Abstract
Paracellular barrier properties of tissues are mainly determined by the composition of claudin heteropolymers. To analyze the molecular organization of tight junctions (TJ), we investigated the ability of claudins (Cld) to form homo- and heteromers. Cld1, -2, -3, -5, and -12 expressed in cerebral barriers were investigated. TJ-strands were reconstituted by claudin-transfection of HEK293-cells. cis-Interactions and/or spatial proximity were analyzed by fluorescence resonance energy transfer inside and outside of strands and ranked: Cld5/Cld5 > Cld5/Cld1 > Cld3/Cld1 > Cld3/Cld3 > Cld3/Cld5, no Cld3/Cld2. Classic Cld1, -3, and -5 but not non-classic Cld12 showed homophilic trans-interaction. Freeze-fracture electron microscopy revealed that, in contrast to classic claudins, YFP-tagged Cld12 does not form homopolymers. Heterophilic trans-interactions were analyzed in cocultures of differently monotransfected cells. trans-Interaction of Cld3/Cld5 was less pronounced than that of Cld3/Cld1, Cld5/Cld1, Cld5/Cld5 or Cld3/Cld3. The barrier function of reconstituted TJ-strands was demonstrated by a novel imaging assay. A model of the molecular organization of TJ was generated.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-011-0680-z) contains supplementary material, which is available to authorized users.
Keywords: Claudins, FRET, Protein–protein interaction, Tight junction
Introduction
Tight junctions (TJ) form a paracellular barrier in epithelia and endothelia. They regulate and limit the paracellular permeation of ions, solutes, and proteins [1]. TJ appear as fusions of the membranes of two neighboring cells (transmission electron microscopy) and as an anastomosing network of strands composed of transmembrane proteins (freeze-fracture electron microscopy) [2]. The tetraspan membrane proteins of the claudin (Cld) family constitute the backbone of TJ by trans- (head to head) and cis- (side by side) interactions [3, 4]. The various claudin subtypes exhibit tissue-specific expression patterns and differ in their barrier properties [5]. For instance, Cld5 tightens the blood–brain barrier against small molecules up to 800 Da [6]; Cld2 forms paracellular pores for monovalent cations; and a complex between Cld16 and Cld19 creates pores for divalent cations [7, 8]. Residues in the first extracellular loop (ECL1) of claudins determine the ability to form paracellular ion pores as well as their charge selectivity [5, 9]. In addition, Gln44 in the ECL1 of human Cld3 prevents heterophilic trans-interactions with Cld4 [10]. Involvement of the ECL2 in trans-interaction between Cld5 molecules of adjacent cells during formation of TJ strands and paracellular barrier development has been demonstrated in vitro [3, 11] and in vivo [12].
Claudins can functionally be divided into barrier-forming, e.g., Cld1, -3, -4, -5, and pore-forming claudins, e.g., Cld2, -7, -10, -16, [5]. Based on sequence homology studies, claudins can be grouped as classic claudins (Cld1–10, -14, -15, -17, -19) with high homology within this group and non-classic claudins (Cld11, -12, -13, -16, -18, -20 to -24) with lower homology [5]. In line with the sequence homology, the classic claudins are likely to share a common helix-turn-helix structure of the ECL2 that is involved in paracellular tightening [3, 11].
Most epithelial and endothelial cells express an assortment of different claudins and it is assumed that they form heteropolymers [13]. However, the pattern of heterophilic compatibility of claudin subtypes is incomplete and the underlying molecular mechanisms are not known. The morphology of TJ-strands in a given tissue depends on the claudin composition [14]. Most epithelia contain continuous strands with intramembranous particles associated with the protoplasmic face (P-face) of the plasma membrane. The very leaky and often fenestrated endothelia of peripheral blood vessels contain discontinuous strands, associated with the exoplasmic face (E-face). In contrast, the very tight brain capillary endothelial cells contain TJ-strands with particles on the P- and E-face [15]. Transfection experiments show that Cld1 and Cld3 form P-face-associated continuous strands where as Cld2, -5, -6, -11 form discontinuous strands partly or fully associated with the E-face. Coexpression of Cld2 with Cld1 or Cld3 increases P-face association and strand continuity [13]. Coexpression of Cld5 with Cld3 correlates with high P-face association and tightness of brain endothelial cells [16, 17]. In summary, freeze fracture and functional data suggest that the combination and stoichiometry of claudin subtypes in heteropolymeric strands defines their P/E-face association and continuity as critical determinants of TJ barrier function.
In this study, homo- and heterophilic interactions between claudins assumed to be expressed in cerebral barriers (Cld1, -2, -3, -5, and -12; [6, 14]) were systematically investigated. The data improve the understanding of the oligomerization of TJ proteins.
Materials and methods
Cell culture, transfection
HEK293 cells (HEK cells) were maintained in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen) supplemented with 10% fetal calf serum, 100 units/ml penicillin, 100 μg/ml streptomycin and 1% l-alanyl-l-glutamine (Invitrogen). Transient transfections of HEK cells were preformed with Lipofectamine 2000 according to the supplier’s recommendations (Invitrogen). Stable lines were selected by 0.5 or 1 mg/ml G418 (Calbiochem). To avoid the development of clonal variations, pools containing different Cld5-YFP-expressing and non-expressing colonies were used. In addition, cells were sorted with FACSVantage SE System (BD Biosciences) to achieve cultures in which >95% of the cells express the CFP/YFP-construct.
Mammalian expression vectors and site-directed mutagenesis
Expression vectors for claudins were based on pECFP-N1/pEYFP-N1 (Clonetech). Plasmids encoding mouse Cld5wt-CFP, Cld5wt-YFP, mutants thereof and corticotropin releasing factor receptor1(CRFR1)-YFP [3], plasmids encoding mouse Cld3Lwt-CFP, Cld3Lwt-YFP, Cld5wt, Cld3wt, Cld3Y147A [18] and CFP-YFP tandem construct [19] have been described previously. Plasmids encoding Cld3-CFP or Cld3-YFP were generated by amplifying full-length Cld3 and introduction of SalI and BamHI restrictions sites before and after the coding sequence by PCR from Cld3Lwt-YFP as template. The products were subcloned in pEYFP-N1 and pECFP-N1, respectively. A plasmid encoding rat Cld12-YFP was generated by amplifying full-length Cld12 without the stop codon and introduction of HindIII and BamHI restrictions sites by PCR from template plasmid (kindly provided by Dr. T. Terasaki, Sendai, Japan). The product was subcloned in pEYFP-N1. Plasmids encoding human Cld1-YFP or Cld2-YFP were generated by amplifying full-length claudins without the stop codon by PCR from pcDNA3.11_Cld1 and pcDNA3.11_Cld2 as templates (kindly provided by Dr. M. Fromm, Berlin, Germany) and subcloned in pEYFP-N1.
Immunocytochemistry and live-cell imaging
Immunocytochemistry was performed as described [3]. For live-cell imaging, transfected cells were transferred to 1 ml DMEM, 10 mM (N-(2-hydroxyethyl)piperazine-N′(2-ethanesulfonic acid) pH 7.5 without phenol red. The plasma membrane was visualized by addition of 20 μl trypan blue, 0.05% in phosphate buffered saline (PBS). Cells were examined with an LSM 510 META system, using an Axiovert 135 microscope equipped with a PlanNeofluar 100×/1.3 objective (Zeiss) [3]. To quantify claudin enrichment at contacts between two claudin-expressing cells, intensity profiles of confocal images of living cells were analyzed using the LSM 510 software (Zeiss). Contacts between two cells were identified by the trypan blue fluorescence peaks. For each cell pair, ≥5 fluorescence intensity profiles were quantified. For cultures of Cld-CFP- or Cld-YFP-expressing cells the enrichment factor (E F) was calculated as: (intensity at contact between two claudin-expressing cells)/(intensity at contact between an expressing and a non-expressing cell × 2) [3]. For cocultures of Cld-CFP- and Cld-YFP-expressing cells the enrichment factor (E F) was calculated as: (intensity at contact between a Cld-CFP- and a Cld-YFP-expressing cells)/(intensity at contact between an expressing and a non-expressing cell). E F > 1.5 (E F of control + 2 SD) was considered as enrichment. In addition, the frequency of enrichments P E (pairs of contacts with E F > 1.5) was determined. Similarly, contact enrichment in fixed and stained cultures was determined using confocal images, intensity profiles, and Axiovision software (Zeiss). Rabbit anti-Cld3 and Cy3-conjugated goat anti-mouse antibodies (Invitrogen) were used for immunocytochemistry.
For determination of the barrier function of claudins, cells were replated on poly-l-lysine-coated glass coverslips, after 2–3 days washed with Hank’s buffered salt solution (HBSS) and incubated with 0.25 μg/ml CellMask™ Deep Red plasma membrane stain (Invitrogen) in HBSS for 5 min. CFP, YFP and CellMask™ f (λ ex = 633 nm, λ em = 650–710 nm) was imaged within 5 min. As a measure for the diffusion barrier preventing CellMask™ labeling, the barrier ratio (Br), was determined using fluorescence intensity profiles as: (CellMask™ intensity at contact between a construct-expressing and a non-expressing cell)/(intensity at contact between two construct-expressing cells). At least 15 pairs of contacts were analyzed for each sample.
FRET (fluorescence resonance energy transfer) analysis by confocal microscopy
Cells were cotransfected with plasmids encoding CFP- and YFP-fusion proteins of claudins or CRFR1. CRFR1-YFP was used as negative control, as this receptor was found to colocalize with Cld5 at cell–cell contacts but does not interact with Cld5 [19]. FRET acceptor photobleaching was performed 3 days after transfection as described [3]. Briefly, CFP and YFP were excited at 458 and 514 nm and detected from 463–495 and 527–634 nm, respectively. Photobleaching of YFP at the area of cell–cell contacts was performed by using 15–25 pulses of the 514-nm argon laser line at 100% intensity. For quantitation, the laser and detector settings were kept constant, and only cells with CFP and YFP intensities below saturation were used. FRET-efficiency (F E) was calculated as F E = (IA − IB) × 100/IA, where IB and IA refer to the CFP intensity before and after photobleaching. In each experiment, the pair Cld5wt-CFP/Cld5wt-YFP was the internal standard. The relative FE was calculated as the FE of a distinct pair of CFP/YFP- fusion proteins divided by F E of Cld5wt-CFP/Cld5wt-YFP. In addition, the FRET efficiency was normalized to a YFP/CFP intensity ratio of 2 (F E − YFP/CFP = 2), because the FRET efficiency was strongly dependent on the YFP/CFP intensity ratio (Fig. S1). For this, logarithmic regression was used to obtain an equation with which F E − YFP/CFP was calculated for YFP/CFP intensity ratio = 2.
FRET analysis by flow cytometry
Cells were cotransfected with plasmids encoding CFP- and YFP-fusion proteins. Two days later, single cell suspensions were prepared by gentle trypsinization, washing, resuspension in PBS containing 5 mM EDTA at 4°C and filtration through a cell strainer (35 μm) before analysis. FRET measurements were performed using a FACSAria II (BD Bioscience) equipped with 405 and 488-nm laser lines. To measure CFP and FRET, cells were excited at 405 nm and fluorescence was detected in the CFP channel with a 450/50 filter, while the FRET-signal was measured with a 525/50 filter. To detect YFP, cells were excited at 488 nm while emission was taken with a 530/30 filter. All dead cells and debris were excluded from the analysis based on forward and sideward scatter gating. To control for autofluorescence and automatic compensation non-transfected and single-transfected cells were included. 50,000 events/sample were analyzed using FlowJo (Treestar) to identify the double-positive transfected cell population and OriginPro (OriginLab) was used to normalize the FRET and YFP values relative to the CFP intensity. A first-order exponential decay regression was performed using the equation {FRET/CFP} = A1 × exp(−{YFP/CFP}/t1) + y0 (A1, t1, yo regression parameter determined by OriginPro). To take into account the dependence of the FRET-efficiency on the acceptor/donor ratio (YFP/CFP), FRET/CFP was calculated for the standardized YFP/CFP intensity ratio of 5. As a measure of the FRET-efficiency the FRET-ratioFC was calculated as (FRET/CFP)sample − (FRET/CFP)Cld5/CRFR1. Cld5/CRFR1 served as internal negative control for unspecific FRET in each experiment.
FRET analysis by spectrofluorometry
Cell suspensions (as described above) were analyzed in a FP-6500 spectrofluorometer (JASCO). CFP-fluorescence was detected with λ excitation = 425 ± 5 nm and λ emission = 475 ± 5 nm, YFP-fluorescence with λ excitation = 490 ± 5 nm and λ emission = 525 ± 5 nm and FRET-Signal with λ excitation = 425 ± 5 nm and λ emission = 525 ± 5 nm. Crosstalk of YFP-fluorescence to the CFP- and FRET-signal was determined experimentally with sample containing YFP or CFP, only (for λ excitation = 425 ± 5 nm and λ emission = 475 ± 5 nm the signal was 2% of the YFP-fluorescence (ε 1 = 0.02) and for λ emission = 525 ± 5 nm the signal was 5% of the YFP-fluorescence (ε 2 = 0.05). As a measure of the FRET-efficiency the FRET-ratioS was calculated as FRET-ratioS = (FRET-signal − ε 2 × YFP-fluorescence)/(CFP-fluorescence − ε 1× YFP-fluorescence).
Fluorescence recovery after photobleaching
Fluorescence recovery after photobleaching (FRAP) was performed on a DM4000 microscope (Leica) equipped with a MicroPoint Laser System (Photonic Instruments), Leica 63X UVI objective, and 37°C heated stage controlled by MetaMorph 7 (MDS). A region of interest was bleached with a nanosecond laser pulse using low intensities and <1 s exposures to minimize phototoxicity. MetaMorph 7 was used for analysis, as described [20, 21].
Freeze-fracture electron microscopy
HEK cells were transfected with claudin-constructs, 3 days later, washed with PBS, fixed with 2.5% glutaraldehyde (electron microscopy grade, Sigma-Aldrich) in PBS for 2 h, washed and processed for freeze-fracture electron microscopy as reported [22].
Statistics
Unless stated otherwise, results are shown as mean ± SEM. Statistical analyses were performed by one-way analysis of variance (ANOVA) and followed by an unpaired Student’s t test. p < 0.02 was taken as significantly different.
Results
The classic claudin1, -3 and -5, but not the non-classic claudin12, are capable of homophilic trans-interactions
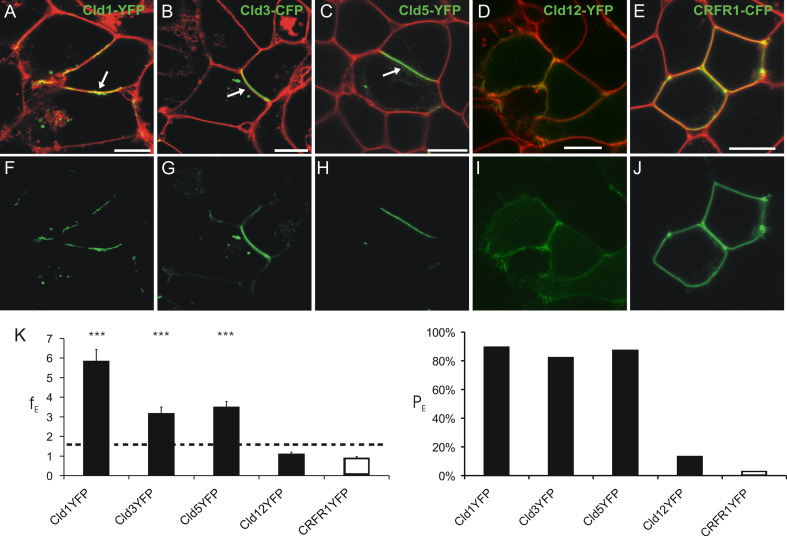
To compare the ability of the blood–brain barrier relevant Cld1, -3, -5, and -12, to form trans-interactions, the enrichment of claudins at contacts between HEK cells transfected with plasmids encoding Cld-YFP was analyzed using a previously established assay [3]. Marked enrichment of Cld1, -3, and -5, but not Cld12 or the negative CRFR1, was shown (Fig. 1). This indicates that the classic Cld1, -3, and -5 [5] but not the non-classic Cld12, have similar abilities for homophilic trans-interaction.
Fig. 1.
Homophilic trans-interaction of claudin (Cld)-1, -3, and -5 but not -12. Confocal microscopy of living HEK cells stably transfected with Cld1-YFP (a, f), Cld3-CFP (b, g), Cld5-YFP (c, h), Cld12-YFP (d, i), and transiently transfected with corticotropin releasing factor receptor 1 (CRFR1)-CFP (g, j). Cld1, -3, -5 but not -12 and CRFR1 (green) are enriched (arrows) at contacts between two claudin-expressing cells indicating trans-interaction. Plasma membrane was visualized with trypan blue (a–e, red). a–e, merge of CFP/YFP and trypan blue; f, h, i, YFP; g, j; CFP. Bar, 10 μm. k Quantification of contact enrichment. For Cld1, Cld3, and Cld5, but not Cld12, the factor of contact enrichment (E F) and the percentage of contacts with E F > 1.5 (PE) are much higher than for the negative control (CRFR1). ***p < 0.001 compared to CRFR1-YFP; E F, mean + SEM; n ≥ 40; dotted line, threshold above trans-interaction is considered
Determinants of trans-interactions in extracellular loop 2 are conserved between claudin5 and claudin3
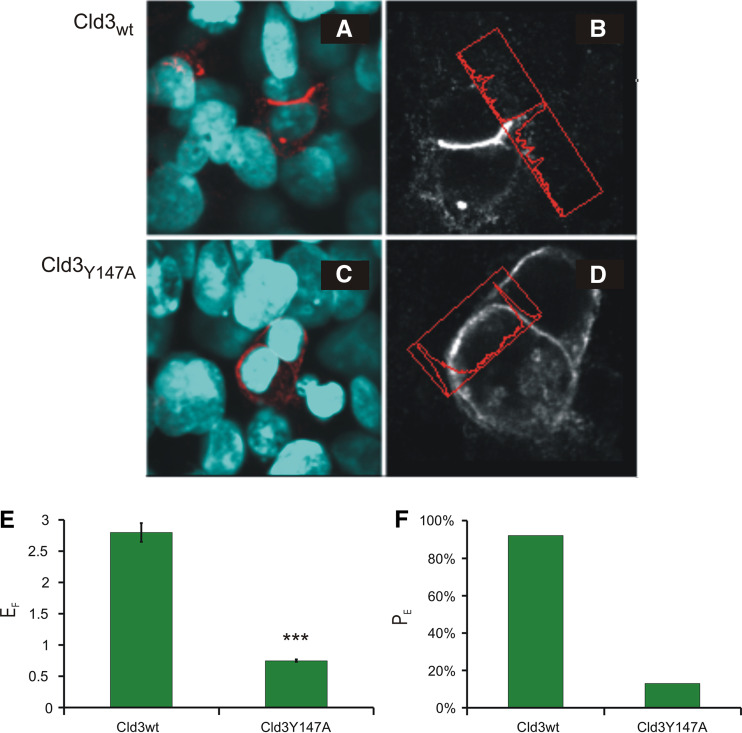
Previously, we have shown that defined residues (e.g., Y148) in the ECL2 of murine Cld5 conserved among classic claudins [5] are involved in homophilic trans-interactions [3]. Therefore, we proposed that homologous residues are involved in homophilic trans-interaction of other classic claudins. To test this, HEK cells were transfected with murine Cld3wt or Cld3Y147A (corresponding to Cld5Y148A) and the development of trans-interactions was assessed by enrichment of Cld3 at contacts between two Cld-expressing cells. In contrast to Cld3wt (Fig. 2a, b), Cld3Y147A (Fig. 2c, d) showed no enrichment at contacts. For Cld3wt an enrichment factor (E F) of 2.8 was determined compared to E F = 0.8 for Cld3Y147A (Fig. 2e). In addition, the percentage of contacts with a E F > 1.5 (P E) was for Cld3wt (91%) much higher than for Cld3Y147A (12%, Fig. 2f). This indicates that Y147A inhibits trans-interaction and, therefore, that the determinants for trans-interaction in the ECL2 are, at least, partially conserved between the classic claudins Cld5 and Cld3.
Fig. 2.
Determinant for trans-interaction in the extracellular loop2 (ECL2) is conserved between Cld5 and Cld3. Three days after transfection of HEK cells with Cld3wt (a, b) or Cld3Y147A (c, d) cells were fixed, stained against Cld3 (red in a, c; white in b, d) and nuclei (blue in a, c) and enrichment at cell–cell contacts quantified using fluorescence intensity profiles (b, d; red box). The factor of contact enrichment (E F) was four times and the percentage of contacts showing an enrichment (PE) 6.5 times higher for Cld3wt than for Cld3Y147A (e, f). This indicates that the substitution Y147A inhibits trans-interaction of Cld3. Mean ± SEM; n = 123 for Cld3wt, n = 213 for Cld3Y147A; ***p < 0.001, compared to Cld3wt
Heterophilic claudin3/claudin5 trans-interactions are weaker than claudin3/claudin1-, claudin5/claudin1-, and homophilic trans-interactions
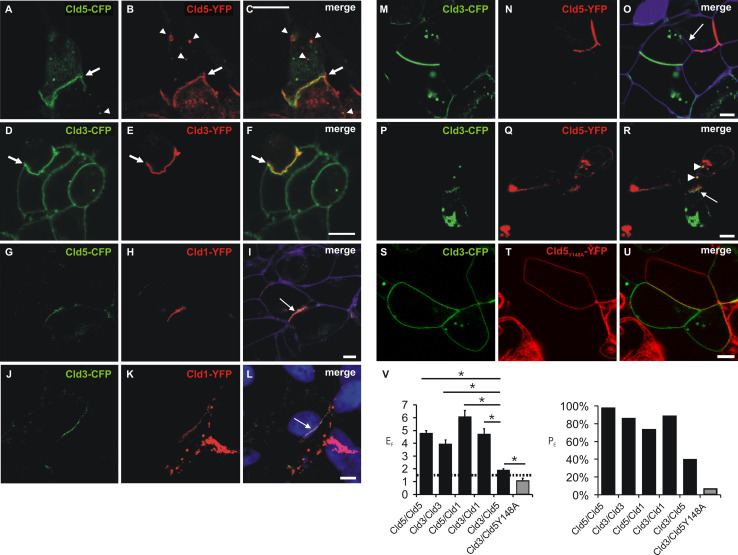
In order to compare the homo- and heterophilic trans-interactions of Cld1, -3, and-5, mixed cocultures of HEK cells monotransfected with Cld-CFP- or Cld-YFP constructs were analyzed by confocal microscopy. Strong coenrichment was detected for Cld5-YFP/Cld5-CFP, Cld3-YFP/Cld3-CFP, Cld1-YFP/Cld5-CFP and Cld1-YFP/Cld3-CFP (Fig. 3a–l). For cocultures of Cld5-YFP/Cld3-CFP (Fig. 3m–r) coenrichment was much less prominent but still greater than for Cld5Y148A-YFP/Cld3-CFP (Fig. 3s–u) that showed no contact enrichment. Quantitative analysis (Fig. 3v) indicates that homophilic Cld3- and Cld5 trans-interactions as well as heterophilic Cld5/Cld1- and Cld3/Cld1 trans-interactions can be formed to a similar extent whereas Cld3/Cld5 trans-interactions are much weaker. In addition, the latter is abolished by the substitution Y148A in Cld5. Despite the fact that the Cld3/Cld5 coenrichment was not prominent, Cld3-CFP-containing vesicles (Fig 3p–r, arrow heads) were detected in Cld5-YFP-expressing cells to an extent similar to Cld5-CFP-containing vesicles in Cld5-YFP-expressing cells and Cld5-YFP-containing vesicles in Cld5-CFP-expressing cells (Fig 3a–c, arrow heads). Thus claudins are, at least partly, internalized together with trans-interacting claudins of a neighboring cell, as reported earlier [23]. Hence, this coendocytosis indicates Cld3/Cld5 trans-interaction.
Fig. 3.
Heterophilic trans-interaction between claudins. Confocal microscopy of cocultures of cells monotransfected with Cld5-CFP/Cld5-YFP (a–c), Cld3-CFP/Cld3-YFP (d–f), Cld5-CFP/Cld1-YFP (g–i), Cld3-CFP/Cld1-YFP (j–l), Cld3-CFP/Cld5-YFP (m–r), or Cld3-CFP/Cld5Y148A-YFP (s–u). CFP- (green) and YFP- fluorescence (red) was imaged (a–u) and, partly, the plasma membrane visualized with trypan blue (i, o; violet) or nuclei stained with DAPI (l, blue). Strong colocalization and coenrichment of claudins at cell contacts (arrows) was detected at homologous (Cld5/Cld5, a–c, Cld3/Cld3, d–f) and heterologous cell contacts for Cld1/Cld5 (g–i) and Cld1/Cld3 (j–l). At contacts between Cld3- and Cld5-expressing cells, coenrichment was found to be weaker and less frequent (example without (m–o, arrow) and with (p–r, arrow) coenrichment). Cld3-CFP containing vesicles (r, arrow heads) were detected in Cld5-YFP-expressing cells similar as Cld5-YFP containing vesicles (a–c, arrow heads) in Cld5-CFP-expressing cells and vice versa. At contacts between Cld3- and Cld5-Y148-expressing cells, no coenrichment was found (s–u). v Quantification of contact coenrichment. For Cld5/Cld5, Cld3/Cld3, Cld1/Cld3 and Cld1/Cld5 the factor of contact enrichment (E F) was significantly higher than for Cld3/Cld5 and for Cld3/Cld5 than for Cld3/Cld5Y148A that showed E F similar to the negative control (Fig. 1). Similar differences were obtained for percentage of contacts with enrichment (PE). Bar, 5 μm. *p < 0.01; n ≥ 35; mean + SEM; dotted line, threshold above trans-interaction is considered
Coexpressed claudins colocalize only partly
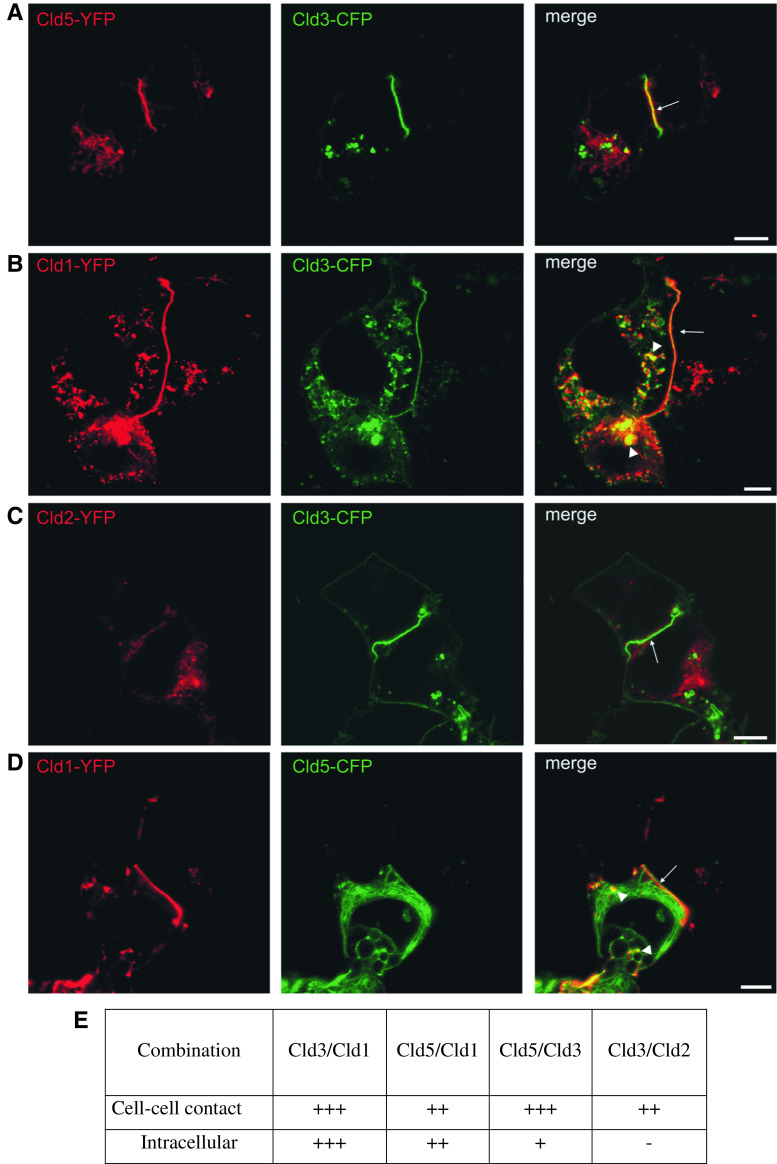
The subcellular distribution of claudins was compared by transient coexpression for pairs of Cld1, -2, -3, and -5 in HEK cells and subsequent analysis by confocal microscopy (Fig. 4). Compared to stable transfection (Fig. 1), the amount of claudins in intracellular compartments was higher. Nevertheless, at contacts between transient transfectants colocalization of all claudin pairs was observed. Strong colocalization in intracellular compartments (arrow heads) was found for Cld3/Cld1 (Fig. 4b), partially for Cld5/Cld1 (Fig. 4d) but not for Cld3/Cld5 and Cld3/Cld2 (Fig. 4a, c). These differences in subcellular distribution indicate subtype-specific differences in capacity for heterophilic cis-interactions, targeting to the plasma membrane and/or internalization of the claudins.
Fig. 4.
Differential colocalization of coexpressed claudin subtypes at cell–cell contacts and intracellular compartments. Pairs of Cld1, -2, -3, and -5 were coexpressed in HEK cells and the subcellular distribution analyzed by confocal microscopy. At contacts between two Cld-expressing cells (arrows) colocalization of Cld3/Cld5 (a), Cld3/Cld1 (b), Cld5/Cld1 (d), and partly Cld3/Cld2 (c) was observed. Strong colocalization in intracellular compartments (arrow heads) was found for Cld3/Cld1 (b), partially for Cld5/Cld1 (d) but not for Cld3/Cld5 (a) and Cld3/Cld2 (c). Bar, 5 μm (e) Summary of the colocalization analysis.−, no; (+)/+, weak; ++/+++, strong colocalization
Analysis of cis-interaction at cell–cell contacts indicates claudin subtype-specific differences
Due to the limited resolution of light microscopy apparent colocalization is not sufficient to demonstrate interaction between claudins. Hence, fluorescence resonance energy transfer (FRET) was applied to investigate homo- and heterophilic cis-interactions between Cld1, -2, -3, and -5 at cell–cell contacts, HEK cells were cotransfected with respective pairs of YFP- and CFP-fusion proteins of claudins. FRET was analyzed at contacts of living cells by acceptor photobleaching [19]. The FRET-efficiency (F E) was strongly dependent on the acceptor/donor (YFP/CFP) ratio but independent of the YFP− or YFP + CFP intensity (measure for expression level) as shown for Cld5-CFP/Cld5-YFP as an example (Fig. S1). This underlines the specificity of the FRET-signal since it indicates a clustered but not random distribution of claudins [24].
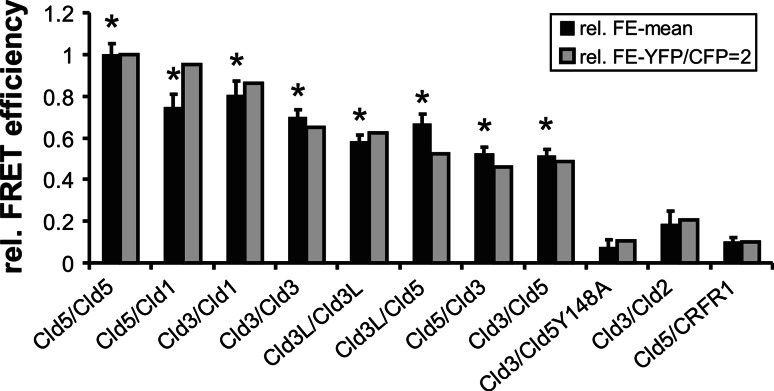
Due to the dependence on YFP/CFP ratios, the F E of different claudin pairs was compared for cells with similar YFP/CFP ratio (Fig. 5, black columns). In addition, the F E was normalized to a YFP/CFP fluorescence intensity ratio of 2 (Fig. 5, gray columns). As shown previously [3], the mean F E for Cld5-CFP/Cld5-YFP was ten times higher than for that of the negative control, Cld5/CRFR1. The normalized FE decreased in the order: Cld5/Cld5, Cld5/Cld1, Cld3/Cld1 > Cld3/Cld3 > Cld3/Cld5. For these, but not for Cld3/Cld2, FE were significantly higher than for the negative control. For Cld3, fusion constructs without and with a flexible linker between Cld3 and CFP/YFP (Cld3L) produced similar results (Fig 5.). This indicates that the lower FE for Cld3/Cld3 and Cld3/Cld5 compared to Cld5/Cld5 are not due to sterical hindrance of the CFP/YFP-tag in Cld3-containing strands. In addition, Cld3-CFP/Cld5-YFP and Cld3-YFP/Cld5-CFP gave similar results. In summary, these data indicate claudin subtype-specific differences in cis-interaction at cell–cell contacts. The absence of FRET between Cld3-CFP and Cld5Y148A-YFP further suggests that the trans-interaction (that is inhibited by the substitution Y148A [3]) is necessary for close proximity between Cld3 and Cld5 at cell–cell contacts.
Fig. 5.
Claudin subtype-specific differences in homo- and heterophilic cis-interactions of claudins at cell–cell contacts. HEK cells were cotransfected with YFP- and CFP-fusion proteins (given as CFP/YFP pairs). Fluorescence resonance energy transfer (FRET) at cell–cell contacts was analyzed in living cells by acceptor photobleaching. Relative FRET-efficiency (rFE, (FEsample/FECld5/Cld5, black columns) and rFE normalized to YFP/CFP fluorescence intensity ratio = 2 (gray columns) using logarithmic regression (see “Methods”). Mean + SEM; n = 20–50; *p < 0.01 compared to Cld5/CRFR1; Cld3L, Cld3 with linker
cis-Interactions between claudins differ in a subtype-specific manner outside of cell–cell contacts
To investigate the cis-interaction of claudins outside of cell–cell contacts FRET was analyzed in cell suspensions by flow cytometry. This technique was applied because thousands of cells of one sample can be analyzed individually. Previous studies have demonstrated the feasibility of detecting FRET by flow cytometry [25, 26]. However, published methods do not consider the dependence of the FRET-efficiency on the acceptor/donor ratio, YFP/CFP (see Fig. S1 and [27, 28]. Hence, we improved the analysis of the flow cytometry data by calculating, as a measure of the FRET-efficiency, the FRET-ratioFC for a standardized YFP/CFP-ratio (see methods and Fig. S2).
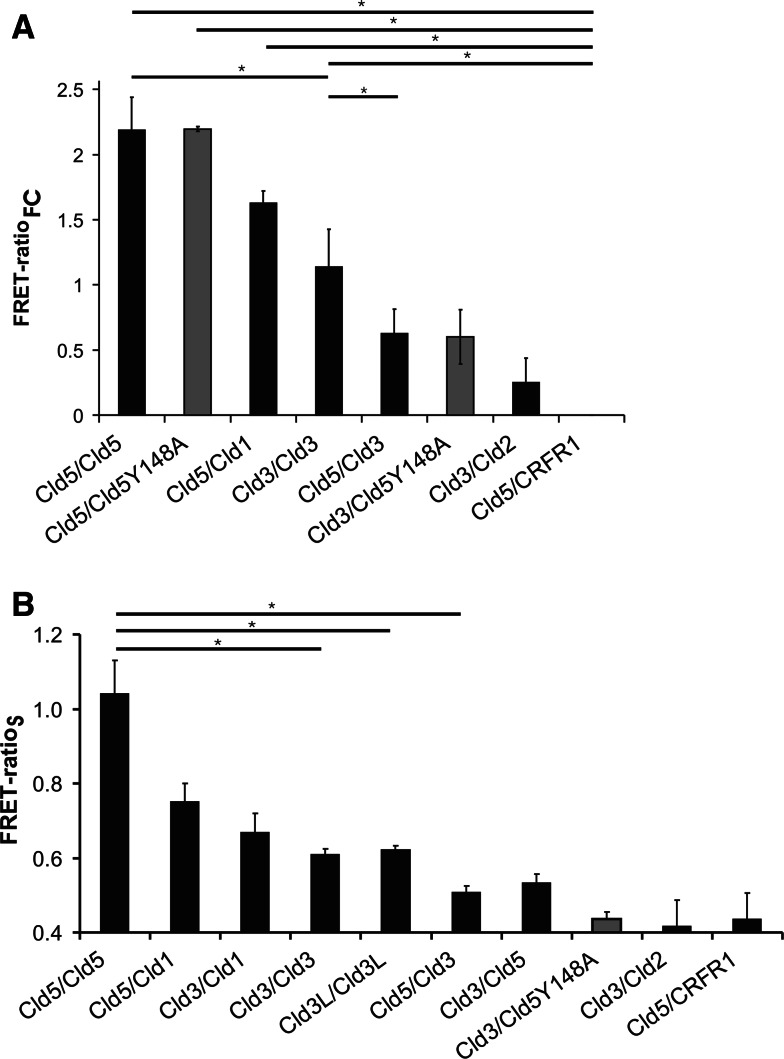
For cells expressing different claudin-pairs, the FRET-efficiency decreased in the order: Cld5/Cld5 > Cld5/Cld1 > Cld3/Cld3 > Cld3/Cld5 (Fig. 6a). The FRET-ratio for Cld3/Cld2 was in the range of the negative control (Cld5/CRFR1). Similar results were obtained by FRET analysis of cell suspensions using spectrofluorometry (Fig. 6b), and of intracellular compartments using confocal microscopy (Fig. S3). In addition, Cld5/Cld5Y148A showed a FRET-ratio similar to that of Cld5/Cld5 and Cld3/Cld5Y148A showed a FRET-ratio similar to that of Cld3-Cld5 (Fig. 6a). Taken together, the data suggest that cis-interactions of claudins outside of cell–cell contacts differ in a subtype-specific manner.
Fig. 6.
Claudin subtype-specific differences in homo- and heterophilic cis-interactions in cell suspension. HEK cells were cotransfected with CFP- and YFP-fusion proteins (e.g., Cld5CFP/Cld5YFP), detached from the culture plate and single cell suspension prepared by pipetting. a FRET-analysis by flow cytometry. As a measure for the FRET-efficiency, the FRET-ratioFC was calculated.*p < 0.02, n = 3–5, for each sample >2,500 coexpressing cells were analyzed. b FRET-analysis by spectrofluorometry. As a measure for the FRET-efficiency, the FRET-ratioS was calculated. In addition to indicated pairs, the FRET-ratioS was for all pairs but not Cld3/Cld2 and Cld3/Cld5Y148A significantly higher than for the negative control (Cld5/CRFR1). *p < 0.01, n = 4–16
Freeze-fracture electron microscopy revealed claudin subtype-specific characteristics of formation of tight junction strands
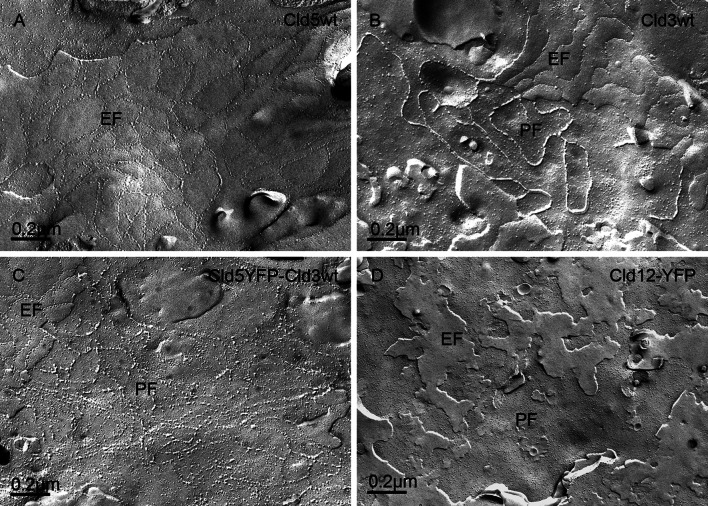
Cld5 strands are known to appear as discontinuous chains of intramembranous particles that are associated with the exoplasmic face (E-face) of the plasma membrane [29]. We reproduced this finding (Fig. 7a) and verified that non-tagged and YFP-tagged Cld5 form strands of similar morphology [3]. Also, we reproduced earlier reports [13] that Cld3, in contrast to Cld5, forms continuous strands with intramembranous particles associated with the protoplasmic face (P-face) of the plasma membrane. In contrast, grooves with very few particles are found on the E-face (Fig. 7b). Coexpression of Cld3 and Cld5 resulted in strands with a mixed phenotype: discontinuous chains of intramembranous particles on the P-face as well as on the E-face (Fig. 7c). In addition, the substitution Y147A in Cld3, that strongly inhibits trans-interaction, abolished the formation of strands. Similarly for Cld12, which showed no strong trans-interactions (Fig. 1), no strands were detected after transfection of HEK cells (Fig 7d). These results demonstrate that trans-interaction is a prerequisite for strand formation. All together, the data demonstrate claudin subtype-specific characteristics of strand formation.
Fig. 7.
Claudin subtype-specific characteristics of formation of tight junction strands. Claudin-transfected HEK cells were analyzed by freeze-fracture electron microscopy. For Cld5wt-expressing cells, discontinuous strands with intramembranous particles associated with the exoplasmic face (E-face) of the plasma membrane were detected (a). For Cld3wt-expressing cells, continuous strands associated with the protoplasmic face (P-face) of the plasma membrane were detected (b). For cells that coexpress Cld5wt-YFP and Cld3wt, intramembranous particles were found on the E- as well as on the P-face (c). For cells transfected with Cld12 (d), Cld3Y147A or non-transfected cells (latter not shown), and for non-transfected cells, no tight junction strands were observed
The mobility of claudins is subtype-specific and affected by trans-interaction
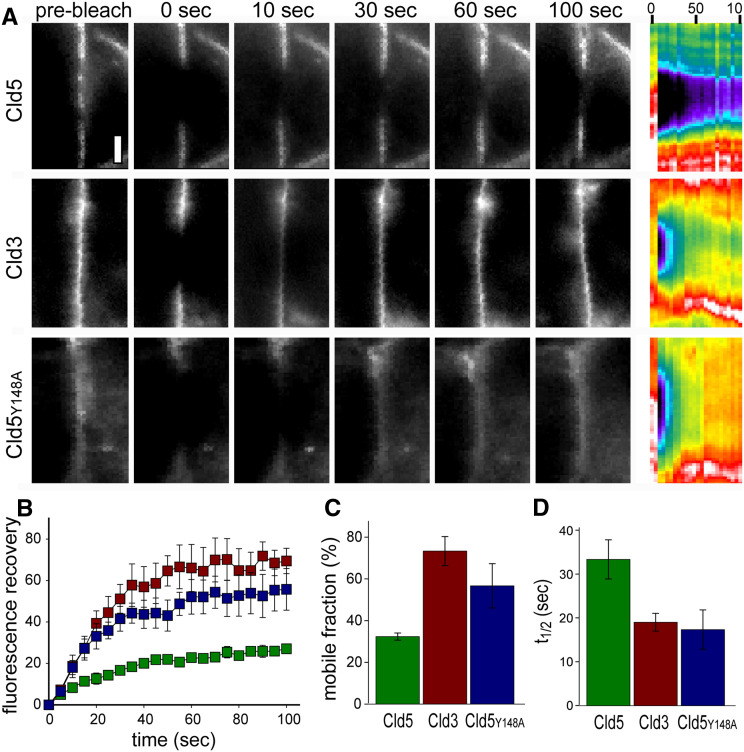
The mobility of claudins was analyzed by fluorescence recovery after photobleaching, FRAP [21]. At cell–cell contacts of HEK cells stably transfected with claudin-YFP constructs, Cld5 showed a mobile fraction of 32 ± 2%, only, whereas Cld3 exhibited a mobile fraction of 73 ± 7% (p < 0.01; Fig. 8a–c). In addition, the t 1/2 of recovery for Cld5 (33 ± 4 s) was nearly twice as long as that of Cld3 (19 ± 2 s; p < 0.01; Fig. 8d). The substitution Y148A in Cld5 resulted in a much higher mobile fraction 57 ± 10% (p < 0.05), and shorter t 1/2 (17 ± 4 s; p < 0.05; Fig. 8c, d) that were not significantly different from mobile fraction and t 1/2 of Cld3. Cld5 was far more mobile within plasma membrane areas away from cell–cell contacts (mobile fraction 61% ± 3%; t 1/2 16 ± 3 s) than at cell–cell contacts (p < 0.02 for each vs. Cld5 at cell contacts). This was similar to FRAP behavior of intracellular Cld5 (mobile fraction 58 ± 3%; t 1/2 18 ± 5 s) as well as Cld5Y148A at cell–cell contacts. In contrast to Cld5, exchange behavior of Cld3 within plasma membrane areas away from cell–cell contacts (mobile fraction 72 ± 4%; t 1/2 18 ± 2 s) was similar to that at cell–cell contacts. These data indicate that trans-interactions of Cld5 at cell–cell contacts and presumably incorporation of Cld5 in strands limits its mobility. The mobility in the plasma membrane is higher for Cld3 than for Cld5. This could be due to a higher amount of nonpolymerized Cld3 in the plasma membrane or due to higher disassembly/assembly rate of the strands for Cld3 relative to Cld5.
Fig. 8.
Mobility of claudins is subtype-specific and affected by trans-interaction. a FRAP behavior of Cld5, Cld3, and Cld5Y148A at areas of cell–cell contact between HEK cells stably transfected with the respective YFP fusion constructs. Individual images are shown as indicated, with the corresponding kymograph at the right. Bar = 2 μm. b Recovery curves for Cld5, Cld3 and Cld5Y148A at areas of cell–cell contact (n = 3 per condition). c Mobile fractions of Cld5, Cld3 and Cld5Y148A at areas of cell–cell contact calculated from recovery data in b (n = 3 per condition). d t 1/2 of Cld5, Cld3, Cld5YY148A recovery at areas of cell–cell contact calculated from data in b (n = 3 per condition)
Claudins form diffusion barriers at cell–cell contacts
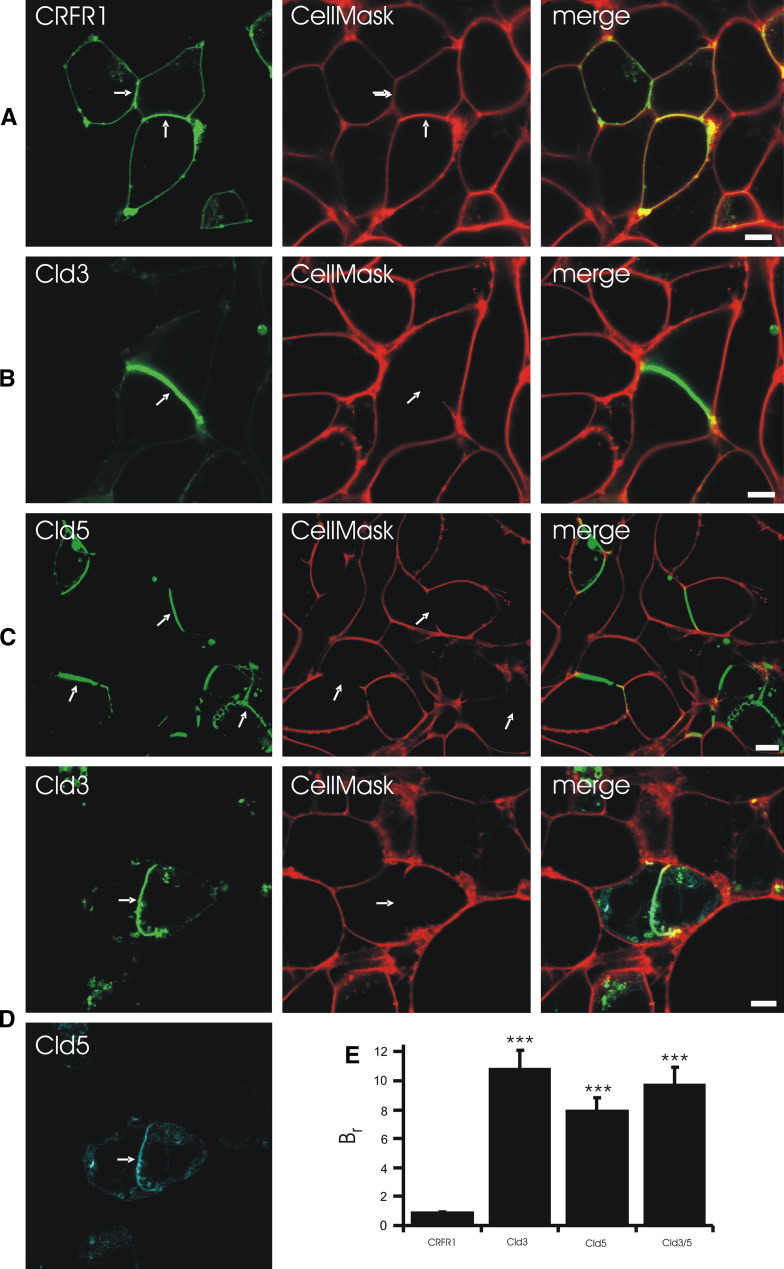
To analyze the barrier function of reconstituted TJ-strands, claudin-expressing HEK cells were incubated with the plasma membrane stain CellMask™ and analyzed by life cell imaging. Incubation with 5 μg/ml for 15 min or with 0.25 μg/ml for 20 min labeled the plasma membrane everywhere. In contrast, incubation with 0.25 μg/ml CellMask for 5 min resulted in a strong labeling of contacts without claudin enrichment but not of contacts with claudin enrichment (Fig. 9, Fig. S5). This was found for contacts between two Cld3-expressing or two Cld5-exppressing but not for CRFR1-expressing cells (Fig. 9a–c). In addition, no clear CellMask™-labeling was found for contacts between two Cld3/Cld5-coexpressing cells (Fig. 9d). This demonstrates that the TJ-strands that are reconstituted by expression of Cld3 and/or Cld5 form a diffusion barrier which CO which inhibits membrane incorporation of an extracellularly administered marker.
Fig. 9.
Claudins form diffusion barrier at contacts between claudin-expressing HEK cells. Cells were incubated with CellMask for 5 min and the labeling of the cell membrane was analyzed by confocal microscopy. a CellMask labeled contacts between CRFR1-CFP(control)-expressing and non-expressing cells as well as contacts between two CRFR1-CFP-expressing cells. At contacts between Cld3-YFP- (b), Cld5-YFP (c) or Cld3-YFP/Cld5-CFP- (d) expressing cells, CellMask labeling was much weaker or absent. e The barrier ratio (Br), as a measure for paracellular barrier preventing CellMask labeling, was determined using confocal intensity profiles. Bar, 5 μm; ***p < 0.001 versus CRFR1; arrows, contacts between cells expressing fusion proteins
Discussion
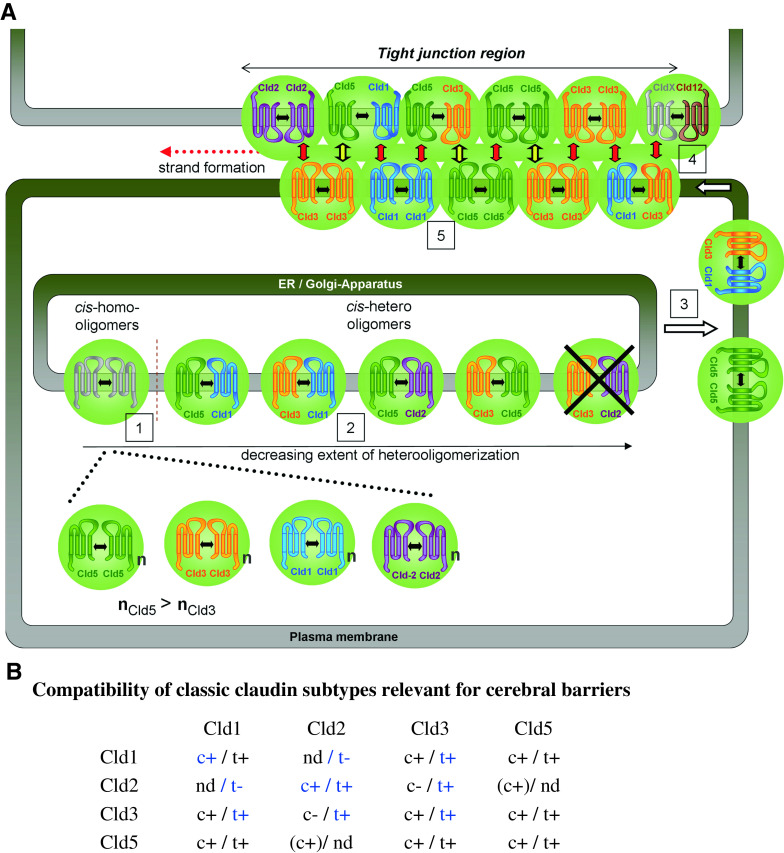
The molecular mechanisms that result in the formation of membranous TJ-strands are not understood. Here, we show claudin subtype-specific characteristics with respect to cis- and trans-interaction as well as formation of TJ-strands. Furthermore, we provide a model for the formation of heteropolymeric strands (Fig. 10).
Fig. 10.
a Model of formation and molecular organization of heteropolymeric claudin strands. Cld1, -2, -3, -5, and -12, assumed to be expressed at cerebral barriers, are shown. Black double arrows, cis-interaction; green spheres, cis-oligomers; n, number of molecules; red arrows, homo- and heterophilic trans-interactions; black/yellow arrows, Cld3/Cld5 trans-interactions; CldX, claudin that incorporates Cld12 by heterophilic trans- or cis-interaction. b Compatibility of classic claudin subtypes relevant for cerebral barriers. c, cis-interaction; t, trans-interaction; +, compatible; −, incompatible. Bracket, detected intracellularly, only; nd, not determined; blue, according to [13, 36, 37], 1–5, see text
Homophilic claudin interactions
Previously, we adapted a cellular reconstitution assay [30] to analyze the formation of TJ-strands on the molecular level. Instead of polarized epithelial or endothelial cells, HEK cells that do not form endogenous TJ were transfected with Cld5-CFP/YFP or mutants thereof to reconstitute defined claudin polymers [3]. Homophilic trans-interaction was detected by enrichment of Cld5 at contacts between two Cld5-expresssing cells. Using this assay, we here describe that Cld1, -3, and -5, which belong to the group of classic claudins [5] show a similar capability for homophilic trans-interaction. This is consistent with earlier studies [10, 13].
In contrast, the non-classic Cld12 did not show considerable homophilic trans-interaction. Consistently, Cld12 did not form homopolymeric strands as do the classic claudins. However, it can not be excluded that the C-terminal YFP-tag prevents strand formation for Cld12 but not for classic claudins. The non-classic claudins show strong sequence differences compared to the classic claudins (e.g., Cld12 contains no aromatic residue corresponding to Y148 in Cld5 and Y147 in Cld3 [5]). Similar to Cld12, the non-classic Cld16 does not form homopolymeric strands [7]. However, the non-classic Cld11 is able to form homopolymeric strands [31]. We show that inhibition of trans-interactions, by removing a conserved aromatic side chain in the ECL2 of classic claudins, diminished strand formation as shown for Cld5 [3] and Cld3 (Figs. 2, 7). In addition, trans-interactions reduce the mobility of Cld5 (Fig 8). Therefore, we conclude that trans-interactions are necessary for strand formation, as discussed earlier [3] and propose that Cld12 and possibly other non-classic claudins lack the ability to form homopolymeric strands due to the lack of ability for homophilic trans-interaction.
Comparison of homo- and heterophilic trans-interactions
To understand the barrier properties of TJ on the molecular level, it is necessary to analyze the heterophilic compatibility of claudins. Heterophilic trans-interactions (heterotypic interactions) have been reported for Cld1/-3, -2/-3, -3/-5 but not for Cld1/-2, -3/-4, -4/-5, -1/-4, -16/-19, -1/-10b and -1/-15 [7, 10, 13, 32]. Heterophilic cis-interactions (heteromeric interactions) have been reported for Cld3 and Cld4 pairs [10] and Cld16 and Cld19 pairs [7]. Heterophilic interactions that could not be clearly distinguished as cis- or trans- were noted for Cld1/2, Cld1/Cld3 and Cld2/Cld3 [13, 33], but the only determinants of cis-interactions identified thus far were G20, Q57, and G123 in human Cld19, which mediate cis-interactions with Cld16 [7].
For the blood–brain barrier Cld1, -3 and -5 are relevant [6, 14]. We found the capability for heterophilic trans-interactions of Cld5/Cld1 and Cld3/Cld1 to be as pronounced as that for homophilic Cld5/Cld5 and Cld3/Cld3 interactions. In contrast, Cld3/Cld5 trans-interactions were much less pronounced, although still significantly stronger than the negative control. The Cld3/Cld1 interaction is consistent with earlier studies [10, 13, 33]. Also similar to our data, the Cld5/Cld5 interaction has been reported to be stronger than Cld3/Cld5 interaction [10]. However, in contrast to our results, the latter authors did not detect trans-interactions between Cld5 and Cld1. This difference could be due to the use of human Cld1 and human Cld5, while we used human Cld1 and mouse Cld5. We also obtained similar results for mouse Cld1 and mouse Cld5 (data not shown). Therefore, although species variation cannot be excluded, the data and fact that sequence similarity between mouse and human is >94% suggest that species effects are unlikely to explain differences of the data. However, the discrepancy could be due to other methodological issues. For example, Daugherty et al. [10] used, in contrast to us, claudin constructs that could bind to proteins containing PDZ-domains, e.g., ZO-1. This could change the subcellular distribution of claudins. Moreover, the use of chemically fixed cells could disturb some interactions. Finally, although the coimmunoprecipitation approach used by Daugherty et al. is a powerful technique, it cannot exclude a loss of interactions between transmembrane proteins after detergent solubilization. In contrast, we analyzed claudins in living cells allowing investigation of interactions under physiological conditions. However, it must also be recognized that our assays do not detect physical association but trace interactions by indirect parameters such as contact-enrichment, TJ-strand formation and FRET.
Claudin subtype-specific cis-interactions
At contacts between claudin-coexpressing cells, colocalization was found for Cld3/Cld5, Cld5/Cld1, Cld3/Cld1 and partly for Cld3/Cld2. However, the different claudin pairs showed striking differences in their intracellular codistribution. Intracellular colocalization was commonly found for Cld1/Cld3, partially for Cld1/Cld5, infrequently for Cld3/Cld5, and very infrequently for Cld2/Cld3. The subtype-specific differences in colocalization suggest that the subcellular distribution is caused by subtype-specific interactions rather than by undirected aggregation. The claudin-containing intracellular compartments did not considerably colocalize with markers for endosomes or lysosomes but partly with those for the endoplasmic reticulum (Fig. S4).
Because of the different extent of colocalization at cell–cell contacts and at intracellular compartments, we performed FRET-analysis independently at and away from cell–cell contacts. At cell–cell contacts and therefore presumably in TJ-strands, cis- and trans-interactions are likely formed. There, FRET-efficiencies decreased in the order Cld5/Cld5, Cld5/Cld1, Cld3/Cld1 > Cld3/Cld3 ≥ Cld3/Cld5 and no FRET for Cld3/Cld2. In single cell suspensions, cell–cell contacts and in turn trans-interactions are assumed to be absent. Those FRET-efficiencies decreased in the order Cld5/Cld5 > Cld5/Cld1 > Cld3/Cld3 > Cld3/Cld5, with no FRET found for Cld3/Cld2. This indicates that cis-interactions between claudin pairs are similar outside and inside TJ-strands.
However, especially for Cld3/Cld5, important differences have been found: First, Cld3 and Cld5 demonstrated marked colocalization at cell–cell contacts, but not outside of contacts. Second, the FRET-efficiency of Cld3/Cld5 at cell contacts is closer to that of Cld3/Cld3 than is the FRET efficiency of Cld3/Cld5 outside of contacts. Finally, Cld3/Cld5-, but not Cld5/Cld5- [3], FRET is abolished by Y148A substitution in Cld5 (inhibition of trans-interactions) at cell contacts, but this substitution has no effect on cis-interactions measured in suspension.
These results indicate that trans-interactions are necessary for the FRET between Cld3 and Cld5. However, this FRET is unlikely to be due to direct energy transfer between trans-interacting Cld3 and Cld5 on opposing plasma membranes because the distance between the respective intracellular CFP- and YFP-tags is too big (>8 nm) for efficient FRET [34]. It is also unlikely that Y148A inhibits Cld3/Cld5 but not Cld5/Cld5 cis-interactions. Hence, the data suggest that the FRET-signal and, in turn, the spatial proximity between Cld3 and Cld5 in one membrane at cell–cell contacts is promoted by homo- and/or heterophilic trans-interactions rather than by direct cis-interactions.
Previously, some heterophilic cis-interactions between claudin subtypes have been analyzed by coimmunoprecipitation after cell lysis [10, 33]. Although this approach has provided important data, potential artifacts might occur due to detergent solubilization. In addition, immunofreeze-fracture microscopy of strands with two different labels was applied [13]. Although this technique is very powerful, the spatial resolution is still limited due to the size of antibodies and gold particles and a potential contribution of trans-interactions to colocalization of claudins in strands has to be considered. An effective split ubiquitin assay in yeast was used to analyze cis-interactions independent of trans-interactions [7]. However, yeast lack mammalian characteristics including contacts between plasma membranes. Hence, cis- and trans-interactions cannot be effectively compared in this system.
We complemented the above-mentioned assays by FRET-experiments with HEK cells within and outside of reconstituted TJ-strands. For an efficient FRET-analysis outside of strands, we here developed a new method based on flow cytometry. Similar FRET techniques have been used before [25, 26]. We improved and optimized the method, especially, with consideration of the strong dependence of the FRET-efficiency on the acceptor/donor ratio and the characteristics of junctional proteins.
Reconstituted TJ-strands form a functional diffusion barrier
Claudins are able to reconstitute TJ-strands in non-polar cells [3, 30]. Nevertheless, these strands do not form a continuous sealing belt around the cells as in epi-/endothelial cells. This is probably due to the lack of cytoplasmic TJ-proteins. Hence, standard Transwell assays to analyze paracellular permeability are hindered in such reconstitution systems. To overcome this limitation we developed a novel, imaging-based assay to demonstrate and quantify barrier functions of reconstituted and molecular defined TJ-strands. We showed that a single claudin (e.g., Cld3 or Cld5) is sufficient to form a diffusion barrier. Whether the diffusion is inhibited in the paracellular space and/or within the membrane might be analyzed by continuative experiments including the novel assay.
Model of formation and molecular organization of heteropolymeric claudin strands
The results obtained and data from the literature [10, 13, 33, 35–37] were combined to propose a model of heteropolymeric strands for Cld1, -2, -3, -5, and -12 that are expressed at cerebral barriers (Fig. 10):
Two to six [33, 35] claudin molecules might form oligomers mediated by cis-interactions in intracellular compartments. This is demonstrated by FRET in cell suspensions of Cld-CFP/Cld-YFP-coexpressing cells in which most of the claudins are present in intracellular compartments. This conclusion is supported by FRET analysis of intracellular compartments using confocal microscopy (Fig. S3).
Differences in the FRET-efficiency suggest that cis-interactions in intracellular compartments are differentially pronounced in the order: Cld5/Cld5 > Cld5/Cld1 > Cld3/Cld1 > Cld3/Cld3 > Cld3/Cld5, and that cis-interactions do not occur between Cld3/Cld2. The higher FRET-efficiency for Cld5- compared to Cld3-homooligomers supports the conclusion that Cld5-cis-homooligomers are sterically different from Cld3-cis-homooligomers, e.g., higher number of molecules (nCld5 > nCld3). This is consistent with differences in oligomer size detected by PFO-PAGE [33]. Evidence for homophilic cis-interaction of Cld1 was recently provided by FRET-assays [37]. Due to strong sequence homology among classic claudins [5], Cld2 is also assumed to form cis-oligomers.
The cis-oligomers are transported to the plasma membrane and diffuse to cell–cell contacts.
At cell–cell contacts, trans-interactions trigger formation of polymeric strands, as proposed earlier [3]. Homophilic trans-interactions are possible for Cld1, -3, -5, and -2 [13], but not for Cld12. Heterophilic Cld3/Cld5 trans-interactions are less pronounced than heterophilic Cld3/Cld1 and Cld5/Cld1 and all homophilic trans-interactions investigated. Nevertheless, heterophilic Cld3/Cld5 trans-interactions (Fig. 10, black/yellow arrows) may be essential to form functional heteropolymers, since these are necessary for Cld3/Cld5 colocalization and FRET. Heterophilic Cld3/Cld2, but not Cld1/Cld2, trans-interactions were described [13].
Aromatic residues in the ECL2 that are conserved between classic claudins (e.g., Y148 in mCld5, Y147 in mCld3) are essential for homo- as well as heterophilic trans-interactions but not for the assembly of cis-oligomers.
-
(5)
In TJ-strands, spatial proximity of claudins in one membrane due to cis-interactions and/or trans-interactions occurs in the order of preference: Cld5/Cld5 > Cld5/Cld1, Cld3/Cld1, Cld3/Cld3 > Cld3/Cld5 > Cld3/Cld2.
In contrast to the classic claudins, the non-classic Cld12 is not able to form homophilic trans-interactions or homopolymeric strands. Probably, heterophilic cis- and/or trans-interactions with another TJ-protein are necessary for the incorporation of Cld12 in strands. The interactions depicted in the model (Fig. 10) are consistent with the data.
Freeze-fracture electron microscopy demonstrated claudin subtype-specific characteristics of TJ-strands. Coexpression of Cld3 and Cld5 resulted in strands with a phenotype that represent a mixture of that of Cld3- (P-face) and Cld5-homopolymers (E-face): discontinuous chains of intramembranous particles on the P-face as well as on the E-face. This resembles TJ in endothelia of the blood–brain barrier in vivo. There, the expression of Cld3 together with Cld5 was reported to correlate with mixed P-/E-face association and high tightness of brain endothelial cell junctions [14, 15, 38]. Here, we demonstrate that this functionally important ultrastructural characteristic is indeed due to the formation of Cld3/Cld5 heteropolymeric strands and does not require other blood–brain barrier-specific factors since we observed it in HEK-cells that do not possess blood–brain barrier properties.
In summary, we provide novel mechanistic insights into the principles that define molecular organization of TJ-strands. This improves the understanding of paracellular barriers and might be used to develop new strategies to improve drug delivery across cerebral barriers, e.g., by modulation of the Cld3/Cld5 interaction.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 1 The FRET-efficiency is strongly dependent on the ratio of acceptor/donor (YFP/CFP) fluorescence intensity (A, R2 = 0.51) but not on the YFP fluorescence intensity (B, R2 = 0.02) or the CFP+YFP fluorescence intensity (C, R2 = 0.02) as a measure of the expression level. Individual data points and non-linear regression curve for Cld5-CFP/Cld5-YFP. (PDF 45 kb)
Supplementary material 2 (A) Flow cytometric expression analysis of HEK cells transfected with different constructs. Non- (mock), CFP- or YFP-transfection were used to account for auto fluorescence. CFP/YFP cotransfection and CFP-YFP tandem were used as FRET negative or positive control, respectively. Cdl5-CFP/CRFR1-YFP and Cld5-CFP/Cld5-YFP were used as pairs of transmembrane proteins shown to be FRET-negative or -positive, respectively, by conventional method (Piontek et al., 2008). On the axes the channels are given. FSC, forward scatter. (B) Visualization of FRET signal by comparison of CFP/YFP - FRET negative -, CFP-YFP tandem - FRET positive - (left panel), Cld5-CFP/CRFR1-YFP - FRET negative - and Cld5-CFP/Cld5-YFP - FRET positive - (right panel). FRET-signal is indicated by left shift. (C) Normalization of FRET and YFP values relative to CFP intensity with the corresponding exponential decay fits (left: CFP/YFP (black dots, green line) and CFP-YFP (red dots, blue line); right: Cld5-CFP/CRFR1-YFP (black dots, green line) and Cld5-CFP/Cld5-YFP (red dots, blue line). The fits were used to calculate the FRET-ratioFC as a measure of FRET- efficiency (see Methods). (PDF 455 kb)
Supplementary material 3 FRET-analysis in intracellular compartments by confocal microscopy. For Cld5-CFP/Cld5-YFP, FRET was significant higher than for Cld3-CFP/Cld3-YFP for which FRET was still higher than for the negative control (CFP-tagged endoplasmic reticulum marker, CFP-ER (BD Biosciences) coexpressed with Cld5-YFP, CFP-ER/Cld5-YFP). 36, 34, and 19 cell–cell contacts were analyzed, respectively; p < 0.001. (PDF 35 kb)
Supplementary material 4 In intracellular compartments claudins did not considerably colocalize with markers for endosomes, Golgi apparatus or lysosomes but partly with that for the endoplasmic reticulum (A, arrow). HEK 293 cells were cotransfected with Cld5-YFP and (A) CFP-ER (marker for endoplasmic reticulum, Clontech), (B) CFP-Endo (marker for endosomes, Clontech) or (C, left) CFP-Golgi (marker for Golgi apparatus, Clontech) and 3 days later analyzed by confocal microscopy. In addition, cells were transfected with Cld5 (C, middle) or Cld3 (C, right) and 3 days later incubated with 50 nM LysoTracker (Invitrogen) in growth medium for 30 min at 37°C, the medium exchanged by DMEM with 10 mM N-(2-hydroxyethyl)piperazine-N’(2-ethanesulfonic acid) pH 7.5 without phenol red and analyzed by confocal microscopy. Bar, 5 µm. (PDF 105 kb)
Supplementary material 5 CellMask labels the apical and basolateral plasma membrane; reconstituted TJs delay the labeling. HEK cells stably transfected with Cld5 were labeled with 0.25 μg/ml CellMask for 5 min (A, B) or 20 min (C, D). After 5 min, CellMask labeling (red) is detected in the apical (ap) and basal (bl) part of the lateral plasma membrane but not at the Cld5-positive (green) area at cell–cell contacts (A,B). In contrast, after 20 min, CellMask (red) colocalizes with claudin-5 (green) at cell–cell contacts (C, D). Merge (A, C); CellMalsk (B, D); arrow, Cld5-enrichment at cell–cell contact; arrowhead, apical and basolateral plasma membrane adjacent to claudin-5; bar, 5 µm. (PDF 272 kb)
Acknowledgments
We gratefully acknowledge the help of Ria Knittel in freeze-fracturing. This work was funded by DFG BL308/7-3, 7-4 and PI 837/2-1 and NIH DK61931.
References
- 1.Angelow S, Ahlstrom R, Yu AS. Biology of claudins. Am J Physiol Renal Physiol. 2008;295:F867–F876. doi: 10.1152/ajprenal.90264.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Staehelin LA. Structure and function of intercellular junctions. Int Rev Cytol. 1974;39:191–283. doi: 10.1016/S0074-7696(08)60940-7. [DOI] [PubMed] [Google Scholar]
- 3.Piontek J, Winkler L, Wolburg H, Muller SL, Zuleger N, Piehl C, Wiesner B, Krause G, Blasig IE. Formation of tight junction: determinants of homophilic interaction between classic claudins. FASEB J. 2008;22:146–158. doi: 10.1096/fj.07-8319com. [DOI] [PubMed] [Google Scholar]
- 4.Morita K, Furuse M, Fujimoto K, Tsukita S. Claudin multigene family encoding four-transmembrane domain protein components of tight junction strands. Proc Natl Acad Sci USA. 1999;96:511–516. doi: 10.1073/pnas.96.2.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krause G, Winkler L, Mueller SL, Haseloff RF, Piontek J, Blasig IE. Structure and function of claudins. Biochim Biophys Acta. 2008;1778:631–645. doi: 10.1016/j.bbamem.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 6.Nitta T, Hata M, Gotoh S, Seo Y, Sasaki H, Hashimoto N, Furuse M, Tsukita S. Size-selective loosening of the blood–brain barrier in claudin-5-deficient mice. J Cell Biol. 2003;161:653–660. doi: 10.1083/jcb.200302070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hou J, Renigunta A, Konrad M, Gomes AS, Schneeberger EE, Paul DL, Waldegger S, Goodenough DA. Claudin-16 and claudin-19 interact and form a cation-selective tight junction complex. J Clin Invest. 2008;118:619–628. doi: 10.1172/JCI33970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hou J, Renigunta A, Gomes AS, Hou M, Paul DL, Waldegger S, Goodenough DA. Claudin-16 and claudin-19 interaction is required for their assembly into tight junctions and for renal reabsorption of magnesium. Proc Natl Acad Sci USA. 2009;106:15350–15355. doi: 10.1073/pnas.0907724106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu AS, Cheng MH, Angelow S, Gunzel D, Kanzawa SA, Schneeberger EE, Fromm M, Coalson RD. Molecular basis for cation selectivity in claudin-2-based paracellular pores: identification of an electrostatic interaction site. J Gen Physiol. 2009;133:111–127. doi: 10.1085/jgp.200810154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daugherty BL, Ward C, Smith T, Ritzenthaler JD, Koval M. Regulation of heterotypic claudin compatibility. J Biol Chem. 2007;282:30005–30013. doi: 10.1074/jbc.M703547200. [DOI] [PubMed] [Google Scholar]
- 11.Piehl C, Piontek J, Cording J, Wolburg H, Blasig IE. Participation of the second extracellular loop of claudin-5 in paracellular tightening against ions, small and large molecules. Cell Mol Life Sci. 2010;67:2131–2140. doi: 10.1007/s00018-010-0332-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J, Piontek J, Wolburg H, Piehl C, Liss M, Otten C, Christ A, Willnow TE, Blasig IE, Abdelilah-Seyfried S. Establishment of a neuroepithelial barrier by Claudin5a is essential for zebrafish brain ventricular lumen expansion. Proc Natl Acad Sci USA. 2010;107:1425–1430. doi: 10.1073/pnas.0911996107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Furuse M, Sasaki H, Tsukita S. Manner of interaction of heterogeneous claudin species within and between tight junction strands. J Cell Biol. 1999;147:891–903. doi: 10.1083/jcb.147.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolburg H, Lippoldt A. Tight junctions of the blood–brain barrier: development, composition and regulation. Vascul Pharmacol. 2002;38:323–337. doi: 10.1016/S1537-1891(02)00200-8. [DOI] [PubMed] [Google Scholar]
- 15.Kniesel U, Risau W, Wolburg H. Development of blood–brain barrier tight junctions in the rat cortex. Dev Brain Res. 1996;96:229–240. doi: 10.1016/0165-3806(96)00117-4. [DOI] [PubMed] [Google Scholar]
- 16.Liebner S, Fischmann A, Rascher G, Duffner F, Grote EH, Kalbacher H, Wolburg H. Claudin-1 and claudin-5 expression and tight junction morphology are altered in blood vessels of human glioblastoma multiforme. Acta Neuropathol. 2000;100:323–331. doi: 10.1007/s004010000180. [DOI] [PubMed] [Google Scholar]
- 17.Wolburg H, Wolburg-Buchholz K, Kraus J, Rascher-Eggstein G, Liebner S, Hamm S, Duffner F, Grote EH, Risau W, Engelhardt B. Localization of claudin-3 in tight junctions of the blood–brain barrier is selectively lost during experimental autoimmune encephalomyelitis and human glioblastoma multiforme. Acta Neuropathol. 2003;105:586–592. doi: 10.1007/s00401-003-0688-z. [DOI] [PubMed] [Google Scholar]
- 18.Winkler L, Gehring C, Wenzel A, Muller SL, Piehl C, Krause G, Blasig IE, Piontek J. Molecular determinants of the interaction between Clostridium perfringens enterotoxin fragments and claudin-3. J Biol Chem. 2009;284:18863–18872. doi: 10.1074/jbc.M109.008623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blasig IE, Winkler L, Lassowski B, Mueller SL, Zuleger N, Krause E, Krause G, Gast K, Kolbe M, Piontek J. On the self-association potential of transmembrane tight junction proteins. Cell Mol Life Sci. 2006;63:505–514. doi: 10.1007/s00018-005-5472-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen L, Weber CR, Turner JR. The tight junction protein complex undergoes rapid and continuous molecular remodeling at steady state. J Cell Biol. 2008;181:683–695. doi: 10.1083/jcb.200711165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu D, Marchiando AM, Weber CR, Raleigh DR, Wang YM, Shen L, Turner JR. MLCK-dependent exchange and actin binding region-dependent anchoring of ZO-1 regulate tight junction barrier function. Proc Natl Acad Sci USA. 2010;107:8237–8241. doi: 10.1073/pnas.0908869107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mack AF, Wolburg H. Growing axons in fish optic nerve are accompanied by astrocytes interconnected by tight junctions. Brain Res. 2006;1103:25–31. doi: 10.1016/j.brainres.2006.04.135. [DOI] [PubMed] [Google Scholar]
- 23.Matsuda M, Kubo A, Furuse M, Tsukita S. A peculiar internalization of claudins, tight junction-specific adhesion molecules, during the intercellular movement of epithelial cells. J Cell Sci. 2004;117:1247–1257. doi: 10.1242/jcs.00972. [DOI] [PubMed] [Google Scholar]
- 24.Wallrabe H, Elangovan M, Burchard A, Periasamy A, Barroso M. Confocal FRET microscopy to measure clustering of ligand-receptor complexes in endocytic membranes. Biophys J. 2003;85:559–571. doi: 10.1016/S0006-3495(03)74500-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Banning C, Votteler J, Hoffmann D, Koppensteiner H, Warmer M, Reimer R, Kirchhoff F, Schubert U, Hauber J, Schindler M (2010) A flow cytometry-based FRET assay to identify and analyse protein–protein interactions in living cells. Plos One 5 [DOI] [PMC free article] [PubMed]
- 26.van Wageningen S, Pennings AH, van der Reijden BA, Boezeman JB, de Lange F, Jansen JH. Isolation of FRET-positive cells using single 408-nm laser flow cytometry. Cytom Part A. 2006;69A:291–298. doi: 10.1002/cyto.a.20254. [DOI] [PubMed] [Google Scholar]
- 27.Di WL, Gu Y, Common JEA, Aasen T, O’Toole EA, Kelsell DP, Zicha D. Connexin interaction patterns in keratinocytes revealed morphologically and by FRET analysis. J Cell Sci. 2005;118:1505–1514. doi: 10.1242/jcs.01733. [DOI] [PubMed] [Google Scholar]
- 28.Amiri H, Schultz G, Schaefer M. FRET-based analysis of TRPC subunit stoichiometry. Cell Calcium. 2003;33:463–470. doi: 10.1016/S0143-4160(03)00061-7. [DOI] [PubMed] [Google Scholar]
- 29.Morita K, Sasaki H, Furuse M, Tsukita S. Endothelial claudin: claudin-5/TMVCF constitutes tight junction strands in endothelial cells. J Cell Biol. 1999;147:185–194. doi: 10.1083/jcb.147.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Furuse M, Sasaki H, Fujimoto K, Tsukita S. A single gene product, claudin-1 or -2, reconstitutes tight junction strands and recruits occludin in fibroblasts. J Cell Biol. 1998;143:391–401. doi: 10.1083/jcb.143.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morita K, Sasaki H, Fujimoto K, Furuse M, Tsukita S. Claudin-11/OSP-based tight junctions of myelin sheaths in brain and sertoli cells in testis. J Cell Biol. 1999;145:579–588. doi: 10.1083/jcb.145.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inai T, Kamimura T, Hirose E, Iida H, Shibata Y. The protoplasmic or exoplasmic face association of tight junction particles cannot predict paracellular permeability or heterotypic claudin compatibility. Eur J Cell Biol. 2010;89:547–556. doi: 10.1016/j.ejcb.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 33.Coyne CB, Gambling TM, Boucher RC, Carson JL, Johnson LG. Role of claudin interactions in airway tight junctional permeability. Am J Physiol Lung Cell Mol Physiol. 2003;285:L1166–L1178. doi: 10.1152/ajplung.00182.2003. [DOI] [PubMed] [Google Scholar]
- 34.Sekar RB, Periasamy A. Fluorescence resonance energy transfer (FRET) microscopy imaging of live cell protein localizations. J Cell Biol. 2003;160:629–633. doi: 10.1083/jcb.200210140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mitic LL, Unger VM, Anderson JM. Expression, solubilization, and biochemical characterization of the tight junction transmembrane protein claudin-4. Protein Sci. 2003;12:218–227. doi: 10.1110/ps.0233903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Itallie CM, Mitic LL, Anderson JM (2010) Claudin-2 forms homodimers and is a component of a high molecular weight protein complex. J Biol Chem [DOI] [PMC free article] [PubMed]
- 37.Harris HJ, Davis C, Mullins JG, Hu K, Goodall M, Farquhar MJ, Mee CJ, McCaffrey K, Young S, Drummer H, Balfe P, McKeating JA. Claudin association with CD81 defines hepatitis C virus entry. J Biol Chem. 2010;285:21092–21102. doi: 10.1074/jbc.M110.104836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liebner S, Kniesel U, Kalbacher H, Wolburg H. Correlation of tight junction morphology with the expression of tight junction proteins in blood–brain barrier endothelial cells. Eur J Cell Biol. 2000;79:707–717. doi: 10.1078/0171-9335-00101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1 The FRET-efficiency is strongly dependent on the ratio of acceptor/donor (YFP/CFP) fluorescence intensity (A, R2 = 0.51) but not on the YFP fluorescence intensity (B, R2 = 0.02) or the CFP+YFP fluorescence intensity (C, R2 = 0.02) as a measure of the expression level. Individual data points and non-linear regression curve for Cld5-CFP/Cld5-YFP. (PDF 45 kb)
Supplementary material 2 (A) Flow cytometric expression analysis of HEK cells transfected with different constructs. Non- (mock), CFP- or YFP-transfection were used to account for auto fluorescence. CFP/YFP cotransfection and CFP-YFP tandem were used as FRET negative or positive control, respectively. Cdl5-CFP/CRFR1-YFP and Cld5-CFP/Cld5-YFP were used as pairs of transmembrane proteins shown to be FRET-negative or -positive, respectively, by conventional method (Piontek et al., 2008). On the axes the channels are given. FSC, forward scatter. (B) Visualization of FRET signal by comparison of CFP/YFP - FRET negative -, CFP-YFP tandem - FRET positive - (left panel), Cld5-CFP/CRFR1-YFP - FRET negative - and Cld5-CFP/Cld5-YFP - FRET positive - (right panel). FRET-signal is indicated by left shift. (C) Normalization of FRET and YFP values relative to CFP intensity with the corresponding exponential decay fits (left: CFP/YFP (black dots, green line) and CFP-YFP (red dots, blue line); right: Cld5-CFP/CRFR1-YFP (black dots, green line) and Cld5-CFP/Cld5-YFP (red dots, blue line). The fits were used to calculate the FRET-ratioFC as a measure of FRET- efficiency (see Methods). (PDF 455 kb)
Supplementary material 3 FRET-analysis in intracellular compartments by confocal microscopy. For Cld5-CFP/Cld5-YFP, FRET was significant higher than for Cld3-CFP/Cld3-YFP for which FRET was still higher than for the negative control (CFP-tagged endoplasmic reticulum marker, CFP-ER (BD Biosciences) coexpressed with Cld5-YFP, CFP-ER/Cld5-YFP). 36, 34, and 19 cell–cell contacts were analyzed, respectively; p < 0.001. (PDF 35 kb)
Supplementary material 4 In intracellular compartments claudins did not considerably colocalize with markers for endosomes, Golgi apparatus or lysosomes but partly with that for the endoplasmic reticulum (A, arrow). HEK 293 cells were cotransfected with Cld5-YFP and (A) CFP-ER (marker for endoplasmic reticulum, Clontech), (B) CFP-Endo (marker for endosomes, Clontech) or (C, left) CFP-Golgi (marker for Golgi apparatus, Clontech) and 3 days later analyzed by confocal microscopy. In addition, cells were transfected with Cld5 (C, middle) or Cld3 (C, right) and 3 days later incubated with 50 nM LysoTracker (Invitrogen) in growth medium for 30 min at 37°C, the medium exchanged by DMEM with 10 mM N-(2-hydroxyethyl)piperazine-N’(2-ethanesulfonic acid) pH 7.5 without phenol red and analyzed by confocal microscopy. Bar, 5 µm. (PDF 105 kb)
Supplementary material 5 CellMask labels the apical and basolateral plasma membrane; reconstituted TJs delay the labeling. HEK cells stably transfected with Cld5 were labeled with 0.25 μg/ml CellMask for 5 min (A, B) or 20 min (C, D). After 5 min, CellMask labeling (red) is detected in the apical (ap) and basal (bl) part of the lateral plasma membrane but not at the Cld5-positive (green) area at cell–cell contacts (A,B). In contrast, after 20 min, CellMask (red) colocalizes with claudin-5 (green) at cell–cell contacts (C, D). Merge (A, C); CellMalsk (B, D); arrow, Cld5-enrichment at cell–cell contact; arrowhead, apical and basolateral plasma membrane adjacent to claudin-5; bar, 5 µm. (PDF 272 kb)