Abstract
Bovine κ-caseinoglycomacropeptide (GMP) is a highly modified peptide from kappa-casein produced during the cheese making process. The chemical nature of GMP makes analysis by traditional proteomic approaches difficult, as the peptide bears a strong net negative charge and a variety of post-translational modifications. In this work, we describe the use of electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry (ESI FT-ICR MS) for the top-down analysis of GMP. The method allows the simultaneous detection of different GMP forms that result from the combination of amino acid genetic variations and post-translational modifications, specifically phosphorylation and O-glycosylation. The different GMP forms were identified by high resolution mass spectrometry in both negative and positive mode and confirmation was achieved by tandem MS. The results showed the predominance of two genetic variants of GMP that occur as either mono or bi-phosphorylated species. Additionally, these four forms can be modified with up to two o-glycans generally sialylated. The results demonstrate the presence of glycosylated, bi-phosphorylated forms of GMP, never describe before.
Introduction
Bovine κ-caseinoglycomacropeptide (GMP) is a sixty-four amino acid peptide formed from κ-casein during the cheese manufacturing process [1]. Chymosin acts upon κ-casein by hydrolyzing the peptide bond between residues Phe105 and Met106 to produce two peptides, para-κ-casein and GMP. Para-κ-casein is the N-terminal fragment, contains no posttranslational modifications (PTMs) and precipitates into the curd during the cheese making process. GMP is the C-terminal fragment of κ-casein and unlike para-κ-casein is a highly modified peptide containing all the PTMs that are found in κ-casein, specifically phosphorylation and O-glycosylation[1]. Because of its hydrophilic character GMP remain in solution and is therefore available for recovery in cheese whey. Research into GMP has gained much interest over the last decade, primarily due to the peptide’s bioactive properties, including regulation of gastric secretions[1], promoting the growth of bifidobacteria[1, 2], decreasing platelet aggregation[3] and adhesion of oral micro-organisms to host cells[4, 5]. Many of these bioactivities have been either directly linked to or are greatly influenced by the modifications on GMP, especially glycosylation[5].
The GMP amino acid sequence is highly conserved with two amino acid differences between the two predominant genetic variants, GMPa (Thr157, Asp169) and GMPb (Ile157, Ala169)[1, 6] (figure 1). Although differing in only in two residues, previous studies have demonstrated that different κ-casein genetic variants greatly affect the coagulation properties of bovine milk[7, 8]. In addition, the sites of phosphorylation and O-glycosylation have been mapped out in GMP through the use of enzymatic digests, Edman sequencing, and liquid chromatography. Phosphorylation in GMP appears to occur primarily at the Ser149 residue, although forms lacking phosphorylation or having two sites of phosphorylation have been previously identified [1, 5, 9]. Unlike the site of phosphorylation, sites of O-glycosylation in GMP vary greatly. It has been reported as many as six potential sites of O-glycosylation in GMP (Thr131, Thr133, Thr135, Thr136, Ser141 and Thr142) [5]. The structures of the O-glycans occupying these sites in GMP have been determined to be predominately core 1 type O-glycans and may contain up to two N-acetylneuraminic acid (NeuAc) residues bonded by either an α2–3 glycosidic bond to a galactose (Gal) residue or by an α2–6 glycosidic bond to an N-acetylgalactosamine (GalNac) residue[1, 5, 9].
Figure 1.
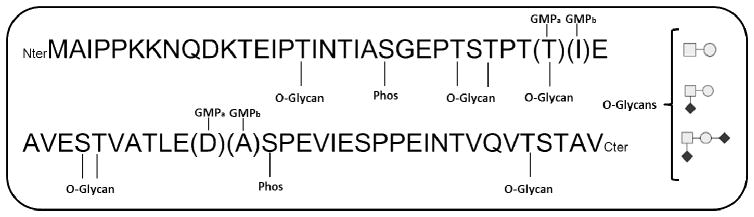
GMP sequence including potential O-glycosilation sites, potential phosphorylation sites as well as the two predominant genetic variants GMPa (Thr157, Asp169) and GMPb (Ile157, Ala169).
Despite the importance of this compound to the industry, there is no rapid and extensive method for characterization. The particular chemistry of GMP has made it a difficult peptide to study by the traditional methods used in proteomics for separations and detection such as polyacrylamide gel electrophoresis (PAGE), UV-VIS spectroscopy or ion chromatography[1]. The amino acid sequence of GMP is devoid of aromatic residues, which limits the use of UV-VIS spectroscopy as a detector in chromatographic separations[1, 6]. Separation and detection of GMP by PAGE is also not practical owing to the GMP molecule bearing a net negative charge except at very low pH values and fails to migrate correctly through the gel, leading to false molecular weight determinations[1, 10]. In addition, the net negative charge of GMP also limits the use of cation exchange chromatography as a means of separation. Advances in both capillary electrophoresis and high–performance anion exchange chromatography have shown promising results for the analysis of GMP [5]. However, both of these methods require either chemical hydrolysis of the glycan followed by introduction of a chemical tag or chemical modifications of the peptide in order to facilitate separation or detection of the glycans [5, 11, 12]. Affinity based methods like lectin affinity chromatography [13] or precipitation of GMP based on its affinity to chitosan [13, 14] have been used to separate the different glycoforms for identification and quantification. Whereas these methods can be successful, they are time intensive and have potential problems with specificity in regards to the isolation and detection of the glycoforms.
In this work, a purified GMP sample from a batch production was studied by direct infusion in an ESI FT-ICR MS. Using both, negative and positive mode and combining them with different fragmentation techniques, all GMP phospho- and glycoforms were properly identified. The top-down methodology described in this study is significatively simple compared with previous approaches requiring little sample preparation other than desalting of the sample. The minimal sample preparation speeds the analysis and reduces potential bias. This method could be potentially used with other large peptides that have undergone extensive post-translational modification.
Experimental
Chemicals and Sample set
Trifluoroacetic acid (TFA) and ammonium acetate were both purchased from Fisher Scientific (Pittsburgh, PA). Acetonitrile (ACN) was purchased from Sigma Aldrich (St. Louis, MO). Water was prepared in house using a Barnsted E-Pure (ThermoScientific, Waltham, MA) water treatment system and was purified to a final resistance of 18 MΩ. The GMP sample used in this study was kindly provided by Davisco Foods International, Inc (Eden Prairie, MN, USA) from their GMP Biopure product line. The sample was 90% pure. The purification procedure can be found under the patent US6800739B2.
Sample preparation
One milligram of the GMP sample was desalted prior to the analysis by using Discovery DSC-8 solid phase extraction columns (SPE) containing 500mg of extraction material (Sigma Aldrich, St. Louis, MO). The extraction was performed according to the method supplied by Michrom Bioresources, Inc. for peptide desalting. The desalted GMP was dried using a centrifugal vacuum device and stored at −20 °C. For the mass spectrometry analysis, the GMP sample was reconstituted in one milliliter of a solution of water:acetonitrile (1:1) spiked with 5 mM ammonium acetate.
Mass Spectrometry
Mass spectrometry experiments were performed on a Varian MS-920 hybrid triple quadrupole Fourier transform ion cyclotron resonance mass spectrometer (FTICR) equipped with an actively shielded 9.4 T superconducting magnet. Briefly, this instrument consists of a standard triple quadrupole mass spectrometer that is capable of passing ions to the FT-ICR portion of the instrument. Analyses were performed in both positive and negative mode. Samples, introduced by direct infusion at a flow rate of 10uL/min, were ionized using a standard electrospray ionization source (ESI). The ESI source conditions were optimized being a needle voltage of −4500 kV, shield voltage of −600 V, drying gas temperature of 300 °C, drying gas pressure of 10 psi and a nebulizing gas pressure of 15 psi. The triple quadrupole was operated in ion guide mode with the ion transmission window set at m/z 1990 and a dwell time of 0.5 seconds. The voltages on the ion optics and ion accumulation times were adjusted as needed to maximize ion transmission to the ICR cell. Ions were excited for detection or removed from the ICR cell during isolation events by application of arbitrary waveforms. All transients were recorded at a digitizer rate of 2 MHz with 2048K samples being taken to give a transient length of 1.049 seconds. Following acquisition, each transient was zero-filled a single time and apodized with a Blackman windowing function. Prior to data collection the FT-ICR was externally calibrated using Agilent ESI Tune mix (Agilent, Santa Clara, CA).
Tandem mass spectrometry experiments were performed using both infrared multiphoton dissociation (IRMPD) and electron capture dissociation (ECD) in the positive mode. Ions of interest were initially mass filtered in the quadrupole side of the instrument and further isolated within the ICR cell by application of an arbitrary waveform. A pulse of nitrogen gas was introduced during the IRMPD event to collisionally cool the product ions and keep them in the path of the IR beam. ECD experiments were performed following isolation by irradiating the ions with electrons generated with a heated rhenium filament. Different charge states were tested and the period of irradiation and the energy of the electrons were optimized to fragment efficiently the ions.
Data analysis
All acquired spectra were analyzed in the Varian Omega FTMS software. All spectra were charge deconvoluted and the average mass for each isotope cluster was determined with the Omega software. ECD fragmentation experiments were analyzed with MS-Seq of Protein Prospector [15]. Masses were allowed 5 ppm and 10 ppm error for the parent ion and the fragments respectively. No complete peptide modifications were included but phosphorylation was allowed as a potential modification at the serine and threonine residues. A non-specific cleavage was used to search against the bovine proteome obtained from Uniprot. Instrument fragmentation was set as ESI-FT-ICR-ECD.
Results and Discussion
High-resolution cationic mode experiments
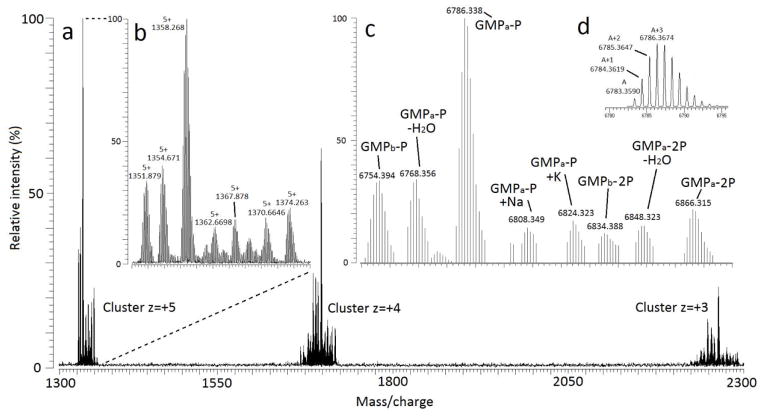
The positive mode ESI FT-ICR MS spectrum of the GMP sample showed several clusters of multiply charged ions (up to charge state +7) spanning the m/z range 900–3500 and varying greatly in intensity. Figure 2a shows charge state from +3 to +5 in the m/z range 1300–2300. While identification of the various forms of GMP is possible in the m/z domain, it is greatly simplified by using the charge deconvoluted data as all of the different forms of GMP will occur at one unique mass rather than occurring at multiple m/z values.
Figure 2.
GMP results in the positive mode; (a) mass spectrum in the 1300–2300 m/z range showing different GMP forms clustering by charge-state; (b) zoom in cluster +5; (c) deconvoluted spectrum; (d) theoretical isotopic pattern of GMPa-P.
The initial analysis of the deconvoluted GMP spectrum in the positive mode (Figure 2c) showed several distinct forms. The more intense signal in the sample corresponds to the phosphorylated GMPa (GMPa-P), whose more abundant isotope (A+3) was detected with a mass of 6786.3382 Da. GMPa-P is the most intense signal in the spectrum being the base peak and roughly three times as abundant as the other genetic variant, phosphorylated GMPb (GMPb-P). Both GMPa-P and GMPb-P were both identified by comparing their deconvoluted experimental masses to theoretical masses. The isotopic pattern of each form of GMP showed good agreement with the theoretical distribution (Figure 2d and Table S2). Both GMPa and GMPb appear in the spectrum in two forms: mono and bi-phosphorylated species (GMPa-2P and GMPb-2P in figure 2c) Dephosphorylated species were not found. Additionally, adducts with sodium and potassium as well as dehydrated species were detected.
Genetic variant confirmation by ECD
Unambiguous identification of the genetic variants A and B was confirmed by ECD tandem-MS. Tandem-MS experiments of quasimolecular ions 970.48+7 m/z and 965.92+7 m/z corresponding with GMPa-P and GMPb-P forms were performed (figures S1a and S1b). The ECD fragmentation results were analyzed with MS-Tag from Protein prospector [15] against the whole bovine proteome. MS-Tag matched 55 ion fragments from the tandem MS spectrum of ion 970.48+7 to GMPa-P phosphorylated at residue S170. Similarly, MS-Tag identified ion 965.92+7 to GMPb-P matching 19 fragments. A table with fragment assignments is shown at the supplementary material (table S1).
High-resolution anionic mode experiments
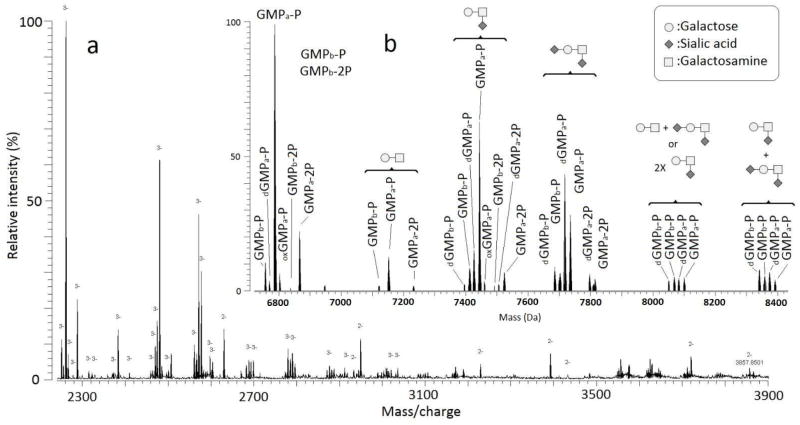
The negative mode ESI FT-ICR MS spectrum of the GMP sample (Figure 3a) showed several clusters of multiply charged ions spanning the m/z range 2000–3500 and varying greatly in intensity. The charge states of these ions were −2 and −3, with the majority of the ions being in the −3 charge state.
Figure 3.
GMP results in the negative mode; (a) mass spectrum in the 2300–3900 m/z range showing different GMP forms with charge states −2/−3; (b) deconvoluted spectrum.
The deconvoluted spectrum in the negative mode (Figure 3b) revealed a larger diversity of GMP forms compared to the positive mode. Again, both GMPa and GMPb are found phosphorylated (GMPa/b-P) and biphosphorylated (GMPa/b-2P) with their corresponding cohort of oxidized and dehydrated forms. No adducts with sodium or potassium were found. The other forms of GMP that were detected in the sample were determined to be GMPa-P and GMPb-P but have been modified with different glycan structures. The presence of sialylated glycans facilitates the detection of GMP glycosylated forms in the anionic mode. GMP is negatively charged even in acidic conditions (pI≈4) [16]. As the sample is infused without chromatographic separation, non-glycosylated GMP competes the glycosylated GMP for the charge during ESI. Ion suppression of glycopeptides by non-glycosylated peptides is a common phenomenon in MS [17, 18]. In the anionic mode on the other hand, the additional negative charge of the sialic acid groups seems to facilitate the glycopeptide ionization.
The identification of the glycosylated forms and the glycans on these species was made by determining the difference in mass between the species that were potentially glycosylated and GMPa-P/GMPbB-P. The addition of each carbohydrate residue (each residue being the carbohydrate minus a water molecule to account for the bond to the peptide) to a peptide causes a distinct increase in the mass, which can be used as a diagnostic mass difference. As an example, the addition of a single N-acetylhexoseamine (HexNAc) residue to GMPa-P will lead to a mass increase of 203.079 Da, while the addition of a disaccharide composed by HexNAc and hexose (Hex) would lead to a mass increase of 365.132 Da. It was determined that the glycosylated forms shown are modified with glycans consisting of HexNAc, Hex, and sialic acid (NeuAc) residues. Some forms of GMPa-P and GMPb-P were observed to contain one or two glycan moieties (Table 1).
Table 1.
Summary of GMP forms found.
| Genetic Variant | Phosphorylation | Glycosylation | Mass observeda,b | Theoretical Massa,b | Mass accuracyc |
|---|---|---|---|---|---|
| A | 1 | - | 6786.3382 | 6786.3674 | 4.3 |
|
|
7151.4574 | 7151.4996 | 5.9 | ||
|
7442.6270 | 7442.5950 | −4.3 | ||
|
7733.6904 | 7733.6896 | 0.1 | ||
|
|
8098.7926 | 8098.8226 | 3.7 | ||
|
|
8389.8903 | 8389.9180 | 3.3 | ||
| 2 | - | 6866.3152 | 6866.3337 | 2.7 | |
|
|
7231.4544 | 7231.4659 | 1.6 | ||
|
7522.5583 | 7522.5613 | 0.4 | ||
|
7813.6685 | 7813.6567 | −1.5 | ||
| B | 1 | - | 6754.3944 | 6754.4140 | 2.9 |
|
|
7119.5775 | 7119.5462 | −4.4 | ||
|
7410.6564 | 7410.6416 | −2 | ||
|
7701.7093 | 7701.7370 | 3.6 | ||
|
|
8066.8490 | 8066.8692 | 2.5 | ||
|
8357.9930 | 8357.9646 | −3.4 | ||
| 2 | - | 6834.3878 | 6834.3803 | −1.1 | |
|
7490.5959 | 7490.6079 | 1.6 | ||
|
7781.6808 | 7781.7033 | 2.9 |
Dalton units.
More abundant isotope (A+3).
ppm.
The smallest glycan that was detected is a monosaccharide of composition HexNAc, with the largest glycan being a tetrasaccharide of composition HexNAcHexNeuAc2. On the glycosylated forms bearing two glycans a mix of the tri- and tetrasaccharide glycans were found. These findings are in agreement with what has previously been published on the glycosylation of GMP [1, 5, 9, 19, 20]. After identification of all forms of GMP that were expected based on literature, several species in the spectra were left unidentified and did not match what has previously been reported. Based on the increase in mass (≈79.9 Da) these species were tentatively assigned as being forms of GMP with two sites of phosphorylation. Doubly phosphorylated GMP has been previously reported, albeit at low abundance and only occurring in non-glycosylated species [9]. In this study, the double phosphorylation were observed for both the non-glycosylated GMP and also for the GMP bearing a single glycan, while the combination of two sites of phosphorylation and two or more sites of glycosylation were not observed. In a more recent study combining electrophoresis and mass spectrometry Jensen et al. [20] identified six different κ-casein isoforms. Otherwise in agreement with our results, bi-phosphorylated glycosylated forms were not found in that study although unidentified low abundance κ-casein forms were present in the gel. Sulfation of a tyrosine residue would also result in a mass increase of 79.9 Da. However, GMP has no tyrosine residues in its sequence; therefore sulfation of the peptide can be eliminated. Sulfation of a glycan structure can also be ruled out as the unidentified species are seen for all forms of GMP, including the non-glycosylated form.
Although the specific biological role of the GMP modifications is not fully understood a number of studies indicated that both phosphorylation and o-glycosylation are involved on the stability of the casein micelles [21, 22]. Briefly, the negative charge that these modifications add to κ-casein enhances the electrostatic repulsion of the micelles preventing their aggregation. In turn, the stability of the casein micelles affects the precipitation of the curd during the cheese making process. In this context, the presence of bi-phosphorylated glycosylated forms of GMP is expected to increase the charge of κ-casein and hence the micelle stability. Nevertheless, a rheological study would be needed to determine the importance of this new GMP form on the precipitation of the curd.
O-glycosylation confirmation by IRMPD
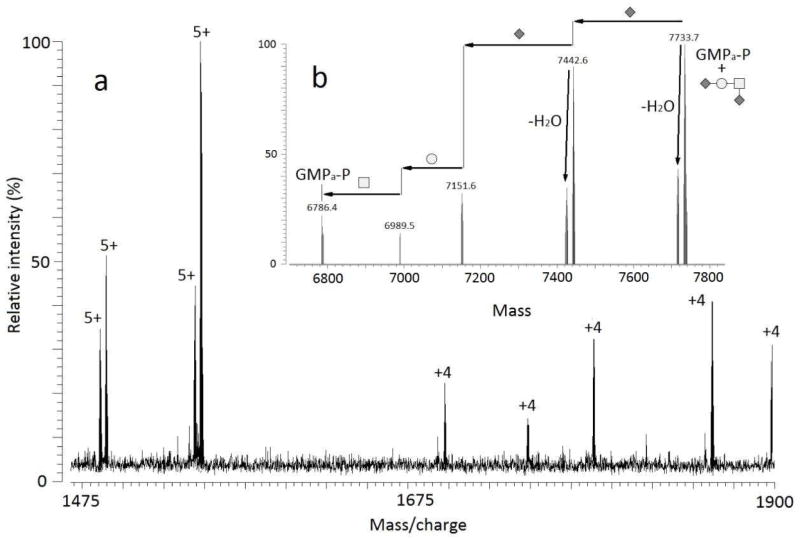
Additional confirmation of the glycan composition was made by performing tandem mass spectrometry experiments on the 7733.6904 Da species that was assigned as a singly glycosylated GMPa-P. This glycoform was isolated as m/z 1547.7381, the 5+ charge state, in the positive mode and was fragmented by IRMPD [19] (Figure 4a). While the corresponding ion-molecule in the negative mode is orders of magnitude more intense, its fragmentation yield only the dehydrated form (data not shown). IRMPD fragmentation of this glycosylated form of GMP in the positive mode was possible by accumulation of 50 spectra. The charge deconvoluted IRMPD spectrum of m/z 1548.1 is shown in Figure 4b.
Figure 4.
IRMPD fragmention of 1547.7381 (5+) (a) raw spectrum, (b) deconvoluted spectrum.
The results of the IRMPD fragmentation clearly support the mass-based glycan assignment of HexNAcHexNeuAc2. The isolated ion fragmented in a manner such that the glycan can be sequenced by looking at the differences in mass between the fragments. It can be seen that the glycan contained two NeuAc residues as there is a loss of 291 Da from the precursor ion and a second loss of 291 Da from the fragment at 7442.6 Da. The final diagnostic loss in the spectrum occurs between 7151.6 Da and 6786.4 Da, a difference in mass of 365 Da, which corresponds to the disaccharide HexNAcHex.
Conclusion
A method for the comprehensive analysis of posttranslational modifications in bovine caseinoglycomacropeptide using electrospray ionization Fourier transform ion cyclotron mass spectrometry has been presented. The method makes use of differences in mass between different species and a combinatorial approach based on diagnostic carbohydrate masses in order to identify the different glycoforms in terms of their glycan composition. Confirmation was obtained by Tandem-MS. Otherwise in agreement with previous studies, glycosylated, bi-phosphorylated forms of GMP, never describe before were detected.
Supplementary Material
Acknowledgments
Funding provided by the National Institute of Health (AT007079, HD061923 and 1U24DK097154) is gratefully acknowledged. Authors thank Davisco Foods International, Inc. (Eden Prairie, MN, USA.) for the bovine GMP sample. A.G. wants to thank the Spanish National Research Council for the economical supply 2009 EST CSIC–1536.
Footnotes
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
References
- 1.Brody EP. Biological activities of bovine glycomacropeptide. Br J Nutr. 2000;84:39–46. doi: 10.1017/s0007114500002233. [DOI] [PubMed] [Google Scholar]
- 2.Proulx M, Gauthier SF, Roy D. Effect of casein hydrolysates on the growth of Bifidobacteria. Le Lait. 1992;72:393–404. [Google Scholar]
- 3.Jolles P, Levy-Toledano S, Fiat AM, Soria C, Gillessen D, Thomaidis A, Dunn FW, Caen JP. Analogy between fibrinogen and casein. Effect of an undecapeptide isolated from kappa-casein on platelet function. Eur J Biochem. 1986;158:379–382. doi: 10.1111/j.1432-1033.1986.tb09764.x. [DOI] [PubMed] [Google Scholar]
- 4.Neeser JR, Grafstrom RC, Woltz A, Brassart D, Fryder V, Guggenheim B. A 23 kDa membrane glycoprotein bearing NeuNAc alpha 2–3Gal beta 1–3GalNAc O-linked carbohydrate chains acts as a receptor for Streptococcus sanguis OMZ 9 on human buccal epithelial cells. Glycobiology. 1995;5:97–104. doi: 10.1093/glycob/5.1.97. [DOI] [PubMed] [Google Scholar]
- 5.Tran NT, Daali Y, Cherkaoui S, Taverna M, Neeser JR, Veuthey JL. Routine o-glycan characterization in nutritional supplements--a comparison of analytical methods for the monitoring of the bovine kappa-casein macropeptide glycosylation. J Chromatogr A. 2001;929:151–163. doi: 10.1016/s0021-9673(01)01176-1. [DOI] [PubMed] [Google Scholar]
- 6.Whitney RM. Proteins in milk. In: Marth EH, editor. Fundamentals of Dairy Chemistry. Van Nostrand Reinhold; New York: 1988. p. 88. [Google Scholar]
- 7.Ikonen T, Ahlfors K, Kempe R, Ojala M, Ruottinen O. Genetic Parameters for the Milk Coagulation Properties and Prevalence of Noncoagulating Milk in Finnish Dairy Cows. J Dairy Sci. 1999;82:205–214. doi: 10.3168/jds.S0022-0302(99)75225-2. [DOI] [PubMed] [Google Scholar]
- 8.Wedholm A, Larsen LB, Lindmark-Månsson H, Karlsson AH, Andrén A. Effect of Protein Composition on the Cheese-Making Properties of Milk from Individual Dairy Cows. J Dairy Sci. 2006;89:3296–3305. doi: 10.3168/jds.S0022-0302(06)72366-9. [DOI] [PubMed] [Google Scholar]
- 9.Minikiewicz P, Slangen CJ, Lagerwerf FM, Haverkamp J, Rollema HS, Visser S. Reversed-phase high-performance liquid chromatographic separation of bovine kappa-casein macropeptide and characterization of isolated fractions. J Chromatogr A. 1996;743:123–135. doi: 10.1016/0021-9673(96)00122-7. [DOI] [PubMed] [Google Scholar]
- 10.Zhang J, Ren Y, Ma Z, Huang B, Cai Z, Li D. Quantitative determination of bovine caseinoglycomacropeptide in infant formulas by ultra-high-performance liquid chromatography-electrospray-ionization mass spectrometry. J Sep Sci. 2011;34:2751–2758. doi: 10.1002/jssc.201100437. [DOI] [PubMed] [Google Scholar]
- 11.El Rassi Z. Recent developments in capillary electrophoresis of carbohydrate species. Electrophoresis. 1997;18:2400–2407. doi: 10.1002/elps.1150181230. [DOI] [PubMed] [Google Scholar]
- 12.Bigge JC, Patel TP, Bruce JA, Goulding PN, Charles SM, Parekh RB. Nonselective and efficient fluorescent labeling of glycans using 2-amino benzamide and anthranilic acid. Anal Biochem. 1995;230:229–238. doi: 10.1006/abio.1995.1468. [DOI] [PubMed] [Google Scholar]
- 13.Koliwer-Brandl H, Siegert N, Umnus K, Kelm A, Tolkach A, Kulozik U, Kuballa J, Cartellieri S, Kelm S. Lectin inhibition assays for the analysis of bioactive milk sialoglycoconjugates. Int Dairy J. 2011;21:413–420. [Google Scholar]
- 14.Casal E, Corzo N, Moreno FJ, Olano A. Selective recovery of glycosylated caseinmacropeptide with chitosan. J Agric Food Chem. 2005;53:1201–1204. doi: 10.1021/jf040256e. [DOI] [PubMed] [Google Scholar]
- 15.Jiménez CR, Huang L, Qiu Y, Burlingame AL. Current Protocols in Protein Science. John Wiley & Sons, Inc; 2001. Searching Sequence Databases Over the Internet: Protein Identification Using MS-Tag. [DOI] [PubMed] [Google Scholar]
- 16.Bjellqvist B, Hughes GJ, Pasquali C, Paquet N, Ravier F, Sanchez JC, Frutiger S, Hochstrasser D. The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis. 1993;14:1023–1031. doi: 10.1002/elps.11501401163. [DOI] [PubMed] [Google Scholar]
- 17.Larsen MR, Højrup P, Roepstorff P. Characterization of Gel-separated Glycoproteins Using Two-step Proteolytic Digestion Combined with Sequential Microcolumns and Mass Spectrometry. Mol Cell Proteomics. 2005;4:107–119. doi: 10.1074/mcp.M400068-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Geng M, Zhang X, Bina M, Regnier F. Proteomics of glycoproteins based on affinity selection of glycopeptides from tryptic digests. J Chromatogr B. 2001;752:293–306. doi: 10.1016/s0378-4347(00)00550-8. [DOI] [PubMed] [Google Scholar]
- 19.Wuhrer M, Catalina MI, Deelder AM, Hokke CH. Glycoproteomics based on tandem mass spectrometry of glycopeptides. J Chromatogr B. 2007;849:115–128. doi: 10.1016/j.jchromb.2006.09.041. [DOI] [PubMed] [Google Scholar]
- 20.Jensen HB, Holland JW, Poulsen NA, Larsen LB. Milk protein genetic variants and isoforms identified in bovine milk representing extremes in coagulation properties. J Dairy Sci. 2012;95:2891–2903. doi: 10.3168/jds.2012-5346. [DOI] [PubMed] [Google Scholar]
- 21.Tyrisevä AM, Elo K, Kuusipuro A, Vilva V, Jänönen I, Karjalainen H, Ikonen T, Ojala M. Chromosomal Regions Underlying Noncoagulation of Milk in Finnish Ayrshire Cows. Genetics. 2008;180:1211–1220. doi: 10.1534/genetics.107.083964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frederiksen PD, Andersen KK, Hammershøj M, Poulsen HD, Sørensen J, Bakman M, Qvist KB, Larsen LB. Composition and effect of blending of noncoagulating, poorly coagulating, and well-coagulating bovine milk from individual Danish Holstein cows. J Dairy Sci. 2011;94:4787–4799. doi: 10.3168/jds.2011-4343. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.