular TRPM6 and TRPM7 expression may lead to blood pressure regulation. This alteration may also contribute to hypomagnesemia and intracellular Mg2+ deficiency in hypertension, diabetes, and cardiovascular diseases (29).
We also observed that polymorphisms in the genes related to magnesium transport, including CLDN19, SLC41A1, and MMGT1, lead to significantly increased T2D risk among AA and HA women with lower magnesium intake, which seems to support the notion that alteration in Mg2+ transport may lead to impaired insulin response and thus increase T2D risk. In low-magnesium media in the kidney cortex of mice maintained on low-magnesium diets, MMGT1 mRNA levels do increase, leading to regulated pathways for Mg2+ transport in the Golgi and post-Golgi organelles of epithelium-derived cells (30). Among women with higher magnesium intake, some carriers of variants in NIPA1 and NIPA2 appear to have a lower T2D risk, which is consistent with a previous observation that links high magnesium to decreased cell surface of NIPA1 (31). Future work to confirm our observations in large multiethnic cohorts or consortia is warranted.
Measurement of magnesium intake estimated by the FFQ is known to work best only to provide a relative ranking of long-term average intake among participants in well-characterized populations. Moreover, magnesium intake assessed by our FFQ has a correlation of 0.7 when validated against dietary records (32, 33). Thus, the magnitude of relation may be underestimated in our study, given that this error is not likely to differ across specific genotypes or the T2D outcome of interest. Some may speculate that magnesium intake may not necessarily reflect the amount of magnesium available to the body; therefore, magnesium concentration may provide further information to investigate whether magnesium differs according to genetic variants for a given magnesium intake. In the Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium, genomic regions that included TRPM6 and CNNM2, CNNM3, and CNNM4 were associated with magnesium concentrations (34). The top polymorphisms appeared to be involved in magnesium transport and homeostasis; it is speculated that they may also modify individual risk for diabetes despite a difference in magnesium intake (35). However, the literature lacks a report of difference in magnesium concentrations according to susceptible genetic variants for a given magnesium intake. Until now, because of feasibility difficulties (assay characteristics, specimen requirement, and costs), no data on magnesium concentrations are available in the WHI cohort. Although blood concentrations of total magnesium are commonly used for defining magnesium deficiency, they are usually within a narrow range under tight homeostatic regulation in the human body and cannot reliably reflect both inter- and intra-individual magnesium status. Thus, there is an urgent need to develop accurate, reliable, and affordable biomarker measures to assess individual magnesium status in population studies, which would provide more informative answers about magnesium status in future studies. Further, previous studies showed that magnesium plays an important role in the synthesis and metabolism of vitamin D, which receptors exist on pancreatic β cells and insulin-sensitive tissues, and which repletion improves glucose and insulin homeostasis in animal models of vitamin D deficiency (36, 37). Given the biological interconnection between magnesium and vitamin D, disentangling the independent and potentially interactive effects of these factors is of critical importance, and further research may provide insights into the mechanism of the development of diabetes through ion channel pathways. Strengths of this study include a well-characterized population of >10,000 minority women with complete genome scans. Although false positives remain a possibility, the candidate gene approach with careful adjustment for multiple comparisons and strong a priori biological information will continue to shed light on the genetic susceptibility of complex diseases. Further, as a complement to single-marker analysis (12), we performed sliding window haplotype analyses which suggested that HA women who were carriers of the haplotypes SLC41A2 rs12582312-rs10861279 and MRS2 rs1056285-rs7738943 were associated with T2D risk.
In conclusion, findings from this national cohort of AA and HA postmenopausal women indicate that genetic variations in several magnesium-related ion channels may affect T2D risk with possible effect modifications by magnesium intake and ethnicity. In particular, among HA women with low magnesium intake, CLDN19, SLC41A1, and MMGT1 were associated with increased risk of T2D. Among HA women with high magnesium intake, the KCNJ11 gene was associated with an increased T2D risk and NIPA2 was associated with a decreased T2D risk. Among AA women with high magnesium intake, the CNNM1 gene was found to have a protective association with T2D. Further confirmation of our observations in WHI-SHARe is clearly warranted in future mechanistic studies.
Supplementary Material
Acknowledgments
We thank Adam Frankish at Wellcome Trust Sanger Institute for help with bioinformatics investigation of our findings. SL designed the research; CBE and SL were involved in the data collection; KHKC and SAC analyzed the data; KHKC, SAC, YS, MC, CBE, W-CHW, and SL participated in the interpretation of findings that led to this manuscript; SAC and SL wrote the first draft. All authors read and approved the final manuscript.
Footnotes
Abbreviations used: AA, African American; ABCC8, ATP-binding cassette, subfamily C, member 8 gene; CLDN16, claudin 16 gene; CLDN19, claudin 19 gene; CNNM1, cyclin mediator 1 gene; CNNM2, cyclin mediator 2 gene; FDR, false discovery rate; FXYD2, FXYD domain-containing ion transport regulator 2 gene; HA, Hispanic American; KATP, ATP-sensitive K+; KCNJ11, potassium inwardly rectifying channel, subfamily J, member 11 gene; MMGT1, membrane magnesium transporter 1 gene; MRS2, mitochondrial RNA splicing 2 gene; NIPA1, nonimprinted in Prader-Willi/Angelman syndrome 1 gene; NIPA2, nonimprinted in Prader-Willi/Angelman syndrome 2 gene; SLC12A3, solute carrier family 12 (anion exchanger), member 3 gene; SLC41A2, solute carrier family 41 (anion exchanger), member 2 gene; SHARe, SNP Health Association Resource; SNP, single nucleotide polymorphism; T2D, type 2 diabetes; TRP, transient receptor potential; TRPM6, transient receptor potential cation channel, subfamily M, member 6 gene; TRPM7, transient receptor potential cation channel, subfamily M, member 7 gene; WHI, Women’s Health Initiative; ZDHHC17, zinc finger, DHHC-type containing 17 gene.
References
- 1.Lopez-Ridaura R, Willett WC, Rimm EB, Liu S, Stampfer MJ, Manson JE, Hu FB. Magnesium intake and risk of type 2 diabetes in men and women. Diabetes Care 2004;27:134–40. [DOI] [PubMed] [Google Scholar]
- 2.Song Y, Hsu YH, Niu T, Manson JE, Buring JE, Liu S. Common genetic variants of the ion channel transient receptor potential membrane melastatin 6 and 7 (TRPM6 and TRPM7), magnesium intake, and risk of type 2 diabetes in women. BMC Med Genet 2009;10:4–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Romero JR, Castonguay AJ, Barton NS, Germer S, Martin M, Zee RY. Gene variation of the transient receptor potential cation channel, subfamily M, members 6 (TRPM6) and 7 (TRPM7), and type 2 diabetes mellitus: a case-control study. Transl Res 2010;156:235–41. [DOI] [PubMed] [Google Scholar]
- 4.Ashcroft FM, Harrison DE, Ashcroft SJ. Glucose induces closure of single potassium channels in isolated rat pancreatic beta-cells. Nature 1984;312:446–8. [DOI] [PubMed] [Google Scholar]
- 5.Gloyn AL, Siddiqui J, Ellard S. Mutations in the genes encoding the pancreatic beta-cell KATP channel subunits Kir6.2 (KCNJ11) and SUR1 (ABCC8) in diabetes mellitus and hyperinsulinism. Hum Mutat 2006;27:220–31 10.1002/humu.20292. [DOI] [PubMed] [Google Scholar]
- 6.Gribble FM, Tucker SJ, Haug T, Ashcroft FM. MgATP activates the beta cell KATP channel by interaction with its SUR1 subunit. Proc Natl Acad Sci USA 1998;95:7185–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Design of the Women's Health Initiative clinical trial and observational study. The Women's Health Initiative Study Group. Control Clin Trials 1998;19:61–109. [DOI] [PubMed] [Google Scholar]
- 8.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thornton T, McPeek MS. ROADTRIPS: case-control association testing with partially or completely unknown population and pedigree structure. Am J Hum Genet 2010;86:172–84. [DOI] [PMC free article] [PubMed]
- 10.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006;38:904–9. [DOI] [PubMed] [Google Scholar]
- 11.Benjamini Y, H. Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 1995;57:289–300.
- 12.Hsu YH, Niu T, Song Y, Tinker L, Kuller LH, Liu S. Genetic variants in the UCP2–UCP3 gene cluster and risk of diabetes in the Women's Health Initiative Observational Study. Diabetes 2008;57:1101–7. [DOI] [PubMed] [Google Scholar]
- 13.R Development Core Team. R: a language and environment for statistical computing [Internet]. Vienna, Austria: R Foundation for Statistical Computing; 2010 [cited 2014 Sep 2]. Available from: http://wwwR-project.org.
- 14.Aguilar-Bryan L, Bryan J. Molecular biology of adenosine triphosphate-sensitive potassium channels. Endocr Rev 1999;20:101–35. [DOI] [PubMed] [Google Scholar]
- 15.Edghill EL, Flanagan SE, Ellard S. Permanent neonatal diabetes due to activating mutations in ABCC8 and KCNJ11. Rev Endocr Metab Disord 2010;11:193–8. [DOI] [PubMed] [Google Scholar]
- 16.Gloyn AL, Cummings EA, Edghill EL, Harries LW, Scott R, Costa T, Temple IK, Hattersley AT, Ellard S. Permanent neonatal diabetes due to paternal germline mosaicism for an activating mutation of the KCNJ11 gene encoding the Kir6.2 subunit of the beta-cell potassium adenosine triphosphate channel. J Clin Endocrinol Metab 2004;89:3932–5. [DOI] [PubMed] [Google Scholar]
- 17.Gloyn AL, Pearson ER, Antcliff JF, Proks P, Bruining GJ, Slingerland AS, Howard N, Srinivasan S, Silva JM, Molnes J, et al. Activating mutations in the gene encoding the ATP-sensitive potassium-channel subunit Kir6.2 and permanent neonatal diabetes. N Engl J Med 2004;350:1838–49. [DOI] [PubMed] [Google Scholar]
- 18.Slingerland AS, Hattersley AT. Mutations in the Kir6.2 subunit of the KATP channel and permanent neonatal diabetes: new insights and new treatment. Ann Med 2005;37:186–95. [DOI] [PubMed] [Google Scholar]
- 19.Gloyn AL, Weedon MN, Owen KR, Turner MJ, Knight BA, Hitman G, Walker M, Levy JC, Sampson M, Halford S, et al. Large-scale association studies of variants in genes encoding the pancreatic beta-cell KATP channel subunits Kir6.2 (KCNJ11) and SUR1 (ABCC8) confirm that the KCNJ11 E23K variant is associated with type 2 diabetes. Diabetes 2003;52:568–72. [DOI] [PubMed] [Google Scholar]
- 20.Zhou D, Zhang D, Liu Y, Zhao T, Chen Z, Liu Z, Yu L, Zhang Z, Xu H, He L. The E23K variation in the KCNJ11 gene is associated with type 2 diabetes in Chinese and East Asian population. J Hum Genet 2009;54:433–5. [DOI] [PubMed] [Google Scholar]
- 21.Alsmadi O, Al-Rubeaan K, Wakil SM, Imtiaz F, Mohamed G, Al-Saud H, Al-Saud NA, Aldaghri N, Mohammad S, Meyer BF. Genetic study of Saudi diabetes (GSSD): significant association of the KCNJ11 E23K polymorphism with type 2 diabetes. Diabetes Metab Res Rev 2008;24:137–40. [DOI] [PubMed] [Google Scholar]
- 22.Doi Y, Kubo M, Ninomiya T, Yonemoto K, Iwase M, Arima H, Hata J, Tanizaki Y, Iida M, Kiyohara Y. Impact of Kir6.2 E23K polymorphism on the development of type 2 diabetes in a general Japanese population: the Hisayama Study. Diabetes 2007;56:2829–33. [DOI] [PubMed] [Google Scholar]
- 23.Gonen MS, Arikoglu H, Erkoc Kaya D, Ozdemir H, Ipekci SH, Arslan A, Kayis SA, Gogebakan B. Effects of single nucleotide polymorphisms in K(ATP) channel genes on type 2 diabetes in a Turkish population. Arch Med Res 2012;43:317–23. [DOI] [PubMed] [Google Scholar]
- 24.Palmer ND, Langefeld CD, Bryer-Ash M, Rotter JI, Taylor KD, Bowden DW. Association of the Kir6.2 E23K variant with reduced acute insulin response in African-Americans. J Clin Endocrinol Metab 2008;93:4979–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics 2010;26:2069–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, Timpson NJ, Perry JR, Rayner NW, Freathy RM, et al. ; Wellcome Trust Case Control Consortium (WTCCC). Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 2007;316:1336–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chavali S, Mahajan A, Tabassum R, Dwivedi OP, Chauhan G, Ghosh S, Tandon N, Bharadwaj D. Association of variants in genes involved in pancreatic beta-cell development and function with type 2 diabetes in North Indians. J Hum Genet 2011;56:695–700. [DOI] [PubMed]
- 28.Kalis M, Bolmeson C, Esguerra JL, Gupta S, Edlund A, Tormo-Badia N, Speidel D, Holmberg D, Mayans S, Khoo NK, et al. Beta-cell specific deletion of Dicer1 leads to defective insulin secretion and diabetes mellitus. PLoS One 2011;6:e29166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yogi A, Callera GE, Antunes TT, Tostes RC, Touyz RM. Vascular biology of magnesium and its transporters in hypertension. Magnes Res 2010;23:S207–15. [DOI] [PubMed] [Google Scholar]
- 30.Goytain A, Quamme GA. Identification and characterization of a novel family of membrane magnesium transporters, MMgT1 and MMgT2. Am J Physiol Cell Physiol 2008;294:C495–502. [DOI] [PubMed] [Google Scholar]
- 31.Goytain A, Hines RM, El-Husseini A, Quamme GA. NIPA1(SPG6), the basis for autosomal dominant form of hereditary spastic paraplegia, encodes a functional Mg2+ transporter. J Biol Chem 2007;282:8060–8. [DOI] [PubMed] [Google Scholar]
- 32.Chacko SA, Song Y, Nathan L, Tinker L, de Boer IH, Tylavsky F, Wallace R, Liu S. Relations of dietary magnesium intake to biomarkers of inflammation and endothelial dysfunction in an ethnically diverse cohort of postmenopausal women. Diabetes Care 2010;33:304–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patterson RE, Kristal AR, Tinker LF, Carter RA, Bolton MP, Agurs-Collins T. Measurement characteristics of the Women's Health Initiative food frequency questionnaire. Ann Epidemiol 1999;9:178–87. [DOI] [PubMed] [Google Scholar]
- 34.Meyer TE, Verwoert GC, Hwang SJ, Glazer NL, Smith AV, van Rooij FJ, Ehret GB, Boerwinkle E, Felix JF, Leak TS, et al. Genome-wide association studies of serum magnesium, potassium, and sodium concentrations identify six loci influencing serum magnesium levels. PLoS Genet 2010;6:e1001045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hruby A, McKeown NM, Song Y, Djousse L. Dietary magnesium and genetic interactions in diabetes and related risk factors: a brief overview of current knowledge. Nutrients 2013;5:4990–5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chacko SA, Song Y, Manson JE, Van Horn L, Eaton C, Martin LW, McTiernan A, Curb JD, Wylie-Rosett J, Phillips LS, et al. Serum 25-hydroxyvitamin D concentrations in relation to cardiometabolic risk factors and metabolic syndrome in postmenopausal women. Am J Clin Nutr 2011;94:209–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deng X, Song Y, Manson JE, Signorello LB, Zhang SM, Shrubsole MJ, Ness RM, Seidner DL, Dai Q. Magnesium, vitamin D status and mortality: results from US National Health and Nutrition Examination Survey (NHANES) 2001 to 2006 and NHANES III. BMC Med 2013;11:187–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.