Abstract
Background: Hundreds of naturally occurring milk peptides are present in term human milk. Preterm milk is produced before complete maturation of the mammary gland, which could change milk synthesis and secretion processes within the mammary gland, leading to differences in protein expression and enzymatic activity, thereby resulting in an altered peptide profile.
Objective: This study examined differences in peptides present between milk from women delivering at term and women delivering prematurely.
Methods: Nano-LC tandem mass spectrometry was employed to identify naturally occurring peptides and compare their abundances between term and preterm human milk samples at multiple time points over lactation. Term milk samples were collected from 8 mothers and preterm milk was collected from 14 mothers. The 28 preterm and 32 term human milk samples were divided into 4 groups based on day of collection (<14, 14–28, 29–41, and 42–58 d).
Results: Preterm milk peptide counts, ion abundance, and concentration were significantly higher in preterm milk than term milk. Bioinformatic analysis of the cleavage sites for peptides identified suggested that plasmin was more active in preterm milk than term milk and that cytosol aminopeptidase and carboxypeptidase B2 likely contribute to extensive milk protein breakdown. Many identified milk peptides in both term and preterm milk overlapped with known functional peptides, including antihypertensive, antimicrobial, and immunomodulatory peptides.
Conclusion: The high protein degradation by endogenous proteases in preterm milk might attenuate problems because of the preterm infant’s immature digestive system. This trial was registered at clinicaltrials.gov as NCT01817127.
Keywords: human milk, peptidomics, premature, term, mass spectrometry, plasmin, carboxypeptidase B2, cytosol aminopeptidase, functional peptide
Introduction
Human milk contains hundreds of naturally occurring peptides derived from intramammary disassembly of milk proteins (1–4) by milk proteases, including plasmin (5), cathepsin D (6), and elastase (7, 8). Many of the released peptides have high homology to known antimicrobial and immunomodulatory peptides (1, 4), and milk peptides as an ensemble kill Escherichia coli and Staphylococcus aureus (1). Preterm milk (from mothers who give birth at <37 wk gestation) has higher protein concentration (9), higher energy content (10), higher lipid concentration (10), an altered FA profile (11), lower lactose (after the first week) (10), and higher sodium, chloride, magnesium, and iron (12) compared to term milk. Chromogenic enzymatic assays show that preterm milk has higher plasmin activity than term milk (13). We previously demonstrated that plasmin is the main protease that hydrolyzes term human milk proteins in the mammary gland (8). We hypothesized that the higher plasmin activity in preterm milk results in increased released peptides compared to term milk with potential biological consequences for the preterm mother and infant.
Ferranti et al. (2) found, via matrix-assisted laser desorption ionization and electrospray MS, >100 peptides originating from αs1-, β-, and κ-casein in milk samples obtained from 1 mother within the first week after premature delivery at 25 wk gestation. A large number of identified peptides in the preterm mother’s milk were also found in 2 term mothers’ milk samples, which suggests that the same enzymatic mechanisms are at play in both preterm and term milk. Armaforte et al. (13) found via 2D-SDS-PAGE, in-gel trypsin digestion, and MS that low molecular weight casein fragments were overexpressed in preterm milk compared to term milk, whereas intact αs1- and β-casein were present at lower concentrations in preterm than term milk. These findings suggest that more degradation of casein occurs in preterm milk than term milk, which coincides with the finding that plasmin activity is higher in preterm milk (13).
In this paper, we report profiles and comparisons of the peptides, both qualitatively and quantitatively, in term and preterm milk samples over lactation with nano-LC tandem MS. We examine the patterns of enzymatic protein degradation in term and preterm milk. Finally, we examine the peptides produced for homology to known functional peptides.
Methods
Sample collection.
Informed consent was obtained from all mothers participating in the study, and the study was approved by the UC Davis Institutional Review Board. Human milk samples were collected from 14 healthy mothers who delivered preterm infants (24–32 wk gestation) and from 8 healthy mothers enrolled in the UC Davis Foods for Health Institute Lactation Study who gave birth to term infants (37–41 wk gestation) (clinicaltrials.gov identifier NCT01817127). Preterm infants were in the neonatal intensive care unit of the UC Davis Medical Center in Sacramento, California. Samples were collected from 2 to 58 d after parturition by pumping on-site or at home with clean electric breast pumps into sterile plastic containers and stored immediately at −20°C. The breast was cleansed with water on a washcloth (no soap or alcohol) before pumping. Samples were transported to UC Davis on ice and then stored at −40°C. In total, 28 preterm and 32 term human milk samples were collected and divided into 4 groups based on day of collection (<14, 14–28, 29–41, and 42–58 d). The number of observations in each day of lactation group for term and preterm samples is shown in Table 1. Specific dates of collection for each mother are shown in Supplemental Table 1. Subject characteristics, including gestational age at birth, maternal age, parity, birth mode, and infant gender are shown in Supplemental Table 2.
TABLE 1.
Number of observations for each lactation stage group for preterm and term milks
| Lactation stage groups |
||||
| <14 d | 14–28 d | 29–41 d | 42–58 d | |
| Gestational age | ||||
| Preterm | 8 | 6 | 8 | 6 |
| Term | 8 | 8 | 8 | 8 |
Sample preparation.
Samples were thawed on ice. Peptides were extracted as previously described (14) with the following modifications. Briefly, 100 μL of each human milk sample was centrifuged at 16,000 × g for 15 min at 4°C. The upper lipid layer was removed and the infranate (skim milk) was collected. The centrifugation procedure was repeated once. One hundred microliters of water and 1 μL of 10-μg/mL peptide standards stock solution (containing equal parts Leu-enkephalin, gonadoliberin, angiotensin I, and neurotensin; Peptide Calibration Standard Set; ProteoChem) were added to 25-μL skim milk for each sample. Then, 505 μL Folch solution was added and samples were centrifuged at 16,000 × g for 15 min at 4°C. The top layer was collected and dried by centrifugal evaporation (MiVac Quattro concentrator; GeneVac). Samples were rehydrated in 100 μL nanopure water and mixed with use of a vortex mixer. Remaining proteins were precipitated with 1:1 (sample to solution) 200-g/L trichloroacetic acid, followed by mixing, centrifugation, and collection of the supernatant. Supernatants were applied to a 96-well C18 solid-phase extraction plate to purify peptides and eluted as described (14). Eluted peptides were dried by vacuum centrifugation and rehydrated in 40 μL nanopure water for quadrupole time-of-flight analysis.
Peptide concentration determination.
The microplate procedure for the Micro BCA Protein Assay Kit (Thermo Scientific) was employed to measure peptide contents of each extracted peptide sample (15). Absorbance measurements were performed with a UV-visible recording spectrophotometer (Spectra Max 340 Microplate Reader Spectrophotometer; Molecular Devices) set to 562 nm. Because color development continues even after cooling to room temperature, all measurements were taken within 10 min. Standard curves were prepared to determine the amount of peptide in each sample. R2 values were >0.99. All samples were measured in duplicate.
Peptide analysis.
Peptide analysis was performed with the Agilent nano-LC chip-cube 6520 quadrupole time-of-flight tandem mass spectrometer. Term milk samples were analyzed in MS-only mode, whereas preterm samples were analyzed in both MS/MS and MS-only modes. The chip employed contained an enrichment and analytical column packed with a C18 stationary phase connected to an electrospray ionization source. The gradient elution solvents were (A) 3% acetonitrile/0.1% formic acid and (B) 90% acetonitrile/0.1% formic acid. The gradient employed was ramped from 0 to 8% B from 0 to 5 min, 8 to 26.5% B from 5 to 24 min, 26.5 to 100% B from 24 to 42 min, followed by 100% B for 10 min, 100 to 0% B from 52 to 55 min, and 100% A for 10 min (to re-equilibrate the column). Capillary voltage was varied within the range of 1900–2000 V to maintain a stable spray. Drying gas was set to a temperature of 325°C and a flow of 3 L/min. MS-only data were collected for positive ions within an m/z window of 500–3000 at a rate of 1 spectrum/s. For MS/MS analysis, data for positive fragment ions was collected within an m/z window of 100–3000, also at a rate of 1 spectrum/s. Isolation width was set to medium (∼4 amu). Collision energy was determined from precursor m/z values with use of a linear equation with a slope of 0.03600 and an intercept of −4.80 [Energy = 0.03600(m/z) − 4.80]. Minimum precursor ion abundance was set to 1000 ion counts (absolute) and 0.01 (relative).
Creation of peptide library.
To create a peptide library, the preterm milk samples were assayed through use of the same method but with MS/MS as described previously (14) [the library of term milk peptides used in the present study was collected previously (4)]. Peptides were identified with use of X!Tandem (16) with parameters described previously (14). The results from X!Tandem for all samples were compiled for all milk samples. Peptide libraries were created as described previously (4). Peptides with e-values ≤0.01 were removed (a 99% confidence level threshold). The MS proteomics data have been deposited to the ProteomeXchange Consortium (http://www.proteomexchange.org) via the PRIDE partner repository (17) with the dataset identifier PXD001079.
Peptide identification.
Peak intensities for each peptide were extracted from the data as described previously (4). The database match employed a mass error tolerance of 20 ppm and a retention time tolerance of 1 min.
Statistical analysis.
The peptides identified were grouped by gestational age (preterm and term) and day of lactation (<14, 15–28, 29–41, and 42–58 d). For statistical analysis of the peptide counts, ion abundances and concentration, and ion abundances grouped by protein, the mixed effect model with repeated measurements was applied. Gestational age and day of lactation were fixed effects, whereas the individual mothers were the random effect. More specifically, to model the number of peptides in each observation as a function of gestational age at birth and lactation stage, a generalized linear mixed-effects model with Poisson distribution was applied. To model the overall mean abundance and concentration of peptides and changes in predicted plasmin activity in each observation as a function of gestational age at birth and lactation stage, a linear mixed-effects model was applied. For examining the peak area of individual peptides, peptides not present in at least 75% of the samples of each group (either 21 preterm samples or the 24 term samples) were omitted. The peak areas of all samples were transformed into natural log scale. A likelihood ratio test in a linear mixed effect model was applied to model the intensity of each peptide as a function of gestational age at birth, the interaction effect, and the lactation stage for preterm and term samples separately. The Benjamini Hochberg False Discovery Rate method was used to correct for multiple comparisons. Unpaired equal variance two-tailed t tests were employed to compare total peptide number, ion abundance, and peptide concentration in term and preterm milk samples, as well as peptide number, ion abundance, and peptide concentration in term and preterm milk samples by protein, lactation stage, and enzyme. Values in the text are presented as means ± SEMs. All statistical analyses were conducted in R software, version 2.15.2. For all the analyses, peptides with P values ≤ 0.05 were deemed significantly different.
Functional annotation.
Identified peptide sequences from the milk samples were searched against a library of known functional milk peptides from the literature (18–21). Peptides in the samples that completely or partially encompassed a known functional peptide were counted for the bioactive peptide table. For partial matches, peptides had to match the functional peptide in the database by at least 80% overlap.
Computational enzymatic analysis.
Peptide sequences and cleavage sites were analyzed with use of an in-house program called PEnTab [available online (22)] to estimate/predict the enzyme activity as described previously (23). The enzyme specificity patterns used in the algorithm for evaluating cleavages are shown in Supplemental Table 3.
Cleavage sites of cytosol aminopeptidase and carboxypeptidase B2 were determined through manual examination of peptide sequences. Peptides that could have been produced by a plasmin cleavage followed by removal of the C-terminal arginine or lysine (24, 25) were interpreted as potential carboxypeptidase B2 products under the condition that another peptide with the C-terminal arginine or lysine present was identified. Because cytosol aminopeptidase has a broad cleavage specificity and can remove single N-terminal amino acids (Uniprot, P28838), the identified peptides were manually examined for groups of peptides that showed a pattern of sequential loss of N-terminal amino acids (illustrated in Table 2). For a peptide within these groups to be considered a product of cytosol aminopeptidase, the presence of a second peptide with 1 additional amino acid added to its N-terminal end was required. Peptides that were produced by N-terminal removal of arginine or lysine were not counted as cytosol aminopeptidase products because the enzyme cannot cleave at the C-end of either of these residues (26).
TABLE 2.
A human milk β-casein peptide cascade resulting from potential cytosol aminopeptidase activity
| Protein | Residues | Sequence |
| β-casein | (20–33) | ESLSSSEESITEYK |
| β-casein | (21–33) | SLSSSEESITEYK |
| β-casein | (22–33) | LSSSEESITEYK |
| β-casein | (23–33) | SSSEESITEYK |
| β-casein | (24–32) | SSEESITEY1 |
| β-casein | (25–33) | SEESITEYK |
Removal of the C-terminal lysine residue in β-casein (24–32) matches to a carboxypeptidase B2 cleavage.
Grouping and visualizing peptide signals.
Peptide sequences and their normalized intensities were aligned over their corresponding proteins of origin with use of an in-house script written in Python (PepEx) (23).
Results
Overall, 445 endogenous peptides were identified with >99% confidence in the term and preterm milk samples. Of the identified peptides, only 1 was unique to term samples and 9 were only in preterm samples. The sequences of all 445 peptides are shown in Supplemental Table 4. All identified peptide sequences are deposited along with the X!Tandem result files and MS/MS raw data in the ProteomeXchange with identifier PXD001079.
Preterm milk contains more peptide sequences.
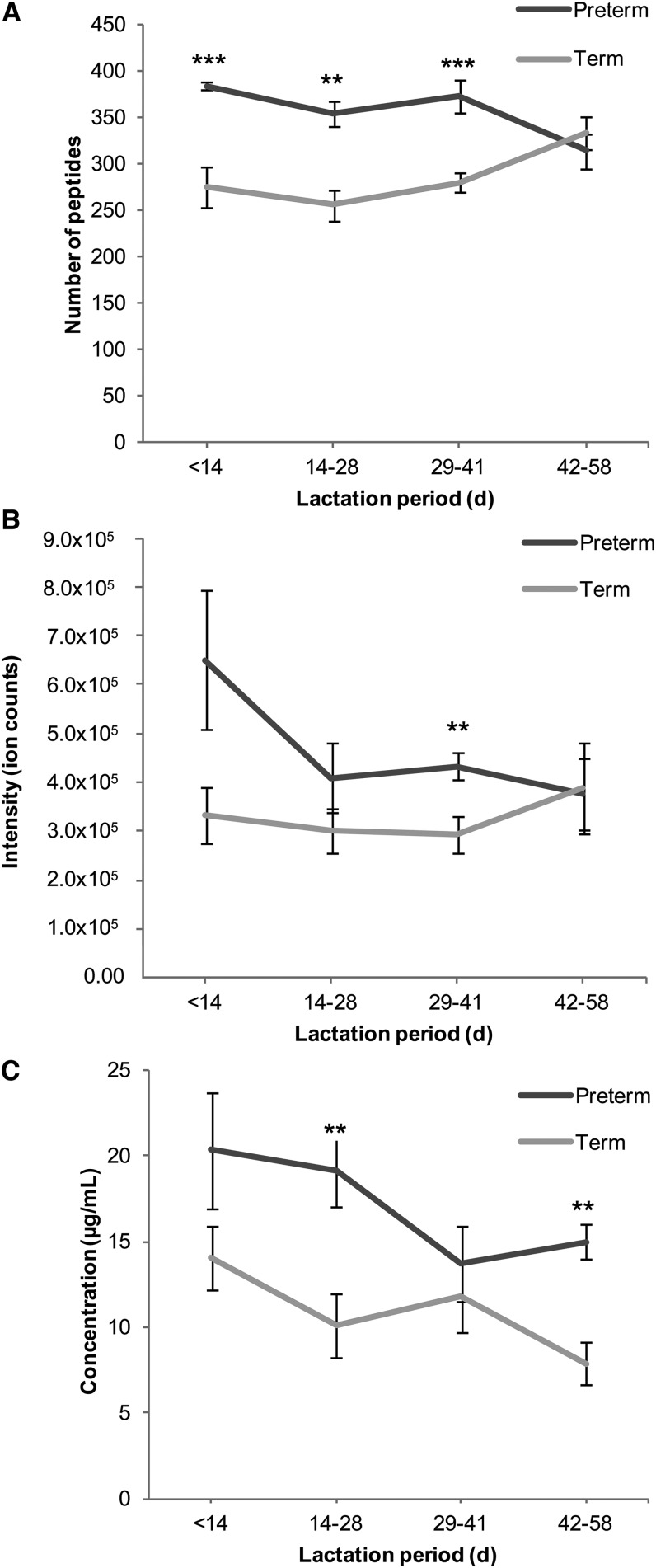
The mean number of peptides in preterm samples was significantly higher than in term samples (359 ± 8.33 vs. 268 ± 9.78, P = 5.41 × 10−7). The peptide counts were significantly higher in the first 3 lactation periods for preterm compared to term milk (Figure 1A).
FIGURE 1.
Number, ion intensities, and concentrations of peptides in preterm and term human milk over lactation time. Number of peptides in preterm and term milk over lactation time (A). Intensities of peptides in preterm and term milk over lactation time (B). Concentrations of peptides in preterm and term human milk over lactation time (C). Values are means ± SEMs. Number of observations (n) in each group is shown in Table 1. Asterisks indicate significant differences between preterm and term milk: **P < 0.01, ***P < 0.001.
A generalized linear mixed-effects model with Poisson distribution was used to calculate the effect of gestational age at birth (preterm and term delivery) and day of lactation on the total number of peptides present in the milk. The total number of peptides was significantly affected by term vs. preterm birth (P = 4.28 × 10−10) (Figure 1). Neither preterm nor term milk showed a significant change over time in the number of peptides present until the last lactation stage. The effect of lactation stage became significant when the last stage was reached (P = 7.57 × 10−5). The interaction of maturation (term vs. preterm birth) and lactation stage also became significant at the last lactation stage (P = 7.56 × 10−13), which was also observed as the crossed line of preterm and term data in the last lactation stage in Figure 1.
Peptide intensity variation.
The mean total peptide intensity (peak area) was higher in preterm milk samples than in term milk samples (477,613 ± 49,300 vs. 329,129 ± 27,800 ion counts, P = <2.00 × 10−16), although deviations from the means were large (Figure 1B). The change over time in the mean intensity of peptides was not statistically significant in preterm or term milks, whereas the interaction term was significant in the last lactation stage (P = 2.86 × 10−5).
A likelihood ratio test in a linear mixed-effects model was used to calculate the effect of gestational age at birth and day of lactation on the abundance of peptides. After omitting peptides not present in at least 75% of the samples of each group (either 21 preterm samples or 24 term samples), 382 peptides remained. Among the 382 peptides, 144 were significantly affected by gestational age at birth, and 65 peptides were significantly affected by the interaction between gestational age at birth and lactation period. The P values of the likelihood ratio tests of these significantly different peptides are shown in the last 2 columns of Supplemental Table 4.
A likelihood ratio test in a linear mixed-effects model was used to calculate the effect of day of lactation on the abundance of individual peptides. Forty-eight and 37 peptides were significantly altered over lactation in preterm milk and term milk samples, respectively (Supplemental Table 5).
Peptide concentration variation.
The mean total peptide concentration was higher in preterm milk samples than in term milk samples (17.1 ± 1.31 vs. 11.0 ± 0.950 μg/mL, P = 3.76 × 10−4). This finding corresponds with the findings that peptide numbers and peptide ion abundances are higher for preterm milks than term milks. The peptide concentrations were significantly higher at 14–28 and 42–58 d of lactation for preterm compared to term milk (Figure 1C). According to the likelihood ratio test of the linear mixed-effect model, the effect of maturation (i.e., term vs. preterm) on peptide concentration is significant (P < 2.20 × 10−16). The change over time in the mean peptide concentration was not statistically significant (P = 5.30 × 10−2). Likewise, there was no significant interaction (P = 0.37).
Protein of origin.
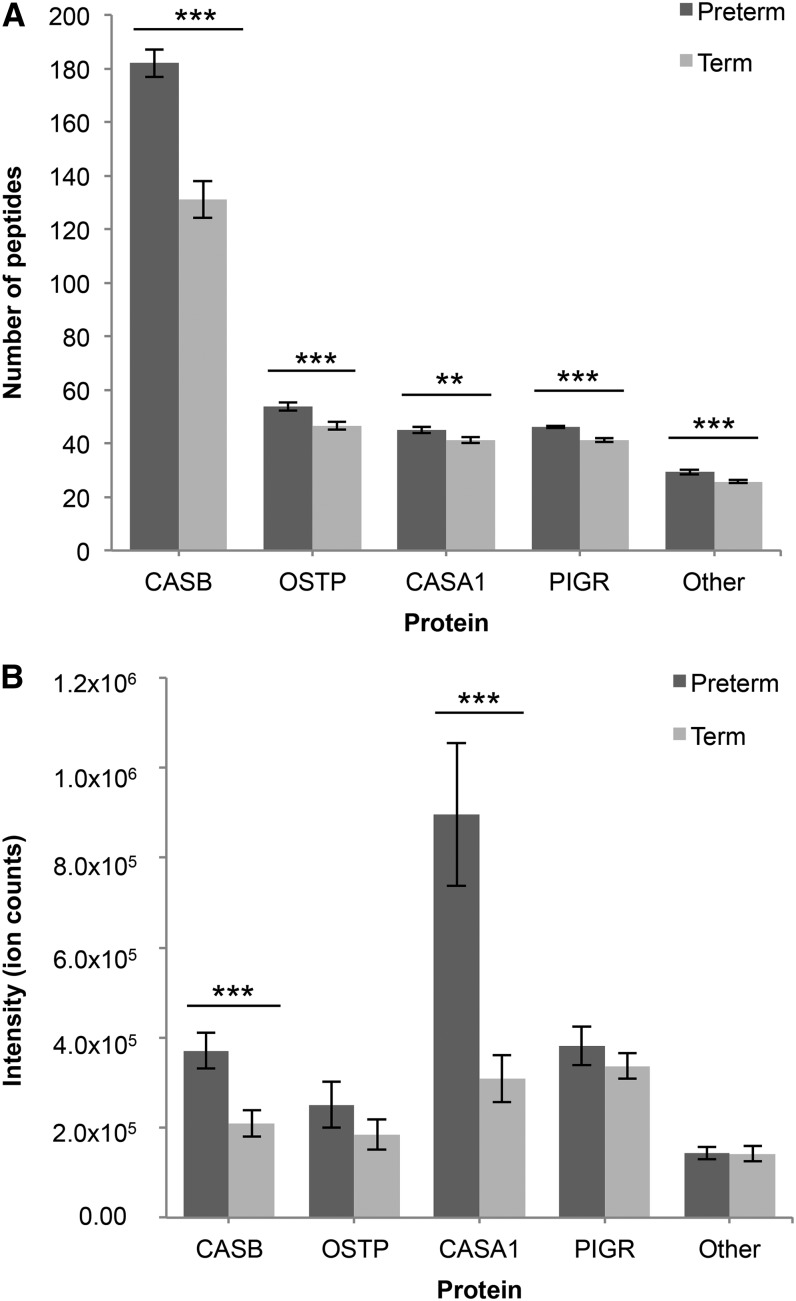
The majority of milk peptides in both term and preterm samples are derived from β-casein (54% by count). Other major protein precursors included osteopontin (15%), αs1-casein (12%), and polymeric immunoglobulin receptor (PIGR) (11%). In total, identified milk peptides were derived from 15 protein precursors. For further analyses, a group called “other” was created that includes 11 proteins for which <12 peptides were derived per protein, and includes κ-casein, butyrophilin subfamily 1 member A1, mucin-1, complement C4-B, perilipin-2, apolipoprotein B receptor, bile salt-activated lipase, fibrinogen α-chain, progonadoliberin-1, neurotensin/neuromedin N, and TNF receptor superfamily member 11B. As reported in previous articles on human milk peptidomics (1, 23), no peptides were identified from the highly abundant whey proteins lactoferrin and α-lactalbumin or the immunoglobulins. The number of peptides released from all 4 of the major protein precursors was significantly greater for preterm milk samples than term milk samples (β-casein, P = 1.05 × 10−7; osteopontin, P = 1.23 × 10−3; αs1-casein, P = 9.30 × 10−3; PIGR, P = 1.80 × 10−6) (Figure 2A).
FIGURE 2.
Number and intensity of peptides in preterm and term human milk by protein of origin. Number of peptides identified in preterm and term milk by protein of origin (A). Intensity of peptides in preterm and term milk by protein of origin (B). Values are means ± SEMs; n = 32 for term milk, n = 28 for preterm milk. Asterisks indicate significant differences between preterm and term milk: **P < 0.01, ***P < 0.001. CASA1, αs1-casein; CASB, β-casein; OSTP, osteopontin; Other, combination of all other protein precursors identified; PIGR, polymeric immunoglobulin receptor.
Of the 4 major peptide-releasing proteins identified, peptides from αs1-casein were most abundant despite β-casein releasing a greater number of peptides (Figure 2B). Peptides from β-casein (P = 1.48 × 10−3) and αs1-casein (P = 4.88 × 10−4) were significantly higher in abundance in preterm milk than in term milk (Figure 2B).
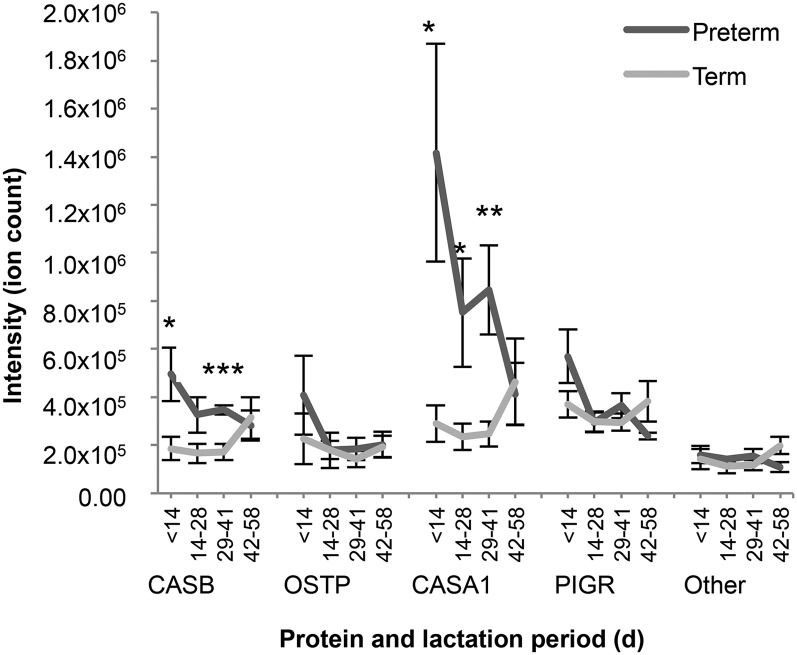
The intensity of PIGR changed significantly over time (P = 4.30 × 10−2), but β-casein, osteopontin, αs1-casein, and the “other” proteins did not change significantly over time in either the term or preterm sample groups (P = 0.63, P = 0.30, P = 0.28, P = 0.69, respectively) (Figure 3). Six peptides with the same peptide sequence (RPKLPLRYPERLQNPSESSEPIPLESREEYMNGMN) but different modifications (1O; 2P 1P; 2O; 1P 2O; 2O 2D; 2P 2O 1D) were responsible for the high intensity of αs1-casein–derived peptides at <14 d of lactation.
FIGURE 3.
Intensity of peptides in preterm and term human milk by protein of origin over lactation time. Values are means ± SEMs. Number of observations (n) in each group is shown in Table 1. Asterisks indicate significant differences between preterm and term milk: *P < 0.05, **P < 0.01, ***P < 0.001. CASA1, αs1-casein; CASB, β-casein; OSTP, osteopontin; Other, combination of all other protein precursors identified; PIGR, polymeric immunoglobulin receptor.
Enzyme degradation patterns.
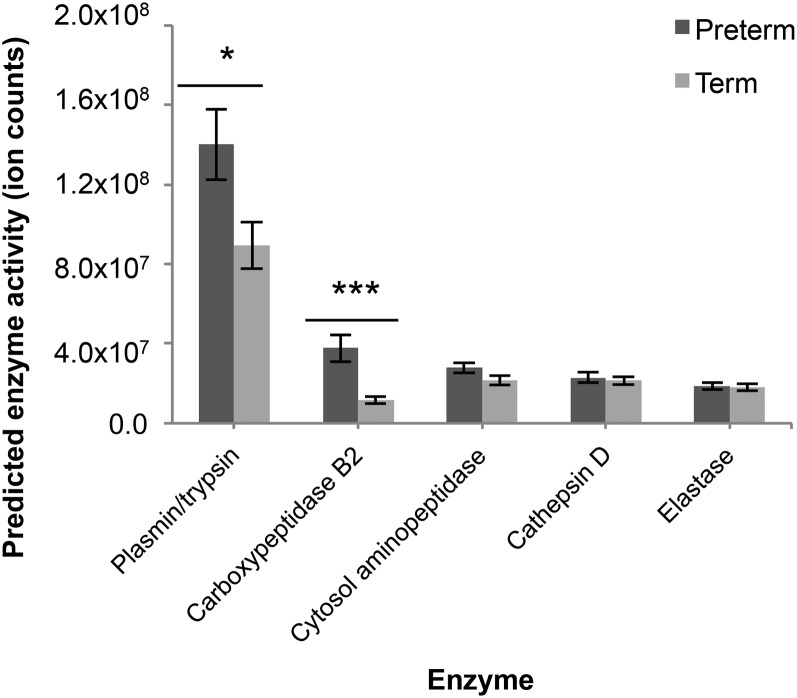
The enzyme degradation pattern in both preterm and term milk was associated with plasmin, cathepsin D, and elastase, as predicted by computational analysis. Through manual examination of the peptide data, many potential cytosol aminopeptidase and carboxypeptidase B2 cleavages were also identified (Figure 4). A recent study identified cytosol aminopeptidase and carboxypeptidase B2 in human milk (16).
FIGURE 4.
Predicted enzyme activity of plasmin, carboxypeptidase B2, cytosol aminopeptidase, cathepsin D, and elastase in preterm and term human milk. Values are means ± SEMs; n = 32 for term milk, n = 28 for preterm milk. Asterisks indicate significant differences between preterm and term milk: *P < 0.05, ***P < 0.001.
To assess carboxypeptidase B2 activity, we identified peptides with C-termini that may have been produced by a plasmin cleavage followed by the removal of the C-terminal lysine or arginine (24, 25). These peptides were considered potential carboxypeptidase B2 products if the results contained a second peptide that had the corresponding lysine or arginine residue still present. A total of 53 peptide cleavage sites were attributed to carboxypeptidase B2. Predicted carboxypeptidase B2 activity was significantly higher in preterm samples (Figure 4).
Cytosol aminopeptidase cleaves single amino acid residues from the N-terminus of peptides with broad specificity (26). Therefore, to determine its activity, we searched the results for groups of peptides that showed sequential removal of N-terminal residues (an example is illustrated in Table 1). Within these groups, a peptide was considered to be a product of cytosol aminopeptidase if its N-terminal sequence displayed the loss of a single amino acid residue that was present at the N-terminus of a different peptide in the group. A total of 18 peptide cascades (a group of peptides differing by sequential loss of single amino acids) attributable by prediction to cytosol aminopeptidase activity were identified, most of which came from β-casein–derived peptides (Supplemental Table 6).
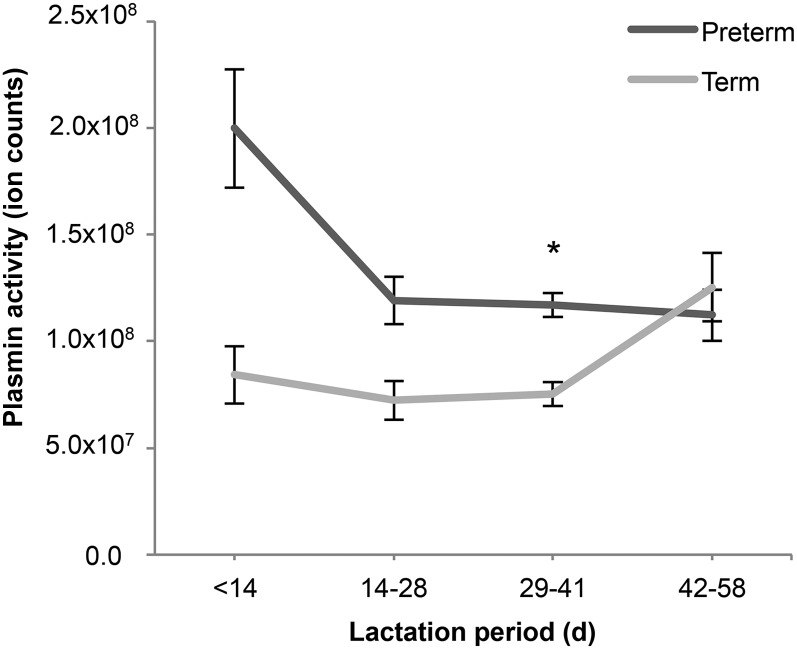
In both term and preterm milk, by computational prediction, the enzyme degradation pattern was most highly associated with plasmin, a protease active in human milk (13). The intensity of peptides ascribed to plasmin cleavage was significantly higher in the preterm milk than in term milk (Figure 4). Changes in predicted plasmin activity over time for the term and preterm groups were not significant (P = 0.51) (Figure 5).
FIGURE 5.
Predicted enzyme activity of plasmin in preterm and term human milk over lactation period. Values are means ± SEMs. Number of observations (n) in each group is shown in Table 1. Asterisk indicates significant difference between preterm and term milk: *P < 0.05.
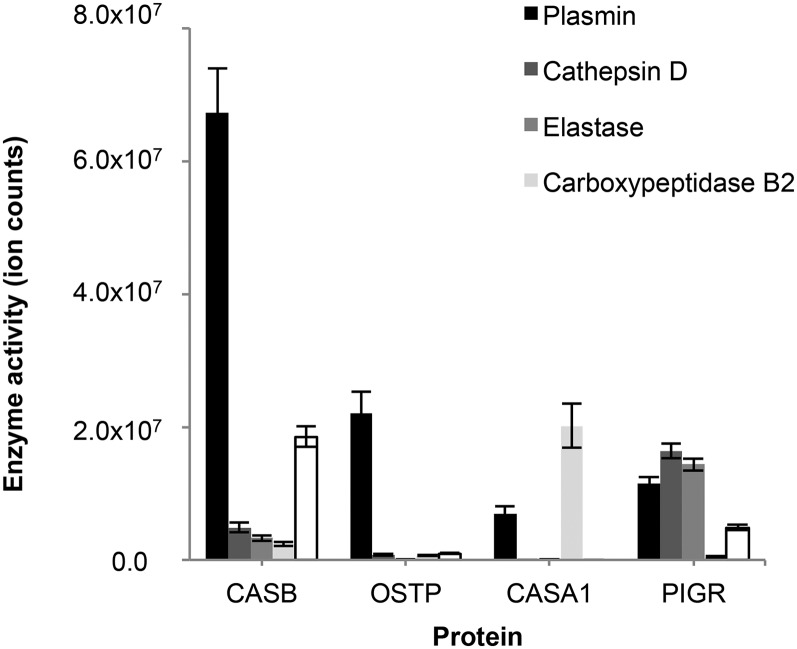
According to our model’s prediction, plasmin participated in the degradation of all 4 major proteins (Figure 6, Supplemental Figure 1). Plasmin was the most prominent enzyme predicted for β-casein and osteopontin degradation. Plasmin, cathepsin D, elastase, and cytosol aminopeptidase were all predicted to play major roles in degradation of PIGR. Predicted carboxypeptidase B2 activity was most evident on αs1-casein.
FIGURE 6.
Predicted activity of enzymes responsible for the endogenous human milk protein degradation based on the sum of peptide intensities. Values are means ± SEMs; n = 60. CASA1, αs1-casein; CASB, β-casein; OSTP, osteopontin; PIGR, polymeric immunoglobulin receptor.
Selectivity of protein digestion.
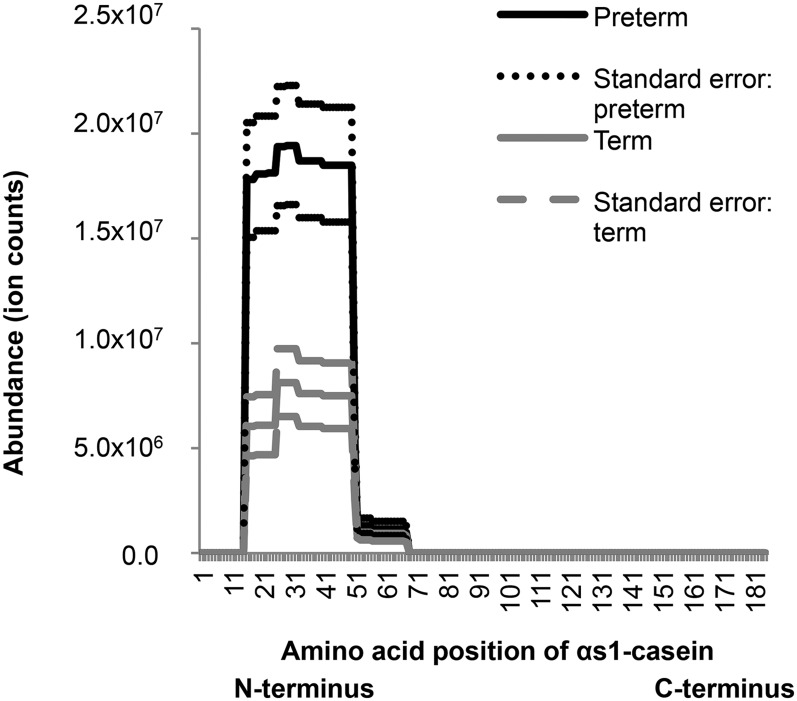
In both term and preterm milk, αs1-casein, β-casein, PIGR, and osteopontin were digested, but other highly abundant proteins including lactoferrin, α-lactalbumin, and immunoglobulins were not digested. The proteolytic maps of αs1-casein (Figure 7), β-casein, PIGR, and osteopontin (Supplemental Figures 2–4) in preterm milk matched the patterns in term milk. For example, αs1-casein was cleaved at the same specific site in the sequence in both term and preterm milk (Figure 7), although the intensity of αs1-casein–derived peptides was higher in preterm than term milk.
FIGURE 7.
Proteolytic map of αs1-casein in preterm and term human milk samples determined by mapping peptide intensities onto the αs1-casein protein sequence. Values are means ± SEMs; n = 32 for term milk, n = 28 for preterm milk.
Comparison of identified peptides to functional database.
Peptides in all samples were compared for sequence matches with an in-house database of known functional peptides. This search revealed that 25.4% of term and 18.9% of preterm milk peptides contained a functional sequence, or a sequence that showed 80% similarity to a functional peptide; 97.1% of these matching peptides were present in both term and preterm milk. The 4 peptides from the functional library that matched the experimental data possess antihypertensive (18), antimicrobial (19, 27), and immunomodulatory (21) functions. Although most of the peptides with high homology to functional peptides were present in both preterm and term milk samples, 45 of these peptides had significant differences in their mean abundances between the preterm and term groups (P < 0.05, Supplemental Table 7).
Discussion
This study showed that the number, ion intensity, and concentration of peptides released was higher in preterm milk than term milk (Figure 1) and that plasmin activity is likely greater in preterm milk than term milk (Figure 4), especially in early lactation. These findings align with a previous study (13) that showed low molecular weight casein fragments to be more abundant in preterm milk (gel electrophoresis), and that plasmin activity was greater in preterm milk (chromogenic assay).
Milk plasmin and plasminogen concentrations are known to vary with the stage of lactation, lactation number (number of delivered births), and somatic cell count (28). Plasmin activity increases as the number of lactations increases (29). During mastitis, the somatic cell count and the endogenous proteolytic activity in milk increase substantially (30, 31). This increased proteolytic activity in mastitis has been hypothesized to be due to an increase in plasminogen transmission from blood to milk (32) and increased protease secretion from somatic cells (33, 34). Tight junctions of the mammary gland are leaky during pregnancy and undergo closure around parturition (35). A previous review suggested that the mammary glands of mothers at <37 wk gestation have leaky tight junctions because of immaturity (36). We hypothesize that incomplete closure of tight junctions in the immature mammary gland of preterm mothers underlies increased leakage of proteases and protease activators into the milk, explaining the higher amounts of peptides and higher predicted plasmin activity in preterm milk compared to term milk that we observed.
Mammary epithelial cells secrete a variety of protease activators and inhibitors that influence proteolytic activity in milk, including α2-macroglobulin and α2-antiplasmin, both of which inhibit the conversion of plasminogen to plasmin, and inter α-trypsin inhibitor, which inhibits trypsin activation (34, 37). These mammary cells also secrete tissue-type and urokinase-type plasminogen activators (38, 39) and kallikrein (39), which cleave plasminogen to plasmin, the active enzyme form (5). The increased proteolytic activity in preterm milk could be due to increased expression of activators or decreased expression of inhibitors in the immature preterm mammary gland.
The majority of peptides, by count, were derived from β-casein and most were predicted to be released by plasmin; however, most of the peptides, by abundance, were derived from αs1-casein, and these peptides matched only to a small degree with plasmin, elastase, and cathepsin D cleavage patterns. Our manual enzyme specificity analysis predicts that αs1-casein degradation is mainly due to carboxypeptidase B2 activity (Figure 6). Forty-nine percent of the identified αs1-casein peptides have a C-terminus that could have been created by a plasmin cleavage followed by the removal of the C-terminal lysine or arginine by carboxypeptidase B2. This sequential cleavage would explain why plasmin activity on αs1-casein appeared to be low because our enzyme analysis program would not recognize a potential plasmin cleavage site if the C-terminal arginine or lysine was subsequently removed by carboxypeptidase B2. The abundance of those peptides that had been acted upon by carboxypeptidase B2 therefore did not contribute to the activity calculation of plasmin. To our knowledge, this study provides the first evidence that carboxypeptidase B2 may be active in human milk. In addition, we demonstrated, via our bioinformatic approach, that the predicted carboxypeptidase B2 activity is higher in preterm milk than term milk.
Potential cytosol aminopeptidase cleavages were mainly observed in peptides derived from β-casein. As shown in Table 1, peptide cascades were used to identify probable instances of aminopeptidase activity. A disadvantage of this approach is that it ignores the possibility that cytosol aminopeptidase may have acted upon a peptide that was not obviously part of a cascade. Because cytosol aminopeptidase can cleave almost any N-terminal residue (all except K or R), it is expected to be active beyond what was identified in this study. However, we chose to use peptide cascades for aminopeptidase activity estimation because the cascades provided the most reliable evidence of sequential removal of single amino acid residues.
Previous data showed that trypsin (7) and prothrombin (40) are also present in milk. Plasmin, trypsin, and thrombin have overlapping specificities and, therefore, could not be differentiated with this method (41). However, to our knowledge, trypsin and thrombin activity in human milk have never been demonstrated. One study (42) found that isolated trypsin from human milk is active against a trypsin substrate (benzoyl-L-arginine p-nitroanilide). The activity of isolated trypsin, however, does not imply activity within milk because milk contains αs1-antitrypsin. The researchers did find degradation of the substrate in a few milk samples; however, because benzoyl-L-arginine p-nitroanilide is also a substrate for plasmin (43), this finding does not provide good evidence for trypsin activity. Therefore, in this paper, cleavages after lysine or arginine were referred to as predicted plasmin cleavages. Some proportion of cleavages attributed here to plasmin may be due to trypsin or thrombin activity. Cathepsins B (40), H (44), and S (40, 44) have been identified in milk, but because there is no evidence for their activity, they were not included in the analysis.
Individual peptide relative quantification.
The peptide peak intensities were extracted based on retention time and mass. Although strict matching criteria were employed (1 min retention time window, 20 ppm mass error), some incorrect assignments are possible. For example, a peptide with a mass similar to the library peptide (<20 ppm difference) that co-elutes with a library peptide may be assigned as the library peptide.
Functional peptides.
Across lactation, a large number of functional peptides were present in both preterm and term milk with actions including antihypertensive, antimicrobial, and immunomodulatory. The release of these peptides is relevant because most milk peptides are inactive within the sequence of the intact parent protein, until they are released, and thus, activated by proteolytic enzymes (45). For example, during the digestion of highly phosphorylated β-caseins, phosphopeptides are formed. By count, 26% of all released peptides were phosphorylated, with more than half of these multiphosphorylated. These peptides keep calcium soluble, thereby facilitating its absorption (46). Phosphopeptides may contribute to the high bioavailability of calcium in human milk. By count, preterm and term peptides showed the same number of matches with known antimicrobial peptides, although the difference in mean abundance between preterm and term samples was statistically significant for 74.3% of the antibacterial matches.
Protein specificity.
As previously shown for term milk (1, 23), many peptides in preterm milk were derived from specific sections of the proteins osteopontin, PIGR, and αs1-casein (Figure 7, Supplemental Figures 3 and 4). Interestingly, the same pattern was present with both term and preterm milk, although abundances were higher in the preterm samples. This specificity in digestion of proteins may be due to properties inherent to these proteins’ structures, as suggested previously (1, 23). Like term milk samples examined previously, preterm milk did not contain protein fragments from the highly abundant whey proteins lactoferrin, α-lactalbumin, and secretory immunoglobulin A. As in previous studies, we hypothesize that the presence of casein-derived peptides in such high numbers is related to the looser, rheomorphic structure of the caseins—which allows less restricted access to cleavage sites by proteases—as opposed to the more rigid, globular structures of milk serum proteins (1). Likewise, we suspect that lactoferrin, α-lactalbumin, and immunoglobulin proteins are protected from degradation in the mammary gland by their structures, including glycosylation state. These whey proteins may also resist digestion because they do not associate with casein micelles, which are known to bind most of the milk plasmin (5). Because these whey proteins have important functions intact—antimicrobial and immunologic functions in the gut to protect the infant from infections (47, 48)—their lack of degradation may be functionally beneficial.
Relation to infant digestion.
The preterm infant, compared with the term infant, is thought to be poorly equipped for the task of protein digestion (45, 49). For example, a previous study showed that gastrointestinal trypsin activity is lower in preterm than term infants in early life (49). Another study showed that gut trypsin activity actually decreases with milk feeding in premature infants (50). The high protein degradation by endogenous proteases in preterm milk might attenuate this lowered overall proteolysis. At first glance, given that premature infants <32 wk gestation rarely survived before modern medicine and supplemental feeding (51, 52), an evolutionary relation between these factors seems unlikely. However, studies in marsupials demonstrating that significant differences between milk from the “premature nipple” compared to the “mature nipple” result in marked differences in growth of the joey (53) support the hypothesis that preterm milk may be better suited to the premature infant. The alternative hypothesis, that term mother’s milk is the gold standard for both term and preterm infants and that preterm milk is the result of a poorly controlled enzymatic disassembly of proteins in the preterm mother’s mammary gland, would suggest the need to develop new methods for processing term donor human milk to preserve milk proteins and peptides for premature infants. Future experiments should focus on a comparison of human milk digestion peptidomes between term and preterm infants to evaluate how closely the bioavailable peptides in the stomach of preterm infants match to the standard of the term infant. Such data could provide insights into methods for improving preterm infant nourishment.
Supplementary Material
Acknowledgments
The authors thank Cora J Dillard for editing the manuscript and Dr. David Rocke for assistance with design of the statistical analysis. DCD, MAU, JBG, and DB designed the research; JTS and MAU designed the clinical sample collection; DCD, AG, and CBL established the peptidomics methodology; DCD and RCR performed the MS analysis; DCD, CJS, RCR, TT, EAP, and KAH analyzed the data; DCD, CJS, and RCR wrote the paper; and DCD had primary responsibility for the final content. All authors read and approved the final manuscript.
References
- 1.Dallas DC, Guerrero A, Khaldi N, Castillo PA, Martin WF, Smilowitz JT, Bevins CL, Barile D, German JB, Lebrilla CB. Extensive in vivo human milk peptidomics reveals specific proteolysis yielding protective antimicrobial peptides. J Proteome Res 2013;12:2295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferranti P, Traisci MV, Picariello G, Nasi A, Boschi V, Siervo M, Falconi C, Chianese L, Addeo F. Casein proteolysis in human milk: tracing the pattern of casein breakdown and the formation of potential bioactive peptides. J Dairy Res 2004;71:74–87. [DOI] [PubMed] [Google Scholar]
- 3.Guerrero A, Dallas DC, Contreras S, Chee S, Parker EA, Sun X, Dimapasoc LM, Barile D, German JB, Lebrilla CB. Mechanistic peptidomics: factors that dictate the specificity on the formation of endogenous peptides in human milk. Mol Cell Proteomics 2014;13:3343–51. [DOI] [PMC free article] [PubMed]
- 4.Dallas DC, Guerrero A, Khaldi N, Borghese R, Bhandari A, Underwood MA, Lebrilla CB, German JB, Barile D. A peptidomic analysis of human milk digestion in the infant stomach reveals protein-specific degradation patterns. J Nutr 2014;144:815–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heegaard CW, Larsen LB, Rasmussen LK, Højberg KE, Petersen TE, Andreasen PA. Plasminogen activation system in human milk. J Pediatr Gastroenterol Nutr 1997;25:159–66. [DOI] [PubMed] [Google Scholar]
- 6.Vĕtvicka V, Vagner J, Baudys M, Tang J, Foundling S, Fusek M. Human breast milk contains procathepsin D–detection by specific antibodies. Biochem Mol Biol Int 1993;30:921–8. [PubMed] [Google Scholar]
- 7.Borulf S, Lindberg T, Mansson M. Immunoreactive anionic trypsin and anionic elastase in human milk. Acta Paediatr Scand 1987;76:11–5. [DOI] [PubMed] [Google Scholar]
- 8.Khaldi N, Vijayakumar V, Dallas DC, Guerrero A, Wickramasinghe S, Smilowitz JT, Medrano JF, Lebrilla CB, Shields DC, German JB. Predicting the important enzyme players in human breast milk digestion. J Agric Food Chem 2014;62:7225–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zucht HD, Raida M, Adermann K, Mägert HJ, Forssmann WG. Casocidin-I: a casein-alpha(s2) derived peptide exhibits antibacterial activity. FEBS Lett 1995;372:185–8. [DOI] [PubMed] [Google Scholar]
- 10.Anderson GH, Atkinson SA, Bryan MH. Energy and macronutrient content of human milk during early lactation from mothers giving birth prematurely and at term. Am J Clin Nutr 1981;34:258–65. [DOI] [PubMed] [Google Scholar]
- 11.Bitman J, Wood L, Hamosh M, Hamosh P, Mehta NR. Comparison of the lipid composition of breast milk from mothers of term and preterm infants. Am J Clin Nutr 1983;38:300–12. [DOI] [PubMed] [Google Scholar]
- 12.Lemons JA, Moye L, Hall D, Simmons M. Differences in the composition of preterm and term human milk during early lactation. Pediatr Res 1982;16:113–7. [DOI] [PubMed] [Google Scholar]
- 13.Armaforte E, Curran E, Huppertz T, Ryan CA, Caboni MF, O'Connor PM, Ross RP, Hirtz C, Sommerer N, Chevalier F. Proteins and proteolysis in pre-term and term human milk and possible implications for infant formulae. Int Dairy J 2010;20:715–23. [Google Scholar]
- 14.Dallas DC, Guerrero A, Parker EA, Garay LA, Bhandari A, Lebrilla CB, Barile D, German JB. Peptidomic profile of milk of Holstein cows at peak lactation. J Agric Food Chem 2014;62:58–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith PK, Krohn RI, Hermanson G, Mallia A, Gartner F, Provenzano M, Fujimoto E, Goeke N, Olson B, Klenk D. Measurement of protein using bicinchoninic acid. Anal Biochem 1985;150:76–85. [DOI] [PubMed] [Google Scholar]
- 16.Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics 2004;20:1466–7. [DOI] [PubMed] [Google Scholar]
- 17.Vizcaíno JA, Côté RG, Csordas A, Dianes JA, Fabregat A, Foster JM, Griss J, Alpi E, Birim M, Contell J. The Proteomics Identifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res 2013;41:D1063–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maruyama S, Nakagomi K, Tomizuka N, Suzuki H. Angiotensin I-converting enzyme inhibitor derived from an enzymatic hydrolysate of casein. II. Isolation and bradykinin-potentiating activity on the uterus and the ileum of rats. Agric Biol Chem 1985;49:1405–9. [Google Scholar]
- 19.Recio I, Visser S. Identification of two distinct antibacterial domains within the sequence of bovine a-s2-casein. Biochim Biophys Acta 1999;1428:314–26. [DOI] [PubMed] [Google Scholar]
- 20.Minervini F, Algaron F, Rizzello C, Fox P, Monnet V, Gobbetti M. Angiotensin I-converting-enzyme-inhibitory and antibacterial peptides from Lactobacillus helveticus PR4 proteinase-hydrolyzed caseins of milk from six species. Appl Environ Microbiol 2003;69:5297–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayes M, Stanton C, Fitzgerald GF, Ross RP. Putting microbes to work: dairy fermentation, cell factories and bioactive peptides. Part II: bioactive peptide functions. Biotechnol J 2007;2:435–49. [DOI] [PubMed] [Google Scholar]
- 22.Parker EA. Peptidomics enzyme estimator. 2014 [cited 2014 Nov 1]. Available from: https://github.com/eparker05/Peptidomics-enzyme-estimator.
- 23.Guerrero A, Dallas DC, Contreras S, Chee S, Parker EA, Sun X, Dimapasoc LM, Barile D, German JB, Lebrilla CB. Mechanistic peptidomics: factors that dictate specificity in the formation of endogenous peptides in human milk. Mol Cell Proteomics 2014;13:3343–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eaton DL, Malloy B, Tsai SP, Henzel W, Drayna D. Isolation, molecular cloning, and partial characterization of a novel carboxypeptidase B from human plasma. J Biol Chem 1991;266:21833–8. [PubMed] [Google Scholar]
- 25.Hendriks D, Wang W, Scharpé S, Lommaert M-P, van Sande M. Purification and characterization of a new arginine carboxypeptidase in human serum. Biochim Biophys Acta 1990;1034:86–92. [DOI] [PubMed] [Google Scholar]
- 26.P28838–Cytosol aminopeptidase. 2014 June 11 [cited 2014 Jul 8]. Available from: http://www.uniprot.org/uniprot/P28838.
- 27.Azuma N, Nagaune S, Ishino Y, Mori H, Kaminogawa S, Yamauchi K. DNA-synthesis stimulating peptides from human β-casein. Agric Biol Chem 1989;53:2631–4. [Google Scholar]
- 28.Politis I, Ng Kwai Hang K, Giroux R. Environmental factors affecting plasmin activity in milk. J Dairy Sci 1989;72:1713–8. [DOI] [PubMed] [Google Scholar]
- 29.Ismail B, Nielsen S. Invited review: plasmin protease in milk: current knowledge and relevance to dairy industry. J Dairy Sci 2010;93:4999–5009. [DOI] [PubMed] [Google Scholar]
- 30.Mansor R, Mullen W, Albalat A, Zerefos P, Mischak H, Barrett DC, Biggs A, Eckersall PD. A peptidomic approach to biomarker discovery for bovine mastitis. J Proteomics 2013;85:89–98. [DOI] [PubMed] [Google Scholar]
- 31.Guerrero A, Dallas DC, Contreras S, Bhandari A, Cánovas A, Islas-Trejo A, Medrano JF, Parker EA, Wang M, Hettinga K, et al. Peptidomic analysis of healthy and subclinically mastitic bovine milk. Int Dairy J 2014 Oct 28 (Epub ahead of print; DOI: 10.1016/j.idairyj.2014.09.006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaartinen L, Sandholm M. Regulation of plasmin activation in mastitic milk: correlation with inflammatory markers and growth of streptococcus agalactiae. Zentralbl Veterinarmed B 1987;34:42–50. [DOI] [PubMed] [Google Scholar]
- 33.Bastian ED, Brown RJ. Plasmin in milk and dairy products: an update. Int Dairy J 1996;6:435–57. [Google Scholar]
- 34.Kelly A, O’Flaherty F, Fox P. Indigenous proteolytic enzymes in milk: a brief overview of the present state of knowledge. Int Dairy J 2006;16:563–72. [Google Scholar]
- 35.Nguyen D-AD, Neville MC. Tight junction regulation in the mammary gland. J Mammary Gland Biol Neoplasia 1998;3:233–46. [DOI] [PubMed] [Google Scholar]
- 36.Anderson G. The effect of prematurity on milk composition and its physiological basis . Fed Proc 1984;43:2438–42. [PubMed] [Google Scholar]
- 37.Lindberg T, Ohlsson K, Weström B. Protease inhibitors and their relation to protease activity in human-milk. Pediatr Res 1982;16:479–83. [DOI] [PubMed] [Google Scholar]
- 38.Korycha-Dahl M, Dumas BR, Chene N, Martal J. Plasmin activity in milk. J Dairy Sci 1983;66:704–11. [Google Scholar]
- 39.Wickramasinghe S, Rincon G, Islas-Trejo A, Medrano JF. Transcriptional profiling of bovine milk using RNA sequencing. BMC Genomics 2012;13:45–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Molinari CE, Casadio YS, Hartmann BT, Livk A, Bringans S, Arthur PG, Hartmann PE. Proteome mapping of human skim milk proteins in term and preterm milk. J Proteome Res 2012;11:1696–714. [DOI] [PubMed] [Google Scholar]
- 41.Christensen B, Schack L, Kläning E, Sørensen ES. Osteopontin is cleaved at multiple sites close to its integrin-binding motifs in milk and is a novel substrate for plasmin and cathepsin D. J Biol Chem 2010;285:7929–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Monti JC, Mermoud A-F, Jolles P. Trypsin in human milk. Experientia 1986;42:39–41. [DOI] [PubMed] [Google Scholar]
- 43.Christensen U, Müllertz S. Mechanism of reaction of human plasmin with α-N-benzoyl-l-arginine-p-nitroanilide: titration of the enzyme. Biochim Biophys Acta 1974;334:187–98. [Google Scholar]
- 44.Palmer DJ, Kelly VC, Smit AM, Kuy S, Knight CG, Cooper GJ. Human colostrum: identification of minor proteins in the aqueous phase by proteomics. Proteomics 2006;6:2208–16. [DOI] [PubMed] [Google Scholar]
- 45.Dallas DC, Underwood MA, Zivkovic AM, German JB. Digestion of protein in premature and term infants. J Nutr Disord Ther 2012;2:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sato R, Noguchi T, Naito H. Casein phosphopeptide (CPP) enhances calcium absorption from the ligated segment of rat small intestine. J Nutr Sci Vitaminol (Tokyo) 1986;32:67–76. [DOI] [PubMed] [Google Scholar]
- 47.Newburg DS, Walker WA. Protection of the neonate by the innate immune system of developing gut and of human milk. Pediatr Res 2007;61:2–8. [DOI] [PubMed] [Google Scholar]
- 48.Lönnerdal B. Bioactive proteins in human milk: mechanisms of action. J Pediatr 2010;156:S26–30. [DOI] [PubMed] [Google Scholar]
- 49.Borgström B, Lindquist B, Lundh G. Enzyme concentration and absorption of protein and glucose in duodenum of premature infants. AMA J Dis Child 1960;99:338–43. [PubMed] [Google Scholar]
- 50.Engberg S, Mansson M, Andersson Y, Jakobsson I, Lindberg T. Trypsin and elastase activity in duodenal juice from preterm infants before and after a meal of human milk. Prenat Neonatal Med 1999;4:466–71. [Google Scholar]
- 51.Arslanoglu S, Moro GE, Ziegler EE. Optimization of human milk fortification for preterm infants: new concepts and recommendations. J Perinat Med 2010;38:233–8. [DOI] [PubMed] [Google Scholar]
- 52.Beck S, Wojdyla D, Say L, Betran AP, Merialdi M, Requejo JH, Rubens C, Menon R, Van Look PF. The worldwide incidence of preterm birth: a systematic review of maternal mortality and morbidity. Bull World Health Organ 2010;88:31–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kwek JH, Iongh RD, Digby MR, Renfree MB, Nicholas KR, Familari M. Cross-fostering of the tammar wallaby Macropus eugenii pouch young accelerates fore-stomach maturation. Mech Dev 2009;126:449–63. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.