Abstract
The potential for personalized sequencing to individually optimize medical treatment in diseases such as cancer and for pharmacogenomic application is just beginning to be realized, and the utility of sequencing healthy individuals for managing health is also being explored. The data produced requires additional advancements in interpretation of variants of unknown significance to maximize clinical benefit. Nevertheless, personalized sequencing, only recently applied to clinical medicine, has already been broadly applied to the discovery and study of disease. It is poised to enable the earlier and more accurate diagnosis of disease risk and occurrence, guide prevention and individualized intervention as well as facilitate monitoring of healthy and treated patients, and play a role in the prevention and recurrence of future disease. This article documents the advancing capacity of personalized sequencing, reviews its impact on disease-oriented scientific discovery and anticipates its role in the future of medicine.
Keywords: cancer genomics, disease discovery, electronic medical record, genomic medicine, individualized therapy, next-generation sequencing, personalized medicine, pharmacogenetics, pharmacogenomics, precision medicine, prevention, whole-exome sequencing, whole-genome sequencing
Background
In the 10 years since the official completion of the Human Genome Project (HGP) [1] technological advances in the speed and scale of sequencing analysis have maintained an accelerating pace. The tools produced by these advances now enable holistic analysis of individual human genomes at a cost and within a timeframe to allow practical and productive application to research questions and, more recently, personalized clinical evaluations.
Targeted, single gene sequence analysis of individual patients has been a clinically applicable diagnostic tool since before completion of the Human Genome Project. However, such testing has always been limited to thoroughly characterized genes, for which a phenotype is recognizable and clinically certified testing is available. Current next-generation sequencing technologies, including whole-exome sequencing (WES) and whole-genome sequencing (WGS) now allow analyses beyond a handful of genes, to include a more comprehensive genetic analysis. These sequencing tools are being actively applied to well-studied, but as yet unconquered diseases such as cancer, where significant advances in understanding the pathophysiology, diagnostics, treatment and surveillance are likely to greatly benefit patients; they are also being applied to the analyses of unsolved diseases in children and adults, and more recently to the analyses of healthy individuals. It is expected that genetic information will play an ever increasingly important role in helping us to better predict, diagnose and treat diseases. Here, we present a perspective on how next-generation sequencing may change pharmacogenomics and medicine as a whole through discovery and treatment of many types of disease and personalized pharmacological intervention.
Cancer genome sequencing
Many of the most innovative tools of biomedical investigation have been based on understanding the various presentations of cancer. Accordingly, soon after the completion of the HGP and the advent of new sequencing technologies, genome sequencing was applied to the analysis of cancer. Cancer has long been recognized as being caused by acquisition of multiple genetic mutations, which are thought to ‘drive’ cells toward uncontrollable growth. Studies have described driver gene versus passenger gene mutations in many forms of cancer [2]. Driver gene mutations are classically defined as mutations that, when they occur in a cell, confer a selective growth advantage and drive the cell’s progression to malignancy [3]. Some driver mutations are inherited at birth (e.g., APC and mutations) whereas others are acquired somatically and may be heavily influenced by environmental exposure.
Personalized sequencing impacts cancer in several ways. The first is cancer cell DNA sequencing. One of the first genomic studies to apply WGS to cancer involved analysis of the DNA of an acute myeloid leukemia patient in which both tumor and normal cells were sequenced [4]. Ten mutations were identified in the tumor DNA and not in the normal DNA. Two of these had been previously described as linked to acute myeloid leukemia and the remaining eight were novel. This proof of principle study put forward WES/WGS as a tool to discover novel mutations and potential therapeutic targets. This effort has now been expanded on a very large scale. One large project is The Cancer Genome Atlas (TCGA), which is systematically analyzing WGS and WES of more than 20 types of human cancer [5]. One of the biggest outcomes of these efforts is the discovery that most cancers are very different from one other, although common mutated pathways can often be observed. For example, in ovarian cancer patients mutations in the BRCA1 and the BRCA2 pathways, affecting homologous recombination, are frequently observed. Moreover, cancer from different tissues of origin can often have the same types of mutations. For example, the EGFR gene, previously known to be commonly mutated in breast cancer patients, is often amplified or mutated in other cancer types. As such, cancers are now being classified based on their genetic changes rather than their tissue of origin.
Another active area in the pathophysiology of cancer is the clonal evolution theory of cancer. In 1976 Peter Nowell posited that cancer develops as differently mutated clonal cells out-compete each other, with the expectation that less fit variant clones die, leaving one clone to comprise the majority of a tumor [6]. However, genomic analyses in recent years have demonstrated that for many cases, there is a significant level of genetic diversity within single tumors, suggesting that tumors are more mosaic, rather than being dominated by a predominant clone [7]. These observations have raised the profile of several aspects of tumoral genetic heterogeneity, and the role heterogeneity plays in diagnosis and treatment of cancer. For example, intratumoral, intermetastatic, intrametastatic and interpatient tumor heterogeneity each impact our efforts to achieve early diagnosis and successful therapeutic intervention [3].
New methods have emerged that use DNA sequencing to monitor cancer progression. Tumor DNA sequencing is rapidly expanding its capacity to produce a clinically relevant tumor profile. This is currently focused on somatic DNA variations, but there is growing effort to analyze RNA expression and DNA methylation patterns. Such information can help determine which signaling pathways are active in tumor cells, which may not have been suggested by histological assessment alone, and thereby suggests therapeutic avenues that would not be uncovered by conventional methods [8].
Cancer sequencing treatment implications
Personalized tumor DNA sequencing can directly impact treatment by identifying mutations that can suggest therapeutic treatments. In some cases the information from DNA sequencing can identify a known cancer target or pathway for which an existing pharmacological treatment is available (often initially used for a cancer involving a different tissue) and sometimes even new potential targets are uncovered. For example, researchers recently found through WES, a loss of function mutation in TSC1 in approximately 5% of advanced bladder cancers. This specific mutation correlated with tumor sensitivity to everolimus, suggesting that this subgroup of bladder cancer patients might benefit from everolimus therapy [9]. Other examples of genome sequencing based clinical interventions include utilization of EGFR kinase inhibitors in cancers with EGFR gene mutations (found in many different types of cancers), and BRAF inhibitors in tumors with BRAF mutations (often found in melanomas) [10,11]. In these situations application of pharmacogenomic principles to individual tumors is critical to determine their susceptibility to these specific drug therapies, as only a fraction of patients will respond to these targeted therapies and treating patients prior to confirming their tumor’s sensitivity would expose patients to drug side effects while allowing their cancers to advance [3]. For example, identification of KRAS alterations in codons 12 or 13, which occurs in approximately 30% of colon cancer patients, suggest some toxicity risk and no particular treatment benefit with EGFR specific antibodies [12]. Despite these advances with clear impact on current patient care, tumor somatic mutation assessment has impacted clinical intervention for a limited number of cancers. Currently, less than 10% of oncology drugs approved by the US FDA have documented molecular predictors of efficacy, and there is tremendous potential for progress in this area [8].
Even as sequencing has led to these advances in cancer therapy, new targets are emerging, such as within the pathways of tumor suppressor genes, which individually can be difficult to impact therapeutically. For example, BRCA1 and BRCA2 gene defects impact downstream DNA repair pathways and make cells more susceptible to drugs that inhibit repair of DNA damage, such as PARP, and clinical trials with this strategy are in underway [13]. Individual cancer genomes have been noted to contain highly variable numbers (often 30–70 mutations) in coded proteins. Each of these changes is foreign to the native immune system and exploiting these changes has been suggested to lead to the development of highly tumor-specific antigens as a powerful tool for cancer directed therapies [3]. This is one example of advances in the development of molecularly targeted cancer therapies enabled by genomic sequencing. This is especially important in the context of the substantial problem of cancer drug resistance, in which resistance causing events in tumors appear to be selected for in a Darwinian fashion [14,15]. The mechanism of cancer drug resistance can be influenced by genetic and histological background of the tumor as well as previously applied treatments [14,16]. One of the strategies to combat drug resistance may eventually include using simultaneous drug combinations [14,17].
Cancer pharmacogenomics
Another major area of impact of cancer genome sequencing is germline sequencing. Germline sequencing enables the estimation of underlying patient risk arising from known alterations causing characterized syndromes of cancer predisposition, such as familial adenomatous polyposis or Li Fraumeni syndrome, which facilitates implementation of prophylactic interventions and screening protocols to optimize early detection. Familial predisposition is estimated to account for up to 10% of melanoma, breast, colon and gastric cancers and up to 25% of ovarian cancer. Among these, testing is available to identify known predisposing genes in approximately 2–3% of colon cancer, 3–5% of gastric cancer, 5–10% of breast cancer, up to 10% of melanoma and up to 25% of endometrial cancer [18–23]. Germline testing can potentially impact an estimated 40,000 new cases of these types of cancer alone [24], in addition to the thousands of family members who benefit from germline sequencing by finding they do not carry the genetic predisposition. While germline sequencing currently impacts a minority of cancer, it is clear that significant potential remains for personal sequencing to discover novel genetic etiologies that account for the thousands of familial and individual cases of cancer for which a molecular etiology remains unclear. For example, WGS and WES analysis of individuals with pancreatic cancer identified segregating variants of the ATM gene, implicating it as a pancreatic cancer predisposition gene [25]. WGS of patients with multiple adenomas and/or colorectal adenocarcinoma (CRC) found mutations in POLE and POLD, identifying them as CRC susceptibility genes [26].
These same technologies are being applied to address pharmacogenomic issues, such as the etiology of chemotherapeutic failure, clinical side effects, drug metabolism or drug resistance. The identification of a germline variant of TPMT was found to result in life-threatening toxicity in patients treated with mercaptopurine (a treatment for acute lymphoblastic leukemia). This led the FDA to recommend genotyping of patients prior to treatment, and reduce the dosage for those with appropriate genotypes [27]. The FDA currently recommends genotyping prior to treatment with other chemotherapeutics, such as irinotecan for CRC [28]. Patient-specific drug metabolism can also lead to inadequate dosing, as in the case of tamoxifen for estrogen receptor positive breast carcinoma. Tamoxifen is metabolized into multiple metabolites, including endoxifen, which is central to treatment efficacy. Germline patient sequencing found variants in the CYP2D6 gene, which are associated with lower serum concentrations of endoxifen due to decreased enzyme activity and lead to risk of drug failure due to inadequate dosing [29]. In addition to the clear potential for cancers to mutate and develop resistance to particular chemotherapeutic interventions [8], germline sequence analysis has found patients whose tumors are inherently resistant. For example, a study of chronic myeloid leukemia patients found a deletion of BCL2-like (also known as BIM) in patients whose cancer treatment was resistant to tyrosine kinase inhibitors. Further analyses confirmed presence of the BIM deletion in the germline of the patients resistant to treatment, and that the proapoptotic domain affected by the deletion was the mechanism of the patients’ inherent resistance to tyrosine kinase inhibitor therapeutics [28]. Each of these circumstances demonstrates the increasingly important role of pretreatment, germline personalized sequencing in successful cancer intervention.
Although the benefit of genetic information is clear, the overall impact of these advances on patient care currently remains limited. To date, approximately 40 FDA approved oncology drugs (Table 1) have been updated to include clinically relevant pharmacogenomic information in their package inserts [30]. Similarly, only approximately 40 known cancer genes have FDA approved drugs, some with multiple drugs per gene target. However, greater than 30 additional cancer genes have experimental drugs under development, which will greatly enhance the impact of genome sequencing in the future [31].
Table 1.
US FDA-approved oncology drugs with package inserts containing pharmacogenetics and pharmacogenomics information.
| Drug | Pharmacogenomic biomarker(s) |
|---|---|
| Ado-trastuzumab emtansine | ERBB2 |
| Afatinib | EGFR |
| Anastrozole | ESR1, PGR |
| Arsenic trioxide | PML/RARA |
| Bosutinib | BCR/ABL1 |
| Brentuximab vedotin | TNFRSF8 |
| Busulfan | Ph chromosome |
| Capecitabine | DPYD |
| Cetuximab | EGFR, KRAS |
| Cisplatin | TPMT |
| Crizotinib | ALK |
| Dabrafenib | BRAF, G6PD |
| Dasatinib | BCR/ABL1 |
| Denileukin diftitox | IL2RA |
| Erlotinib | EGFR |
| Everolimus | ERBB2, ESR1 |
| Exemestane | ESR1 |
| Fluorouracil | DPYD |
| Fulvestrant | ESR1 |
| Ibritumomab tiuxetan | MS4A1 |
| Imatinib | KIT, BCR/ABL1, PDGFRB, FIP1L1/PDGFRA |
| Irinotecan | UGT1A1 |
| Lapatinib | ERBB2 |
| Letrozole | ESR1, PGR |
| Mercaptopurine | TPMT |
| Nilotinib | BCR/ABL1, UGT1A1 |
| Obinutuzumab | MS4A1 |
| Ofatumumab | MS4A1 |
| Omacetaxine | BCR/ABL1 |
| Panitumumab | EGFR, KRAS |
| Pazopanib | UGT1A1 |
| Pertuzumab | ERBB2 |
| Ponatinib | BCR-ABL T315I |
| Rasburicase | G6PD |
| Rituximab | MS4A1 |
| Tamoxifen | ESR1, PGR, F5, F2 |
| Thioguanine | TPMT |
| Tositumomab | MS4A1 |
| Trametinib | BRAF |
| Trastuzumab | ERBB2 |
| Tretinoin | PML/RARA |
| Vemurafenib | BRAF |
F2: Prothrombin; F5: Factor V Leiden; Ph: Philadelphia.
Data taken from [32].
Cancer sequencing & surveillance
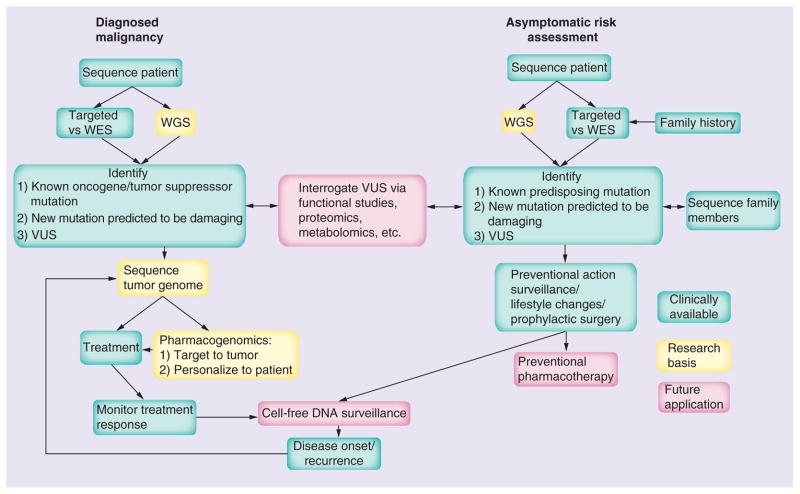
Following intervention for a diagnosed malignancy, such as CRC, personalized sequencing will likely also play a role in guiding ongoing surveillance for recurrence. With one WGS costing the approximate equivalent of one to two colonoscopies [33], post-treatment identification of known genetics susceptibilities can inform individualized management and follow-up protocols. An area of promise for future monitoring of remission and recurrence is that of cell-free DNA (cfDNA) sequencing through which it is possible to detect cancer mutations in body fluids such as blood, urine and stool, which enables monitoring of response to treatment and tumor evolution [34]. Developed and clinically applied in the realm of prenatal diagnostics, the analysis of cell-free fetal DNA, as collected from maternal plasma collection, has recently lead to non-invasive WGS of a fetus at 18.5 weeks gestation, enabled in part by the capacity to distinguish maternal from fetal DNA sequence [35]. The observation of significant levels of tumor DNA in the blood of patients with cancer has led to the idea of monitoring tumor cfDNA levels as a marker of disease [36]. The presence of tumor specific mutations offers an additional opportunity for cfDNA analysis, with post-treatment sequencing of cfDNA having the potential to identify those mutations unique to the eradicated primary tumor, as a marker of remission and sentinel indicator of recurrence [36]. In light of the advances made in cfDNA analysis in the prenatal realm to the point of WGS of an actively growing fetus, it seems reasonable to envision a future ability to obtain WGS of a patient tumor at an early stage of recurrence, as a noninvasive form of surveillance that potentiates early re-intervention. It is crucial to emphasize here that surveillance strategies require tests with high sensitivity and specificity, the establishment of which will require large randomized control trials. This is necessary to avoid the potential harm from false positives or ambiguous results. Overall, this could lead personalized sequencing to become an integral part of the full spectrum of clinical cancer care, including risk assessment, prevention, disease screening and diagnostics, personalized pharmacogenomic-based therapy, and post-therapy surveillance (Figure 1).
Figure 1. Personalized sequencing in cancer: current and future medical applications.
This includes applications that are currently clinically available, those applied in research protocols and those envisioned for the future, such as preventional management.
WES: Whole-exome sequencing; WGS: Whole-genome sequencing; VUS: Variants of unknown significance.
Sequencing the unknown: rare diseases
Multiple genome sequencing studies have already uncovered novel relationships for genetic variants with monogenetic Mendelian disorders and complex diseases [37–42]. Approximately 7000 well-defined Mendelian disorders are currently known, of which the corresponding allelic variants underlying fewer than half of these monogenic disorders have been discovered, and the etiology of many monogenic diseases is still unknown [42,43]. Furthermore, genome sequencing enables us to decipher the causes and even guide treatment of an ever-growing number of ‘mystery’ diseases, of which many cluster in families but can also involve individual probands, such as Charcot–Marie–Tooth neuropathy, Miller’s syndrome, and dopa (3,4-dihydroxyphenylalanine)-responsive dystonia [37,41,42]. Sequencing has shown the potential to provide a solution in cases where there is an initial inability to make a clinical diagnosis of the disease and in rare cases has been shown to subsequently direct a course of treatment [44].
A notable example was reported by Worthey et al. A male infant presented with proctitis. This progressed to pancolitis, which was concerning for a Crohn’s disease-like affliction. The severity of disease was suggestive of underlying immune dysfunction, however, substantial clinical evaluation was unable to determine a definitive diagnosis. Utilization of WES in this patient identified over 16,000 variants which, after further analysis observed a novel mutation in the X-linked inhibitor of the apoptosis gene, not previously connected with Crohn’s disease but known to be involved in the pro-inflammatory response [45]. Functional analyses confirmed the deleterious nature of the mutation and the diagnosis of X-linked inhibitor of apoptosis deficiency. Indicated treatment was hematopoetic cell transplant, after which the patient experienced resolution of the symptoms of colitis [45]. Another example is described by Bainbridge et al., in two fraternal twins afflicted by clinical symptoms of dystonia whose diagnostic evaluation was unrevealing until one of the twins experienced symptomatic improvement with L-dopa treatment, at which time they were diagnosed with dopa-responsive dystonia (DRD), based on this clinical response. Even with L-dopa treatment, the patients continued to experience a combination of mild tremor, dystonic posturing, unsteady gait, dysphonia and bradykinesia [46]. The twins’ DNA, as well as their parents and an unaffected sibling, were subjected to WGS and, after shared mutation analysis, filtering and genetic annotation, three genes with significant nonsynonymous mutations were found, one of which, SPR, had been previously associated with DRD. SPR encodes an enzyme important to the generation of BH4, a cofactor for dopamine and serotonin. Functional studies confirmed the deleterious impact of the compound heterozygous SPR mutations found in the patients, and their treatment was modified to include a serotonin precursor, which is recommended in patients with DRD due to SPR mutations. Two weeks after therapeutic modification, the patients both experienced symptomatic improvement including increased ability to participate in athletic activities at school [46]. Though a minority of cases result in successful treatment interventions, even a diagnosis without a current therapy provides a family with important information regarding a patient’s prognosis, medical management and allows for informed family planning.
Challenges of sequence interpretation
Despite these clear successes, many challenges make these successes less frequent than is desirable. In one of the above examples, multiple individuals in addition to the probands underwent WGS to facilitate filtering of the thousands of identified variants, which are not relevant to the clinical question in the proband. However in many clinical scenarios, only a single proband is under evaluation and, even with sequencing of both of the proband’s parents, thousands of variants will segregate in a fashion that makes it difficult to unequivocally identify the causative variant. Filtering of identified variants is also dependent on the clarity of the phenotype, as candidate gene lists are developed based on known disease gene associations. If a patient’s phenotype is too broadly defined or nonspecific, then the identification of likely candidate genes, from thousands of sequence variants, is significantly complicated. The clarity of a patient’s phenotype may also be difficult to describe in the setting of monogenic conditions with decreased penetrance or variable expressivity. Finally, once a promising candidate is distilled from the filtering process, there is no standardized approach to functionally verify that the causative genetic mutation has been ascertained. In the examples noted, in vitro functional analyses were performed to obtain supportive evidence, though the true confirmation of the diagnosis was observed in the patients’ response to genomic sequence based treatment. Unfortunately, there remain many genetic diseases without a known treatment, for which these means of confirmation is unavailable.
Sequencing & the potential for discovery
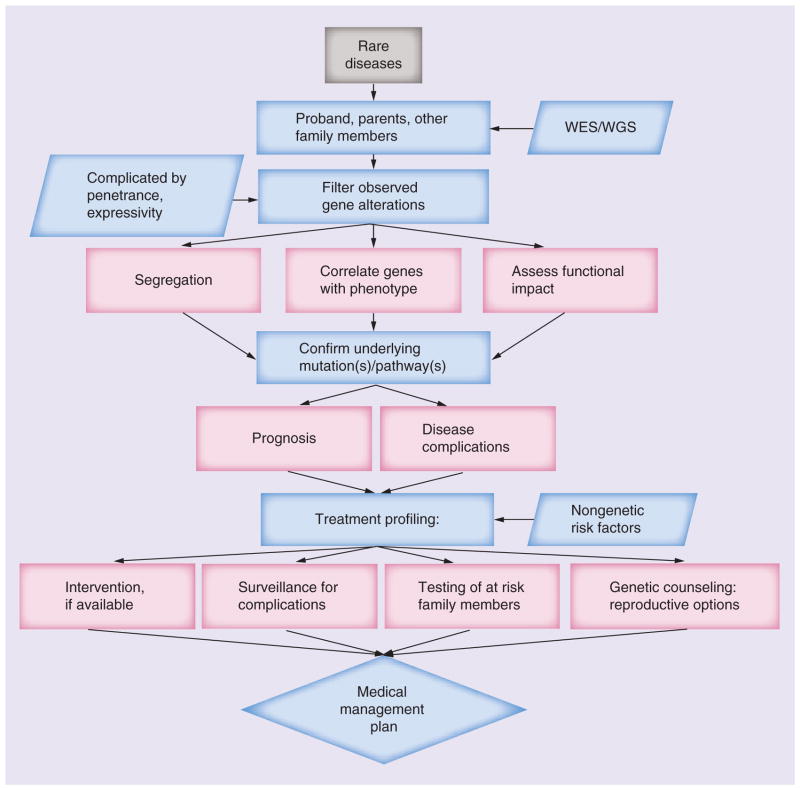
Nevertheless, the application of WES/WGS appears well suited to the elucidation of genetic diseases of Mendelian inheritance, as outlined in Figure 2. It is a powerful approach to the discovery of novel causative genes underlying Mendelian disorders where conventional strategies have failed. Even in conditions where conventional approaches are expected to find the genetic etiology, WES/WGS provides a means to accelerate discovery [47]. WES in particular is anticipated to accelerate the discovery of genes causing rare Mendelian disorders as: many known alleles of these conditions disrupt protein-coding sequences; a large fraction of rare protein impacting variants are predicted to have deleterious affects; and the exome represents an enriched genomic subset in which to search for these alterations with large effect sizes providing the opportunity to capture nearly all of the protein-coding gene rare alleles present in a sample [47]. This includes diagnostic application to pediatric patients with rare diseases, like the examples already described. There is also potential to impact other inherited disease, such as the wide range of inherited cardiovascular diseases. Nonsyndromic cardiomyopathies such as dilated cardiomyopathy (DCM), hypertrophic cardiomyopathy, arrhythmogenic right ventricular dysplasia and left ventricular noncompaction, among others, have been attributed to mutations in over 40 genes [48]. Among these known cardiomyopathy genes, it is estimated that the specific genetic cause is identified in as many as 65% of familial hypertrophic cardiomyopathy cases, 50% of arrhythmogenic right ventricular dysplasia cases and 30% of DCM cases; however, lower identification rates are achieved in sporadic cases. Of the remaining undiagnosed cases of familial cardiomyopathy, application of WES/WGS has identified rare variants, for example, in the DCM gene TTN, with 25% of familial cases demonstrating potentially causative variants [48]. With such an extensive, and growing, collection of genes accounting for the various inherited cardiomyopathies, many of which overlap between categories, personalized sequencing is a promising tool for streamlining of diagnostics, via both WES/WGS as well as targeted sequencing panels. It also demonstrates ongoing promise in delineating the underlying rare variants responsible for those cases of heritable cardiomyopathy yet to be elucidated.
Figure 2. Investigation of rare diseases: potential and pitfalls.
As the obstacles facing WES/WGS in the diagnosis of rare diseases are overcome, they can ultimately guide medical management.
WES: Whole-exome sequencing; WGS: Whole-genome sequencing.
Clinical personalized sequencing
The process of performing WES began to be offered on a clinical basis in 2011. There are currently a number of CLIA-certified laboratories that offer WES as a diagnostic test for patients with a phenotype for which an underlying molecular etiology has not yet been defined. While WES can be performed on the patient only, various sites offer, and recommend, testing of family trios, including the patient and both parents, to facilitate subsequent sequence interpretation. In all cases of clinically available WES, identified mutations thought to be of clinical relevance are confirmed via Sanger sequencing prior to being reported. The output of clinical WES includes disruptive mutations to which the patient’s phenotype is attributable, mutations which appear unrelated to the observed phenotype, and variants of unknown significance (VUS).
There can also be mutations and incidental findings discovered during WGS and WES studies that predispose to conditions unrelated to the original indication, the reporting of which has been addressed by the American College of Medical Genetics and Genomics (ACMG) and remains a subject of ongoing evaluation [49]. Though early estimates of the success of WES/WGS in diagnosing rare disease have been as high as 50% [50], recent studies suggest the success rate to be closer to 20–30% [51,52], with chief obstacles being efficient and accurate clinical interpretation of the genomic variants [53] and the fact that many genes have yet to be associated with a specific disorder, an obstacle WES itself will help to overcome. Nevertheless, it is probably just a matter of time until pharmacogenomic sequencing studies experience similar successful discoveries in the realm of rare Mendelian diseases.
Sequencing the ‘healthy’
Future steps will involve integration of established disease variants into clinical decision-making for asymptomatic, healthy individuals. Several pilot projects have been published where sequenced genomes from single individuals were annotated for known genetic risk factors [54–56]. The Varimed database, which contains published knowledge on hundreds of thousands of genetic variations in relation to thousands of traits, formed the reference for annotation with the Risk-OGram algorithm [55]. Other examples of annotation databases are: Online Mendelian Inheritance in Man (OMIM) [57,58], the Human Gene Mutation Database (HGMD) [59,60], NCBI ClinVar [57,61] the European Genome-phenome Archive [62], dbGaP [63,64] and the GWAS catalog [65,66]. Ashley et al. provided the first example where a patient with a family history of vascular disease and early sudden death was clinically assessed, including the patient’s full genome sequence, to provide risk prediction for coronary artery disease and screening for causes of sudden cardiac death [54]. The ‘Snyderome’ paper revealed that genome sequencing can be used to assess various medical risks, direct the monitoring of specific diseases (in this study, aplastic anemia and Type 2 diabetes) and successfully guide lifestyle interventions and pharmacotherapy [55]. The subject carried a TERT mutation, predicted to be damaging, which has been associated with aplastic anemia [67]. However, measurements of telomere length suggested little or no decrease in telomere length and a modest increase in numbers of cells with short telomeres. Importantly, the patient and his mother share the same mutation but neither exhibit symptoms of aplastic anemia, indicating that this mutation does not always result in disease and is likely context specific in its effects. This illustrates that previously reported statistically significant associations of genetic variants with diseases may have imperfect positive predictive values. The subject was predicted to have significantly elevated risk levels for hypertriglyceridemia and diabetes, including associated variants in GCKR (homozygous) [68], KCNJ11 (homozygous) [69] and TCF7 (heterozygous) [70]. Consistent with the elevated hypertriglyceridemia risk, triglycerides were found to be high (321 mg/dl) at the beginning of the study and these levels were reduced (81–116 mg/dl) after regularly taking simvastatin (20 mg/day). Although the subject lacked many known factors associated with diabetes (nonsmoker, normal BMI) and for that reason usually would not have been screened, monitoring of glucose levels and glycated hemoglobin revealed the onset of Type 2 diabetes during study follow-up as diagnosed by the subject’s physician. Interestingly, the participant possessed two genotypes in the LPIN1 and SLC22A1 genes associated with favorable responses to two diabetic drugs (rosiglitazone and metformin). Nonetheless, after dramatic changes in diet, exercise and ingestion of low doses of acetylsalicylic acid, gradual decreases in glucose and glycated hemoglobin levels were observed and no auxiliary pharmacological agents were prescribed.
Pharmacogenomic sequencing
In addition to drugs relevant to diabetes, the subject described by Chen et al. had pharmacogenomic variants such as that of VKORC1 (C/T) associated with a low maintenance dose of warfarin and CYP2C19, which has been associated with increased risk of bleeding on standard doses of clopidogrel. There were also variants associated with slow metabolism of codeine, increased risk of neurological adverse events and Stevens–Johnson syndrome with carbamazepine and increased risk of adverse effects with methotrexate, among others [55]. The subject described by Ashley et al. similarly carried the VKORC1 variant (C/T) for low warfarin maintenance dose, and variants in CYP4F2 associated with reduced warfarin dosing, ADRB1 suggesting favorable response to atenolol, HMGCR associated with favorable response to statins, and CDKN2A/B suggesting reduced likelihood of response to metformin and troglitazone, among others [54]. In each case, these findings could impact the choice or dosing of medications in these individuals, should any of the impacted drugs be indicated in future medical management. In both cases, the pharmacogenomics variants were annotated based on the Pharmacogenomics Knowledge Base (PharmGKB), a publicly available web-based knowledge base [71]. It contains data from approximately 2500 variants, from which approximately 650 are specifically related to drug response phenotypes, each of which are assigned levels of evidence through literature review by database curators [54]. It represents one of the most up to date sources of human genetic variation as relevant to drug response. There are a number of databases accumulating pharmacogenomic information, including PharmaADME [72,73], the human cytochrome P450 (CYP) allele nomenclature web-site [72,74], the human arylamine N-acetyltransferase (NAT) gene nomenclature website [72,75], Pharmacogenetics of Membrane Transporters (PMT) database [72,76], Transporter Database (TP-search) [72,77], the UDP-glucuronosyltransferase (UGT) Allele Nomenclature Page [72,78], and PACdb [79,80], among others. The information compiled by these and other sources is anticipated to play an ever growing role in guiding patient care in conjunction with personal sequencing. While these pilot studies were performed in ostensibly healthy individuals, similar sequence analysis has clear potential relevance in individuals for whom any one of the above mentioned drugs may be indicated for a known medical condition.
The described pilots in single individuals should be replicated in greater numbers, potentially leading the way towards more specific upstream screening for risk factors and diseases. Additional genetic variants are known and have been validated to be of potential clinical relevance, such as the Val174Ala allele in the SLCO1B1 gene for statin-induced myopathy [81,82]. HLA B*5701 has been associated with slow or nonprogression of HIV infection and with hypersensitivity reactions to abacavir [83–85]. Therefore, most treatment guidelines recommend that upon considering administration of abacavir, patients should be tested for the presence of this allele, and that those who are positive should not receive the drug. Since the widespread introduction of HLA B*5701 testing, the incidence of hypersensitivity reactions in those receiving abacavir has dropped substantially [86]. Recently, it was shown in a large cohort that HLA B*5701-positive patients were more likely to achieve viral suppression than negative patients on a nonabacavir regimen and less likely to experience viral rebound [87]. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines have been published regarding the use of CYP2D6 and CYP2C19 genotyping test results to modify patient dosing of tricyclic antidepressants, such as amitriptyline and nortriptyline [88]. The effect of an individual patient’s CYP genotype on metabolism of these tricyclic agents can be taken into consideration at initial dosing, in an effort to maximize the efficacy in utilizing these medications for such indications as depression, obsessive compulsive disorder, migraine prophylaxis and neuropathic pain management while minimizing the associated anticholinergic, CNS and cardiac adverse effects [88]. With these and other examples of pharmacogenomic indications, the FDA currently includes pharmacogenomic information in the drug labels of approximately 100 approved drugs (Table 2), in addition to the 40 oncology drugs mentioned previously (Table 1).
Table 2.
US FDA-approved drugs with package inserts containing pharmacogenetics and pharmacogenomics information.
| Drug | Disease type | Pharmacogenomic biomarker(s) |
|---|---|---|
| Abacavir | Infectious diseases | HLA-B |
| Amitriptyline | Psychiatry | CYP2D6 |
| Aripiprazole | Psychiatry | CYP2D6 |
| Atomoxetine | Psychiatry | CYP2D6 |
| Atorvastatin | Endocrinology | LDLR |
| Azathioprine | Rheumatology | TPMT |
| Belimumab | Autoimmune diseases | BTG3 |
| Boceprevir | Infectious diseases | IFNL3 |
| Carbamazepine | Neurology | HLA-B, HLA-A |
| Carglumic acid | Metabolic disorders | NAGS |
| Carisoprodol | Rheumatology | CYP2C19 |
| Carvedilol | Cardiology | CYP2D6 |
| Celecoxib | Rheumatology | CYP2C9 |
| Cevimeline | Dermatology | CYP2D6 |
| Chloroquine | Infectious diseases | G6PD |
| Chlorpropamide | Endocrinology | G6PD |
| Citalopram | Psychiatry | CYP2C19, CYP2D6 |
| Clobazam | Neurology | CYP2C19 |
| Clomipramine | Psychiatry | CYP2D6 |
| Clopidogrel | Cardiology | CYP2C19 |
| Clozapine | Psychiatry | CYP2D6 |
| Codeine | Anesthesiology | CYP2D6 |
| Dapsone | Dermatology, infectious diseases | G6PD |
| Desipramine | Psychiatry | CYP2D6 |
| Dexlansoprazole | Gastroenterology | CYP2C19, CYP1A2 |
| Dextromethorphan and quinidine | Neurology | CYP2D6 |
| Diazepam | Psychiatry | CYP2C19 |
| Doxepin | Psychiatry | CYP2D6 |
| Drospirenone and ethinyl estradiol | Neurology | CYP2C19 |
| Eltrombopag | Hematology | F5, SERPINC1 |
| Esomeprazole | Gastroenterology | CYP2C19 |
| Fluorouracil | Dermatology | DPYD |
| Fluoxetine | Psychiatry | CYP2D6 |
| Flurbiprofen | Rheumatology | CYP2C9 |
| Fluvoxamine | Psychiatry | CYP2D6 |
| Galantamine | Neurology | CYP2D6 |
| Glimepiride | Endocrinology | G6PD |
| Glipizide | Endocrinology | G6PD |
| Glyburide | Endocrinology | G6PD |
| Iloperidone | Psychiatry | CYP2D6 |
| Imipramine | Psychiatry | CYP2D6 |
| Indacaterol | Pulmonary | UGT1A1 |
| Isosorbide and hydralazine | Cardiology | NAT1–2 |
| Ivacaftor | Pulmonary | CFTR |
| Lansoprazole | Gastroenterology | CYP2C19 |
| Lenalidomide | Hematology | del (5q) |
| Lomitapide | Endocrinology | LDLR |
| Mafenide | Infectious diseases | G6PD |
| Maraviroc | Infectious diseases | CCR5 |
| Methylene blue | Hematology | G6PD |
| Metoclopramide | Gastroentrology | CYB5R1–4 |
| Metoprolol | Cardiology | CYP2D6 |
| Mipomersen | Endocrinology | LDLR |
| Modafinil | Psychiatry | CYP2D6 |
| Mycophenolic acid | Transplantation | HPRT1 |
| Nalidixic acid | Infectious diseases | G6PD |
| Nefazodone | Psychiatry | CYP2D6 |
| Nitrofurantoin | Infectious diseases | G6PD |
| Nortriptyline | Psychiatry | CYP2D6 |
| Omeprazole | Gastroenterology | CYP2C19 |
| Pantoprazole | Gastroenterology | CYP2C19 |
| Paroxetine | Psychiatry | CYP2D6 |
| PEG-3350 | Gastroenterology | G6PD |
| Peginterferon alfa-2b | Infectious diseases | IFNL3 |
| Pegloticase | Rheumatology | G6PD |
| Perphenazine | Psychiatry | CYP2D6 |
| Phenytoin | Neurology | HLA-B |
| Pimozide | Psychiatry | CYP2D6 |
| Prasugrel | Cardiology | CYP2C19 |
| Pravastatin | Endocrinology | LDLR |
| Primaquine | Infectious diseases | G6PD |
| Propafenone | Cardiology | CYP2D6 |
| Propranolol | Cardiology | CYP2D6 |
| Protriptyline | Psychiatry | CYP2D6 |
| Quinidine | Cardiology | CYP2D6 |
| Quinine sulfate | Infectious diseases | G6PD |
| Rabeprazole | Gastroenterology | CYP2C19 |
| Rifampin, isoniazid and pyrazinamide | Infectious diseases | NAT1–2 |
| Risperidone | Psychiatry | CYP2D6 |
| Rosuvastatin | Endocrinology | LDLR |
| Simeprevir | Infectious diseases | IFNL3 |
| Sodium nitrite | Antidotal therapy | G6PD |
| Sofosbuvir | Infectious diseases | IFNL3 |
| Succimer | Hematology | G6PD |
| Sulfamethoxazole and trimethoprim | Infectious diseases | G6PD |
| Telaprevir | Infectious diseases | IFNL3 |
| Terbinafine | Infectious diseases | CYP2D6 |
| Tetrabenazine | Neurology | CYP2D6 |
| Thioridazine | Psychiatry | CYP2D6 |
| Ticagrelor | Cardiology | CYP2C19 |
| Tolterodine | Genitourinary | CYP2D6 |
| Tramadol | Analgesic | CYP2D6 |
| Trimipramine | Psychiatry | CYP2D6 |
| Valproic acid | Neurology | POLG, NAGS, CPS1, ASS1, OTC, ASL, ABL2 |
| Velaglucerase alfa | Metabolic disorders | GBA |
| Venlafaxine | Psychiatry | CYP2D6 |
| Voriconazole | Infectious diseases | CYP2C19 |
| Vortioxetine | Neurology | CYP2D6 |
| Warfarin | Cardiology, hematology | CYP2C9, VKORC1, PROC |
F2: Prothrombin; F5: Factor V Leiden.
Data taken from [32].
Information derived from personalized genome sequencing could point to undeveloped or concealed monogenic/oligogenic phenotypes where lifestyle and pharmacological interventions may minimize disease risks and future complications. Additionally, genome sequencing may provide evidence supporting the elevated risk for, or diagnosis of, complex disorders such as Alzheimer’s disease and prostate cancer, where genome sequencing has contributed novel findings [38–40]. Where multiple pharmacological options are available, the pharmacogenomic profile may guide more effective and less deleterious treatment decisions. To this end are more pharmacogenomic studies needed, as the associated variants form the basis for prediction of treatment response. It may become of increasing interest to invest in drug-response sequencing studies within clinical trials. Though it may seem unattractive to the pharmaceutical industry to identify those individuals genetically prone to adverse effects, this information can provide opportunities for the development of drugs with application to a broader population and drugs uniquely effective for significant individual cohorts, both in terms of tolerance and drug metabolism related individualized dosing.
Personal sequencing & family history
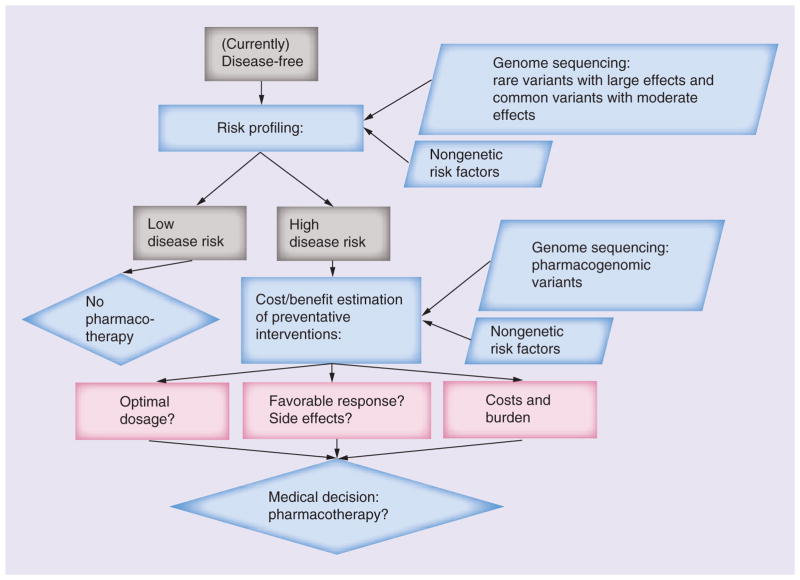
The patient interest and demand for personal sequencing seems poised to grow, for example in those individuals with a known family history of a particular condition as well as healthy, proactive individuals curious to learn about their genomic health and corresponding disease risks. Indeed on-demand genetic testing has already been commercially available for some time, including personal sequencing. As such, we anticipate increasing examples, such as those described above, of sequence analysis in individuals with subclinical or nondisease status, some having traditional risk factors (such as family history), which could contribute to improved prediction of who will not and who will ultimately develop disease and allow preventive measures to potentially avert disease in some (Figure 3).
Figure 3. Profiling rare diseases.
Issues and pipeline for personal genome sequencing in disease risk profiling of the ‘healthy’ state.
Understandably, there remains a significant amount of trepidation in the clinical community regarding genome sequencing in individuals as an adjunctive screen for risk of various types of disease. However, information obtained via a family history has long been an accepted and critical part of an individual’s clinical evaluation as a potent predictor of risk for certain diseases. WES/WGS represents a potentially more accurate means of determining what portion of the family history is specifically relevant to a particular individual’s health, and should be seen as a powerful supplement to the clinical family history.
Limitations of personalized sequencing
Nevertheless, WES and WGS are often performed with relatively low read depth, which results in data of insufficient quality to be directly used in clinical practice. Consequently, they can return thousands or tens of thousands of false-positive variants, necessitating validation by a separate platform such as Sanger sequencing or targeted amplicon sequencing with high read depth. As an example, for the Snyderome a variety of technologies and platforms manufactured by different distributors were applied to achieve deep sequencing. WGS by Complete Genomics (CA, USA; 35 nt paired end; 150-fold total coverage) and Illumina (CA, USA; 100 nt paired end; 120-fold total coverage). WES by Agilent (CA, USA), Illumina (CA, USA) and Nimblegen (WI, USA) at 80- to 100-fold coverage; crossvalidation with Illumina Omni1-Quad genotyping arrays (99.3% sensitivity); stringent data quality control and calling criteria; RNA sequencing by Illumina HiSeq with high depth; Sanger sequencing of randomly selected variants (36/36 single nucleotide variations validated; 14/15 indels validated).
In the majority of cases of clinically available WES, identified mutations thought to be of clinical relevance are confirmed via an independent platform prior to being reported with Sanger sequencing being the predominantly applied confirmatory platform. Sanger sequencing is held by many as the gold standard [89], while some have suggested conventional Sanger may no longer be the gold standard [90]. This appears in part related to the observation that some variants identified by WES/WGS are not confirmable by Sanger sequencing [91–93]. It should be noted that no platform is perfect as each has its own systematic weaknesses [90,94]. Nevertheless, while one clinical laboratory has begun to forgo Sanger confirmation for WES/WGS variants identified above a specified quality threshold, Sanger sequencing remains a relevant technique for validation of variants found by WES/WGS, as the systematic errors associated with each are different [95].
It is also important to realize that previously reported associations of genetic variants with disease may have suboptimal positive- and negative-predictive values. Some variants have been evaluated in this context with sufficient sample sizes in independent studies, but for many associations these statistics remain to be adequately determined. More research is needed to support high-quality evidence-based genomic medicine.
Sequencing & VUS: benefit-to-harm ratio
As application of personal sequencing expands, WES/WGS will yield an abundance of data including VUS [96,97]. Inadequate in silico prediction algorithms and incomplete penetrance are among the factors complicating clinical interpretation of these findings [44]. A ‘binning system’ has been proposed by which genetic variants can be ‘triaged’ in the clinical diagnostic setting to help address the field’s limited, though growing, understanding about most genetic variants, to facilitate focused attention on those variants demonstrated to have clinical implications [98]. Additional in vitro investigations, such as RNA expression and proteomic analyses, will be needed to confidently disregard a VUS or establish its association with a condition. As more sequencing data are becoming available, variants previously designated disease causing, benign or of uncertain significance are being reclassified. This theoretically adds to healthcare costs and the practical and mental burden of patients tested; these aspects need to be taken into consideration and require further investigation.
Ethical discussions surrounding DNA sequencing are ongoing and confidentiality of patient health information has become a serious issue [99]. Patients, but also the general public, should be properly educated about genome sequencing, its applications and limitations, enabling them to make informed health decisions. Informed consent and data sharing agreements should include clauses specifying to whom a patient wishes to grant access to his or her genomic data, while taking into consideration the individuals for whom the patient’s genomic data represents actionable clinical information. While these issues remain to be resolved, we are convinced that correct application of information resulting from personal sequencing will prove to be cost effective with a favorable benefit-to-harm ratio.
Sequencing patients: integration into clinical care
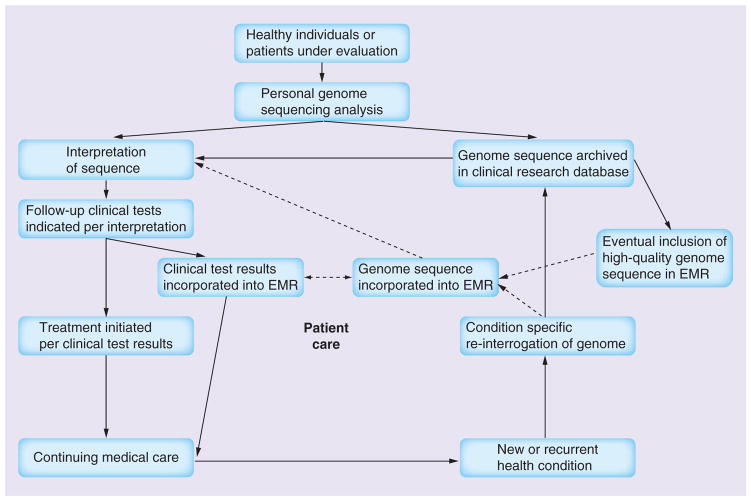
The genome is believed to be relatively stable throughout life and it will gradually become more attractive to retain data resulting from genome sequencing for future use. The cost–effectiveness of personalized exome and genome sequencing will improve with time, as the costs of sequence data generation, processing and storage decline and we will learn how to best utilize this information. For clinical care, it may become relevant to integrate genomic information into medical health records. Any information to be integrated into the electronic medical record (EMR) needs to be of high quality and accuracy, and the current quality of genomic sequence does not meet that level of rigor. An interim strategy would be to establish a clinical research database in which the full genome sequence and downstream analyses are stored and selected results from established, clinically related follow-up tests would get into the EMR (Figure 4). The selection criteria would require regular updating per advancing scientific understanding and the patients’ sequences would need to be regularly re-evaluated, gradually enriching the genomic information in the EMR. A pharmacogenomic example could be that automated alerts would notify the treating physician of genetically determined sensitivity to certain medications or suboptimal metabolism requiring drug dose adjustment, such as the Snyderome related variants impacting clopidogrel and warfarin efficacy [55].
Figure 4. Incorporation of the personal genome sequence into healthcare.
As genome sequence quality improves, future direct incorporation into the EMR will be possible, as indicated by the dashed arrows.
EMR: Electronic medical record.
Studies have also been designed for genotype-driven scans of the interaction of numerous nongenetic factors. A group of investigators lead by Atul Butte devised a design where a set of well-established genetic variants are screened for interactions with a great variety of nongenetic factors, termed ‘environment-wide association study’ [100–102]. One of the published studies found an association between SNP genotypes and β-carotenes, where β-carotenes seem to negate certain detrimental genetic effects and prevent genetically predisposed persons from getting Type 2 diabetes [101]. Whether β-carotene administration to persons with risk genotypes for Type 2 diabetes could actually prevent them from getting the disease remains to be proven. In the future it may even be possible to isolate cells from patients known to carry detrimental alleles and test responses to different compounds in vitro, which may support eventual medical decision-making.
Obstacles to EMR implementation
While personalized sequencing provides us with a wealth of genomic data including single nucleotide variations or SNPs and structural variations, it is key to keep in mind the current limitations of the technology, such as read depth and data quality, when interpreting the results. Confirmation by another reliable platform with high read depth remains necessary. Moreover, there is additive value in combining genomic sequencing information with RNA sequencing, transcriptomic, proteomic and metabolomic data. These omics analyses yield independent information about dynamic changes in health and disease states and are critical to correct interpretation of genomic variation and its clinical application [55,103,104].
Many of these clinical applications for personal sequencing remain in the near and distant future, as we continue to unravel the mysteries of the human genome [97]. As our understanding grows through discovery and validation studies the accuracy of medical genetic sequencing will improve. This will increase the need for carefully curated databases with well-corroborated genetic variants and reference genomes to support sequence interpretation. Analytical validation and evaluation studies with adequate study sample sizes and performed in different population groups are necessary to translate findings from the research realm into clinically validated tests. While the clinical applications of personal sequencing and validated medically actionable variants remains limited, their growing application to the practice of medicine is anticipated to accelerate.
Conclusion
Personalized sequencing has fast evolved to become a tool broadly applied in the study of disease and of increasing value in medical application to the diagnosis and treatment of disease. There is a growing need for improved methods, both in silico and in vitro, to predict the clinical impact of VUS identified in the process of large-scale sequencing. The quality of the currently generated WES and WGS is also in need of improvement, if it is to be incorporated in the future into an individual’s EMR as a reference.
Even with these obstacles, personalized sequencing has already begun to demonstrate its applicability to the practice of medicine. With it we have begun to better understand the etiologies of rare diseases and long studied diseases, such as cancer. Successes have been most evident in the field of rare Mendelian disorders where a single variant is sought to explain the phenotype in a patient, and can guide disease management by establishing the diagnosis. It demonstrates utility in refining and expanding our current diagnostic capacity such as through individual tumor DNA sequencing. Germline sequencing has also produced actionable information able to indicate lifestyle changes for an individual at risk for diabetes, as well as guiding pharmacologic interventions for the treatment of cancer, attuned to both tolerance of the patient and effectiveness of therapy, in a step towards personalized medicine. The application of personalized sequencing to clinically healthy individuals awaits the replication of recent studies in larger cohorts, but has the potential to be a medically valuable application in the not too distant future. As implementation of personalized sequencing on a large-scale is becoming progressively achievable, and accuracy of interpretation is significantly improved, we expect a transformation of healthcare in its current form.
Future perspective
With the rapid advances that have taken place in personalized sequencing over the past 5 years, it is difficult to predict its overall impact on the field of medicine in the coming 5–10 years, though we expect its impact to be significant. For example, while current WES data quality and interpretation remain currently inadequate to the task, there exists the future possibility that personal sequencing technology might be applied in clinically healthy individuals, both with and without known predispositions to disease, as a screening method to detect preclinical conditions, such as cancer, and facilitate pre-emptive treatment, with the potential to abrogate progression of otherwise clinically undetectable disease. It should be noted that, while we feel the current sequence quality and associated interpretative capacity require significant advancement prior to this type of clinical application, asymptomatic adult whole-exome screening tests are already being offered by CLIA-certified facilities (i.e., ‘adult screening exome sequencing’) [105], emphasizing the urgent need for advancement in sequence quality and interpretive acuity. Where WES is currently available on a clinical basis, we would predict WGS to be clinically available in 5–10 years’ time, as an evaluation that is routinely ordered by physicians. As broader utilization of EMRs takes hold in the practice of medicine and the quality of WGS continues to improve, we envision an individual’s genomic sequence becoming archived as an accessible part of their EMR in as little as 10 years’ time, for physicians to reference as a part of patient care.
In this era of medicine where medical practitioners include intensivists who treat diseases at their critical extremity and interventionalists who utilize invasive techniques, often at significant expense and morbidity, to ameliorate the complications of advanced sequelae of preventable disease, we envision a future in which personalized genomic sequencing enables the emergence of a new breed of medical practitioner: the preventional geneticist. Personalized sequencing portends a future for medicine where specialized health-care providers, preventional geneticists, carefully interpreting and applying an individual’s genomic profile can foresee their potential for various major diseases, such as diabetes and cancer. This information can then be used by preventional geneticists, possibly prior to clinical onset of these conditions, to institute surveillance, lifestyle changes and even preemptive pharmacogenomic-based therapeutics, with the potential to delay disease sequelae, and ultimately prevent disease onset in its entirety, defeating disease. Personalized sequencing represents a first major step toward this revolutionary future for medicine.
Executive summary.
Background
Personalized sequencing has advanced in scale, speed and affordability to become a powerful tool in the study of individuals and their diseases.
Cancer genome sequencing
The diagnosis and management of various forms of cancer has benefited from two forms of personalized sequencing: tumor DNA and germline. These have enabled the beginning of individualized therapy of cancer.
Sequencing the unknown: rare diseases
Rare and ‘mystery’ diseases as targets for personalized sequencing have yielded etiologies for previously undiagnosed diseases, in rare cases capacitating effective treatment.
Sequencing the ‘healthy’
At least one study analyzing the genomic sequence of a clinically healthy individual found a predisposition to diabetes, pharmacological interventions to which the individual would favorably respond, and allowed lifestyle changes to prevent the disease’s onset.
Several obstacles impede the current clinical application of personalized sequencing, one of which is the technological limitations of sequencing accuracy, where much progress is needed to allow transition to clinical medicine.
Interpretation of the genomic sequence currently remains a significant impediment to clinical applications. While many approaches are under development, new in silico and in vitro strategies are critical to understanding genomic data with acuity sufficient for clinical decision-making.
Variant(s) of unknown significance uncovered during sequencing will require additional interrogation via RNA expression, proteomic, metabolomic and other functional analyses, to facilitate accurate classification as benign and disease causing.
Sequencing patients: integration into clinical care
An individual’s personalized sequence may eventually become a valuable part of their electronic medical record, to be referenced periodically in the identification and management of disease.
Future perspective
Personalized sequencing represents a major step toward a revolutionary future of disease treatment, prediction and prevention in the practice of medicine.
Acknowledgments
The authors thank the members of the Snyder laboratory and our collaborators in the study of applications for genome sequencing in personalized medicine.
Footnotes
Financial & competing interests disclosure
ED Esplin holds the Tashia and John Morgridge Endowed Post-doctoral Fellowship and is supported by the Child Health Research Institute and the Stanford CTSA grant UL1 TR000093. This work was supported by NIH grants (NIH National Center for Advancing Translational Sciences [NCATS] Grant UL1 TR000093 to ED Esplin and NIH National Institute of Diabetes and Digestive and Kidney Diseases [NIDDK] Grant U54 DK102556 to MP Snyder. MP Snyder serves as founder and consultant for Personalis, and is a member of the scientific advisory board of GenapSys and a consultant for Illumina. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
For reprint orders, please contact: reprints@futuremedicine.com
References
Papers of special note have been highlighted as:
• of interest;
•• of considerable interest
- 1.Collins FS, Green ED, Guttmacher AE, Guyer MS. A vision for the future of genomics research. Nature. 2003;422(6934):835–847. doi: 10.1038/nature01626. [DOI] [PubMed] [Google Scholar]
- 2.Bozic I, Antal T, Ohtsuki H, et al. Accumulation of driver and passenger mutations during tumor progression. Proc Natl Acad Sci USA. 2010;107(43):18545–18550. doi: 10.1073/pnas.1010978107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3••.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. Overview of genome sequencing’s impact on cancer research. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4•.Ley TJ, Mardis ER, Ding L, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456(7218):66–72. doi: 10.1038/nature07485. Early study comparing whole-genome sequencing (WGS) of patient’s tumor cell and normal cell DNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garraway LA, Lander ES. Lessons from the cancer genome. Cell. 2013;153(1):17–37. doi: 10.1016/j.cell.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 6.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194(4260):23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 7.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McLeod HL. Cancer pharmacogenomics: early promise, but concerted effort needed. Science. 2013;339(6127):1563–1566. doi: 10.1126/science.1234139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kreso A, O’Brien CA, Van Galen P, et al. Variable clonal repopulation dynamics influence chemotherapy response in colorectal cancer. Science. 2013;339(6119):543–548. doi: 10.1126/science.1227670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7(3):169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 11.Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364(26):2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amado RG, Wolf M, Peeters M, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26(10):1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 13.Irshad S, Ashworth A, Tutt A. Therapeutic potential of PARP inhibitors for metastatic breast cancer. Expert Rev Anticancer Ther. 2011;11(8):1243–1251. doi: 10.1586/era.11.52. [DOI] [PubMed] [Google Scholar]
- 14.Lord CJ, Ashworth A. Mechanisms of resistance to therapies targeting BRCA-mutant cancers. Nat Med. 2013;19(11):1381–1388. doi: 10.1038/nm.3369. [DOI] [PubMed] [Google Scholar]
- 15.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481(7381):306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ashworth A, Lord CJ, Reis-Filho JS. Genetic interactions in cancer progression and treatment. Cell. 2011;145(1):30–38. doi: 10.1016/j.cell.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 17.Rottenberg S, Jaspers JE, Kersbergen A, et al. High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proc Natl Acad Sci USA. 2008;105(44):17079–17084. doi: 10.1073/pnas.0806092105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gabree M, Seidel M. Genetic testing by cancer site: skin. Cancer J. 2012;18(4):372–380. doi: 10.1097/PPO.0b013e3182624664. [DOI] [PubMed] [Google Scholar]
- 19.Shannon KM, Chittenden A. Genetic testing by cancer site: breast. Cancer J. 2012;18(4):310–319. doi: 10.1097/PPO.0b013e318260946f. [DOI] [PubMed] [Google Scholar]
- 20.Senter L. Genetic testing by cancer site: colon (nonpolyposis syndromes) Cancer J. 2012;18(4):334–337. doi: 10.1097/PPO.0b013e31826094b2. [DOI] [PubMed] [Google Scholar]
- 21.Jasperson KW. Genetic testing by cancer site: colon (polyposis syndromes) Cancer J. 2012;18(4):328–333. doi: 10.1097/PPO.0b013e3182609300. [DOI] [PubMed] [Google Scholar]
- 22.Chun N, Ford JM. Genetic testing by cancer site: stomach. Cancer J. 2012;18(4):355–363. doi: 10.1097/PPO.0b013e31826246dc. [DOI] [PubMed] [Google Scholar]
- 23.Weissman SM, Weiss SM, Newlin AC. Genetic testing by cancer site: ovary. Cancer J. 2012;18(4):320–327. doi: 10.1097/PPO.0b013e31826246c2. [DOI] [PubMed] [Google Scholar]
- 24.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 25.Roberts NJ, Jiao Y, Yu J, et al. ATM mutations in patients with hereditary pancreatic cancer. Cancer Discov. 2012;2(1):41–46. doi: 10.1158/2159-8290.CD-11-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palles C, Cazier JB, Howarth KM, et al. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet. 2013;45(2):136–144. doi: 10.1038/ng.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paugh SW, Stocco G, McCorkle JR, Diouf B, Crews KR, Evans WE. Cancer pharmacogenomics. Clin Pharmacol Ther. 2011;90(3):461–466. doi: 10.1038/clpt.2011.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28••.Wheeler HE, Maitland ML, Dolan ME, Cox NJ, Ratain MJ. Cancer pharmacogenomics: strategies and challenges. Nat Rev Genet. 2013;14(1):23–34. doi: 10.1038/nrg3352. Overview of pharmacogenomic applications to cancer treatment with relevant examples. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Binkhorst L, Van Gelder T, Mathijssen RH. Individualization of tamoxifen treatment for breast carcinoma. Clin Pharmacol Ther. 2012;92(4):431–433. doi: 10.1038/clpt.2012.94. [DOI] [PubMed] [Google Scholar]
- 30.Weng L, Zhang L, Peng Y, Huang RS. Pharmacogenetics and pharmacogenomics: a bridge to individualized cancer therapy. Pharmacogenomics. 2013;14(3):315–324. doi: 10.2217/pgs.12.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wheeler DA, Wang L. From human genome to cancer genome: the first decade. Genome Res. 2013;23(7):1054–1062. doi: 10.1101/gr.157602.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32•.Table of Pharmacogenomic Biomarkers in Drug Labeling. www.fda.gov/drugs/scienceresearch/researchareas/pharmacogenetics/ucm083378.htmOnline resource providing pharmacogenomics information in all US FDA-approved drug labels.
- 33.Sharaf RN, Ladabaum U. Comparative effectiveness and cost–effectiveness of screening colonoscopy vs. sigmoidoscopy and alternative strategies. Am J Gastroenterol. 2013;108(1):120–132. doi: 10.1038/ajg.2012.380. [DOI] [PubMed] [Google Scholar]
- 34.Dunn BK, Jegalian K, Greenwald P. Biomarkers for early detection and as surrogate endpoints in cancer prevention trials: issues and opportunities. Recent Results Cancer Res. 2011;188:21–47. doi: 10.1007/978-3-642-10858-7_3. [DOI] [PubMed] [Google Scholar]
- 35.Kitzman JO, Snyder MW, Ventura M, et al. Noninvasive whole-genome sequencing of a human fetus. Sci Transl Med. 2012;4(137):137ra176. doi: 10.1126/scitranslmed.3004323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benesova L, Belsanova B, Suchanek S, et al. Mutation-based detection and monitoring of cell-free tumor DNA in peripheral blood of cancer patients. Anal Biochem. 2013;433(2):227–234. doi: 10.1016/j.ab.2012.06.018. [DOI] [PubMed] [Google Scholar]
- 37.Bainbridge MN, Wiszniewski W, Murdock DR, et al. Whole-genome sequencing for optimized patient management. Sci Transl Med. 2011;3(87):87re83. doi: 10.1126/scitranslmed.3002243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gudmundsson J, Sulem P, Gudbjartsson DF, et al. A study based on whole-genome sequencing yields a rare variant at 8q24 associated with prostate cancer. Nat Genet. 2012;44(12):1326–1329. doi: 10.1038/ng.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jonsson T, Atwal JK, Steinberg S, et al. A mutation in APP protects against Alzheimer’s disease and age-related cognitive decline. Nature. 2012;488(7409):96–99. doi: 10.1038/nature11283. [DOI] [PubMed] [Google Scholar]
- 40.Jonsson T, Stefansson H, Steinberg S, et al. Variant of TREM2 associated with the risk of Alzheimer’s disease. N Engl J Med. 2013;368(2):107–116. doi: 10.1056/NEJMoa1211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lupski JR, Reid JG, Gonzaga-Jauregui C, et al. Whole-genome sequencing in a patient with Charcot-Marie-Tooth neuropathy. N Engl J Med. 2010;362(13):1181–1191. doi: 10.1056/NEJMoa0908094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ng SB, Buckingham KJ, Lee C, et al. Exome sequencing identifies the cause of a Mendelian disorder. Nat Genet. 2010;42(1):30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peltonen L, Perola M, Naukkarinen J, Palotie A. Lessons from studying monogenic disease for common disease. Hum Mol Genet. 2006;15(Spec 1):R67–R74. doi: 10.1093/hmg/ddl060. [DOI] [PubMed] [Google Scholar]
- 44.Kohane IS. (Mis)treating the pharmacogenetic incidentalome. Nat Rev Drug Discov. 2012;11(2):89–90. doi: 10.1038/nrd3659. [DOI] [PubMed] [Google Scholar]
- 45•.Worthey EA, Mayer AN, Syverson GD, et al. Making a definitive diagnosis: successful clinical application of whole exome sequencing in a child with intractable inflammatory bowel disease. Genet Med. 2011;13(3):255–262. doi: 10.1097/GIM.0b013e3182088158. Early example of whole-exome sequencing (WES) application to diagnose a rare disease and guide effective medical treatment. [DOI] [PubMed] [Google Scholar]
- 46.Bainbridge MN, Wiszniewski W, Murdock DR, et al. Whole-genome sequencing for optimized patient management. Sci Transl Med. 2011;3(87):87re83. doi: 10.1126/scitranslmed.3002243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bamshad MJ, Ng SB, Bigham AW, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011;12(11):745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 48.Norton N, Li D, Hershberger RE. Next-generation sequencing to identify genetic causes of cardiomyopathies. Curr Opin Cardiol. 2012;27(3):214–220. doi: 10.1097/HCO.0b013e328352207e. [DOI] [PubMed] [Google Scholar]
- 49•.Green RC, Berg JS, Grody WW, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Am Coll Med Genet Genomics. 2013;15(7):565–574. doi: 10.1038/gim.2013.73. Current clinical guidelines for communication of results, including incidental findings, of WES and WGS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Need AC, Shashi V, Hitomi Y, et al. Clinical application of exome sequencing in undiagnosed genetic conditions. J Med Genet. 2012;49(6):353–361. doi: 10.1136/jmedgenet-2012-100819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang Y, Muzny DM, Reid JG, et al. Clinical whole-exome sequencing for the diagnosis of Mendelian disorders. N Engl J Med. 2013;369(16):1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Atwal PS, Brennan ML, Cox R, et al. Clinical whole-exome sequencing: are we there yet? Genet Med. 2014 doi: 10.1038/gim.2014.10. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 53.Dewey FE, Grove ME, Pan C, et al. Clinical interpretation and implications of whole-genome sequencing. JAMA. 2014;311(10):1035–1045. doi: 10.1001/jama.2014.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54••.Ashley EA, Butte AJ, Wheeler MT, et al. Clinical assessment incorporating a personal genome. Lancet. 2010;375(9725):1525–1535. doi: 10.1016/S0140-6736(10)60452-7. First sequencing of a healthy individual’s genome to predict risk of disease and identify pharmacogenomic profile. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55••.Chen R, Mias GI, Li-Pook-Than J, et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell. 2012;148(6):1293–1307. doi: 10.1016/j.cell.2012.02.009. First example of personalized sequencing combined with omics analyses, which predicted onset of diabetes in an individual and enabled successful intervention. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gupta R, Ratan A, Rajesh C, et al. Sequencing and analysis of a South Asian-Indian personal genome. BMC Genomics. 2012;13:440. doi: 10.1186/1471-2164-13-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peterson TA, Doughty E, Kann MG. Towards precision medicine: advances in computational approaches for the analysis of human variants. J Mol Biol. 2013;425(21):4047–4063. doi: 10.1016/j.jmb.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Online Mendelian Inheritance in Man. www.omim.org.
- 59.Stenson PD, Mort M, Ball EV, Shaw K, Phillips A, Cooper DN. The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet. 2014;133(1):1–9. doi: 10.1007/s00439-013-1358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.The Human Gene Mutation Database. www.hgmd.org.
- 61.ClinVar. www.ncbi.nlm.nih.gov/clinvar.
- 62.European Genome-Phenome Archive. www.ebi.ac.uk/ega.
- 63.Mailman MD, Feolo M, Jin Y, et al. The NCBI dbGaP database of genotypes and phenotypes. Nat Genet. 2007;39(10):1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.dbGap. www.ncbi.nlm.nih.gov/gap.
- 65.Hindorff LA, Sethupathy P, Junkins HA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA. 2009;106(23):9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Welter D, MacArthur J, Morales J, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yamaguchi H, Calado RT, Ly H, et al. Mutations in TERT, the gene for telomerase reverse transcriptase, in aplastic anemia. N Engl J Med. 2005;352(14):1413–1424. doi: 10.1056/NEJMoa042980. [DOI] [PubMed] [Google Scholar]
- 68.Vaxillaire M, Cavalcanti-Proenca C, Dechaume A, et al. The common P446L polymorphism in GCKR inversely modulates fasting glucose and triglyceride levels and reduces Type 2 diabetes risk in the DESIR prospective general French population. Diabetes. 2008;57(8):2253–2257. doi: 10.2337/db07-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hani EH, Boutin P, Durand E, et al. Missense mutations in the pancreatic islet beta cell inwardly rectifying K+ channel gene (KIR6.2/BIR): a meta-analysis suggests a role in the polygenic basis of Type II diabetes mellitus in Caucasians. Diabetologia. 1998;41(12):1511–1515. doi: 10.1007/s001250051098. [DOI] [PubMed] [Google Scholar]
- 70.Erlich HA, Valdes AM, Julier C, Mirel D, Noble JA. Evidence for association of the TCF7 locus with Type I diabetes. Genes Immun. 2009;10(Suppl 1):S54–S59. doi: 10.1038/gene.2009.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.PharmGKB. Pharmacogenomics. Knowledge. Implementation. www.pharmgkb.org.
- 72.Sim SC, Altman RB, Ingelman-Sundberg M. Databases in the area of pharmacogenetics. Hum Mutat. 2011;32(5):526–531. doi: 10.1002/humu.21454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.PharmaADME. www.pharmaadme.org.
- 74.The Human Cytochrome P450 (CYP) Allele Nomenclature Database. www.cypalleles.ki.se.
- 75.University of Louisville School of Medicine: Department of Pharmacology and Toxicology. http://n-acetyltransferasenomenclature.louisville.edu.
- 76.Pharmacogenetics of Membrane Transporters Database. http://pharmacogenetics.ucsf.edu.
- 77.TP-Search. www.tp-search.jp.
- 78.UGT1A and UGT2B haplotypes and SNPs tables. www.pharmacogenomics.pha.ulaval.ca/sgc/ugt_alleles.
- 79.Gamazon ER, Duan S, Zhang W, et al. PACdb: a database for cell-based pharmacogenomics. Pharmacogenet Genomics. 2010;20(4):269–273. doi: 10.1097/FPC.0b013e328337b8d6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.PACdb. www.pacdb.org.
- 81.Group SC, Link E, Parish S, et al. SLCO1B1 variants and statin-induced myopathy – a genomewide study. N Engl J Med. 2008;359(8):789–799. doi: 10.1056/NEJMoa0801936. [DOI] [PubMed] [Google Scholar]
- 82.Feng Q, Wilke RA, Baye TM. Individualized risk for statin-induced myopathy: current knowledge, emerging challenges and potential solutions. Pharmacogenomics. 2012;13(5):579–594. doi: 10.2217/pgs.12.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mallal S, Nolan D, Witt C, et al. Association between presence of HLA-B*5701, HLA-DR7, and HLA-DQ3 and hypersensitivity to HIV-1 reverse-transcriptase inhibitor abacavir. Lancet. 2002;359(9308):727–732. doi: 10.1016/s0140-6736(02)07873-x. [DOI] [PubMed] [Google Scholar]
- 84.Hughes AR, Mosteller M, Bansal AT, et al. Association of genetic variations in HLA-B region with hypersensitivity to abacavir in some, but not all, populations. Pharmacogenomics. 2004;5(2):203–211. doi: 10.1517/phgs.5.2.203.27481. [DOI] [PubMed] [Google Scholar]
- 85.Mallal S, Phillips E, Carosi G, et al. HLA-B*5701 screening for hypersensitivity to abacavir. N Engl J Med. 2008;358(6):568–579. doi: 10.1056/NEJMoa0706135. [DOI] [PubMed] [Google Scholar]
- 86.Waters LJ, Mandalia S, Gazzard B, Nelson M. Prospective HLA-B*5701 screening and abacavir hypersensitivity: a single centre experience. AIDS. 2007;21(18):2533–2534. doi: 10.1097/QAD.0b013e328273bc07. [DOI] [PubMed] [Google Scholar]
- 87.UK Collaborative HIV Cohort Study Steering Committee. HLA B*5701 status, disease progression, and response to antiretroviral therapy. AIDS. 2013;27(16):2587–2592. doi: 10.1097/01.aids.0000432613.95455.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hicks JK, Swen JJ, Thorn CF, et al. Clinical Pharmacogenetics Implementation Consortium guideline for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants. Clin Pharmacol Ther. 2013;93(5):402–408. doi: 10.1038/clpt.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Macconaill LE. Existing and emerging technologies for tumor genomic profiling. J Clin Oncol. 2013;31(15):1815–1824. doi: 10.1200/JCO.2012.46.5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ulahannan D, Kovac MB, Mulholland PJ, Cazier JB, Tomlinson I. Technical and implementation issues in using next-generation sequencing of cancers in clinical practice. Br J Cancer. 2013;109(4):827–835. doi: 10.1038/bjc.2013.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Linderman MD, Brandt T, Edelmann L, et al. Analytical validation of whole exome and whole genome sequencing for clinical applications. BMC Med Genomics. 2014;7(1):20. doi: 10.1186/1755-8794-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.McCourt CM, McArt DG, Mills K, et al. Validation of next generation sequencing technologies in comparison to current diagnostic gold standards for BRAF, EGFR and KRAS mutational analysis. PLoS ONE. 2013;8(7):e69604. doi: 10.1371/journal.pone.0069604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chin EL, Da Silva C, Hegde M. Assessment of clinical analytical sensitivity and specificity of next-generation sequencing for detection of simple and complex mutations. BMC Genet. 2013;14:6. doi: 10.1186/1471-2156-14-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Esteban-Jurado C, Garre P, Vila M, et al. New genes emerging for colorectal cancer predisposition. World J Gastroenterol. 2014;20(8):1961–1971. doi: 10.3748/wjg.v20.i8.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Strom SP, Lee H, Das K, et al. Assessing the necessity of confirmatory testing for exome-sequencing results in a clinical molecular diagnostic laboratory. Genet Med. 2014;16(7):510–515. doi: 10.1038/gim.2013.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Henrikson NB, Burke W, Veenstra DL. Ancillary risk information and pharmacogenetic tests: social and policy implications. Pharmacogenomics J. 2008;8(2):85–89. doi: 10.1038/sj.tpj.6500457. [DOI] [PubMed] [Google Scholar]
- 97.Westbrook MJ, Wright MF, Van Driest SL, et al. Mapping the incidentalome: estimating incidental findings generated through clinical pharmacogenomics testing. Genet Med. 2013;15(5):325–331. doi: 10.1038/gim.2012.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98•.Berg JS, Khoury MJ, Evans JP. Deploying whole genome sequencing in clinical practice and public health: meeting the challenge one bin at a time. Genet Med. 2011;13(6):499–504. doi: 10.1097/GIM.0b013e318220aaba. Discussion of strategies for incorporation of personal sequence into clinical healthcare delivery. [DOI] [PubMed] [Google Scholar]
- 99.Gymrek M, McGuire AL, Golan D, Halperin E, Erlich Y. Identifying personal genomes by surname inference. Science. 2013;339(6117):321–324. doi: 10.1126/science.1229566. [DOI] [PubMed] [Google Scholar]
- 100.Patel CJ, Chen R, Butte AJ. Data-driven integration of epidemiological and toxicological data to select candidate interacting genes and environmental factors in association with disease. Bioinformatics. 2012;28(12):i121–i126. doi: 10.1093/bioinformatics/bts229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Patel CJ, Bhattacharya J, Butte AJ. An environment-wide association study (EWAS) on Type 2 diabetes mellitus. PLoS ONE. 2010;5(5):e10746. doi: 10.1371/journal.pone.0010746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Patel CJ, Cullen MR, Ioannidis JP, Butte AJ. Systematic evaluation of environmental factors: persistent pollutants and nutrients correlated with serum lipid levels. Int J Epidemiol. 2012;41(3):828–843. doi: 10.1093/ije/dys003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Snyder M, Weissman S, Gerstein M. Personal phenotypes to go with personal genomes. Mol Syst Biol. 2009;5:273. doi: 10.1038/msb.2009.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Snyder M, Du J, Gerstein M. Personal genome sequencing: current approaches and challenges. Genes Dev. 2010;24(5):423–431. doi: 10.1101/gad.1864110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Medical Genetics Test Details. www.bcm.edu/research/medical-genetics-labs/test_detail.cfm?testcode=1605.