Abstract
Discoidin domain receptor (DDR) 1 and 2 are transmembrane receptors that belong to the family of receptor tyrosine kinases (RTK). Upon collagen binding, DDRs transduce cellular signaling involved in various cell functions, including cell adhesion, proliferation, differentiation, migration, and matrix homeostasis. Altered DDR function resulting from either mutations or overexpression has been implicated in several types of disease, including atherosclerosis, inflammation, cancer, and tissue fibrosis. Several established inhibitors, such as imatinib, dasatinib, and nilotinib, originally developed as Abelson murine leukemia (Abl) kinase inhibitors, have been found to inhibit DDR kinase activity. As we review here, recent discoveries of novel inhibitors and their co-crystal structure with the DDR1 kinase domain have made structure-based drug discovery for DDR1 amenable.
Keywords: DDR1 kinase, inhibitors, structure-based drug discovery, comparative modeling
Introduction
DDR1 and DDR2 are RTKs comprising an extracellular Discoidin (DS) homology domain that encompasses the collagen-binding site, a DS-like domain that contributes to collagen-induced receptor activation, an extracellular juxtamembrane region that contains N- and O-glycosylation sites and matrix metalloproteinase cleavage sites [1]. In addition, DDRs have a single transmembrane helix, an intracellular juxtamembrane regulatory region upstream of a cytoplasmic tyrosine kinase domain [2]. The DDR family comprises two distinct members, DDR1 and DDR2. DDR1 has five isoforms, whereas DDR2 has a single one [2].
Upon activation by binding of fibrillar collagens I–III, V, or networking-forming collagen IV, DDR1 undergoes phosphorylation and initiates various downstream signaling pathways. Multiple tyrosine residues within the intracellular juxtamembrane region and tyrosine kinase domain of DDR1 can be phosphorylated and recruit proteins, such as ShcA, SHP-2, and the p85 subunit of phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K) [3–6]. DDR1 stimulates several signaling pathways in a context- and cell type-dependent manner. For example, DDR1 activates estrogen receptor kinase (ERK) signaling in vascular smooth muscle cells [7], but inhibits ERK in mesangial cells [8], and has no effect on ERK activation in T47D breast cancer cells [6]. In addition, DDR1 modulates signaling pathways initiated by other matrix receptors (e.g., integrins) [9], cytokines [e.g., transforming growth factor (TGF)-β] [10], and transmembrane receptors (e.g., Notch1) [11]. Interaction of DDR1 with various receptors is important for the regulation of cell survival, migration, and differentiation in development and pathological conditions [5, 9, 12, 13].
Our understanding of the role of DDR1 in development, tissue homeostasis, and disease has been significantly enhanced by availability of DDR1-deficient mice. These mice have defects in mammary gland morphogenesis and inability of blastocysts to implant properly in the uterine wall [14]. In contrast to these findings, DDR1 ablation has been shown to have a beneficial role in various mouse models of fibrotic diseases, including atherosclerosis [15], pulmonary fibrosis [16], and renal fibrosis [13]. Thus, inhibiting DDR1 might be a promising therapeutic strategy for fibrotic diseases.
The DDR1 kinase domain
The DDR1 intracellular kinase domain shares the typical structure of other kinase domains (Figure 1). However, how DDR1 kinase is activated upon collagen binding is poorly understood. It is thought that the process is fundamentally different from the accepted paradigm of ligand-induced RTK dimerization. Unlike typical RTKs, DDR1 exists as a preformed dimer and, following collagen binding, undergoes receptor oligomerization and internalization, and is phosphorylated unusually slowly. A recent study showed that collagen binding to DDR1 fails to induce a major conformational change that could explain kinase activation, and instead proposed that collagen-induced receptor oligomerization might be responsible for kinase activation [17]. In support of this hypothesis, events that reduce receptor oligomerization, such as antibodies that bind to DS-like domain or enforced covalent receptor dimerization at residues within the DS-like domain, reduce DDR1 phosphorylation and activation. However, mutation of Asn211, a conserved glycosylation site within the DS-like domain, results in ligand-independent activation of DDR1, enhanced receptor dimerization, and internalization, suggesting that, in addition to receptor clustering, ligand-induced internalization also contributes to receptor activation [18].
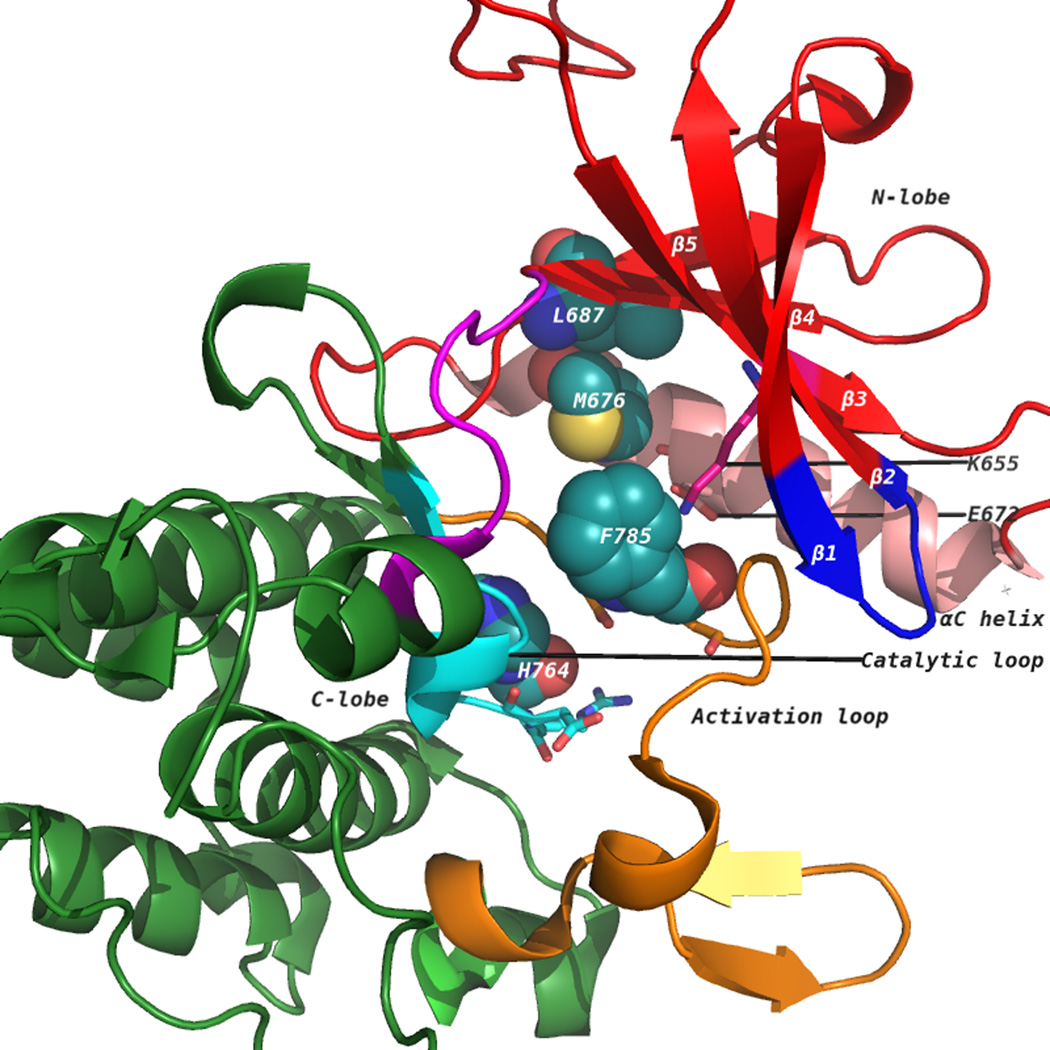
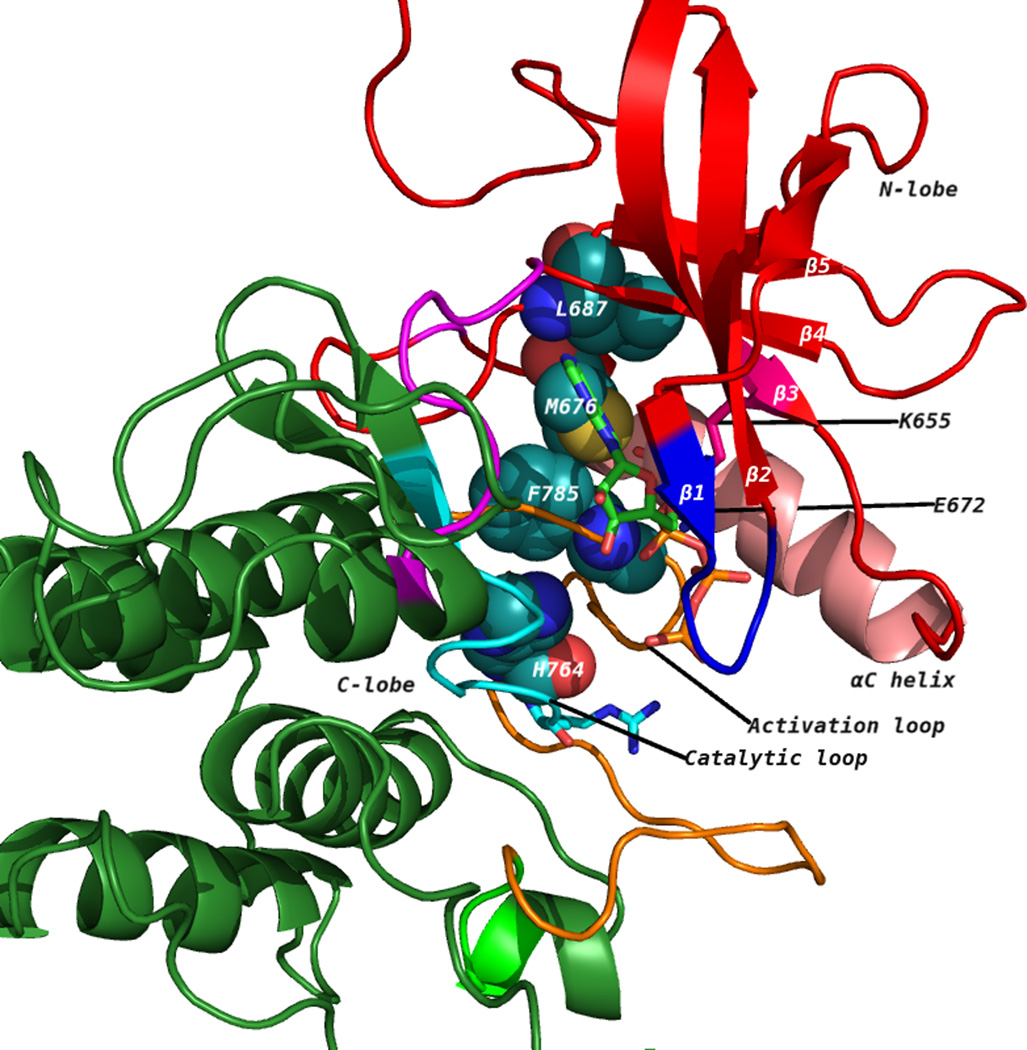
Figure 1.
(A) The discoidin domain receptor 1 (DDR1) kinase domain (3ZOS) has a characteristic bilobal architecture. The image shows the DFG-out (F785 belongs to the DFG motif in the catalytic loop) or the ‘inactive’ state. The N-terminal lobe contains five β strands (1–5, in red) and a universally conserved helix called αC (in pink). The C-terminal lobe is primarily helical (in green). The catalytic loop is in cyan, whereas the activation loop is in orange. The disrupted hydrophobic spine that is characteristic of inactive kinases is shown as spheres. (B) An active DDR1 conformation homology model (DFG-in) showing intact hydrophobic spines in spheres. ATP occupies the cleft between the N-lobe and C-lobe. The ATP-binding pocket is bound by a glycine-rich loop and C-terminal hinge region.
Collagen binding to DDR1 induces a slow receptor tyrosine autophosphorylation of multiple tyrosine residues, including Tyr792, Tyr796, and Tyr797, in the activation loop, which is likely to cause the kinase domain to switch from the inactive to the active state [19]. The active state satisfies chemical restraints that allow the transfer of the γ-phosphate of ATP to the hydroxyl group of tyrosine on the loop, which enables recruitment of substrate proteins. Here, we refer to the residues of the DDR1 kinase domain [Protein Data Bank (PDB): 3ZOS] in relation to their corresponding residues in protein kinase A (PKA, PDB: 1ATP) and the kinase domain of Abl tyrosine kinase. Positions indicated in italics and normal in square brackets are PKA and Abl kinase residue numbers, respectively, equivalent to those in DDR1.
Kinase domain structure and important residues
The tyrosine kinase domain comprises an N-terminal (N-lobe) and a C-terminal (C-lobe) lobe [20]. Figure 1A shows the kinase domain of DDR1 (PDB: 3ZOS) [21]. The N-lobe comprises a five-stranded β-sheet and a prominent α- helix, called αC helix, whereas the C-lobe is mostly helical [20]. The ATP binding pocket lies in the cleft between the two lobes and sits beneath a highly conserved glycine-rich loop, which is between the β1 and β2 strands [20] (Figure 1A). In the active conformation (Figure 1B), this loop positions the γ-phosphate of ATP for catalysis and a conserved valine, Val624 [Val57, Val254] makes a hydrophobic contact to the base of ATP. As with other domains, the DDR1 kinase domain contains the highly conserved DFG and HRD (YRD in case of PKA) motifs on the activation and catalytic loops, respectively. Asp784 [Asp184, Asp381] of the Asp-Phe-Gly (DFG) motif forms polar contacts with all three ATP phosphates. The phenylalanine of Asp784-Phe785-Gly786 [Asp184-Phe185-Gly186, Asp381-Phe382-Gly383] makes hydrophobic contacts with the Met676 [Leu95, Met290] of the αC helix and the histidine of the conserved His764-Arg765-Asp766 [Tyr164-Arg165-Asp166, His361-Arg362-Asp363] motif. The His-Arg-Asp (HRD) motif of DDR1 and Abl [Tyr-Arg-Asp (YRD) in PKA] is part of the ‘activation loop’ that provides a platform for peptide substrate binding. Phosphorylation of tyrosine residues within the activation loop is required to support a configuration that enables binding and phosphorylation of substrate protein. A conserved glutamate residue Glu672 [Glu91, Glu286] located on the αC helix forms an ion pair with the Lys655 [Lys72, Lys271] side chain that coordinates the α- and β-phosphates of ATP. In several active kinases, αC makes direct contact with N-terminal region of activation loop, with its conformation often linked to the DFG motif. Although there is no experimental structure of DDR1 in its active conformation, it is expected that this feature is preserved.
The C-lobe comprises mostly α-helices that surround a central β-sheet and serves as a docking site for substrate proteins. The residues in the interface of C-lobe with N-lobe are involved in catalytic machinery associated with the transfer of phosphate from ATP. Typically, disruption of interactions within the N-lobe and between the two lobes immobilizes kinase activity.
Overall, the DDR1 kinase domain has a typical kinase domain structure and adequate sequence and/or structure similarity with other kinase domains, as shown in Figure 2, which allow homology modeling of the active and inactive conformations.
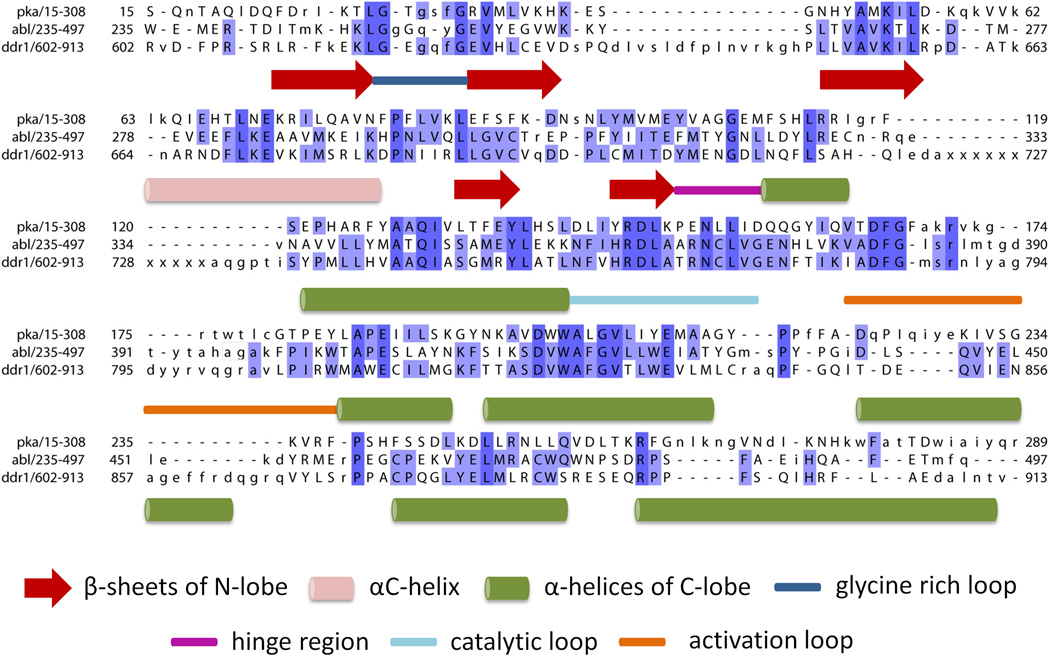
Figure 2.
The structure and sequence alignment of protein kinase A [PKA; Protein Data Bank (PDB): 1ATP-E)], Abl kinase (PDB: 2HYY-A), and discoidin domain receptor 1 (DDR1; PDB: 3ZOS-A). The PDB structures were aligned using PDBEFOLD [37]. The sequence limits are as defined in respective PDB. Missing regions in the DDR1 PDB entry are denoted by ‘x’ and the intensity of blue color indicates sequence conservation. Structural elements are described associated with the sequences.
In addition to conformational changes to the activation loop, αC-helix position and orientation of catalytic residues, conserved spatial arrangement patterns of residues have been identified in the active kinase conformation. Four residues in the ATP-binding site link together N and C lobes of the kinase domain [22, 23]. Residues His764, Phe785, Met676, and Leu687 [Tyr164, Phe185, Leu95, and Leu106; His361, Phe382, Met290, and Leu301] form the so-called ‘hydrophobic spine’. An intact conformation of the hydrophobic spine is essential for maintaining the active state conformation of kinase domain, whereas disruption of the arrangement leads to inactive conformation. The hydrophobic spine supports the relative orientation of the two lobes as a hinge for interconversion of the open and closed conformations required for binding ATP and releasing ADP [23, 24]. Figure 1A and 1B illustrate the hydrophobic spines in inactive conformation of DDR1 (PDB: 3ZOS) and active DDR1 conformation (homology model), respectively.
Targeting DDR1 for inhibition
Given that DDR1 has a key role in pathological conditions, including atherosclerosis, cancer, inflammation, and fibrosis, blocking DDR1-mediated downstream signaling by inhibiting the transfer of γ-phosphate of ATP to the hydroxyl group of the tyrosine kinase on protein substrates is an appealing strategy to prevent DDR1 activation.
DDR1 inhibitors reported so far are ATP competitive inhibitors that bind to either the active (type-1 inhibitors) or inactive (type-2 inhibitors) conformations, preventing transfer of the terminal phosphate group of ATP to the protein substrate. Screening for inhibitory activity against a panel of kinases identified imatinib [25], nilotinib [26], dasatinib [26], and bafetinib [27] as DDR1 inhibitors (Figure 3). Day et al. reported inhibition of DDR1 by imatinib, nilotinib, and dasatinib with IC50 values of 43± 2.4 nM, 3.7± 1.2 nM and 1.35± 0.2 nM, respectively [28]. However, these inhibitors are not selective, because they were originally designed to target Abl kinase. Sun et al. identified (3-(2-(3-(morpholinomethyl)phenyl)thieno [3,2-b]pyridin-7-ylamino) phenol (LCB 03-0110 in Figure 3) as a potent inhibitor of both DDR1 and DDR2 along with several other tyrosine kinases [29]. Recently, Gao et al. identified a series of 3-(2-(pyrazolo[1,5-a]pyrimidin-6-yl) ethynyl)benzamides as potent DDR1 inhibitors, the most potent of which (7rh and 7rj in Figure 3) have IC50 values of 6.8 and 7.0 nM, respectively [30]. Kim et al. reported two inhibitors, DDR1-IN-1 and DDR1-IN-2 (Figure 3), which exhibit an IC50 of 105 nM and 47 nM, respectively [31].
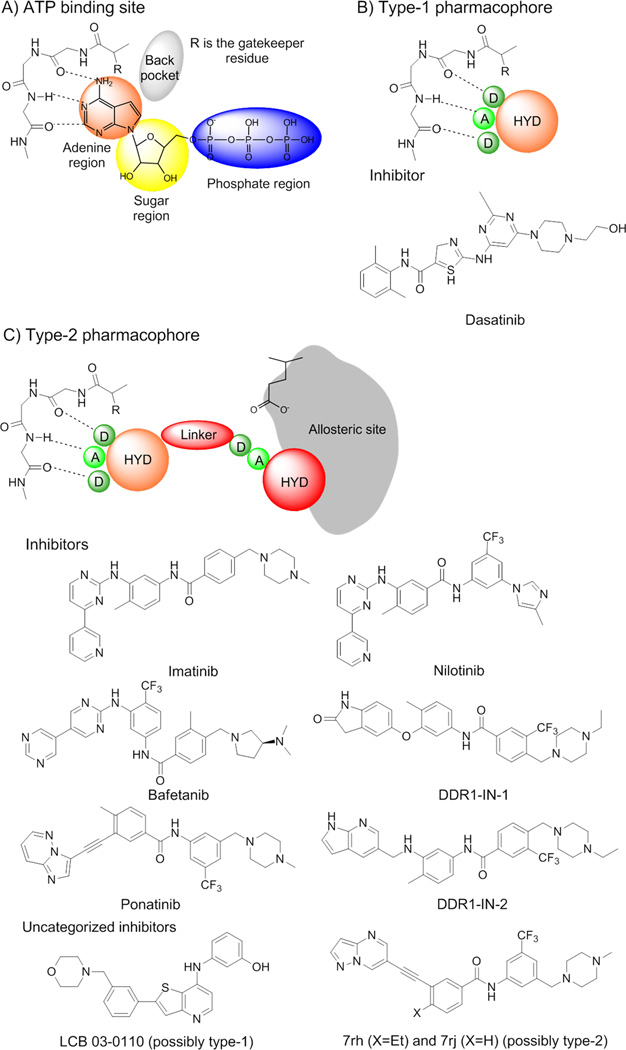
Figure 3.
(A) ATP binding site region and ATP interactions with the hinge residues of a kinase domain. Hydrogen bonds are represented by dashed lines. (B) Type-1 kinase inhibitors mimic the binding of the adenine moiety of ATP and lock the kinase domain in its active-state conformation. The type-1 inhibitor pharmacophore is shown representing the potential hydrogen bonds with the hinge region. (C) Type-2 kinase inhibitors lock the kinase domain in the inactive-conformation by leveraging the ATP binding site as well as the allosteric site that is accessible in the inactive state. The pharmacophore is shown representing the interactions with the hinge region and the allosteric site present in the ‘DFG-out’ conformation. Hydrogen bond donors are represented by circles labeled ‘D’, hydrogen bond acceptors by circles labeled (A). The larger circles labeled ‘HYD’ indicate hydrophobic moieties. The moiety that occupies the adenine ring region is colored in orange. The allosteric site is represented in gray. Adapted from [33].
DDR1–inhibitor complexes
Dasatinib is a type-1 inhibitor that targets kinase domains in the active form and is characterized by an open conformation of the activation loop (see below). Type-1 inhibitors bind the ATP site by mimicking the interaction of the adenine ring with the ‘hinge’ residues of protein. Even though there is no co-crystal structure of the DDR1– dasatinib complex, it is expected that dasatinib will bind in the so-called ‘open conformation’ of the DDR1 kinase domain, which is characterized by a ‘DFG-in’ configuration of the conserved triad DFG at the beginning of activation loop (Figure 1B).
By contrast, imatinib and nilotinib are type-2 inhibitors that bind to and stabilize an inactive kinase form that is characterized by ‘DFG-out’ conformation (see below). The ‘DFG-out’ motif opens an additional cavity, a hydrophobic allosteric site that, in addition to the ATP binding pocket, is targeted by type-2 inhibitors (Figure 1A).
Type-1 inhibitors
Generally, type-1 inhibitors bind to the ATP site (Figure 3A) by mimicking the interactions of the adenine moiety. Figure 3B shows the type-1 kinase pharmacophore, which comprises a hydrogen bond acceptor, two hydrogen bond donors, and a hydrophobic moiety. Type-1 inhibitors typically form one to three hydrogen bonds with kinase hinge residues and some hydrophobic interactions with residues that occupy the region around the adenine ring of ATP [32]. Below, we discuss a homology model of DDR1 in complex with a type-1 inhibitor that illustrates these important interactions.
Type-2 inhibitors
Recently, the inactive conformation of DDR1 bound to imatinib was reported in the PDB (PDB: 4BKJ) [21]. The co-crystal structure of DDR1-IN-1 with DDR1 kinase (PDB: 4CKR) suggests a comparable binding mode to that of imatinib, suggesting that it is a type-2 inhibitor that locks the kinase in the inactive ‘DFG-out’ conformation [31]. Type-2 inhibitors leverage the ATP binding pocket as well as an allosteric site created by a conformational change of the activation loop. The conformational change moves the phenylalanine residue (Phe785 [Phe185, Phe382]) more than 10 Å from its position in its kinase active conformation, creating a hydrophobic site adjacent to the ATP binding pocket. The co-crystal structure of DDR1 (PDB: 4BKJ) and imatinib displays the frequently observed hydrogen-bond interactions with the residues in the allosteric site (Figure S1).
Type-2 inhibitors target additional allosteric sites
Type-1 inhibitors have high cross-reactivity within the kinase family because of a high degree of sequence and structural similarity in their ATP binding site [33]. In general, type-1 inhibitors tend to be promiscuous, because they usually target well-conserved active kinase binding sites [33]. However, type-1 inhibitors have the advantage of inhibiting kinases that have acquired mutations resistant to type-2 inhibitors [34]. Type-2 inhibitors tend to be more selective because the inactive ‘DFG-out’ kinase conformation allows additional interactions between the inhibitor and specific, not-well-conserved exposed hydrophobic sites within the kinase domain (Figure 3C). A third class of inhibitors has been identified that target either the catalytically active (‘DFG-in’ and αC-helix-in) or inactive (‘DFG-out’ and αC-helix-out) kinases by leveraging a hydrophobic back cavity [33]. The back cavity is accessible in kinases that have a small gatekeeper residue, which is the first residue of the hinge connecting the C-lobe and N-lobe. The small gatekeeper residue, Thr701 [Met120, Thr315] in DDR1 (PDB:3ZOS), will allow the design of selective and potent binders that engage with the back pocket that becomes available because of the small side chain.
An intact conformation of the regulatory spine is essential for kinase activity. Efforts have been made to develop non-ATP competitive (i.e., truly allosteric inhibitors that target the regulatory spine). In this context, ARQ197 [35], a non-ATP competitive inhibitor of met proto-oncogene (c-Met), is currently in Phase III clinical trials for non-small-cell lung cancer [24]. The co-crystal structure of c-Met and ARQ197 reveals disrupted interactions between Met1131, Leu1142, His1202, and Phe1223 in the regulatory spine [24]. The inhibitor has shown exceptional exclusivity against a panel of 230 human kinases, of which only four are inhibited to any significant degree [35]. Eathiraj et al. created a generalized computational model of this inactive kinase conformation that was successfully applied to identify a series of fibroblast growth factor receptor (FGFR) TRK inhibitors [36]. Being tyrosine kinases, the novel mode of kinase inhibition is pertinent to DDR kinases and can be explored for identifying selective inhibitors.
DDR1 models for structure-based drug discovery
The sequence similarity in kinase domains has allowed homology modeling of DDR1. Day et al. [28] used homology models based on multiple templates to describe inhibition of DDR1 by imatinib, nilotinib, and dasatinib. Fu et al. [2] built two homology models of DDR1 based on the ‘DFG-in’ (active) and ‘DFG-out’ (inactive) conformations of the DFG motif. For the present review, we developed homology models for DDR1 using the ROSETTA macromolecular modeling suite [37].
DFG-out conformation
Homology models for the ‘DFG-out’ conformation were built to assess the accuracy of ROSETTA when compared with crystal structure PDB: 3ZOS. In addition, the highly variable DFG loop demands development of multiple models for docking studies aimed at finding inhibitors. The models were developed using three templates, each of which was in the DFG-out conformation and had been crystallized with different ligands: (i) FMS tyrosine kinase (PDB: 3BEA); (ii) TRKB kinase (PDB: 4AT5); and (iii) KIT kinase domain (PDB: 4HVS). The top 50 models from each template were used for docking the type-2 inhibitor imatinib. A hierarchical docking protocol (Figure S2 in the supplementary material online) was used to identify favorable homology models based on the ability to recover native poses for the ligand of interest. Models were clustered on the basis of root-mean-square deviation (RMSD) of docked imatinib to native imatinib pose. The top 1% models were taken from each cluster that recovered the native imatinib-binding pose within 0.2 Å and recovered side chain conformations (details in the supplementary material online). The resulting four homology models had the DFG-loop in the ‘out’ conformation with missing hydrophobic spines. Important residues that were discussed previously were recovered well. The structures were used for further docking studies. Meanwhile, the structure was also determined experimentally (PDB: 4BKJ), which enabled us to compare our model in a blind experiment (Figure S3A in the supplementary material online). The best-scoring homology models achieved a RMSD of 2.5 Å to PDB: 4BKJ. Ligand docking into DDR1-inactive homology models yielded docking poses that had a RMSD of 0.2 Å to the experimental imatinib pose in PDB: 4BKJ (Figure S4A in the supplementary material online) and was successful in the recovery of side chain conformations in the binding pocket (Figure S3B in the supplementary material online).
DFG-in conformation
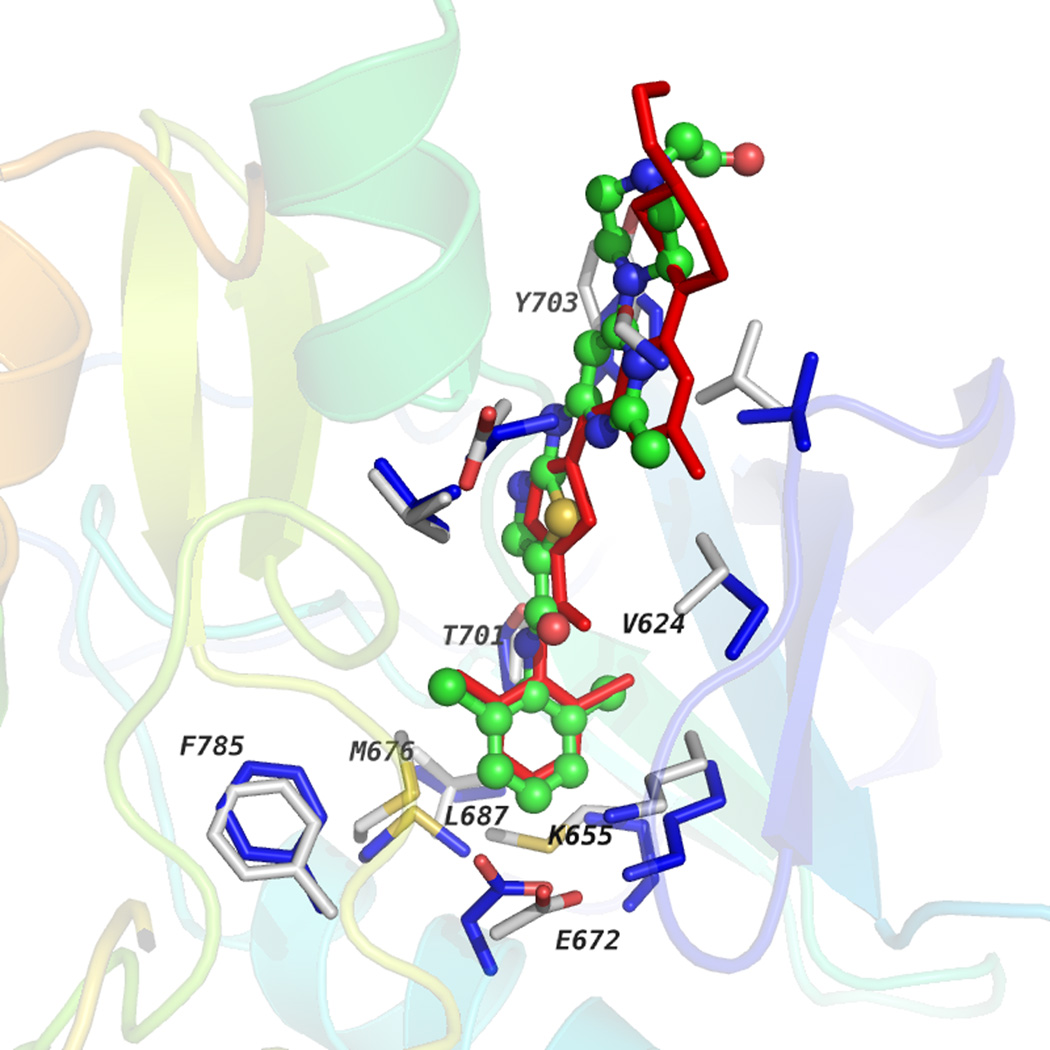
These results gave us confidence in creating a model for DDR1 in the ‘DFG-in’ conformation. Three ‘DFG-in’ conformation template structures bound to different ligands [(i) fibroblast growth factor receptor (FGFR; PDB: 2PVF); (ii) RET kinase (PDB: 2X2L); and (iii) FGFR kinase (PDB: 3C4F)] were used to create homology models using the hierarchical docking protocol represented in Figure S2 in the supplementary material online. Five models were obtained after clustering and model selection as explained above for ‘DDR-out’ conformation. Hydrophobic spines described previously were recovered intact (Figure 1B), whereas Thr701 [Thr315] served as gatekeeper residue as in Abl kinase. Docking dasatinib into five different homology models recovered the binding pose of dasatinib reported in the Abl kinase domain (PDB: 2GQG) within a RMSD of 0.18 Å (Figure S4B in the supplementary material online). Figure 4 shows the dasatinib-docked pose in a multicolored ball-and stick-model and, in red, is its native pose co-crystallized with the Abl kinase domain. The docked model recovered the position of the benzene group of dasatinib deep in the ATP-binding cleft. The hydrogen bond between the nitrogen of the carboxamide group and Thr315 (PDB: 2GQG) was observed in the docked model with Thr701 of DDR1. In the binding pocket, side-chain orientations of residues Glu672 [Glu286], Lys655 [Lys271], Val624 [Val256], and Tyr703 [Phe317] were captured as in the experimental structure. Successful recovery of the binding pose of dasatinib and residue side-chain recovery in the binding pocket suggests that these homology models could be used for high-throughput ligand-docking experiments to identify binders.
Figure 4.
Discoidin domain receptor 1 (DDR1) homology model complexed with dasatinib is shown along with the co-crystal structure of Abl kinase and dasatinib (PDB: 2GQG). Dasatinib docked into the DDR1-active state homology model is shown in the multicolored ball-and stick-model, whereas the native binding pose of dasatinib in 2GQG is shown in the red stick model. Residues in the binding pocket for DDR1 and Abl kinase are shown in gray or blue sticks, respectively. Residues that interact with ligands are labeled along with those that form the hydrophobic spine.
Concluding remarks and future perspectives
The availability of experimental structures as well as high-quality homology models makes DDR1 amenable for the application of structure-based drug discovery methods. Flexible docking with ROSETTALIGAND into crystal structures would improve docking accuracy. In addition, as the number of known DDR1 inhibitors grows [25–27,29–31,38], ligand-based drug discovery methods, such as quantitative structure–activity relation (SAR) models, could be developed. The discovery of inhibitors of DDR1 kinase activity might be beneficial in several pathological conditions where DDR1 has been identified as a potential therapeutic target.
Supplementary Material
Highlights.
DDR1 is a potential therapeutic target as it is implicated in organ fibrosis and several cancers.
DDR1 kinase domain has a typical structure.
Several important residues involved in function are conserved as in other kinase domains.
Homology modeling and docking studies of DDR1 demonstrates possibilities for computer aided drug discovery.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Teaser: Recent inhibitor-bound crystal structures and several novel DDR1 inhibitors allow application of computer-aided drug discovery methods.
References
- 1.Fu HL, et al. Shedding of discoidin domain receptor 1 by membrane-type matrix metalloproteinases. J. Biol. Chem. 2013;288:12114–12129. doi: 10.1074/jbc.M112.409599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fu HL, et al. Discoidin domain receptors: unique receptor tyrosine kinases in collagen-mediated signaling. J. Biol. Chem. 2013;288:7430–7437. doi: 10.1074/jbc.R112.444158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vogel W, et al. The discoidin domain receptor tyrosine kinases are activated by collagen. Mol. Cell. 1997;1:13–23. doi: 10.1016/s1097-2765(00)80003-9. [DOI] [PubMed] [Google Scholar]
- 4.Koo DH, et al. Pinpointing phosphotyrosine-dependent interactions downstream of the collagen receptor DDR1. FEBS Lett. 2006;580:15–22. doi: 10.1016/j.febslet.2005.11.035. [DOI] [PubMed] [Google Scholar]
- 5.Wang CZ, et al. A discoidin domain receptor 1/SHP-2 signaling complex inhibits alpha2beta1-integrin-mediated signal transducers and activators of transcription 1/3 activation and cell migration. Mol. Biol. Cell. 2006;17:2839–2852. doi: 10.1091/mbc.E05-11-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.L’Hote CG, et al. Functional analysis of discoidin domain receptor 1: effect of adhesion on DDR1 phosphorylation. FASEB J. 2002;16:234–236. doi: 10.1096/fj.01-0414fje. [DOI] [PubMed] [Google Scholar]
- 7.Lu KK, et al. Collagen stimulates discoidin domain receptor 1-mediated migration of smooth muscle cells through Src. Cardiovasc. Pathol. 2011;20:71–76. doi: 10.1016/j.carpath.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 8.Curat CA, Vogel WF. Discoidin domain receptor 1 controls growth and adhesion of mesangial cells. J. Am. Soc. Nephrol. 2002;13:2648–2656. doi: 10.1097/01.asn.0000032419.13208.0c. [DOI] [PubMed] [Google Scholar]
- 9.Xu H, et al. Discoidin domain receptors promote alpha1beta1- and alpha2beta1-integrin mediated cell adhesion to collagen by enhancing integrin activation. PLoS ONE. 2012;7:e52209. doi: 10.1371/journal.pone.0052209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roarty K, Serra R. Wnt5a is required for proper mammary gland development and TGF-beta-mediated inhibition of ductal growth. Development. 2007;134:3929–3939. doi: 10.1242/dev.008250. [DOI] [PubMed] [Google Scholar]
- 11.Kim HG, et al. DDR1 receptor tyrosine kinase promotes prosurvival pathway through Notch1 activation. J. Biol. Chem. 2011;286:17672–17681. doi: 10.1074/jbc.M111.236612. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Shintani Y, et al. Collagen I-mediated up-regulation of N-cadherin requires cooperative signals from integrins and discoidin domain receptor 1. J. Cell Biol. 2008;180:1277–1289. doi: 10.1083/jcb.200708137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borza CM, Pozzi A. Discoidin domain receptors in disease. Matrix Biol. 2013 doi: 10.1016/j.matbio.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vogel WF, et al. Discoidin domain receptor 1 tyrosine kinase has an essential role in mammary gland development. Mol. Cell Biol. 2001;21:2906–2917. doi: 10.1128/MCB.21.8.2906-2917.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franco C, et al. Discoidin domain receptor 1 (ddr1) deletion decreases atherosclerosis by accelerating matrix accumulation and reducing inflammation in low-density lipoprotein receptor-deficient mice. Circ. Res. 2008;102:1202–1211. doi: 10.1161/CIRCRESAHA.107.170662. [DOI] [PubMed] [Google Scholar]
- 16.Avivi-Green C, et al. Discoidin domain receptor 1-deficient mice are resistant to bleomycin-induced lung fibrosis. Am. J. Respir. Crit. Care Med. 2006;174:420–427. doi: 10.1164/rccm.200603-333OC. [DOI] [PubMed] [Google Scholar]
- 17.Xu H, et al. Normal activation of discoidin domain receptor 1 mutants with disulphide cross-links, insertions or deletions in the extracellular juxtamembrane region: mechanistic implications. J. Biol. Chem. 2014 doi: 10.1074/jbc.M113.536144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu HL, et al. Glycosylation at Asn211 regulates the activation state of the discoidin domain receptor 1 (DDR1) J. Biol. Chem. 2014;289:9275–9287. doi: 10.1074/jbc.M113.541102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rabiller M, et al. Proteus in the world of proteins: conformational changes in protein kinases. Arch. Pharm. 2010;343:193–206. doi: 10.1002/ardp.201000028. [DOI] [PubMed] [Google Scholar]
- 20.Johnson LN, et al. Active and inactive protein kinases: structural basis for regulation. Cell. 1996;85:149–158. doi: 10.1016/s0092-8674(00)81092-2. [DOI] [PubMed] [Google Scholar]
- 21.Canning P, et al. Structural mechanisms determining inhibition of the collagen receptor DDR1 by selective and multi–targeted type II kinase inhibitors. J. Mol. Biol. 2014 doi: 10.1016/j.jmb.2014.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor SS, Kornev AP. Protein kinases: evolution of dynamic regulatory proteins. Trends Biochem. Sci. 2011;36:65–77. doi: 10.1016/j.tibs.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kornev AP, Taylor SS. Defining the conserved internal architecture of a protein kinase. Biochim. Biophys. Acta. 2010;1804:440–444. doi: 10.1016/j.bbapap.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Norman RA, et al. Structural approaches to obtain kinase selectivity. Trends Pharmacol. Sci. 2012;33:273–278. doi: 10.1016/j.tips.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 25.Bantscheff M, et al. Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat. Biotechnol. 2007;25:1035–1044. doi: 10.1038/nbt1328. [DOI] [PubMed] [Google Scholar]
- 26.Rix U, et al. Chemical proteomic profiles of the BCR-ABL inhibitors imatinib, nilotinib, and dasatinib reveal novel kinase and nonkinase targets. Blood. 2007;110:4055–4063. doi: 10.1182/blood-2007-07-102061. [DOI] [PubMed] [Google Scholar]
- 27.Rix U, et al. A comprehensive target selectivity survey of the BCR-ABL kinase inhibitor INNO-406 by kinase profiling and chemical proteomics in chronic myeloid leukemia cells. Leukemia. 2010;24:44–50. doi: 10.1038/leu.2009.228. [DOI] [PubMed] [Google Scholar]
- 28.Day E, et al. Inhibition of collagen-induced discoidin domain receptor 1 and 2 activation by imatinib, nilotinib and dasatinib. Eur. J. Pharmacol. 2008;599:44–53. doi: 10.1016/j.ejphar.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 29.Sun X, et al. LCB 03-0110, a novel pan-discoidin domain receptor/c-Src family tyrosine kinase inhibitor, suppresses scar formation by inhibiting fibroblast and macrophage activation. J. Pharmacol. Exp. Ther. 2012;340:510–519. doi: 10.1124/jpet.111.187328. [DOI] [PubMed] [Google Scholar]
- 30.Gao M, et al. Discovery and optimization of 3-(2-(Pyrazolo[1,5-a]pyrimidin-6-yl)ethynyl)benzamides as novel selective and orally bioavailable discoidin domain receptor 1 (DDR1) inhibitors. J. Med. Chem. 2013;56:3281–3295. doi: 10.1021/jm301824k. [DOI] [PubMed] [Google Scholar]
- 31.Kim HG, et al. Discovery of a potent and selective DDR1 receptor tyrosine kinase inhibitor. ACS Chem. Biol. 2013 doi: 10.1021/cb400430t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y, Gray NS. Rational design of inhibitors that bind to inactive kinase conformations. Nat. Chem. Biol. 2006;2:358–364. doi: 10.1038/nchembio799. [DOI] [PubMed] [Google Scholar]
- 33.Zuccotto F, et al. Through the ‘gatekeeper door’: exploiting the active kinase conformation. J. Med. Chem. 2010;53:2681–2694. doi: 10.1021/jm901443h. [DOI] [PubMed] [Google Scholar]
- 34.Eglen RM, Reisine T. The current status of drug discovery against the human kinome. Assay Drug Dev. Technol. 2009;7:22–43. doi: 10.1089/adt.2008.164. [DOI] [PubMed] [Google Scholar]
- 35.Munshi N, et al. ARQ 197, a novel and selective inhibitor of the human c-Met receptor tyrosine kinase with antitumor activity. Mol. Cancer Ther. 2010;9:1544–1553. doi: 10.1158/1535-7163.MCT-09-1173. [DOI] [PubMed] [Google Scholar]
- 36.Eathiraj S, et al. Discovery of a novel mode of protein kinase inhibition characterized by the mechanism of inhibition of human mesenchymal-epithelial transition factor (c-Met) protein autophosphorylation by ARQ 197. J. Biol. Chem. 2011;286:20666–20676. doi: 10.1074/jbc.M110.213801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaufmann KW, et al. Practically useful: what the ROSETTA protein modeling suite can do for you. Biochemistry. 2010;49:2987–2998. doi: 10.1021/bi902153g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davis MI, et al. Comprehensive analysis of kinase inhibitor selectivity. Nat. Biotechnol. 2011;29:1046–1051. doi: 10.1038/nbt.1990. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.