SUMMARY
Chromobox homolog 7 (CBX7) plays an important role in gene transcription in a wide array of cellular processes, ranging from stem cell self-renewal and differentiation to tumor progression. CBX7 functions through its N-terminal chromodomain (ChD), which recognizes tri-methylated lysine 27 of histone 3 (H3K27me3), a conserved epigenetic mark that signifies gene transcriptional repression. In this study, we report discovery of small molecules that inhibit CBX7ChD binding to H3K27me3. Our crystal structures reveal the binding modes of these molecules that compete against H3K27me3 binding through interactions with key residues in the methyl-lysine binding pocket of CBX7ChD. We further show that a lead compound MS37452, de-represses transcription of Polycomb repressive complex target gene p16/CDKN2A by displacing CBX7 binding to the INK4A/ARF locus in prostate cancer cells. These small molecules have the potential to be developed into high-potency chemical modulators that target CBX7 functions in gene transcription in different disease pathways.
INTRODUCTION
Polycomb group (PcG) proteins were first identified in Drosophila as silencers of transcription of the bithorax gene complex (BX-C), a group of homeotic (Hox) genes responsible for controlling segmentation (Lewis, 1978). PcG proteins also play a fundamental role in multicellular organism development and cancer progression (Sauvageau and Sauvageau, 2010). PcG proteins function in two main complexes: Polycomb repressive complex 2 (PRC2) deposits a repressive histone H3 lysine 27 methylation (H3K27me3) mark by EZH2; Polycomb repressive complex 1 (PRC1) recognizes H3K27me3 through chromodomain (ChD)-containing Chromobox homolog (CBX) proteins and ubiquitinates H2AK119 by RING1A/B. However, the precise mechanism by which PcG proteins are recruited to repress gene transcription is not fully understood (Simon and Kingston, 2009).
A key PRC1 protein, CBX7 was first identified in functional cDNA screening, designed to extend cell lifespan of normal human prostate epithelial cells by repressing the Ink4a/Arf locus (Gil et al., 2004). This locus encodes key regulators of both retinoblastoma tumor suppressor (Rb) and p53 (Sherr, 2001, 2006); p14Arf/p53- and p16Ink4a/Rb-dependent impairment of cell growth upon CBX7 knockdown highlights CBX7’s role in tumorigenesis (Bernard et al., 2005). CBX7 is also important in stem cell self-renewal and differentiation (Klauke et al., 2013; Morey et al., 2012).
Similar to other CBX proteins, CBX7 is characteristic of an N-terminal ChD responsible for recognition of the repressive mark, H3K27me3. The methyl-lysine binding pocket (also called the ‘aromatic cavity’) is formed by three aromatic residues (Phe11, Trp32, and Trp35 in CBX7), which are highly conserved among all ChD proteins (Bernstein et al., 2006; Kaustov et al., 2011). Mutations of these residues cause CBX7 dissociation from chromatin and alter lifespan of prostate epithelial cells, underscoring the importance of ChD/H3K27me3 binding for CBX7 activity (Gil et al., 2004; Morey et al., 2012; Yap et al., 2010). Thus, we postulated that small molecule disruption of CBX7ChD binding to H3K27me3 would inhibit the transcriptional activity of CBX7 and result in de-repression of its target genes. Such small molecules could be used to fine-tune a balance between stem cell self-renewal and differentiation, and also be developed into potential new therapeutics for cancer treatment. Despite the functional importance of CBX7 in gene regulation, only recently have macrocyclic calixarenes (Tabet et al., 2013) and peptidomimetics (Simhadri et al., 2014) been shown as CBX7ChD antagonists, but no small-molecule chemical inhibitors have been reported for CBX7ChD or other CBX ChDs. Here, we report discovery and characterization of small molecule chemical modulators of the CBX7ChD, and demonstrate that a lead compound, MS37452, induces transcriptional de-repression of p16/CDKN2A by disrupting CBX7ChD binding to H3K27me3 at the INK4A/ARF locus in PC3 prostate cancer cells.
RESULTS AND DISCUSSION
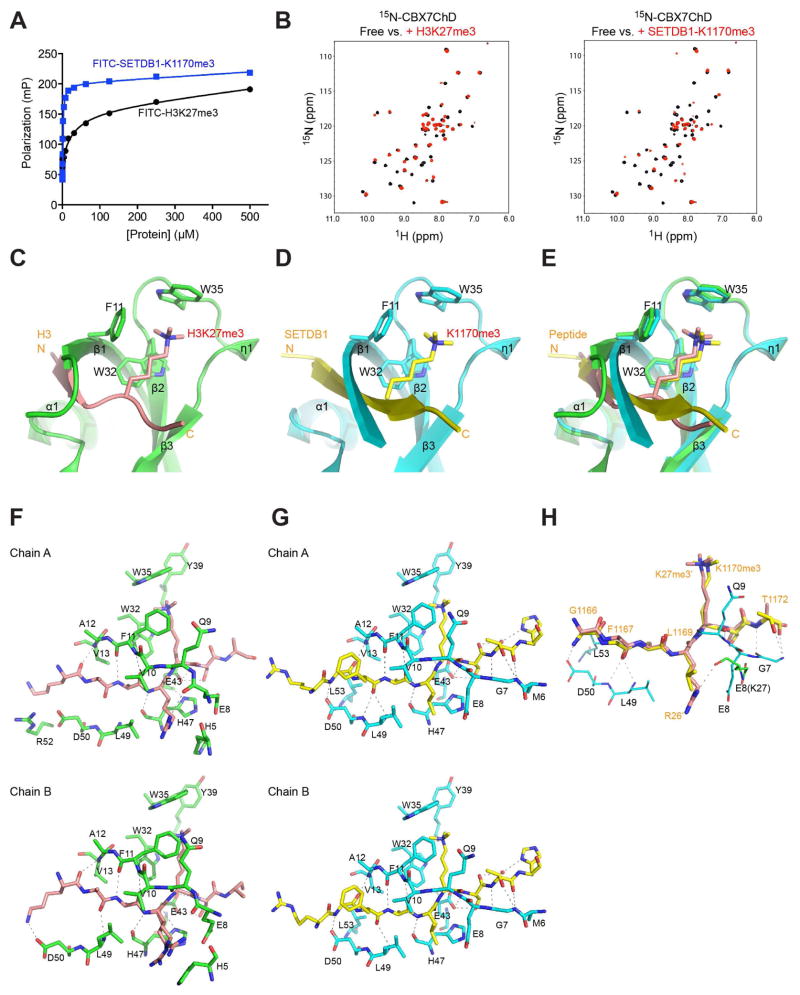
While an H3K27me3 peptide is a well-established biological ligand for CBX7ChD (Bernstein et al., 2006; Yap et al., 2010), its binding affinity is modest (Kd = 27.7 μM) (Figure 1A; Table S1), making it less ideal as a probe for high-throughput chemical screening. To address this issue, we evaluated another tri-methylated-lysine peptide of SETDB1 at K1170 (SETDB1K1170me3) reported to bind to CBX7ChD with higher affinity (Kaustov et al., 2011). Despite similar core sequences of ARKme3S and ALKme3S for H3K27me3 and SETDB1, respectively, the SETDB1 peptide binds to CBX7ChD with much higher affinity (Kd = 1.3 μM) (Figure 1A). The enhanced binding of the SETDB1 peptide is supported by 2D 1H-15N-HSQC (heteronuclear single quantum coherence) NMR spectra (Figure 1B), and also confirmed by a fluorescence anisotropy binding study (see Figure S2J), thus making SETDB1-1170me3 an attractive probe for HTS.
Figure 1. Structural analysis of H3K27me3 and SETDB1 recognition by CBX7ChD.
(A) Measurement of CBX7ChD binding to lysine-methylated H3 and SETDB1 peptides using a fluorescence anisotropy assay.
(B) 2D 1H-15N-HSQC spectra of 15N-CBX7ChD in free (black) or in the presence of H3K27me3 (red, left panel), or SETDB1-1170me3 peptide (red, right panel).
(C) Crystal structure of CBX7ChD bound to an H3K27me3 peptide. CBX7ChD is depicted in green, while H3K27me3 is depicted in salmon.
(D) Crystal structure of CBX7ChD bound to the SETDB1-K1170me3 peptide.
(E) Structural overlay of CBX7ChD bound to the H3K27me3 or SETDB1-K1170me3 peptide.
(F–G) Analysis of CBX7ChD bound to H3K27me3 or SETDB1-K1170me3, respectively. Only residues involved in hydrogen bond or hydrophobic interactions are shown.
(H) Comparison of SETDB1 and H3K27me3 binding to CBX7ChD.
See also Figure S1, and Tables S1 and S2.
To determine the molecular basis of CBX7ChD binding to these lysine-methylated peptides, we solved 1.45 Å- and 1.60 Å-resolution crystal structures of CBX7ChD bound to a H3K27me3 or SETDB1-K1170me3 peptide, respectively (Figure 1C, 1D; Table S2). In the methyl-lysine binding aromatic cavity, the indole of Trp35 recognizes three methyl groups of Kme3, while Phe11 and Trp32 form the base to interact with the acyl side chain of the methylated-lysine (Figure 1E, Figure S1A–S1C). The H3K27me3 peptide binds across the protein with residues Ala24-Ala25-Arg26-K27me3 forming a 2-strand anti-parallel β-sheet with the N-terminal Val10-Ala12 of the protein (Figure 1F). The CBX7ChD has an expanded interface for the SETDB1 peptide with a longer N-terminal β-strand comprised of Gly7-Ala12 (Figure 1D, 1G). This anti-parallel β-sheet is enforced by an extended network of intermolecular hydrogen bond interactions involving protein residues Gly7 and Asp50, as well as hydrophobic interactions with Leu53 that are absent in the H3K27me3-bound structure (Figure 1H). These findings explain the molecular basis for the enhanced affinity of CBX7ChD for SETDB1-K1170me3 over H3K27me3.
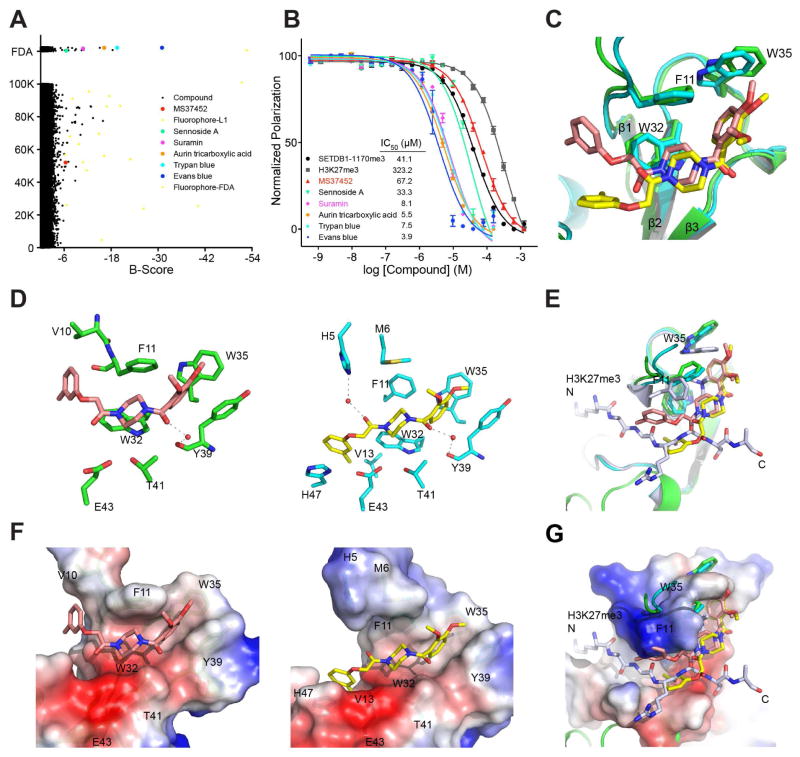
We next conducted high-throughput screening of a library of 2,560 FDA-approved drug molecules and an L1 library of 100,160 compounds selected from commercial sources to identify inhibitors for the CBX7ChD using a fluorescence anisotropy binding assay with a FITC-labeled SETDB1-K1170me3 peptide as an assay probe (Supplemental Information). B-score < −6 was set as the criterion to select initial hits, of which 16 were identified from the FDA library and 40 from the L1 library (Figure 2A). Of these initial hits, 5 hits from the FDA and 1 hit from the L1 library were confirmed as binders of the CBX7ChD by using 2D 1H-15N HSQC spectra (Figures S2A–H and S3) and the fluorescence anisotropy binding assay (Figure 2B).
Figure 2. Discovery of small molecule inhibitors of CBX7ChD.
(A) Hits identified from HTS and confirmed by 2D 1H-15N-HSQC spectra. Compounds from L1 (100K compounds) and FDA (2600 compounds) libraries were plotted according to B-score. All hits with B-score < −6 were further tested by 2D 1H-15N-HSQC spectra, of which the confirmed hits are highlighted with colored dots.
(B) Measurement of CBX7ChD binding to small molecules in a fluorescence polarization assay using FITC-labeled SETDB1-K1170me3 peptide as an assay probe. IC50 was determined using GraphPad Prism.
(C) Superimposition of crystal structures of CBX7ChD in complex with MS37452 in cis or trans conformation.
(D) Analysis of CBX7ChD binding to cis-MS37452 or trans-MS34752. Only residues involved in hydrophobic or H-bond interactions are shown.
(E) Structural comparison of the CBX7ChD bound to MS37452 or H3K27me3 peptide.
(F) Electrostatic potential surface representation of CBX7ChD bound to cis-MS37452 (left) or trans-MS37452 (right).
(G) Superimposition of the structures of CBX7ChD bound to H3K27me3 peptide or MS37452 illustrating conformational change of the Kme3 binding aromatic cage.
See also Figures S1 and S2, and Tables S2 and S3.
MS37452 was the only chemical hit confirmed from the L1 library with binding affinity (Kd) of 28.90 ± 2.71 μM to the CBX7ChD, as determined by NMR titration (Figure S2I). MS37452 disrupts CBX7-H3K27me3 or H3K9me3 binding with Ki of 43.0 and 55.3 μM, respectively, as shown in a fluorescence anisotropy binding assay (Figure S2J).
We then solved a 2.14 Å-resolution crystal structure of the CBX7ChD-MS37452 complex (Table S2). Strikingly, MS37452 adapts two rotamer conformations. The di-methoxyl-benzene and piperazine moieties of MS37452 are bound in the same orientation interacting with Phe11, Trp32 and Trp35 in the aromatic cavity, or sandwiched between Phe11 and Trp32, respectively, whereas the methylbenzene moiety adopts a “cis” or “trans” confirmation with respect to the di-methoxyl-benzene, hinged at the carbonyl that connects the methylbenzene to the piperazine ring (Figure 2C). Notably, in the “trans” conformation, the methylbenzene is positioned in the ChD similarly as H3K27me3 and SETDB1-K1170me3, forming interactions with His47, Val13 and Glu43 (Figure 2D–2G). On the other hand, in the “cis” confirmation, the methylbenzene swings upwards into the protein structure, disrupting the N-terminal β-strand and compromising the stability of the complex (Figure 2D left). The “trans” conformation is secured by water-mediated hydrogen bonding of the ligand’s two carbonyl groups with protein residues His5 (from His-tag) and Tyr39 (Figure 2D right). In either conformation, Trp35 is pushed outward to accommodate the di-methoxyl-benzene moiety within the aromatic cavity (Figure 2E, 2G). Point mutations of Phe11, Trp35, or Tyr39 to Ala almost completely abolished CBX7ChD-MS37452 binding (Figure S2I). These results confirmed that the aromatic cage residues and Tyr39 are critical to the CBX7ChD-MS37452 interaction.
We next examined the selectivity of MS37452 for different CBXChDs. MS37452 binds to a sub-group of CBXChDs, including those of CBX2, CBX4, CBX6, and CBX8, but shows almost no binding to ChDs of CBX1, CBX3 and CBX5 that are also known as heterochromatin 1 (HP1) β, γ, and α (Figures S1B, S2). As determined by HSQC titration, MS37452 has approximately a 3-fold and at least 10-fold weaker affinity for CBX4 or CBX2/6/8 than for CBX7, respectively (Figure S2I). The difference in MS37452 affinity between these two sub-groups is most likely due to Trp35, which is replaced by a Phe in the HP1 ChDs (see Figure S1C). Phe has a smaller side chain than Trp, and is not sufficient to accommodate two methoxyls when the aromatic cage residues adopt an extended conformation upon MS37452 binding (Figure S1D). The difference between CBX7 and CBX2/4/6/8 is possibly due to the collective role of Tyr39, Val13, and His47 (see Figure S1C). Specifically, Tyr39 is substituted to Asp in CBX2/4/6/8, which results in loss of aromatic stacking interaction with the di-methoxyl-benzene ring of MS37452. Val13 in CBX7 and CBX4 corresponds to an Ala in CBX2/6/8, which decreases hydrophobic interactions with the methylbenzene ring of MS37452. The presence of Val13 in both CBX4 and CBX7 explains their comparable binding affinity to MS37452. Finally, the presence of Asn in CBX2/4/6/8 instead of His47 in CBX7 contributes to loss of imidazole ring interactions with methylbenzene ring of MS37452.
We also collected structure-activity relationship (SAR) data for MS37452 with chemical analogs either available in the L1 library or synthesized (Supplemental Information and Table S3). The analogs that contain Cl, Br or no methyl group substitutions on the methylbenzene ring bind to the CBX7ChD with reduced affinity in the range of B-score < −3 but > −6. Analogs lacking one of the two methoxyls (or switching their positions) almost abolishes binding, indicating that the size and position of the two methoxyls are critical for maintaining MS37452’s interaction with Trp35 of the protein. However, we did not find a compound with affinity greater than MS37452. Collectively, these results indicate that both the di-methoxyl-benzene and methylbenzene moieties contribute to overall binding. The former is secured by its interaction with the aromatic cavity residues and Tyr39, whereas the latter is bound in a similar conformation as the methylated lysine, sandwiched between β1 and α1.
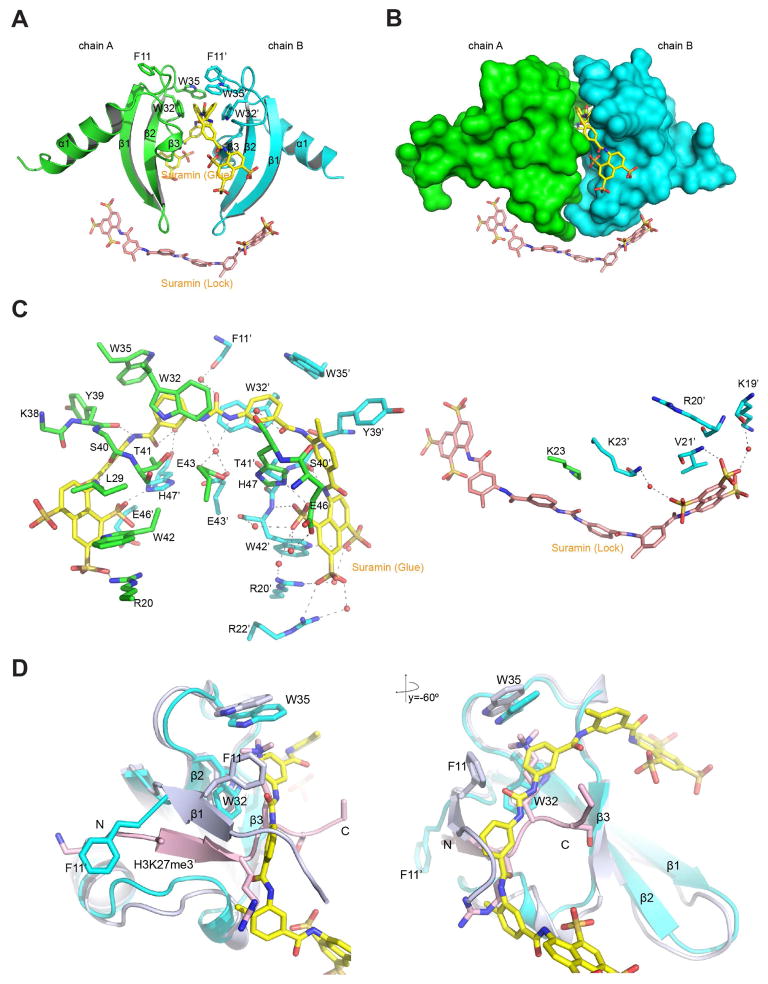
Among the hits identified and confirmed from the FDA library (Figure 2A, Figure S3) are Sennoside A, Suramin, Aurin tricarboxylic acid, Trypan blue and Evans blue. They bind to the CBX7ChD with affinities (IC50) of 33.3, 8.1, 5.5, 7.5 and 3.9 μM, respectively (Figure 2B). Unlike MS37452, these compounds share common features: relatively large in molecular weight, symmetric, and containing poly-hydroxyl groups. Their binding to CBX7ChD also induces severe line broadening in NMR HSQC spectra, indicating ligand binding-induced conformational exchange of the protein (Figure S3). We extended our analysis on suramin, which is colorless, water soluble, and shown to induce protein dimerization (Lima et al., 2009).
We solved a 1.63 Å-resolution crystal structure of the CBX7ChD-Suramin complex, which strikingly reveals two suramin molecules bound to two CBX7ChD molecules in a unit cell (Figure 3A and Table S2). One suramin molecule lies along the cleft/tunnel between the two β-sheets of each CBX7ChD and glues the two domains together, thus referred as “suramin-glue” (Figure 3A, 3B, yellow). The other suramin resides along the plane created by the two β-sheets, orthogonal to suramin-glue (Figure 3A, 3B, salmon), and locks the two CBX7ChD molecules in the region of Lys19 to Gly24, where the β-sheets start to turn, thus named as “suramin-lock”. For the tunnel where suramin-glue lies, Trp32 and Trp35 from two CBX7ChD molecules form the hydrophobic binding pocket. In contrast, Phe11 either rotates (chain B) or points outside the binding tunnel (chain A), thus its aromatic ring does not contribute to binding (Figure 3A–D). A large number of direct or water-mediated hydrogen bonds are established at an extensive interface between suramin and CBX7ChD (Figure 3C). The urea and four amide groups form hydrogen bonds on the bottom surface of the binding tunnel. The sulfonate groups of the suramin-glue interact with a stretch of charged residues Arg20, Glu46 and His47 to facilitate the binding via electrostatic interactions. Suramin-lock only involves interactions with basic residues from chain B, namely Lys19 and Lys23 (Figure 3C, right). In terms of how these crystals are packed, CBX7ChD molecules lie in a 3D network weaved by suramin, and they form a compact 2:2 complex.
Figure 3. Structural analysis of Suramin binding to CBX7ChD.
(A) Crystal structure of a CBX7ChD dimer bound to suramin-glue (yellow) and suramin-lock (salmon) molecules.
(B) Surface presentation of CBX7ChD bound to suramin.
(C) Detailed analysis of the interactions between CBX7ChD and suramin.
(D). Structural comparison of CBX7ChD bound to suramin or H3K27me3 peptide. Chain A and suramin-glue from the CBX7ChD-suramin complex and Chain AC from CBX7ChD-H3K27me3 complex were aligned.
Aside from the methyl-lysine-based peptide inhibitors of the CBXChDs, several methyl-lysine-binding antagonists, particularly against the MBT (malignant brain tumor) domain, have been reported (James et al., 2013). MBTs have a wider methyl-lysine binding pocket and bind preferentially to H3K27me1 or me2, instead of me3. Notably, the aromatic cage of L3MBTL3 is comprised of four residues, Phe387, Phe405, Trp408 and Tyr412, which correspond to Phe11, Trp32, Trp35 and Tyr39 of CBX7, respectively (Figure S1D). L3MBTL3 also contains a critical Asp381, which together with the aromatic cage residues creates a full pocket to accommodate the amine moiety of UNC1215, a potent MBT inhibitor (James et al., 2013). In contrast, aromatic cage residues and Tyr39 of CBX7ChD provide a half-closed pocket for MS37452 binding. Even considering the fact that other residues contribute to MS37452 binding, the methyl-lysine binding pocket of CBX7-MS37452 is still open. In other words, the amine group of UNC1215 binds deep into the pocket in L3MBTL3 while MS37452 lies along the surface groove of CBX7ChD, thus explaining a major difference in affinity between these two methyl-lysine reader domain inhibitors. Additionally, similar to the mode adopted by suramin in binding to CBX7ChD, UNC1215 bridges different domains of L3MBT3 when bound to the protein, thus resulting in a major increase in binding affinity as compared to UNC669 or similar compounds with different linkers (James et al., 2013).
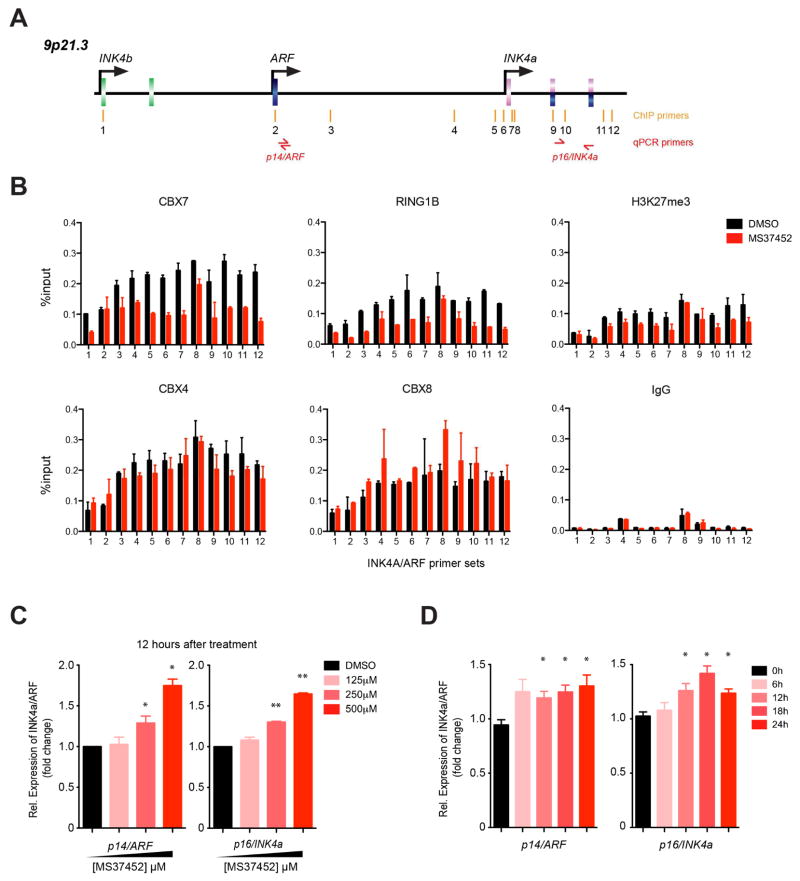
Given that suramin interacts with different proteins, including thrombin and pyruvate kinase (Lima et al., 2009; Morgan et al., 2011), we focused our study of small molecule modulation of CBX7ChD activity in transcriptional repression with MS37452. Prior research showed that mutation or deletion of CBX7ChD results in a decrease in binding of CBX7 to the INK4A/ARF (CDKN2A) locus at multiple sites (Figure 4A) (Bracken et al., 2007). We indeed observed that human PC3 prostate cancer cells treated with MS37452 (250 μM) for 2 hours showed reduced CBX7 occupancy across the INK4A/ARF locus, as determined by chromatin immunoprecipitation (ChIP) (Figure 4B). We further examined INK4A/ARF transcript levels in PC3 cells using quantitative PCR upon dose- and time-dependent treatment of MS37452 (Figure 4C, 4D). After treatment with MS37452 for 12 hours, transcription increased about 25% and 60% for 250 μM and 500 μM of MS37452, respectively, as compared to the DMSO control.
Figure 4. Modulation of CBX7 binding to INK4A/ARF locus by MS37452.
(A) Scheme illustrating the genomic organization of the INK4a/ARF/INK4b gene loci color-coded in pink, blue and green for p16/INK4a, p14/ARF, and p15/INK4b, respectively. Roman numerals and arrows (red) denote sites of quantitative ChIP and PCR primers, respectively, as previously reported (Bracken et al., 2007).
(B) MS37452 reduces CBX7 occupancy at different positions along the INK4a/ARF locus in PC3 cells after 2-hour treatment of MS37452 (250 μM). Plot represented data from one independent experiment (as an example) and error bars donated standard deviation (SD) of technical repeat. Independent ChIP data were listed in Figure S4.
(C), (D) MS37452 induces gene transcriptional de-repression at the INK4a/ARF locus, in a concentration- and time-dependent manner in PC3 cells. Messenger RNA levels of p14/ARF and p16/INK4a were tested using SYBR qPCR after MS37452 treatment. Data were plotted from at least three independent experiments and error bars donated standard error of the mean (SEM).
We further observed reduced occupancy of RING1B, and H3K27me3 at the INK4A/ARF locus by MS37452, but much less, if any change of CBX4 or CBX8 (Figure 4B). MS37452 has much weaker affinity to CBX4ChD or ChX8ChD than to CBX7ChD, consistent with weaker reduction of the former association with the INK4A/ARF locus. As a PRC1 component, RING1B has been shown to interact with CBX7 and play a critical role in CBX7 deposition at chromatin (Morey et al., 2013; Wang et al., 2010). Therefore a reduced RING1B association is consistent with reduced CBX7 occupancy after the compound treatment. The observed reduction of H3K27me3 matches the findings of a previous study showing localized reduction in H3K27me3 signal when CBX7 was silenced (Maertens et al., 2009) without a change of global H3K27me3 level (Figure S4C). Based on these ChIP and mRNA transcription results, we concluded that MS37452 can disrupt CBX7 binding to the INK4A/ARF locus, subsequently resulting in transcriptional de-repression of its target genes p14/ARF and p16/INK4a in PC3 cells.
EXPERIMENTAL PROCEDURES
Procedures for protein expression and purification, biochemical binding assay, and high-throughput chemical screening, protein structural analysis using NMR and X-ray crystallography, as well as cellular analysis of effects of compound treatment in gene transcription are reported in detailed in the Supplemental Information.
Supplementary Material
SIGNIFICANCE.
In this study, we discovered small molecules that inhibit CBX7ChD binding to lysine-methylated peptides with affinity better than its biological target, H3K27me3. One hit, suramin, bridges two CBX7ChD molecules from two perpendicular directions, and another hit, MS37452, directly competes against H3K27me3 binding by occupying both the methyl-lysine binding aromatic cage and an internal portion of the protein. We further demonstrated that MS37452 blocks CBX7 binding to its target INK4/ARF gene locus, and induces de-repression of p14/ARF and p16/INK4a in human PC3 cells. Collectively, our structure-guided discovery and characterization of small molecules for the CBX7ChD demonstrate the promise and feasibility of developing selective chemical inhibitors of this important epigenome reader domain protein. Given the fundamental role of CBX7 in PRC-directed transcriptional repression of the Ink4a/Arf locus in biology and disease, such new inhibitors can potentially be used to provide senescence control, cancer prevention and stem cell lineage specification.
CBX7 represses gene transcription through histone H3 lysine 27 methylation.
Small molecules compete methylated H3 lysine 27 binding to CBX7 chromodomain.
Chemical inhibition of CBX7 chromodomain de-represses p16/CDKN2A in PC3 cells.
The Polycomb group protein CBX7 functions to regulate gene transcriptional repression via its chromodomain binding to methylated lysine 27 of histone H3. Ren et al. report a small molecule that inhibits the CBX7 chromodomain and de-represses target gene p16/CDKN2A in PC3 prostate cancer cells.
Acknowledgments
We thank X6A Workbench 2012 and RapiData 2013 for the X-ray crystallography training both theoretically and experimentally, and beamline members for technical help in data collection at National Synchrotron Light Source (NSLS) of Brookhaven National Laboratory. We thank G. Zhang, Q. Zhang, K.-L. Cheung, G. Lu, L. Peng, S.D. Li, R. Sharma, and Y.F. Sun for technical discussion and advice. The X6A beamline at the Brookhaven National Laboratory is supported by the National Institute of General Medical Sciences (GM0080) and NSLS is funded by DOE (DE-AC02-98CH10886). This work is supported in part by the research grants from National Institutes of Health (to M.-M.Z.).
Footnotes
Supplemental Information includes detailed experimental methods, four figures and four tables and can be found with this article online.
AUTHOR CONTRIBUTIONS
C.R. and M.-M. Z. conceived and designed the experiments for the study. C.R. and K.M. performed HT chemical screen. J.L. and S.G.S. performed the chemical synthesis. C.R. and A.N.P. determined the crystal structures with assistance from J.J. and V.S., C.R. and L.Z. conducted the biochemical study. C.R. carried out the molecular and cell biology experiments with M.W. assistance. C.R. and M.-M.Z. wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bernard D, Martinez-Leal J, Rizzo S, Martinez D, Hudson D, Visakorpi T, Peters G, Carnero A, Beach D, Gil J. CBX7 controls the growth of normal and tumor-derived prostate cells by repressing the Ink4a/Arf locus. Oncogene. 2005;24:5543–5551. doi: 10.1038/sj.onc.1208735. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Duncan EM, Masui O, Gil J, Heard E, Allis CD. Mouse Polycomb Proteins Bind Differentially to Methylated Histone H3 and RNA and Are Enriched in Facultative Heterochromatin. Mol Cell Biol. 2006;26:2560–2569. doi: 10.1128/MCB.26.7.2560-2569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken AP, Kleine-Kohlbrecher D, Dietrich N, Pasini D, Gargiulo G, Beekman C, Theilgaard-Monch K, Minucci S, Porse BT, Marine JC, et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 2007;21:525–530. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil J, Bernard D, Martínez D, Beach D. Polycomb CBX7 has a unifying role in cellular lifespan. Nat Cell Biol. 2004;6:67–72. doi: 10.1038/ncb1077. [DOI] [PubMed] [Google Scholar]
- Hughes RM, Wiggins KR, Khorasanizadeh S, Waters ML. Recognition of trimethyllysine by a chromodomain is not driven by the hydrophobic effect. Proc Natl Acad Sci USA. 2007;104:11184–11188. doi: 10.1073/pnas.0610850104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James LI, Barsyte-Lovejoy D, Zhong N, Krichevsky L, Korboukh VK, Herold JM, MacNevin CJ, Norris JL, Sagum CA, Tempel W, et al. Discovery of a chemical probe for the L3MBTL3 methyllysine reader domain. Nat Chem Biol. 2013;9:184–191. doi: 10.1038/nchembio.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaustov L, Ouyang H, Amaya M, Lemak A, Nady N, Duan S, Wasney GA, Li Z, Vedadi M, Schapira M, et al. Recognition and specificity determinants of the human cbx chromodomains. J Biol Chem. 2011;286:521–529. doi: 10.1074/jbc.M110.191411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauke K, Radulovic V, Broekhuis M, Weersing E, Zwart E, Olthof S, Ritsema M, Bruggeman S, Wu X, Helin K, et al. Polycomb Cbx family members mediate the balance between haematopoietic stem cell self-renewal and differentiation. Nat Cell Biol. 2013;15:353–362. doi: 10.1038/ncb2701. [DOI] [PubMed] [Google Scholar]
- Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- Lima LMTR, Becker CF, Giesel GM, Marques AF, Cargnelutti MT, Neto MD, Monteiro RQ, Verli H, Polikarpov I. Structural and thermodynamic analysis of thrombin:suramin interaction in solution and crystal phases. Bba-Proteins Proteom. 2009;1794:873–881. doi: 10.1016/j.bbapap.2009.03.011. [DOI] [PubMed] [Google Scholar]
- Maertens GN, El Messaoudi-Aubert S, Racek T, Stock JK, Nicholls J, Rodriguez-Niedenfuhr M, Gil J, Peters G. Several distinct polycomb complexes regulate and co-localize on the INK4a tumor suppressor locus. PLoS One. 2009;4:e6380. doi: 10.1371/journal.pone.0006380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey L, Aloia L, Cozzuto L, Benitah SA, Di Croce L. RYBP and Cbx7 define specific biological functions of polycomb complexes in mouse embryonic stem cells. Cell reports. 2013;3:60–69. doi: 10.1016/j.celrep.2012.11.026. [DOI] [PubMed] [Google Scholar]
- Morey L, Pascual G, Cozzuto L, Roma G, Wutz A, Benitah SA, Di Croce L. Nonoverlapping functions of the Polycomb group Cbx family of proteins in embryonic stem cells. Cell Stem Cell. 2012;10:47–62. doi: 10.1016/j.stem.2011.12.006. [DOI] [PubMed] [Google Scholar]
- Morgan HP, McNae IW, Nowicki MW, Zhong WH, Michels PAM, Auld DS, Fothergill-Gilmore LA, Walkinshaw MD. The Trypanocidal Drug Suramin and Other Trypan Blue Mimetics Are Inhibitors of Pyruvate Kinases and Bind to the Adenosine Site. Journal of Biological Chemistry. 2011;286:31232–31240. doi: 10.1074/jbc.M110.212613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvageau M, Sauvageau G. Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell. 2010;7:299–313. doi: 10.1016/j.stem.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ. The INK4a/ARF network in tumour suppression. Nat Rev Mol Cell Biol. 2001;2:731–737. doi: 10.1038/35096061. [DOI] [PubMed] [Google Scholar]
- Sherr CJ. Divorcing ARF and p53: an unsettled case. Nat Rev Cancer. 2006;6:663–673. doi: 10.1038/nrc1954. [DOI] [PubMed] [Google Scholar]
- Simhadri C, Daze KD, Douglas SF, Quon TT, Dev A, Gignac MC, Peng F, Heller M, Boulanger MJ, Wulff JE, et al. Chromodomain Antagonists That Target the Polycomb-Group Methyllysine Reader Protein Chromobox Homolog 7 (CBX7) J Med Chem. 2014 doi: 10.1021/jm401487x. [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Mechanisms of Polycomb gene silencing: knowns and unknowns. Nature reviews Molecular cell biology. 2009:1–12. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- Tabet S, Douglas SF, Daze KD, Garnett GA, Allen KJ, Abrioux EM, Quon TT, Wulff JE, Hof F. Synthetic trimethyllysine receptors that bind histone 3, trimethyllysine 27 (H3K27me3) and disrupt its interaction with the epigenetic reader protein CBX7. Bioorg Med Chem. 2013;21:7004–7010. doi: 10.1016/j.bmc.2013.09.024. [DOI] [PubMed] [Google Scholar]
- Wang Z, Song J, Milne TA, Wang GG, Li H, Allis CD, Patel DJ. Pro isomerization in MLL1 PHD3-bromo cassette connects H3K4me readout to CyP33 and HDAC-mediated repression. Cell. 2010;141:1183–1194. doi: 10.1016/j.cell.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou MM. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.