Abstract
The influence of the position of the amino acid proline in polypeptide sequences is examined by a combination of ion mobility spectrometry-mass spectrometry (IMS-MS), amino acid substitutions, and molecular modeling. The results suggest that when proline exists as the second residue from the N-terminus (i.e., penultimate proline), two families of conformers are formed. We demonstrate the existence of these families by a study of a series of truncated and mutated peptides derived from the 11-residue peptide Ser1-Pro2-Glu3-Leu4-Pro5-Ser6-Pro7-Gln8-Ala9-Glu10-Lys11. We find that every peptide from this sequence with a penultimate proline residue has multiple conformations. Substitution of Ala for Pro residues indicates that multiple conformers arise from the cis- trans isomerization of Xaa1–Pro2 peptide bonds as Xaa–Ala peptide bonds are unlikely to adopt the cis isomer, and examination of spectra from a library of 58 peptides indicates that ~80% of sequences show this effect. A simple mechanism suggesting that the barrier between the cis-and trans-proline forms is lowered because of low steric impedance is proposed. This observation may have interesting biological implications as well, and we note that a number of biologically active peptides have penultimate proline residues.
Keywords: Peptide conformation, Penultimate proline, Proline Cis-Trans isomerization, Ion mobility spectrometry
Introduction
While examining the influence of the position of different amino acid residues in polypeptide sequences on peptide conformation, we find evidence that proline in the second position from the amino-terminus (i.e., penultimate proline) gives rise to two types of peptide families. This penultimate proline has a propensity to induce two types of stable conformations, having mostly similar populations. Prior work has noted the existence of a penultimate proline in a large number of similarly sized signaling peptides [1–4]. It has been proposed that proline in this position protects the small amino acid chain against further enzymatic degradation [1–3]. The evidence presented here that two populations of conformers are favored suggests that penultimate proline may play another role involving multiple structures that may influence activity and binding.
Cis-trans isomerization of peptide bonds is one mechanism that can lead to large changes in the overall structure of peptides and proteins [5–8]. Such isomers are most commonly associated with proline, primarily because of the 20 natural amino acids proline is unique in that it has a secondary amine at the backbone nitrogen atom [9, 10]. This makes it possible for proline to adopt the cis isomer of Xaa–Pro peptide bonds (where Xaa is any amino acid) at a higher frequency than Xaa–Xnp peptide bonds (where Xnp is any non-proline amino acid) [9, 10]. Although most previous studies classify proline residues either as cis or trans configurations, an increasing number of systems appear to adopt structures that utilize both the cis and trans isomers, and the two forms may have different functions [11]. Thus, it is important to understand the fundamental factors that influence the ability of Xaa–Pro bonds to adopt both cis and trans isomers.
In this study, we use a combination of electrospray ionization (ESI) [12], ion mobility spectrometry (IMS) [13–17], and mass spectrometry (MS) to examine the conformation of a range of peptide sequences. The mobility of an ion through a buffer gas depends upon the shape of the ion, and thus is an effective means of delineating large differences in conformation. When combined with ESI and MS, it is possible to sample different conformer populations directly from solution [18–25]. Because of the ability to separate conformations that arise from both cis and trans configurations, IMS-MS is an appealing technique to analyze cis-trans isomerization of proline-containing peptides. Previous IMS-MS studies have established the important role that proline plays in the conformations of polypeptide ions [26–29]. Pierson et al. have shown by IMS-MS that proline residues are important in establishing multiple conformations of the nonapeptide bradykinin [27].
In this paper, we primarily examine the influence of proline position on the distribution of conformations observed by IMS-MS for the peptide Ser1-Pro2-Glu3-Leu4-Pro5-Ser6-Pro7-Gln8-Ala9-Glu10-Lys11—a tryptic peptide found in several proteins across different organisms. For example, it is located at residues 271–281 in the CLPB Protein in Homo sapiens [30]. We are interested in studying this peptide in detail because it has multiple peaks in the ion mobility distribution of the [SPELPSPQAEK + 2H]2+ ion. Examination of this peptide (and related shorter sequences formed as by-products of the synthesis) motivated us to analyze a library of doubly charged peptides, which was available in our laboratory; these studies show that ~80% of all sequences that contain a penultimate proline adopt multiple conformers. Thus, it appears to be a common feature, in at least some families of sequences. We propose a simple mechanism in which the barrier of the system is lower because of decreased steric impedance associated with the penultimate position to explain the results. Overall, these findings expand upon previous studies [26, 27] and help to illuminate the more complex role that proline plays in establishing peptide conformations.
Experimental
Ion Mobility Spectrometry-Mass Spectrometry
IMS-MS experiments were performed on a home-built, ~2-meter instrument described in detail previously [31, 32]. Peptides were electrosprayed from 49.5:49.5:1.0 water:acetonitrile:formic acid solutions. Ions were produced by electrospray ionization with a Triversa Nanomate (Advion Bioscience, Inc., Ithaca, NY, USA). Ions are stored in a Smith-geometry hourglass-shaped ion funnel [33]. Packets of ions are periodically pulsed into the drift tube, which is operated with 3.00±0.02 Torr He buffer gas and an electric field of 10 V·cm−1. Ions exit the drift tube through a differentially pumped region and are analyzed by an orthogonal geometry two-stage reflectron time-of-flight mass spectrometer in a nested fashion [34].
Calculating Collision Cross Sections
Drift time (tD) distributions can be converted to collision cross section (Ω) distributions according to [35],
| (1) |
where ze is the charge of the ion. Kb is Boltzmann’s constant. T and P are the temperature and pressure, respectively. E and L are the electric field and length of the drift tube, respectively. N is the neutral number density at STP. For the instrument used in this study, collision cross sections can be calculated two ways. First, we can measure the time it takes the ions to traverse the first region of the drift tube from the source gate to the selection gate at the middle funnel and use Equation 1 directly. Alternatively, we can use the time required for ions to traverse the entire drift tube. However, the ion funnels are not operated with a uniform electric field. Thus, we must calibrate measurements to well-studied systems such as bradykinin. Cross section values obtained from both methods typically agree within 1%. This allows for the conversion of drift time distributions to accurate collision cross section distributions.
In order to compare collision cross section distributions of peptides in which Ala residues have been substituted for Pro residues, we need to account for the intrinsic size difference between the side chains. Previous efforts have focused on developing intrinsic size parameters (ISPs) to provide insight into the average contribution each amino acid residue has to the collision cross section of peptide ions [36–38]. From this work, it was determined that the substitution of Ala for Pro residues leads to peptides with smaller cross sections on average. To account for this intrinsic difference in size, we have shifted our collision cross section distributions by 3.5 Å2 for each Pro → Ala substitution. This shift in cross section is in good agreement with work by Pierson et al. in which triply charged bradykinin distributions were shifted by 2.5 Å2 for each Ala substitution. It is worth mentioning that previous work suggests ISPs are dependent on charge state [36–38]. Furthermore, the 3.5 Å2 correction per Pro → Ala substitution yields more precisely aligned distributions for the peptides analyzed in this study and both values lead to the same cis and trans isomer assignments.
Peptide Synthesis
Peptides were synthesized by Fmoc solid-phase synthesis on an Apex 396 peptide synthesizer (AAPPTec, Louisville, KY, USA) similar to the method described previously [27]. Fmoc side-chain protected amino acids and Wang-type polystyrene resins were used (Midwest Biotech, Fischers, IN, USA). Twenty percent piperidine in dimethylformamide was used for Nα deprotections. 1,3-Diisopropylcarbodiimide/6-chloro-1-hydroxybenzotriazole or 3-(diethoxyphosphoryloxy)-1,2,3-benzotriazin-4(3H)-one was used as the coupling reagent. Peptides were cleaved from the resin with a trifluoroacetic acid:triisopropylsilane:methanol solution at a 18:1:1 ratio, and 5% 2,2′-(ethylenedioxy)diethanethiol was added for methionine-containing peptides. Peptides were washed and precipitated in diethyl ether. Precipitated products were dried down with a vacuum manifold before reconstituting in H2O and finally the ESI solvent.
Molecular Modeling
Molecular modeling simulations were completed by a simulated annealing procedure with the Insight II software package (Accelrys, Inc., San Diego, CA, USA) similar to simulations described previously [26]. Simulations were performed on the [SPQAEK + 2H]2+ion with charges assigned to the N-terminus and the Lys side chain. Simulated annealing was completed on two initial starting structures that differ by geometry of the Ser1-Pro2 peptide bond (cis or trans). Ions were heated from 298 to 500 K over 5 ps, equilibrated at 500 K for 10 ps, cooled to 298 K over 5 ps, and equilibrated at 298 K for 30 ps. The extensible systematic force field (ESFF) and a dielectric of 1 were used; 100 trial geometries were obtained from each run. Collision cross sections were calculated using the trajectory method in Mobcal [39].
Results and Discussion
IMS-MS of SPELPSPQAEK and Truncated Peptides
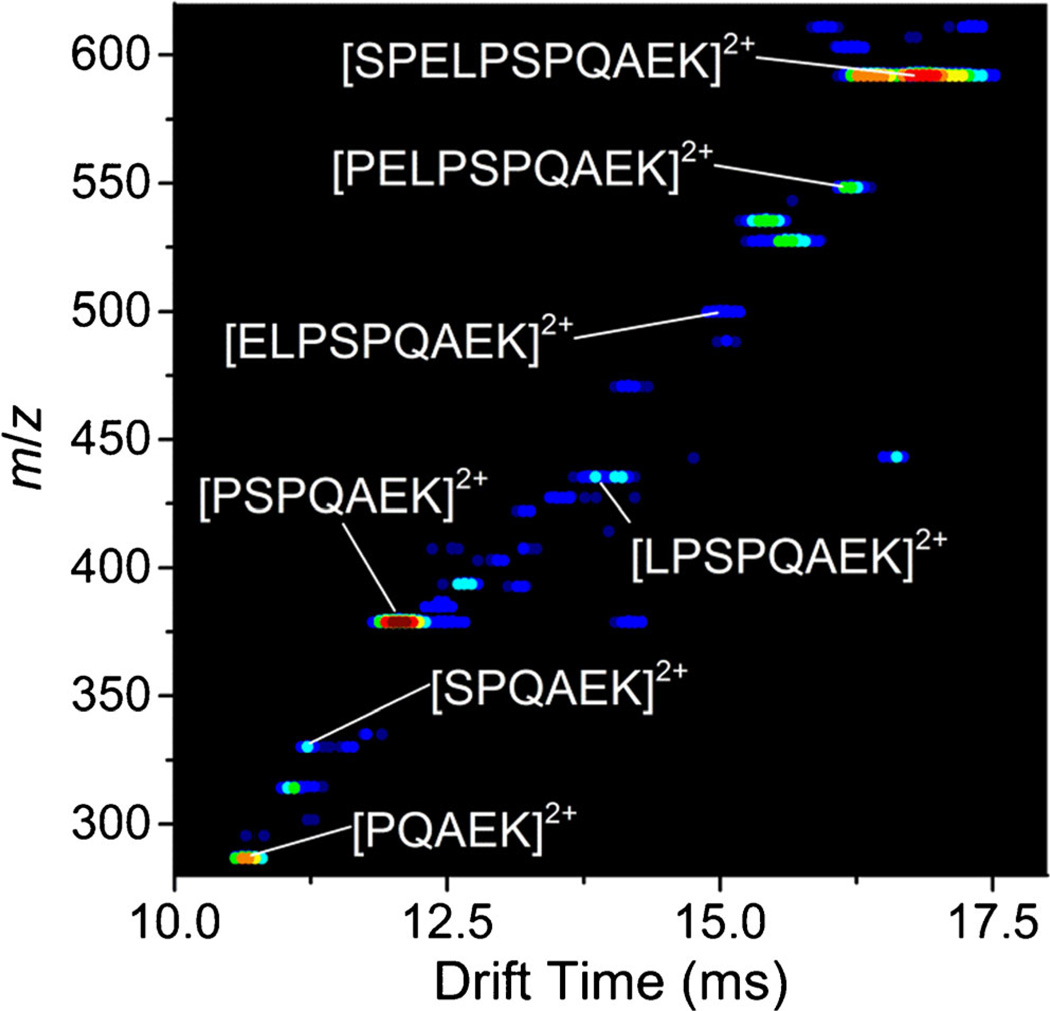
Figure 1 shows the two-dimensional IMS-MS plot of ions produced by ESI of the crude product from the synthesis of the peptide SPELPSPQAEK. In addition to the [SPELPSPQAEK + 2H]2+ ion, we observe the truncated series of peptides (e.g., [PELPSPQAEK + 2H]2+) derived from the full-length sequence in relatively high abundance. Truncated peptide sequences result from the inefficiency of peptide synthesis. Although inefficient synthesis is generally thought to be problematic, this makes it possible to obtain IMS-MS measurements for many unique peptide sequences from a single synthesis. From this series of truncated peptides, we observed an interesting trend—as single amino acid residues are added from the 5 residue peptide to the full-length sequence, the distribution of conformers for each peptide is markedly different. That is, some peptides have multiple conformers and others do not. Upon this discovery, we synthesized each individual truncated peptide derived from the SPELPSPQAEK separately to probe this observation in detail. It is worth mentioning that distributions obtained from the two different syntheses are nearly identical.
Figure 1.
Two-dimensional IMS-MS plot of crude synthesis product of the peptide SPELPSPQAEK electrosprayed from 49.5:49.5:1 water:acetonitrile:formic acid. Doubly charged [M + 2H]2+ truncated sequences of the full length peptide are labeled
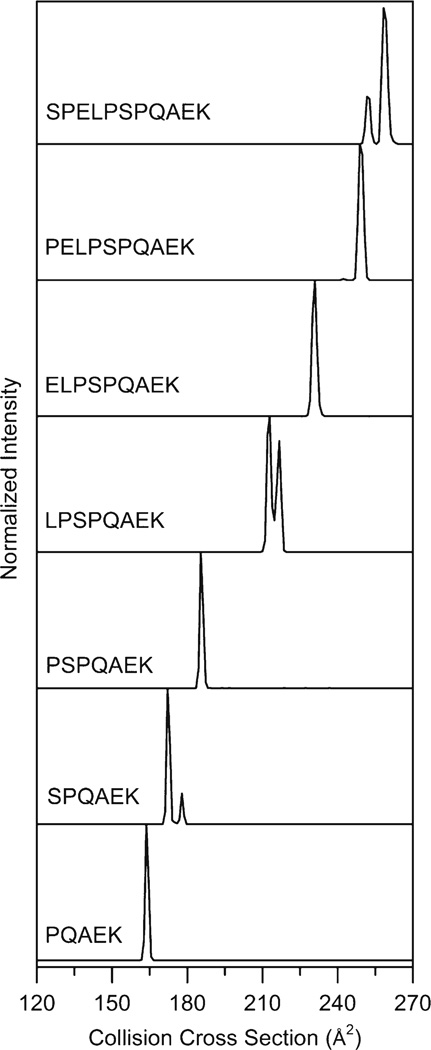
Figure 2 shows the collision cross section distributions for the peptide [SPELPSPQAEK + 2H]2+ ion and truncated peptides. The distribution has two peaks at 252 and 258 Å2 with normalized abundances of 23% and 77%, respectively. Based on previous studies of proline-containing peptides by ion mobility, one likely explanation for the multiple conformations is that one of the Xaa–Pro bonds adopts both the cis and trans isomer [26, 27]. However, it is difficult to assign what residue or region of residues in the peptide sequence are important to observing multiple conformer families from the cross section distribution alone. Thus, we will consider the truncated versions of the peptide to examine how the distributions of ions change as a function of length and amino acid composition.
Figure 2.
Collision cross section distributions for doubly charged peptide [M + 2H]2+ ions (where M = SPELPSPQAEK, PELPSPQAEK, ELPSPQAEK, LPSPQAEK, PSPQAEK, SPQAEK, and PQAEK). Distributions were obtained from IMS-MS measurements by integrating the drift bins for a narrow m/z window for each ion of interest
Collision cross section distributions for the truncated peptides [PELPSPQAEK + 2H]2+, [ELPSPQAEK + 2H]2+, [LPSPQAEK + 2H]2+, [PSPQAEK + 2H]2+, [SPQAEK + 2H]2+, and [PQAEK + 2H]2+ are displayed in Figure 2. It is apparent that the removal of a single amino acid from the N-terminus dramatically influences the distribution of conformers. The distribution of [PELPSPQAEK + 2H]2+ is markedly different than the full-length sequence. Although both features contain multiple peaks, the minor feature for the [PELPSPQAEK + 2H]2+ ion is less than 1% normalized abundance with the major feature at 249 Å2. This trend of observing different distributions for each subsequent truncation is observed across all sequences. The distribution for the [ELPSPQAEK + 2H]2+ ion is dominated by a single peak at 231 Å2. The removal of the Glu residue from the N-terminus leads to two highly abundant peaks for the [LPSPQAEK + 2H]2+ ions at 213 and 217 Å2. Once again, the removal of the N-terminal amino acid changes the distribution as only a single peak at 185 Å2 is observed for the [PSPQAEK + 2H]2+ ion. The distribution for the [SPQAEK + 2H]2+ ion has two features located at 172 and 178 Å2. Finally, the [PQAEK + 2H]2+ ion has a single peak at 164 Å2.
It is interesting that we do not observe a specific length at which all subsequent peptides have similar conformer distributions for the sequences analyzed in this study. This suggests that each amino acid residue influences the conformation, and small modifications such as the addition of a single amino acid to the N-terminus can lead to global conformational changes. Distributions oscillate between being dominated by a single peak and multiple peaks as the sequence is extended. The [SPELPSPQAEK + 2H]2+, [LPSPQAEK + 2H]2+, and [SPQAEK + 2H]2+ ion distributions have multiple conformations observed in relatively high abundance, whereas the [PELPSPQAEK + 2H]2+, [ELPSPQAEK + 2H]2+, [PSPQAEK + 2H]2+, and [PQAEK + 2H]2+ ion distributions are dominated by a single peak.
One commonality between the sequences with multiple features, [SPELPSPQAEK + 2H]2+, [LPSPQAEK + 2H]2+, and [SPQAEK + 2H]2+, is the presence of a Pro2 residue. However, when the N-terminal residue is removed such that Pro is at position 1, the distribution is dominated by a single peak. This suggests that the cis-trans isomerization of Xnp1–Pro2 peptide bonds allows for multiple conformer families and not the more centrally positioned residues. It is important to point out that the cis-trans isomerization of proline occurs at the peptide bond preceding proline (i.e., Xaa–Pro). Therefore, Pro1 residues are unable to adopt both cis and trans isomers and, consequently, multiple conformers.
IMS-MS Analysis of Pro Substituted Peptides
The analysis of the truncated peptides has shown that three sequences, [SPELPSPQAEK + 2H]2+, [LPSPQAEK + 2H]2+, and [SPQAEK + 2H]2+, have multiple conformer families at relatively high abundance. Thus, we will focus on determining the amino acid residues that are important to stabilizing multiple conformations for these sequences. This work was inspired by previous IMS-MS studies by Pierson et al. in which Ala residues were substituted for Pro residues in the nonapeptide bradykinin (BK) [27]. Substituting Ala for Pro residues likely fixes the Xaa–Ala peptide bond in the trans isomer. It was shown that the cis-trans isomerization of proline residues are important for triply charged [BK + 3H]3+ ion conformations and specific Xaa–Pro bonds were assigned as cis or trans for different conformations.
SPELPSPQAEK Pro → Ala Substitutions
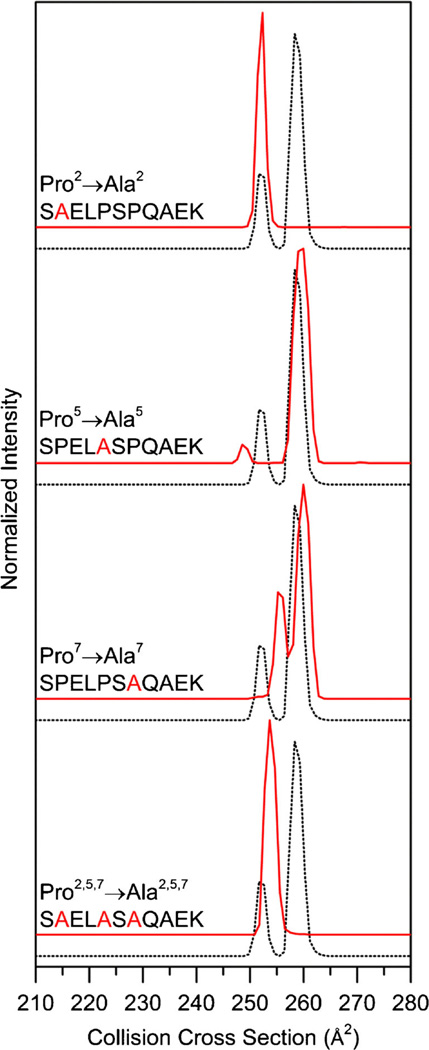
Figure 3 shows the collision cross section distributions for the Pro → Ala analogues of the [SPELPSPQAEK + 2H]2+ ions. Throughout this manuscript, cross section distributions for Pro → Ala substituted peptides have been corrected for the size difference between proline and alanine as explained above. The cross section distribution for the Pro2 → Ala substitution is dominated by a single peak at 252 Å2. This suggests that the cis-trans isomerization of Ser1–Pro2 peptide bond of [SPELPSPQAEK + 2H]2+ ion is crucial for the conformation at 258 Å2. Furthermore, it supports the results from the truncated series that suggest Pro2 residues are important for multiple conformers.
Figure 3.
Collision cross section distributions for Pro2 → Ala, Pro5 → Ala, Pro7 → Ala, and Pro2,5,7 → Ala substituted peptides from the [SPELPSPQAEK + 2H]2+ ions. Substituted distributions are shown in red and the natural sequence is shown as a dotted trace for ease of comparison. Collision cross section distributions for substituted peptides are adjusted for the size difference between Ala and Pro as explained in text
In order to verify that the N-terminal region of the peptide leads to multiple structures, we have analyzed the Pro5 and Pro7 substituted peptides. Both [SPELASPQAEK + 2H]2+ and [SPELPSAQAEK + 2H]2+ distributions have multiple peaks (Figure 3). The collision cross sections of the major peak for both the Ala5 and Ala7 peptides is 260 Å2. The conformers at 260 Å2 are less than 1% larger than the major peak at 258 Å2 for the peptide [SPELPSPQAEK + 2H]2+ ion. Although we observe a peak near 258 Å2 when both Pro5 and Pro7 are substituted, this peak is not observed for the Pro2 substitution. Therefore, we assign the conformation at 258 Å2 as cis-Pro2, trans-Pro5, trans-Pro7. A summary of cis-trans isomer assignments and populations can be found in Table 1.tgroup
Table 1.
Proline Cis-Trans Isomer Assignments
| Sequence | Collision cross section (Å2) | Normalized abundance (%)a | Pro2 | Pro4 | Pro5 | Pro7 |
|---|---|---|---|---|---|---|
| SPELPSPQAEK | 258 | 76.9 | Cis | - | Trans | Trans |
| SPELPSPQAEK | 252 | 23.1 | Trans | - | Trans | Trans |
| LPSPQAEK | 213 | 57.0 | Cis | Trans | - | - |
| LPSPQAEK | 217 | 43.0 | Trans | Trans | - | - |
| SPQAEK | 172 | 83.4 | Cis | - | - | - |
| SPQAEK | 178 | 16.6 | Trans | - | - | - |
Peak abundances are the integrated area for each respective peak normalized to the total area for each collision cross section distribution shown in Figure 2.
As mentioned above, both the distributions for the Pro5 → Ala and Pro7 → Ala sequences have minor features. Although our data suggests that the Pro5 and Pro7 residues are not the sites key to having multiple peaks, they dramatically influence the distribution of conformations. The minor features for the Ala5 substituted peptide is 248 Å2. The distribution for the Ala7 substituted peptide has two minor features at 251 and 255 Å2. In addition to the difference in the number of peaks and their respective cross sections, the populations are also different. Several previous studies have focused on the local environment surrounding proline residues with significant attention to the identity of the residue N-terminally adjacent to proline [40, 41]. A recent study suggests that the nonlocal environment influences cis-Pro residues in proteins found in the Protein Databank [42]. Our results suggest that distal residues influence the population of cis-trans isomers for peptide ions. One possible explanation for this is that intramolecular interactions that stabilize the structure of the peptide change upon substituting Ala for Pro. The backbone nitrogen atoms of proline residues are unable to participate in intramolecular H-bonding because of the lack of hydrogen bond donor on the backbone nitrogen. However, Ala residues are able to form intramolecular H-bonds.
We have performed the triple substitution, Pro2,5,7 → Ala, as this peptide should represent the all trans peptide. The cross section distribution for the triply substituted peptide has a single peak located at 254 Å2, less than 1% larger than the peak at 252 Å2 for the peptide [SPELPSPQAEK + 2H]2+ ion (Figure 3). Therefore, we have confirmed the conformer at 252 Å2 for the [SPELPSPQAEK + 2H]2+ ion as the trans configuration at Pro2, Pro5, and Pro7 (Table 1).
LPSPQAEK Pro → Ala Substitutions
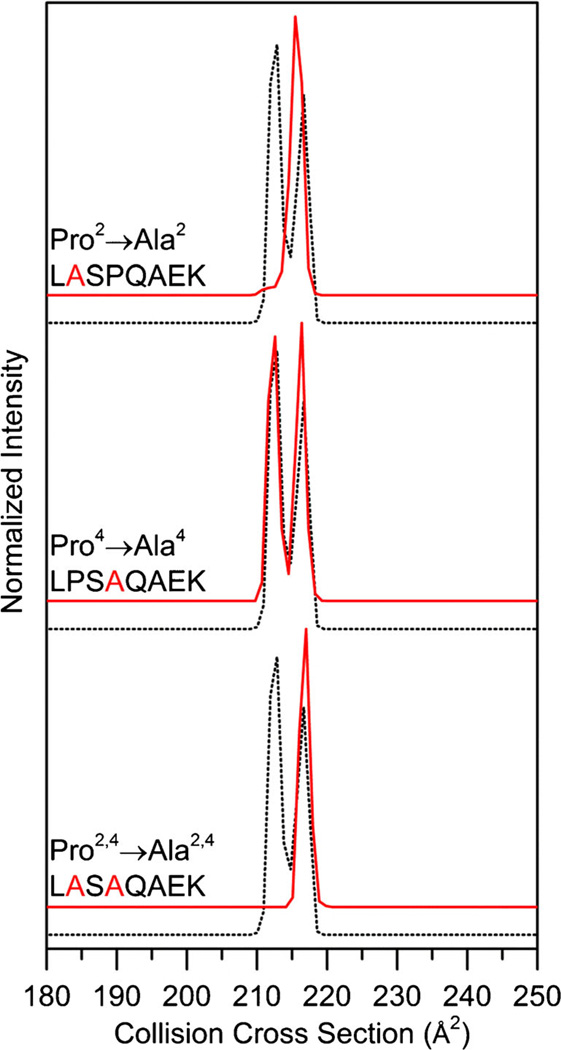
Figure 4 shows the collision cross section distribution for the Pro → Ala substituted peptides for the [LPSPQAEK + 2H]2+ ions. Upon Pro2 → Ala substitution, we observe a distribution that is dominated by a single conformation at 215 Å2. Although a minor feature is observed at 212 Å2, it comprises less than 1% of the distribution. In contrast, the Pro4→Ala [LPSAQAEK + 2H]2+ distribution is strikingly similar to the [LPSPQAEK + 2H]2+ ion distribution, having two peaks at 213 and 216 Å2 of nearly equal abundance. This suggests that the Pro2 residue is important to observing multiple conformer families of nearly equal abundance. The conformer at 213 Å2 is assigned as cis-Pro2, trans-Pro4 (Table 1). Due to the presence of a peak ~217 Å2 for both the Pro2→ Ala and Pro4→ Ala distributions, this conformer is assigned as trans-Pro2 and trans-Pro4 (Table 1).
Figure 4.
Collision cross section distributions for Pro2 → Ala, Pro4 → Ala, and Pro2,4 → Ala substituted peptides from the [LPSPQAEK + 2H]2+ ions. Substituted distributions are shown in red and the natural sequence is shown as a dotted trace for ease of comparison. Collision cross section distributions for substituted peptides are adjusted for the size difference between Ala and Pro as explained in text
In order to confirm the cis-trans isomer assignment, we have analyzed the doubly substituted peptide Pro2,4 → Ala. As shown in Figure 4, the cross section distribution for the doubly substituted Pro2,4 → Ala peptide has a single peak at 217 Å2. [LASAQAEK + 2H]2+ likely represents the all-trans peptide. This provides further evidence that conformer at 217 Å2 for [LPSPQAEK + 2H]2+ ions is trans-Pro2, trans-Pro4 (Table 1).
SPQAEK Pro → Ala Substitutions
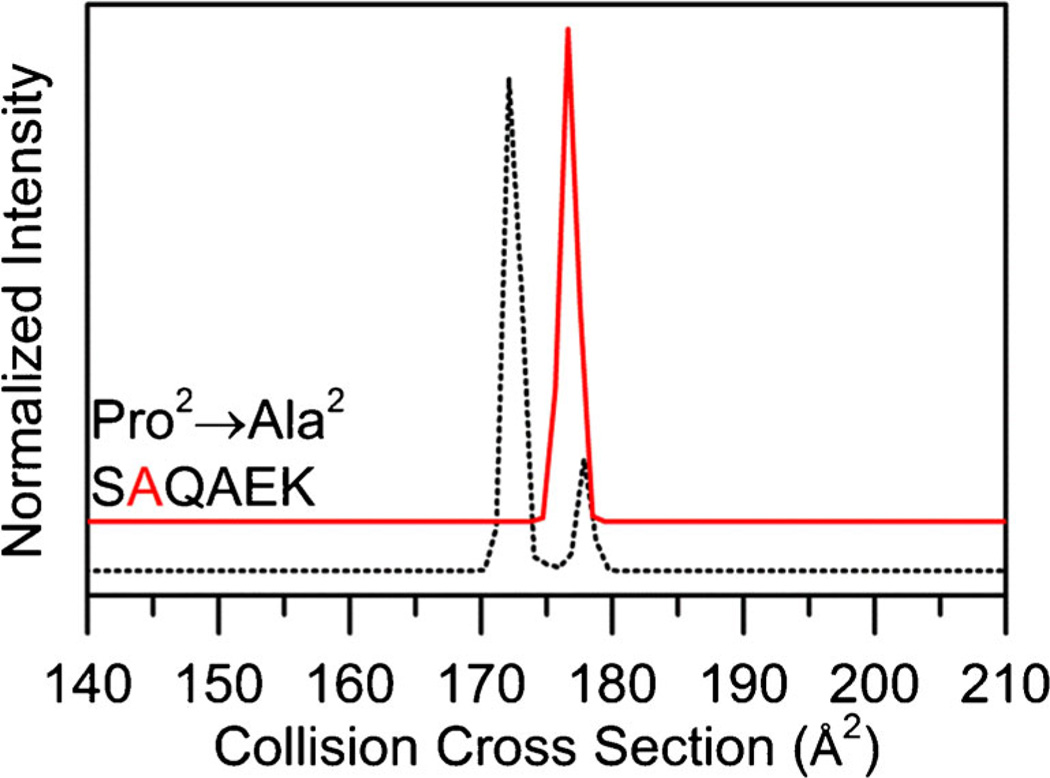
Figure 5 shows the cross section distribution for the Pro2 → Ala substituted peptide [SAQAEK + 2H]2+. Upon substitution, the distribution has a single peak at 177 Å2. This peak is aligned with the minor feature located at 178 Å2 of the [SPQAEK + 2H]2+ distribution. Because this peptide contains a single Pro residue, the assignment is relatively straightforward. The Ser1–Pro2 peptide bond for the conformer at 178 Å2 is assigned as trans because a similar conformer is observed for the [SAQAEK + 2H]2+, which likely has a trans-Ser1–Ala2 peptide bond. The conformer at 172 Å2 is assigned as a cis-Pro2 peptide bond because it is not observed in the [SAQAEK + 2H]2+ distribution (Table 1).
Figure 5.
Collision cross section distributions for Pro2 → Ala from the [SPQAEK + 2H]2+ ions. Substituted distributions are shown in red and the natural sequence is shown as a dotted trace for ease of comparison. Collision cross section distributions for substituted peptides are adjusted for the size difference between Ala and Pro as explained in text
Examination of a Penultimate Proline-Containing Peptide Library
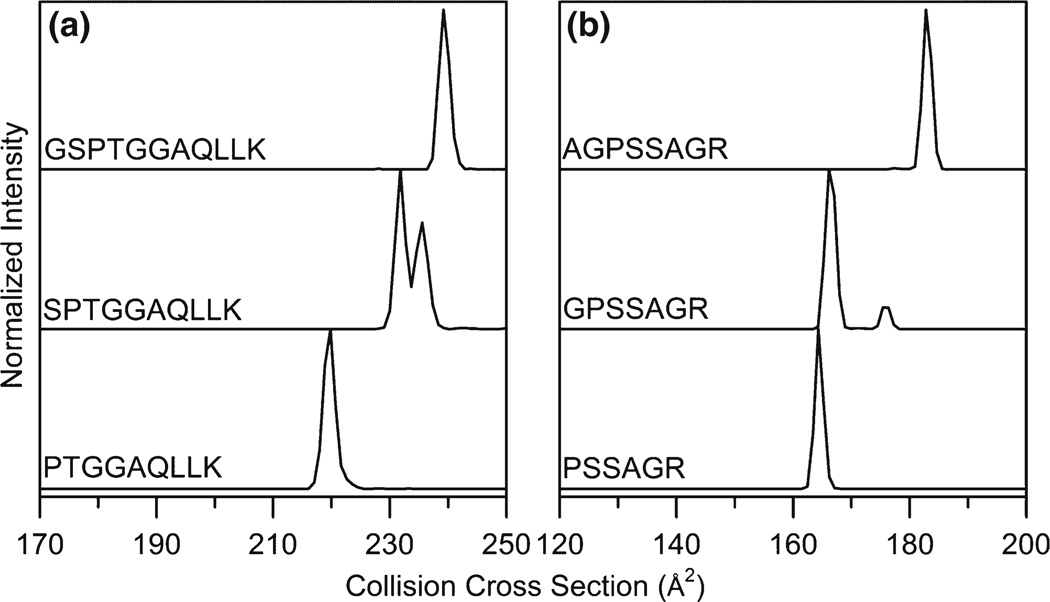
In order to determine if the trend of peptides with a penultimate proline residue adopting multiple conformers is widely observed, ion mobility distributions were measured for 52 additional [M + 2H]2+ peptides with a penultimate proline. IMSMS data was obtained from a library of synthesized peptides in a similar method to that shown in Figure 1, including both truncated and full-length sequences. Figure 6 shows collision cross section distributions for several [M+ 2H]2+ peptides from this dataset. In both examples, we observe a single peak in the mobility distribution for the Pro1 peptide, multiple peaks of considerable abundance for the Pro2 peptides, and a distribution that is dominated by a single peak for the Pro3 peptides.
Figure 6.
Collision cross section distributions for a series of [M + 2H]2+ peptides containing Pro1, Xaa1-Pro2, and Xaa1-Xaa2-Pro3 sequences
If we consider all penultimate proline-containing peptides examined in this study, 46 of 58 (79%) peptide sequences have multiple conformations in the IMS distribution. The complete list of sequences is listed in Supplementary Information (Table S1). Previous IMS-MS measurements by Counterman et al. found that 57% of proline-containing peptides had multiple features in the IMS-MS distribution [26]. It is worth mentioning that the previous measurement included Pro2 peptides. This suggests that penultimate proline-containing peptides adopt multiple conformations at a relatively high frequency across a range of peptide lengths and sequence compositions.
Investigation into Cis- versus Trans-Stabilizing Intramolecular Interactions by Molecular Modeling
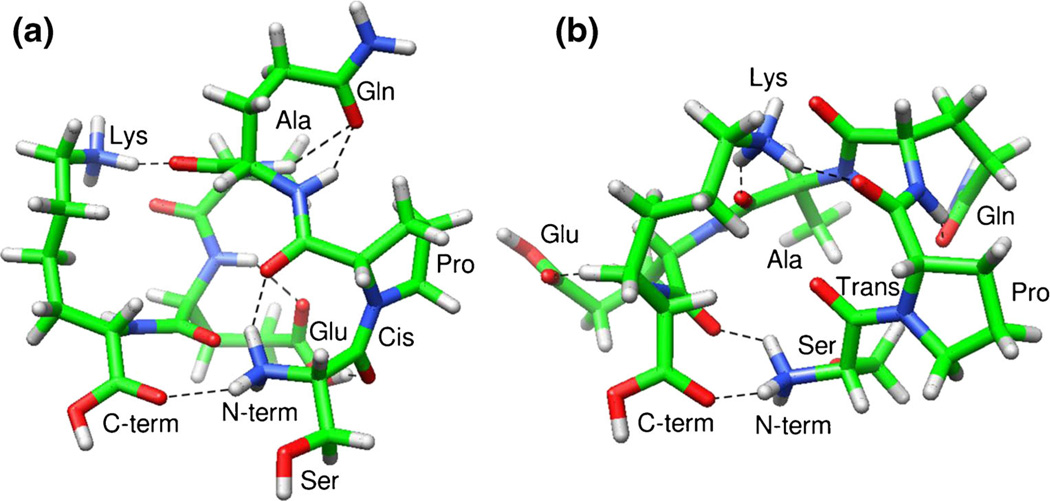
Preliminary molecular modeling simulations were performed to provide insight into the differences in the intramolecular interactions that exist when Xaa-Pro bonds adopt the cis versus trans configuration. Simulations were performed on the [SPQAEK + 2H]2+ ion. This peptide was chosen because it has two well-resolved peaks in the cross section distribution, contains a single proline residue at position two, and is relatively small. Therefore, it provides an excellent model to probe the intramolecular interactions that stabilize both the cis and trans geometries of proline when located at the second position as shown in the experimental results.
Figure 7 shows two representative low-energy structures obtained from molecular modeling simulations. We selected the lowest energy structure for the cis-Pro isomer and a relatively low energy structure with similar deviation from the experimental results for the trans-Pro isomer. Collision cross section values are within 2.0% of the cross sections measured by IMS-MS; the modeled cis-Pro (Figure 7a) and trans-Pro (Figure 7b) conformers have calculated cross sections of 175 and 181 Å2, respectively. Several general similarities exist between the two modeled structures—the Gln, Glu, and Lys side chains interact with the peptide backbone and the N- and C-terminus interact with each other. However, one key difference appears to be the disparity in the interactions with the oxygen atom of the second carbonyl group of the Pro2 residue. The cis-Pro peptide has an interaction between the protonated N-terminus and the carbonyl oxygen of Pro2. Conversely, the trans-Pro peptide has an interaction between the protonated Lys side chain and the carbonyl oxygen of Pro2. Therefore, the interaction of the protonated N-terminus and the carbonyl of the Pro2 side chain may stabilize the cis geometry of the Ser1– Pro2 peptide bond. This may also explain why we did not observe similar distributions for conformations after an amino acid is added to the N-terminus such that proline is in the third position. Upon addition of amino acids to the N-terminus, this interaction may become less energetically favorable and a single conformation may become preferred.
Figure 7.
Representative structures of the [SPQAEK + 2H]2+ ions with Ser1–Pro2 peptide bond in the cis (a) and trans (b) forms obtained from molecular modeling simulations. Amino acid side chains, terminal regions, and the geometry of the Ser1–Pro2 bond are labeled for clarity. H-bonds determined using the recommended relaxed constrains in Chimera are represented as dotted lines [43]
Simple Mechanistic Consideration
In addition to the intramolecular interactions that are key to penultimate proline residues adopting multiple conformations mentioned above, it is worthwhile to consider potential mechanistic explanations for the preference of penultimate proline residues to adopt multiple conformations. One likely explanation is that the low steric impedance of rotating the Xaa1–Pro2 bond leads to a relatively low barrier between the cis and trans isomers. This allows the peptide to rotate more freely between the cis and trans isomers, making the tail region more flexible to adopt multiple conformers. For more internally located proline residues, the barrier to rotate larger regions of the peptide surrounding Xaa–Pro bonds may be increased compared with the terminal region. Also, intramolecular interactions surrounding the peptide may more effectively lock the Xaa–Pro peptide bond into a single isomer. We cannot rule out that a similar effect would be observed for proline residues located at the C-terminus. However, we have only studied tryptic peptides to date; therefore, all sequences analyzed terminate in Lys or Arg residues.
Conclusions
IMS-MS techniques and molecular modeling were used to examine the conformation families in a series of truncated and Pro → Ala substituted analogues of the peptide [SPELPSPQAEK + 2H]2+ ion. Our results are in good agreement with previous IMS-MS studies that suggest proline residues are important for multiple conformations [26, 27]. However, for this system, all proline residues are not equally important. We find a preference for Pro2 residues to adopt both the cis and trans isomer, resulting in multiple conformer families observed by IMS-MS. We find this trend to be true across a variety of peptide sequences; ~80% of all penultimate proline-containing peptides analyzed have multiple stable conformations. This is not meant to imply that only penultimate proline residues can adopt multiple conformations; previous IMS-MS studies have observed Pro residues located at other positions in peptides with multiple conformations [26, 27]. Rather, we suggest that this position may have an intrinsic ability to stabilize both cis-and trans-Xaa1-Pro2 isomers. This is likely due to low steric impedance of the N-terminal region, making this region of the peptide flexible and allowing for isomerization between the cis and trans forms of Xaa1–Pro2 peptide bonds.
Finally, it is interesting to consider the biological implications of Pro2 establishing multiple conformations. Proline is commonly found in the second position in many biological systems, including neuro- and vasoactive peptides [2, 4]. Although the high frequency of Pro2 residues is often hypothesized to stem from the ability of Pro residues to protect the N-terminal region from enzymatic degradation [1–3], it is possible that if Xaa1–Pro2 peptide bonds have a propensity to adopt both cis and trans isomers, this may have additional implications in peptide signaling.
Supplementary Material
Acknowledgments
The authors thank David Smiley and the DiMarchi research group at Indiana University for assistance with peptide synthesis. This work is supported by a grant from the NIH (R01 GM103725).
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s13361-014-1049-y) contains supplementary material, which is available to authorized users.
References
- 1.Falk K, Rötzschke O, Stevanović S, Jung G, Rammensee H-G. Pool sequencing of natural HLA-DR, DQ, DP ligands reveals detailed peptide motifs, constraints of processing, and general rules. Immunogenetics. 1994;39:230–242. doi: 10.1007/BF00188785. [DOI] [PubMed] [Google Scholar]
- 2.Vanhoof G, Goossens F, De Meester I, Hendriks D, Scharpé S. Proline motifs in peptides and their biological processing. FASEB J. 1995;9:736–744. [PubMed] [Google Scholar]
- 3.Nelson CA, Vidavsky I, Viner NJ, Gross ML, Unanue ER. Amino-terminal trimming of peptides for presentation on major histocompatibility complex class II molecules. Proc. Natl. Acad. Sci. U. S. A. 1997;94:628–633. doi: 10.1073/pnas.94.2.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Severini C, Improta G, Falconieri-Erspamer G, Salvadori S, Erspamer V. The tachykinin peptide family. Phamacol. Rev. 2002;54:285–322. doi: 10.1124/pr.54.2.285. [DOI] [PubMed] [Google Scholar]
- 5.Sarkar P, Reichman C, Saleh T, Birge RB, Kalodimos CG. Proline cis-trans isomerization controls autoinhibition of a signaling protein. Mol. Cell. 2007;25:413–426. doi: 10.1016/j.molcel.2007.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brazin KN, Mallis RJ, Fulton DB, Andreotti AH. Regulation of the tyrosine kinase Itk by the peptidyl-prolyl isomerase cyclophilin A. Proc. Natl. Acad. Sci. U. S. A. 2002;99:1899–1904. doi: 10.1073/pnas.042529199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lummis SCR, Beene DL, Lee LW, Lester HA, Broadhurst RW, Dougherty DA. Cis-trans isomerization at a proline opens the pore of a neurotransmitter-gated ion channel. Nature. 2005;438:248–252. doi: 10.1038/nature04130. [DOI] [PubMed] [Google Scholar]
- 8.Torbeev VY, Hilvert D. Both the cis-trans equilibriumand isomerization dynamics of a single proline amide modulate B2-microglobulin amyloid assembly. Proc. Natl. Acad. Sci. U. S. A. 2013;110:20051–20056. doi: 10.1073/pnas.1310414110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stewart DE, Sarkar A, Wampler JE. Occurrence and role of cis peptide bonds in protein structures. J. Mol. Biol. 1990;214:253–260. doi: 10.1016/0022-2836(90)90159-J. [DOI] [PubMed] [Google Scholar]
- 10.MacArthur MW, Thornton JM. Influence of proline residues on protein conformation. J. Mol. Biol. 1991;218:397–412. doi: 10.1016/0022-2836(91)90721-h. [DOI] [PubMed] [Google Scholar]
- 11.Andreotti AH. Native state proline isomerization: an intrinsic molecular switch. Biochemistry. 2003;42:9515–9524. doi: 10.1021/bi0350710. [DOI] [PubMed] [Google Scholar]
- 12.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989;246:64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 13.Clemmer DE, Jarrold MF. Ion mobility measurements and their applications to clusters and biomolecules. J. Mass Spectrom. 1997;32:577–592. [Google Scholar]
- 14.Wyttenbach T, Kemper PR, Bowers MT. Design of a new electrospray ion mobility mass spectrometer. Int. J. Mass Spectrom. 2001;212:13–23. [Google Scholar]
- 15.McLean JA, Ruotolo BT, Gillig KJ, Russell DH. Ion mobility-mass spectrometry: a new paradigm for proteomics. Int. J. Mass Spectrom. 2005;240:301–315. [Google Scholar]
- 16.Bohrer BC, Merenbloom SI, Koeniger SL, Hilderbrand AE, Clemmer DE. Biomolecule analysis by ion mobility spectrometry. Annu. Rev. Anal. Chem. 2008;1:293–327. doi: 10.1146/annurev.anchem.1.031207.113001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanu AB, Dwivedi P, Tam M, Matz L, Hill HH., Jr Ion mobility-mass spectrometry. J. Mass Spectrom. 2008;43:1–22. doi: 10.1002/jms.1383. [DOI] [PubMed] [Google Scholar]
- 18.Ruotolo BT, Giles K, Campuzano I, Sandercock AM, Bateman RH, Robinson CV. Evidence for macromolecular protein rings in the absence of bulk water. Science. 2005;310:1658–1661. doi: 10.1126/science.1120177. [DOI] [PubMed] [Google Scholar]
- 19.Ruotolo BT, Robinson CV. Aspects of native proteins are retained in vacuum. Curr. Opin. Chem. Biol. 2006;10:402–408. doi: 10.1016/j.cbpa.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 20.Bleiholder C, Dupuis NF, Wyttenbach T, Bowers MT. Ion mobility-mass spectrometry reveals a conformational conversion from random assembly to B-sheet in amyloid fibril formation. Nat. Chem. 2011;3:172–177. doi: 10.1038/nchem.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pierson NA, Chen L, Valentine SJ, Russell DH, Clemmer DE. Number of solution states of bradykinin from ion mobility and mass spectrometry measurements. J. Am. Chem. Soc. 2011;133:13810–13813. doi: 10.1021/ja203895j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen L, Chen S-H, Russell DH. An experimental study of the solvent-dependent self-assembly/disassembly and conformer preferences of gramicidin. A. Anal. Chem. 2013;85:7826–7833. doi: 10.1021/ac401370t. [DOI] [PubMed] [Google Scholar]
- 23.Silveira JA, Fort KL, Kim D, Servage KA, Pierson NA, Clemmer DE, Russell DH. From solution to the gas phase: stepwise dehydration and kinetic trapping of Substance P reveals the origin of peptide conformations. J. Am. Chem. Soc. 2013;135:19147–19153. doi: 10.1021/ja4114193. [DOI] [PubMed] [Google Scholar]
- 24.Wyttenbach T, Pierson NA, Clemmer DE, Bowers M. Ionmobility analysis of molecular dynamics. Annu. Rev. Phys. Chem. 2014;65:175–196. doi: 10.1146/annurev-physchem-040513-103644. [DOI] [PubMed] [Google Scholar]
- 25.Shi L, Holliday AE, Shi H, Zhu F, Ewing MA, Russell DH, Clemmer DE. Characterizing intermediates along the transition from polyproline I to polyproline II using ion mobility spectrometry-mass spectrometry. J. Am. Chem. Soc. 2014;136:12702–12711. doi: 10.1021/ja505899g. [DOI] [PubMed] [Google Scholar]
- 26.Counterman AE, Clemmer DE. Cis-trans signatures of proline-containing tryptic peptides in the gas phase. Anal. Chem. 2002;74:1946–1951. doi: 10.1021/ac011083k. [DOI] [PubMed] [Google Scholar]
- 27.Pierson NA, Chen L, Russell DH, Clemmer DE. Cis-trans isomerizations of proline residues are key to bradykinin conformations. J. Am. Chem. Soc. 2013;135:3186–3192. doi: 10.1021/ja3114505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schenk ER, Ridgeway ME, Park MA, Leng F, Fernandez-Lima F. Isomerization kinetics of AT hook decapeptide solution structures. Anal. Chem. 2014;86:1210–1214. doi: 10.1021/ac403386q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Warnke S, Baldauf C, Bowers MT, Pagel K, von Helden G. Photo-dissociation of conformer-selected ubiquitin ions reveals site-specific cis/trans isomerization of proline peptide bonds. J. Am. Chem. Soc. 2014;136:10308–10314. doi: 10.1021/ja502994b. [DOI] [PubMed] [Google Scholar]
- 30.Wu CH, Yeh L-SL, Huang H, Arminski L, Castro-Alvear J, Chen Y, Hu Z, Kourtesis P, Ledley RS, Suzek BE, Vinayaka CR, Zhang J, Barker WC. The protein information resource. Nucleic Acids Res. 2003;31:345–347. doi: 10.1093/nar/gkg040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koeniger SL, Merenbloom SI, Valentine SJ, Jarrold MF, Udseth HR, Smith RD, Clemmer DE. An IMS-IMS analogue of MS-MS. Anal. Chem. 2006;78:4161–4174. doi: 10.1021/ac051060w. [DOI] [PubMed] [Google Scholar]
- 32.Merenbloom SI, Koeniger SL, Valentine SJ, Plasencia MD, Clemmer DE. IMS-IMS and IMS-IMS-IMS/MS for separating peptide and protein fragment ions. Anal. Chem. 2006;78:2802–2809. doi: 10.1021/ac052208e. [DOI] [PubMed] [Google Scholar]
- 33.Tang K, Shvartsburg AA, Lee H-N, Prior DC, Buschbach MA, Li F, Tolmachev AV, Anderson GA, Smith RD. High-sensitivity ion mobility spectrometry/mass spectrometry using electrodynamic ion funnel interfaces. Anal. Chem. 2005;77:3330–3339. doi: 10.1021/ac048315a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoaglund CS, Valentine SJ, Sporleder CR, Reilly JP, Clemmer DE. Three-dimensional ion mobility/TOFMS analysis of electrosprayed biomolecules. Anal. Chem. 1998;70:2236–2242. doi: 10.1021/ac980059c. [DOI] [PubMed] [Google Scholar]
- 35.Mason EA, McDaniel EW. Transport properties of ions in gases. New York: Wiley; 1988. pp. 1–29. [Google Scholar]
- 36.Valentine SJ, Counterman AE, Clemmer DE. A database of 660 peptide ion cross sections: use of intrinsic size parameters for bona fide predictions of cross sections. J. Am. Soc. Mass Spectrom. 1999;10:1188–1211. doi: 10.1016/S1044-0305(99)00079-3. [DOI] [PubMed] [Google Scholar]
- 37.Srebalus-Barnes CA, Clemmer DE. Assessing intrinsic side chain interactions between i and i +4 residues in solvent-free peptides: a combinatorial gas-phase approach. J. Phys. Chem. A. 2003;107:10566–10579. [Google Scholar]
- 38.Dilger JM, Valentine SJ, Glover MS, Ewing MA, Clemmer DE. A database of alkali metal-containing peptide cross sections: influence of metals on size parameters for specific amino acids. Int. J. Mass Spectrom. 2012;330/332:35–45. [Google Scholar]
- 39.Mesleh MF, Hunter JM, Shvartsburg AA, Schatz GC, Jarrold MF. Structural information from ion mobility measurements: effects of the long-range potential. J. Phys. Chem. 1996;100:16082–16086. [Google Scholar]
- 40.Reimer U, Scherer G, Drewello M, Kruber S, Schutkowski M, Fischer G. Side-chain effects on peptidyl-prolyl cis/trans isomerization. J. Mol. Biol. 1998;279:449–460. doi: 10.1006/jmbi.1998.1770. [DOI] [PubMed] [Google Scholar]
- 41.Pal D, Chakrabarti P. Cis peptide bonds in proteins: residues involved, their conformations, interactions, and locations. J.Mol. Biol. 1999;294:271–288. doi: 10.1006/jmbi.1999.3217. [DOI] [PubMed] [Google Scholar]
- 42.Wathen B, Jia Z. Local and nonlocal environments around cis peptides. J. Proteome Res. 2008;7:145–153. doi: 10.1021/pr0704027. [DOI] [PubMed] [Google Scholar]
- 43.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.