Abstract
The TET2 DNA dioxygenase regulates cell identity and suppresses tumorigenesis by modulating DNA methylation and expression of a large number of genes. How TET2, like most other chromatin modifying enzymes, is recruited to specific genomic sites is unknown. Here we report that WT1, a sequence-specific transcription factor, is mutated in a mutually exclusive manner with TET2, IDH1 and IDH2 in acute myeloid leukemia (AML). WT1 physically interacts with and recruits TET2 to its target genes to activate their expression. The interaction between WT1 and TET2 is disrupted by multiple AML-derived TET2 mutations. TET2 suppresses leukemia cell proliferation and colony formation in a manner dependent on WT1. These results provide a mechanism for targeting TET2 to specific DNA sequence in the genome. Our results also provide an explanation for the mutual exclusivity of WT1 and TET2 mutations in AML and suggest an IDH1/2-TET2-WT1 pathway in suppressing AML.
Keywords: WT1, TET2, Mutual exclusive mutations, AML
INTRODUCTION
The recently identified TET (ten-eleven translocation) family of proteins, which includes TET1, TET2, and TET3 in mammalian cells, catalyze three sequential oxidation reactions: converting 5-methylcytosine (5mC) first to 5-hydroxymethylcytosine (5hmC), then 5-formylcytosine (5fC), and finally 5-carboxylcytosine (5caC) (He et al., 2011; Ito et al., 2011; Tahiliani et al., 2009). A subsequent decarboxylation of 5caC, by either a thymine-DNA glycosylase (TDG) or other DNA repair enzymes, could then lead to DNA demethylation (Kohli and Zhang, 2013). Genetic studies in mutant mice have linked the function of Tet genes to various biological pathways, including zygotic, embryonic and perinatal development (Dawlaty et al., 2013; Gu et al., 2011), differentiation of haematopoietic cells (Ko et al., 2011; Li et al., 2011; Moran-Crusio et al., 2011; Quivoron et al., 2011), and induced pluripotent stem cell (iPSC) reprogramming (Costa et al., 2013; Doege et al., 2012). Such diverse and complex roles are consistent with the binding of TET proteins and the distribution of their catalytic products, 5hmC, 5fC and 5caC throughout the genome (Shen et al., 2013; Song et al., 2013; Williams et al., 2011; Wu et al., 2011). However, it is unclear how TET proteins bind to specific locus in the genome.
Pathologically, TET2 is frequently mutated in hematopoietic malignancies of both myeloid, in particular acute myeloid leukemia (AML, ~15 – 20%), and lymphoid lineages such as angioimmunoblastic T-cell lymphoma (AITL, ~30 – 40%) (Delhommeau et al., 2009; Quivoron et al., 2011; Tefferi et al., 2009). In a subset of AML with wild-type TET2 gene, TET2 enzyme is also catalytically inactivated by D-2-hydroxyglutarate (D-2-HG), an oncometabolite produced by mutated isocitrate dehydrogenase 1 and 2 (IDH1 and IDH2) (Chowdhury et al., 2011; Xu et al., 2011), which occurs in about 20% of AMLs in a mutually exclusive manner with TET2 mutations (Figueroa et al., 2010). In addition to IDH1, IDH2 and TET2, wilm’s tumor gene, WT1, is also frequently mutated in AML (2013; Fernandez-Mercado et al., 2012; Liang et al., 2013; Patel et al., 2012; Rocquain et al., 2010; Welch et al., 2012). WT1 encodes a sequence-specific zinc-finger transcription factor involved in the control of organ development and cell differentiation, in particular nephrogenesis and haematopoiesis, and in tumor suppression by regulating the expression of genes involved in different cellular pathways (Huff, 2011; Rivera and Haber, 2005). In an effort to determine how mutations of WT1 contribute to the development of AML, we noted that the WT1 gene is mutated in AML in a mutually exclusive manner with that targeting TET2, IDH1, and IDH2. This pattern of TET2 and WT1 mutation in AML led us to hypothesize that WT1 and TET2 may function in the same pathway in suppressing AML.
RESULTS
WT1, TET2, IDH1 and IDH2 are mutated mutually exclusively in AML
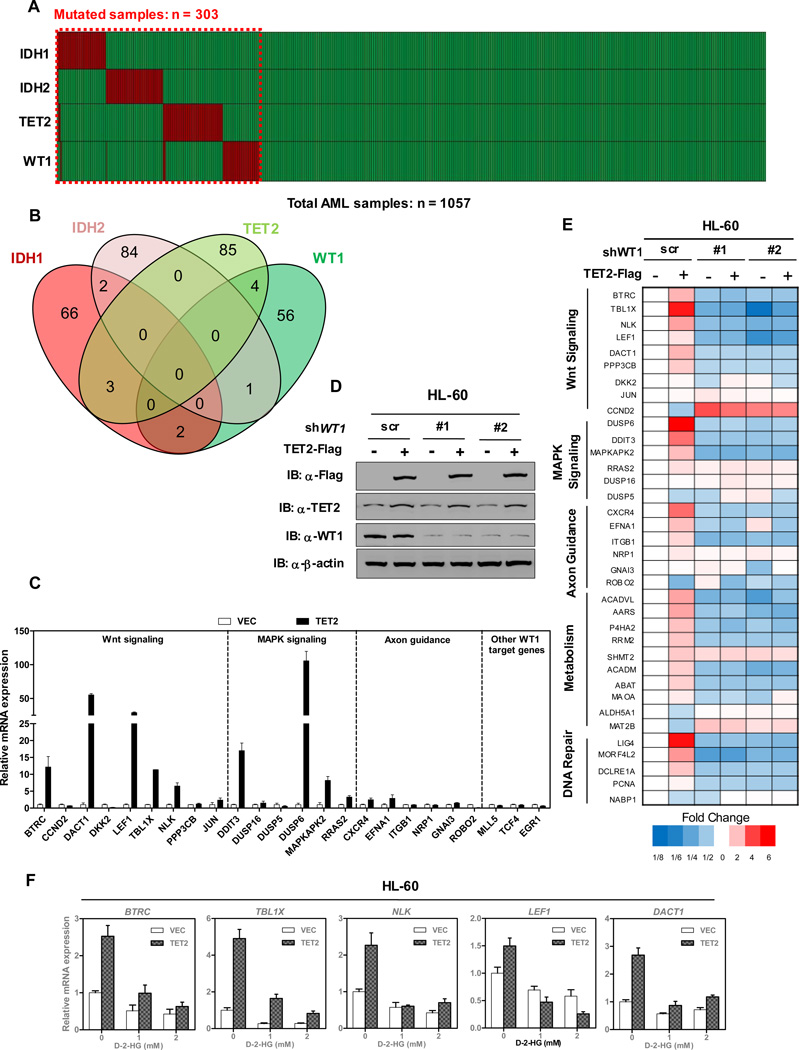
Somatic mutations targeting IDH1, IDH2, TET2 and WT1 genes occur frequently in AML. We carried out a meta-analysis of a total of 1,057 AML cases where all four genes have been sequenced from six separate studies. 303 cases (28.7%) carried mutations targeting at least one of the four genes. As previously reported, the mutations targeting IDH1, IDH2, and TET2 genes occur mutually exclusively (Figueroa et al., 2010). Notably, the mutations targeting WT1 also occur in a mutually exclusive pattern with those targeting IDH1, IDH2 or TET2 in AML (Figures 1A and 1B). The mutual exclusive mutation patterns of TET2 and WT1 led us to hypothesize that TET2 and WT1 may function in the same pathway.
Figure 1. TET2 activates WT1 target genes.
(A) Somatic variants in IDH1/2, TET2 and WT1 were identified in total 1057 AML cases, in which 303 cases carried at least one mutation. Data were collected from six different studies (2013; Fernandez-Mercado et al., 2012; Liang et al., 2013; Patel et al., 2012; Rocquain et al., 2010; Welch et al., 2012)
(B) Overlap of IDH1/2, TET2 and WT1 mutations in 303 mutated cases of AML.
(C) Flag-tagged full-length TET2 was overexpressed in HEK293T cells, and the mRNA expression of indicated WT1-target genes was determined by qRT-PCR.
(D) Overexpression of TET2 in stable HL-60 cells with or without WT1 knockdown. HL-60 cells were transduced with retrovirus expressing different shRNAs against WT1 and retrovirus expressing Flag-tagged full-length TET2. The expression of WT1 and TET2 proteins was determined by western blot.
(E) Stable HL-60 cells were generated as described in (D), and the mRNA expression of WT1 target genes was determined by qRT-PCR.
(F) Stable HL-60 cells overexpressing TET2-Flag or empty vector control were treated with the indicated concentrations of cell-permeable D-2-HG for 12 hrs. mRNA expression of WT1-target genes was determined by qRT-PCR.
Shown are average values of triplicated results with standard deviation (S.D.).
See also Figure S1.
TET2 activates WT1 target genes
To determine the functional significance of mutual exclusive mutation patthern between WT1 and TET2, we first determined whether WT1 and TET2 could affect the expression of each other. It is known that two alternative splice sites result in four major isoforms of WT1: different in the inclusion or exclusion of exon 5 (Ex5+/Ex5−) and three amino acid residues (Lysine-Threonine-Serine, KTS) encoded by exon 9. We found that overexpression of either the (+KTS) or the (−KTS) isoform of WT1 led to up-regulation of BTRC and TBL1X, two known direct WT1-target genes (Kim et al., 2009). In contrast, ectopic expression of either WT1 isoform did not affect the gene expression of TET2 in various cultured cells and vice versa (Figures S1A and S1B). Next, we examined whether TET2, as a broad epigenetic modifier, can modulate WT1 target gene expression. We found that ectopic expression of TET2 in HEK293T cells resulted in the activation of a number of WT1-target genes, including those involved in the Wnt signaling, MAPK signaling and axon guidance pathway (Figure 1C). In addition, we also found that the effect of TET2 on activating WT1-target gene expression was dependent on the catalytic activity of TET2, as expression of TET2 catalytic inactive mutant (CM) did not up-regulate the expression of WT1-target genes (Figures S1C and S1D). Moreover, co-overexpression of TET2 and WT1 in HEK293T cells synergistically activates the expression of WT1 target genes in a dose-dependent manner (Figures S1E and S1F). Furthermore, we utilized three short hairpin RNAs (shRNAs) against WT1 to knock-down its expression in HEK293T cells (Figure S1G). We found that WT1 depletion almost completely abrogated the effect of TET2-mediated activation of WT1-target genes (Figures S1H and S1I), suggesting that the function of TET2 in activating WT1-target gene is dependent on WT1.
Given that the mutations of WT1 and TET2 occur frequently in AML, we then stably infected human AML HL-60 leukemic cells with retroviral vectors expressing Flag-tagged human full-length TET2 (Figure 1D). We found that overexpression of TET2 indeed resulted in the activation of a number of WT1-target genes in HL-60 cells (Figure 1E). When we utilized two different shRNAs against WT1 to deplete its expression in HL-60/TET2-Flag cells, we found that the TET2-induced upregulation of WT1-target gene expression was broadly suppressed by WT1 depletion (Figure 1E), further supporting the notion that TET2 activates WT1-target genes in a manner dependent on WT1. Notably, when the HL-60/TET2-Flag cells were treated with cell-permeable D-2-HG, which is produced by mutant IDH1 and IDH2 and inhibits the activity of TET enzymes (Xu et al., 2011), we found that D-2-HG effectively inhibited the effect of TET2 on activating WT1-target gene expression (Figure 1F). Moreover, overexpression of wild-type IDH1 in HL-60/TET2-Flag cells activated WT1-target genes, while overexpression of R132C mutant IDH1 inactivated WT1-target genes (Figures S1J and S1K). These results further support the notion that the ability of TET2 in activating WT1-target gene expression requires its catalytic activity. It is also consistent with the observation that IDH1, IDH2, TET2 and WT1 genes are mutated in AML in a mutually exclusive manner.
WT1 physically binds to TET2
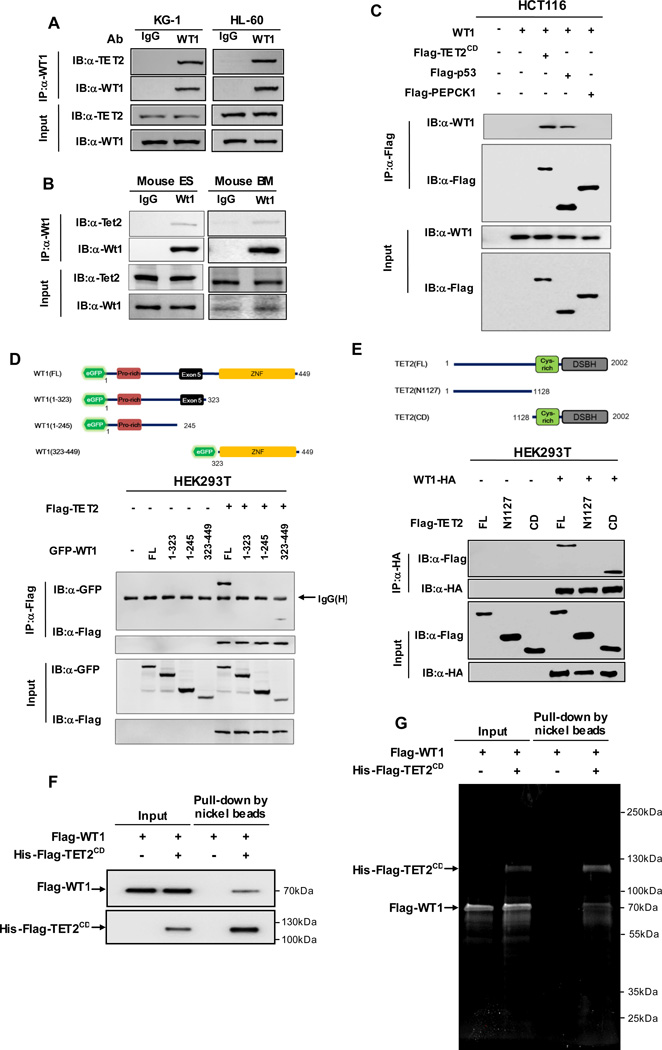
How does TET2 selectively induce expression of WT1-target genes? An appealing model is that the sequence specific transcription factor WT1 directly binds to TET2 and recruits TET2 to its target genes. To test this hypothesis, we first examined the protein expression of TET2 and WT1 in various cell types. We found that TET2 and WT1 proteins were expressed at a readily detectable level in both HL-60 and KG-1 cells, and were low in HEK239T and HCT116 cells (Figure S2A). Next, we determined the endogenous WT1-TET2 association by coupled immunoprecipitation and western blotting (IP-western) in HL-60 and KG-1cells (Figure 2A), as well as mouse embryonic stem cells and primary bone marrow cells (Figure 2B). This experiment demonstrated that the WT1-TET2 association was readily detected in these four cell lines, suggesting that TET2 and WT1 interact with each other and may be broadly involved in cell regulation beyond hematopoietic lineages.
Figure 2. TET2 directly binds to WT1.
(A) Endogenous WT1 protein was immunoprecipitated from two human AML cell lines (i.e. KG-1 and HL-60), following by western blot to detect TET2. Normal rabbit IgG was used as a negative control.
(B) Endogenous Wt1 protein was immunoprecipitated from mouse ES cells or bone marrow (BM) cells, following by western blot to detect Tet2. Normal rabbit IgG was used as a negative control.
(C) HCT116 cells were transiently transfected with plasmids expressing indicated genes. Protein-protein interaction was examined by IP-western using indicated antibodies.
(D) Wild-type WT1 and its deletion mutants, as shown in the schematic illustration, were co-expressed with Flag-TET2 in HEK293T cells. Protein-protein interaction was examined by IP-western using indicated antibodies.
(E) Wild-type TET2 and its deletion mutants, as shown in the schematic illustration, were co-expressed with WT1-HA in HEK293T cells. Protein-protein interaction was examined by IP-western using indicated antibodies.
(F–G) Recombinant Flag-6xHis-TET2CD (200 ng) and Flag-WT1 (400 ng) proteins purified from baculovirus infected Sf9 cells were incubated together. Nickel beads were then added and bound proteins were eluted with imidazole and resolved by SDS-PAGE. WT1-TET2CD binding was examined by either western blot (F) or SYPRO Ruby staining (G).
See also Figure S2.
To further determine the specificity of WT1-TET2 interaction, we ectopically expressed WT1 and the TET2 CD domain (TET2CD) that contains both the Cys-rich and the DSBH domains in HCT116 cells. p53, which has previously been shown to bind WT1 (Maheswaran et al., 1995; Maheswaran et al., 1993), and phosphoenolpyruvate carboxykinase (PEPCK1) were included as a positive and negative control, respectively. This experiment demonstrated the retention of WT1 in the immunoprecipitates of TET2CD (Figure 2C). Furthermore, we found that neither exon 5 nor KTS is required for WT1 to bind with either TET2CD or full-length TET2 (Figure S2B). Interestingly, the interaction of WT1 is specific to TET2, as evidenced by the observation that neither the (Ex5−/+KTS) nor the (Ex5+/−KTS) isoform of WT1 bound to the CD domain or full-length of TET1 (Figures S2C and S2D).
To define the binding regions between WT1 and TET2, we constructed a series of deletion mutants of both proteins. We found that TET2 binds to the zinc finger domain (residue 323–449), but not the N-terminal region (residue 1–323) of WT1 (Figure 2D). Similar mapping experiments showed that WT1 binds to the CD domain, but not the N-terminal region (residue 1 to 1127) of TET2 (Figure 2E). More importantly, in vitro studies with purified recombinant proteins demonstrated a direct interaction between human WT1 and the CD domain of human TET2 proteins at 1:1 stoichiometry (Figures 2F and 2G). Collectively, these data demonstrate that WT1 and TET2 physically interact with each other, with the zinc finger domain of WT1 and the CD domain of TET2 being primarily responsible for their association.
WT1 recruits TET2 to its target genes
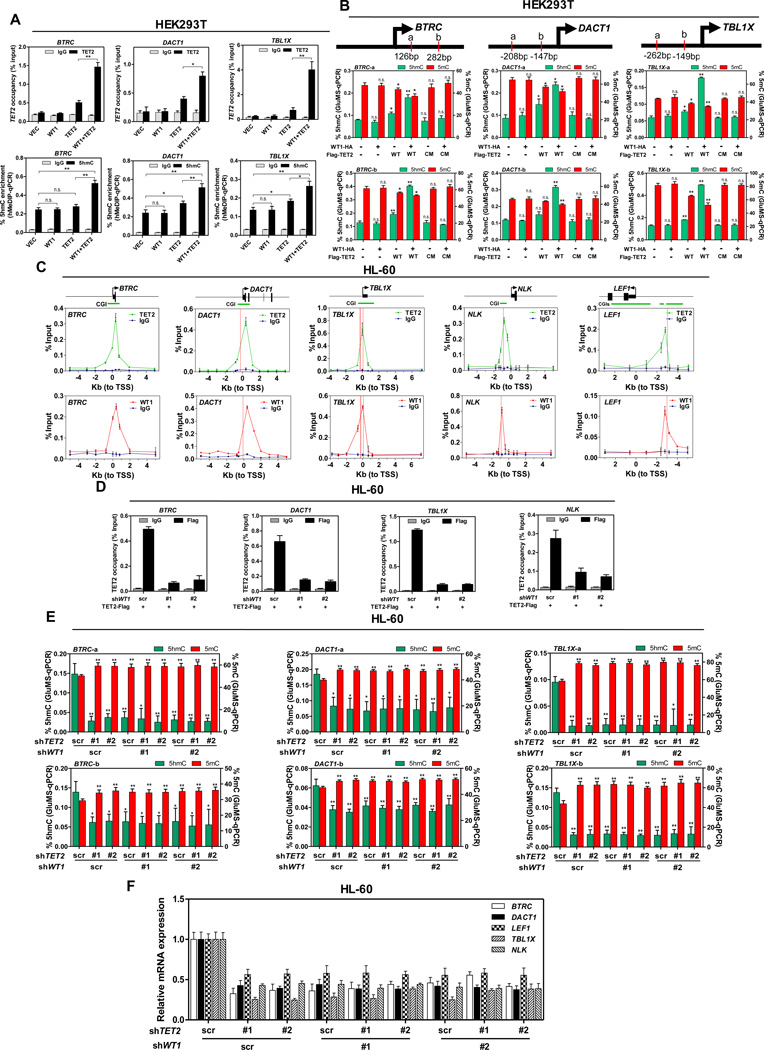
If TET2 is recruited by WT1 to specific DNA sequence, then TET2 should bind with the promoters of WT1-target genes in a manner dependent on WT1. Supporting this hypothesis, coupled chromatin immunoprecipitation and quantitative PCR (ChIP-qPCR) analysis demonstrated the binding of ectopically expressed TET2 to transcription start sites (TSSs) and CpG islands (CGIs) of WT1-target genes (i.e., BTRC, DACT1, and TBL1X) in HEK293T cells (Figure 3A). Notably, the TET2 occupancy on the promoters of WT1-target genes was substantially increased by the co-expression of WT1 in HEK293T cells (Figure 3A). Hydroxymethylated DNA immunoprecipitation quantitative PCR (hMeDIP-qPCR) analysis was used to track the 5hmC change in the CpG-rich regions of WT1-target gene promoters. We found that co-expression of WT1 and TET2 in HEK293T cells significantly increased 5hmC levels at the promoter regions of BTRC (p = 0.0058), DACT1 (p = 0.0015), and TBL1X (p = 0.0071) (Figure 3A). To more accurately quantify the 5hmC and 5mC status at the specific locus within WT1-target gene promoters, we performed glucosylated hydroxymethyl-sensitive qPCR (GluMS-qPCR), which enables single-base level determination of the methylation status of CCGG sites in the genome (Xu et al., 2012). Our data demonstrated that co-expression of WT1 and TET2 in HEK293T cells led to a significant increase in 5hmC and a contaminant decrease in 5mC at particular nucleotide loci (marked as ‘a’ and ‘b’) near TSS of WT1-target genes (Figure 3B). In addition, we also found in the promoter regions of WT1-target genes at the sites where 5hmC was increased by TET2 overexpression that ectopic expression of TET2 led to increased levels of the activating histone marker H3K4me3, and that the increase in H3K4me3 was further enhanced by co-overexpressing TET2 with WT1 (Figure S3A). This is in line with previous studies showing that in mouse ES cells gene promoters that are high in 5hmC are enriched in H3K4me3, whereas those enriched in 5mC are depleted of H3K4me3 marks (Ficz et al., 2011). In contrast to WT1 overexpression, ChIP–qPCR analysis showed that WT1 depletion reduced the TET2 occupancy on the promoters of WT1-target genes to near the background levels in HEK293T cells, but had little effect on TET2 occupancy on the promoters of TMEM116 and CDKL4 (Figure S3B), which are known TET2 target genes independent of WT1 (Deplus et al., 2013).
Figure 3. TET2 is recruited by WT1 to its target genes.
(A) Flag-TET2 was transiently expressed either singularly or with WT1-HA in HEK293T cells. The occupancy of Flag-TET2 at the promoter regions of WT1-target genes was determined by ChIP-qPCR (upper), and the 5hmC enrichment at the Flag-TET2 binding sites was determined by hMeDIP-qPCR (lower). Mouse IgG and rabbit IgG were included as negative controls for ChIP-qPCR and hMeDIP-qPCR, respectively.
(B) Flag-tagged wild-type (WT) TET2 or its catalytic inactive mutant (CM) of TET2 was transiently overexpressed either singularly or with HA-WT1 in HEK293T cells. The site-specific 5hmC and 5mC levels were determined by using GluMS-qPCR. a, b indicate MspI/HapII recognition sites of each target gene. Arrow denotes promoter orientation.
(C) Stable HL-60 cells with knockdown of endogenous TET2 and put-back of Flag-tagged TET2 were generated as described in Figure S3C. The occupancy of TET2-Flag and endogenous WT1 on the promoter regions of indicated WT1-target genes was determined by ChIP-qPCR. Mouse IgG and rabbit IgG were included as negative controls. Arrow denotes promoter orientation, CGI (green line) indicates CpG islands, red dash line indicates WT1 binding motif.
(D) Stable HL-60 cells in (C) were transduced with retrovirus expressing different shRNAs against WT1. The occupancy of TET2-Flag on the promoter regions of indicated WT1-target genes was determined by ChIP-qPCR. Mouse IgG was included as negative control.
(E) HL-60 cells were transduced with retrovirus expressing different shRNAs against WT1 and/or TET2 as described in Figure S3F. Site-specific levels of 5hmC and 5mC were determined by using GluMS-qPCR.
(F) Stable HL-60 cells in (E) were examined for the mRNA expression of indicated WT1-target genes, as determined by qRT-PCR.
Shown are average values of triplicated results with standard deviation (S.D.). * denotes P < 0.05 for the indicated comparison;** denotes P < 0.01 for the indicated comparison; n.s. = not significant.
See also Figure S3.
We then performed ChIP-qPCR analysis to determine the TET2 occupancy on the promoters of WT1-target genes in HL-60 cells. Because of the lack of a TET2 antibody suitable for ChIP analysis of endogenous TET2 occupancy, we generated stable HL-60 cells in which endogenous TET2 was depleted and Flag-TET2 was expressed at a level similar to that of endogenous TET2 protein (Figure S3C). Of 17 direct WT1-targets genes from five different signaling pathways, we found that WT1 and TET2 bind to all of them at the CpG-rich promoter regions (Figures 3C and S3D). Moreover, the TET2 occupancy on the promoters of WT1-target genes was significantly decreased to near the background levels by the deletion of endogenous WT1 in these stable HL-60 cells expressing Flag-TET2 (Figures 3D and S3E). In addition, we also generated stable HL-60 cells with retroviral vectors encoding different shRNAs against WT1 and/or TET2 (Figure S3F). We found that knockdown of either WT1 or TET2 led to significantly decreased 5hmC levels and increased 5mC levels at specific nucleotide loci (marked as ‘a’ and ‘b’) in WT1-target genes (e. g, BTRC, DACT1, and TBL1X), but no additive effect was found upon the co-depletion of both genes (Figure 3E). In accord, knockdown of either gene led to down-regulation of WT1-target genes, but no additive effect was observed upon the co-depletion of both genes (Figure 3F). Taken together, these results support the idea that WT1 recruits TET2 to specific genomic sites to regulate gene expression.
TET2 inhibits leukemia cell proliferation in a WT1-dependent manner
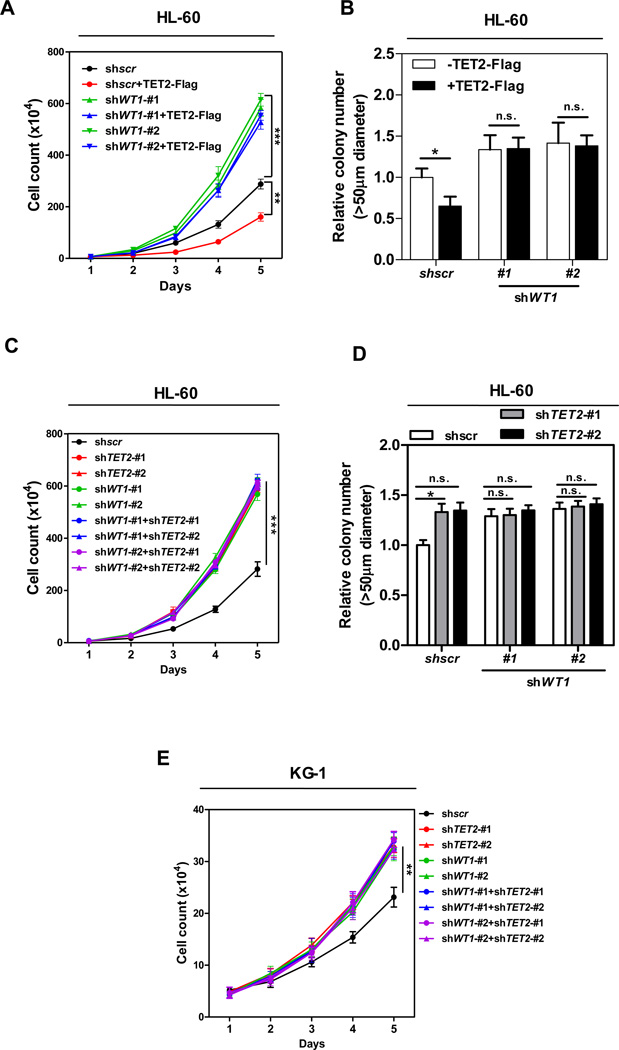
To investigate the physiological relevance of the WT1-TET2 interaction, we determined the functional interaction of WT1 and TET2 in controlling leukemia cell proliferation and colony formation. We first examined the effect of ectopically expressed TET2 in the presence and absence of WT1. The expression of TET2 and knockdown efficiency of WT1 were verified by direct immunoblotting (Figure 1D). Consistent with tumor suppression function of TET2, in multiple independent experiments, HL-60 cells stably overexpressing Flag-tagged TET2 displayed slower proliferation (Figure 4A) and reduced clonogenicity (Figure 4B) as compared to control cells transfected with the empty vector. Notably, the inhibitory effects of TET2 on leukemic cell proliferation and colony formation were abolished by WT1 knockdown.
Figure 4. TET2 inhibits leukemia cell proliferation in a WT1-dependent manner.
(A–B) Cell proliferation (A) and colony formation (B) of stable HL-60 cells overexpressing full-length TET2 with or without WT1 knockdown were determined by cell counting and colony-forming assay, respectively.
(C–D) Cell proliferation (C) and colony formation (D) of stable HL-60 cells with knockdown of WT1 and/or TET2 were determined by cell counting and colony-forming assay, respectively.
(E) Cell proliferation of stable KG-1 cells with knockdown of WT1 and/or TET2 was determined by cell counting.
Shown are average values of triplicated results with standard deviation (S.D.). *denotes the p < 0.05, **denotes the p < 0.01, and ***denotes the p < 0.001 for the indicated comparison. n.s.= not significant.
See also Figure S4.
We then determined the effect of TET2 and/or WT1 knockdown inHL-60 cells. The knockdown efficiency of TET2 and WT1 was determined by qRT-PCR (Figure S3F). We found that the proliferation and colony formation of HL-60 cells were stimulated by the depletion of either TET2 or WT1 gene, but not additively by the co-depletion of both genes (Figures 4C and 4D). We stably infected another human AML cell line, KG-1 cells, with retroviral vectors encoding different shRNAs against WT1 and/or TET2 (Figure S4A). In line with the observations in HL-60 cells, knockdown of either TET2 or WT1 gene in KG-1 cells led to downregulation of WT1-target genes, and again no additive effect was found upon the co-depletion of both genes (Figure S4B). Cell proliferation was stimulated by the knockdown of either TET2 or WT1 gene in KG-1 cells, but not additively by the co-depletion of both genes (Figure 4E). Collectively, these results suggest that TET2 and WT1 function in the same pathway to inhibit leukemia cell proliferation and colony formation.
Multiple recurrent AML-derived mutations in TET2 disrupt its WT1 binding
Besides alterations causing truncation, frame-shift, insertions or deletions, a large number of TET2 missense mutations have been reported in AML and only a few of them have been functionally characterized (Ko et al., 2010). To seek evidence supporting the WT1-mediated TET2 recruitment and explore the pathological significance of their mutual exclusive mutation patterns in AML, we examined the effect on the WT1 binding by a panel of AML-derived mutations targeting the CD domain of TET2 (TET2CD) where WT1 binds. Notably, of total 28 AML-derived TET2 mutants we tested, at least seven, D1242V, G1256D, R1302G, A1505T, P1617H, G1861R and T1884A, severely reduced or completely abolished interaction with Flag-tagged WT1 and their occupancy on WT1 target genes in HEK293T cells (Figures S5A and S5B). Importantly, these WT1-binding defective TET2 mutants were incapable of binding with endogenous WT1 protein in HL-60 cells (Figure 5A). None of these 7 mutations targets well-defined α-KG- or Fe2+-binding residues. To directly examine whether these mutations affect the catalytic activity of TET2, we performed in vitro TET2 activity assay using genomic DNA as substrate and immunopurified TET2CD as the enzyme. The wild-type TET2CD converted all 5mC and 5hmC to 5caC as seen by the loss of both 5mC and 5hmC after reaction, while the catalytic mutant (CM) TET2CD that harbors two mutations disrupting binding with Fe2+(He et al., 2011; Ito et al., 2011; Tahiliani et al., 2009) and control HA precipitates from cells transfected with the empty vector had no detectable activity (Figure 5B). Of seven WT1-binding deficient TET2 mutants we have characterized, six (D1242V, G1256D, R1302G, A1505T, G1861R, and T1884A) exhibited apparent catalytic activity in converting 5mC and 5hmC to 5caC and one mutant (P1617H) displayed reduced catalytic activity (Figure 5B). In addition, we also found that AML-derived, WT1-binding deficient mutants, A1505T and P1617H, retained their catalytic activity in oxidizing 5mC, leading to a reduction in 5mC, although less active when compared with wild-type TET2 (Figure S5D). Furthermore, in vivo TET2 activity assay confirmed that these WT1-binding defective TET2 mutants retained the catalytic function to produce 5hmC, but their activity was reduced compared to wild-type TET2 (Figure S5E). In HEK293T cells, overexpression of these WT1-binding defective TET2 mutants did not increase 5hmC in the CpG-rich regions of WT1-target gene promoters (data not shown). As a result, overexpression of these WT1-binding defective TET2 mutants failed to activate WT1-target genes in HEK293T cells (Figure S5C). We conclude that these AML-derived mutations inactivate the function of TET2 mainly by disrupting its binding with WT1 instead of inactivating the catalytic activity of TET2.
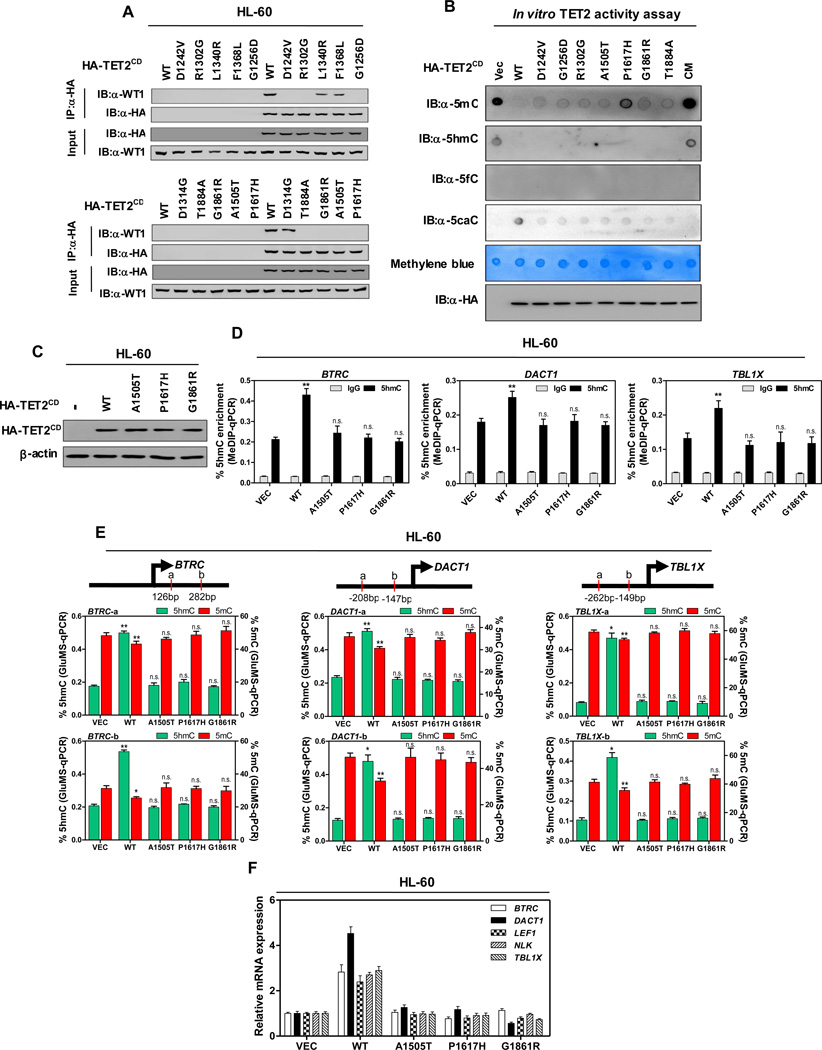
Figure 5. AML-derived mutations in TET2 disrupt WT1 binding.
(A) HL-60 cells were transiently transfected with vectors encoding HA-tagged wild-type TET2CD, or AML-derived WT1-binding defective TET2 mutants as indicated, and the ectopically expressed TET2 proteins were immunoprecipitated, following by western blot to detect endogenous WT1.
(B) In vitro TET2 catalytic activity assay. Genomic DNA were isolated from human monocyte-derived macrophage (MDM) cells, sonicated and incubated with immunopurified HA-tagged wild-type and mutants human TET2 at 37°C for 2 hrs. After termination of the reaction, a fraction of reaction mixture from each reaction was subjected to dot-blot assay using the antibodies specific for 5mC, 5hmC, 5fC and 5caC. See Experimental Procedures for more details. The amount of DNA in each reaction was examined by methylene blue staining. CM refers to a catalytic mutant of TET2 that harbors two mutations disrupting the binding with Fe2+
(C) HL-60 cells were transduced with lentiviral vectors encoding HA-tagged wild-type TET2CD, or AML-derived WT1-binding defective TET2CD mutants as indicated, and the expression of ectopic TET2CD proteins was determined by western blot.
(D–F) Stable HL-60 cells were generated as described above in (C). The 5hmC enrichment at TET2 binding sites at the promoter regions of indicated WT1-target genes was determined by hMeDIP-qPCR (D), rabbit IgG was included as negative control. Moreover, the site-specific levels of 5hmC and 5mC were determined by using GluMS-qPCR (E), and the mRNA expression of indicated WT1-target genes was determined by qRT-PCR (F).
Shown are average values of triplicated results with standard deviation (S.D.). *denotes the p < 0.05 and **denotes the p < 0.01 for the indicated comparison. n.s.= not significant.
See also Figure S5.
To further demonstrate that AML-derived, WT1-binding defective TET2 mutants specifically lose their ability to regulate WT1-target genes, we stably infected HL-60 cells with lentiviral vectors encoding the wild-type TET2CD or three TET2 mutants (A1505T, P1617H or G1861R). Both wild-type TET2 and its mutants were expressed at a similar level in HL-60 cells as verified by direct immunoblotting (Figure 5C). By performing hMeDIP-qPCR analysis, we found that overexpression of wild-type TET2CD, but not WT1-binding defective TET2 mutants in HL-60 cells, significantly increased 5hmC levels in the CpG-rich regions of WT1-target gene promoters (Figure 5D). Moreover, GluMS-qPCR analysis revealed that these WT1-binding defective TET2 mutants failed to increase 5hmC levels and decrease 5mC levels at specific loci near TSS of WT1-target genes in HL-60 cells (Figure 5E). As a result, overexpression of the wild-type TET2CD, but not AML-derived WT1-binding defective TET2 mutants, induced the expression of WT1-target genes in HL-60 cells (Figure 5F), suggesting a functional dependency of TET2 on binding with WT1 to regulate the expression of WT1-target genes in leukemia cells.
AML-derived, WT1 binding-defective TET2 mutants fail to inhibit leukemia cell proliferation
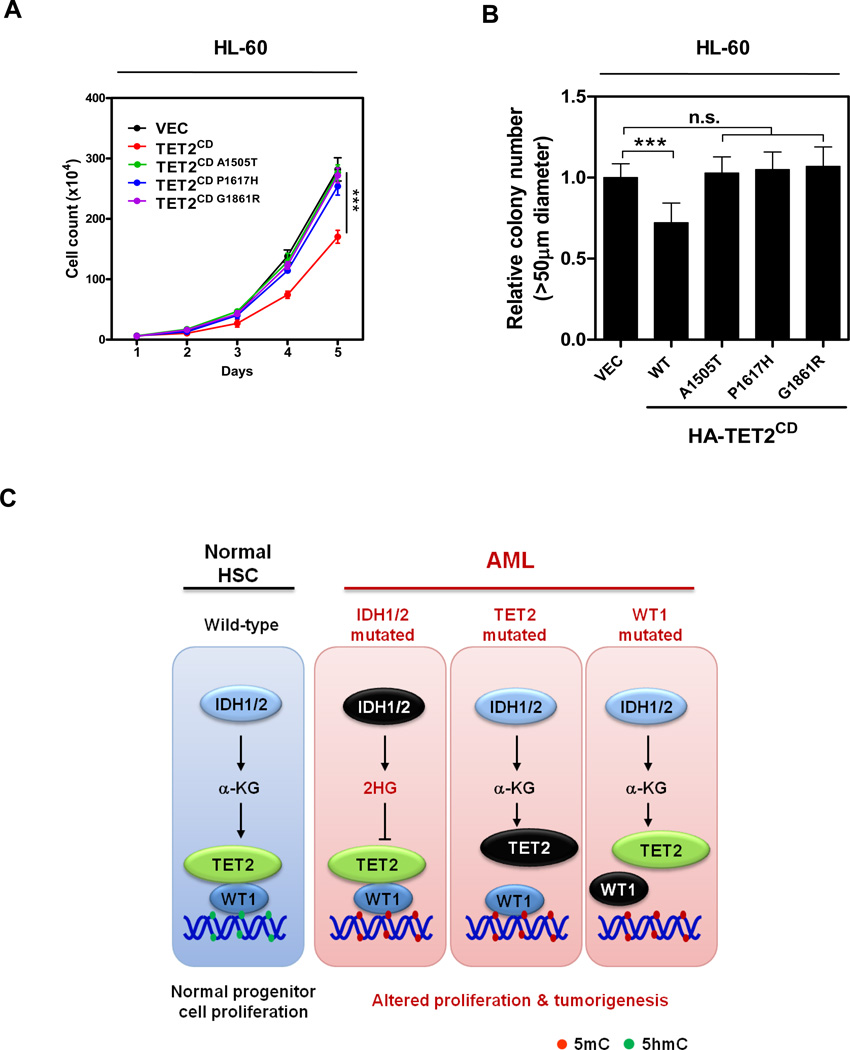
Finally, we investigated the functional consequence of AML-derived, WT1-binding defective mutations in TET2. We found that HL-60 cells stably overexpressing the wild-type TET2CD, but not three TET2 mutants (A1505T, P1617H or G1861R), displayed decreased proliferation and impaired clonogenicity as compared to control cells transfected with the empty vector (Figures 6A and 6B). Taken together, our results suggest that TET2 negatively regulates leukemia cell proliferation in a WT1-dependent manner and that this tumor suppressor function of TET2 is inactivated by multiple AML-associated TET2 mutations that lost its binding with WT1 (Figure 6C).
Figure 6. AML-derived WT1 binding-defective TET2 mutants fail to suppress leukemia cell proliferation.
(A–B) Cell proliferation (A) and colony formation (B) of stable HL-60 cells expressing wild-type or WT1 binding-defective mutants of HA-TET2CD were determined by cell counting and colony-forming assay, respectively.
(C) A schematic illustration of the IDH1/2-TET2-WT1 pathway in AML suppression. See also Table S2.
DISCUSSION
Like most epigenetic modifying enzymes (Smith and Shilatifard, 2010), the TET DNA dioxygenases do not appear to bind to specific DNA sequences by themselves. Therefore, these general chromatin modifying enzymes must be recruited to specific targets in the genome by other factors, presumably sequence specific DNA binding proteins, to regulate specific cellular processes. At present, little is known about how the recruitment of TET is achieved. In this study, we demonstrate that WT1 binds directly to TET2 and recruits TET2 to specific genomic sites to regulate the expression of WT1 target genes. Our results suggest a mechanism—binding with a sequence-specific transcription factor—by which the TET2 DNA methylhydroxylating enzyme is recruited to specific sequences in the genome, thereby converting 5mC to 5hmC at specific nucleotides in specific target genes. This mechanism is supported by three lines of evidence: First, TET2 physically interacts with WT1. Second, TET2 binds to the promoters of genes that are known to be bound and regulated by WT1 and importantly the binding of TET2 to these WT1-target genes depends on the presence of WT1 protein. Third, the binding of TET2 to WT1-target gene is functional, as seen by the increase of 5hmC and the concomitant decrease of 5mC at the same sites and eventual transcription activation of its target genes. The functional relevance of WT1-recrutied TET2 is further supported by the finding that TET2 inhibits leukemia cell proliferation in a WT1-dependent manner.
The pathological significance of WT1-TET2 interaction is evidenced by the finding that this interaction is disrupted by multiple recurrent mutations in TET2 associated with AML, resulting in the loss of ability of TET2 to bind to and activate the expression of WT1-target genes while retaining the catalytic activity of TET2. The WT1-TET2 physical association and dependency of TET2 on WT1 to bind and activate WT1-target genes provide an explanation for the mutual exclusive pattern of mutations of these two genes in AML. Loss-of-function mutations targeting WT1 would have the same consequence as loss-of-function mutations of TET2 in disrupting the regulation of WT1-target genes. There is no advantage for mutating both genes in the same cell for the AML development. Likewise, mutations in IDH1 or IDH2, which produce 2-HG and causes functional inactivation of TET2 enzyme, would also block the activation of WT1-target genes that are normally activated by the recruitment of TET2, as shown by the inhibition of TET2-mediated WT1-target gene expression in cells treated with D-2-HG (Figure 1F) and in cells overexpressing IDH1 R132C mutant (Figure S1K). This result provides an explanation for the mutually exclusive mutations between IDH1, IDH2, TET2 and WT1 in AML. Together with the fact that mutations targeting IDH1 and IDH2 share the same biochemical consequences of inactivating their normal activity in producing α-KG and gaining of function in producing 2-HG, our results offer a molecular explanation for the mutually exclusive pattern of mutations targeting these four genes, IDH1/2, TET2 and WT1 in AML. Hence, these results suggest a linear IDH1/2-TET2-WT1 pathway in the suppression of AML (Figure 6C).
Of note, in addition to AML, both WT1 and TET2 genes were also found to be mutated in other types of cancer, such as bladder urothelial carcinoma, breast invasive carcinoma, colorectal adenocarcinoma, kidney renal clear cell carcinoma, liver hepatocellular carcinoma, lung adenocarcinoma, melanoma, uterine corpus endometrioid carcinoma (Table S2). Although the size of these tumor samples harboring either TET2 or WT1 mutation is too small for conducting a statistical analysis, we note that mutations targeting TET2 and WT1 genes rarely/never occur in the same tumor except for colorectal adenocarcinoma, suggesting their mutual exclusive pattern and a functional interaction in most cases. It is thus possible that the function of the IDH1/2-TET2-WT1 pathway is not limited to hematopoietic cells and may be broadly involved in suppressing other types of tumors.
EXPERIMENTAL PROCEDURES
Please refer to “Supplemental Experimental Procedures” for more detailed information about antibodies, plasmids, cell lines, cell culture and cell transfection, immunoprecipitation and western blotting, RNA isolation and quantitative real-time (qRT) PCR, protein concentration determination, colony formation assay, in vitro and in vivo TET2 activity assays.
Generation of stable WT1 and/or TET2 knock-down cell pools
To generate stable WT1 knock-down cell pools in HEK293T cells, shRNA targeting WT1 gene was constructed and the shRNA targeting sequence is shown in Table S1. The shRNA construct was co-transfected with vectors expressing the gag and vsvg genes into HEK293T cells using a two-plasmid packaging system as previously described (Chen et al., 2013). Retroviral supernatant was harvested 36 hrs after transfection, and mixed with 8 µg/mL polybrene to increase the infection efficiency. Cells were infected with the retrovirus and selected in 1 µg/ml puromycin for 1 week.
To generate stable WT1 and/or TET2 knock-down cell pools in HL-60 and KG-1 cells, shRNA targeting TET2 gene was constructed in pMKO.1-hyg vector and the shRNA targeting sequences are shown in Table S1. The virus was produced as described above. Retroviral supernatant was harvested 36 hrs after transfection, and mixed with 8 µg/mL polybrene to increase the infection efficiency. WT1-knocking down stable cells were infected with the retrovirus and selected in 200 µg/ml hygromycin B for 1 week.
Lentiviral transduction
For lentivirus production, the HA-tagged TET2CD domain expressing vectors were co-transfected with packaging (psPAX2) and envelope (pMD2.G) vectors into HEK293T cells. Lentivirus was harvested from the supernatant at 24 hrs post transfection, and mixed with 8 µg/mL polybrene to increase the infection efficiency. HL-60 cells were infected with the lentivirus and selected in 1 µg/ml puromycin for 1 week.
Baculovirus and pull-down assay
Flag-TET2CD baculovirus was generated by transfecting Sf9 cells with pDEST10-His-Flag-TET2CD by Cellfectin II (Invitrogen) for 72 hrs and amplified twice to get the P3 stock. Sf9 cells were cultured in Sf-900™ II SFM (Invitrogen) at 27°C, infected with P3 stock of Flag-TET2CD baculovirus for 72 hrs, cells were harvested and centrifuged for 15 min at 3000 rpm, 4°C. Cell were then lysed in a NP-40/Triton lysis buffer (40 mM Tris-HCl/pH7.5, 300 mM NaCl, 0.2% NP-40, 0.4% Triton X-100, 5 mM DTT, protease inhibitors) for 1 hr. Recombinant proteins were affinity purified Flag beads (Sigma) for 4 hrs at 4°C. The beads were washed three times with the wash buffer (40 mM Tris-HCl/pH7.5, 300 mM NaCl, 0.2% NP-40, 1 mM DTT, 10% Glycerol) and eluted with the Flag peptide (Sigma), then stored at −80°C.
For the pull-down assay, 400 ng purified recombinant Flag-WT1 (Creative Biomart) and 200 ng recombinant His-Flag-TET2CD were mixed with nickel beads (Qiagen) in the wash buffer described above at 4°C overnight. The beads were collected by centrifugation at 2,000 rpm for 3 min at 4°C and bound proteins were eluted with 300 mM imidazole and subjected to SDS-PAGE. Protein-protein interaction was examined either by Western-blot with Flag antibody or SYPRO Ruby staining (Invitrogen).
Chromatin Immunoprecipitation (ChIP)-qPCR assays
ChIP-qPCR assays were performed as described previously (Lan et al., 2007). Briefly, Cells were crosslinked with 1% paraformaldehype and sonicated. Solubilized chromatin was immunoprecipitated with antibodies against WT1, Flag, HA, H3K4me3 or negative control IgG. Antibody-chromatin complexes were pulled-down using protein A-sepharose (Millipore), washed and then eluted. After cross-link reversal and proteinase K (TaKaRa) treatment, immunoprecipitated DNA was extracted with phenol-chloroform, ethanol precipitated. The DNA fragments were further analyzed by quantitative PCR
MeDIP-qPCR and hMeDIP-qPCR analyses
Genomic DNA from cells was prepared using a phenol-chloroform method. The MeDIP and hMeDIP assays were performed as previously described (Ito et al., 2010; Mohn et al., 2009). Briefly, genomic DNA was denatured and then immunoprecipitated with anti-5mC, anti-5hmC antibody or IgG control antibody and protein G magnetic Dynabeads (Invitrogen). After washing for three times, beads were treated with proteinase K for 4 hours. DNA was prepared with phenol-chloroform and precipitated by using cold ethanol, and pulled-down DNA was analyzed by qPCR. All the primers were listed in supplemental Table S1.
GluMS-qPCR analysis
The 5hmC and 5mC levels in TET2 binding regions containing MspI/HpaII sites were measured by a restriction enzyme-based assay (EpiMark kit, NEB) (Kinney et al., 2011). Genomic DNA was treated with or without T4 Phage β-glucosyltransferase and then digested by MspI, HpaII or no enzyme. The MspI- and HpaII-resistant fractions were quantified by qPCR and normalized to the mock digestion. Primers used for GluMS-qPCR are listed in Supplementary Table S1.
Statistical Analysis
Statistical analyses were performed with a two-tailed unpaired Student's t-test. All data shown represent the results obtained from triplicated independent experiments with standard errors of the mean (mean ± S.D.). The values of p<0.05 were considered statistically significant.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the members of the Fudan MCB laboratory for discussions and support throughout this study. We also thank the Biomedical Core Facility of Fudan University for sustain research excellence. This work was supported by the 973 Program (No. 2012CB910303 to D.Y.; No. 2012CB910101 to K.L.G.), the NSFC grant (No. 81372198 to D. Y.), the NSFC Program of International Cooperation and Exchanges (No. 81120108016 to LXQ, Y.X.), and the Shanghai "Phosphor" Science Foundation, China (No. 14QA1400600 to D.Y.). This work was also supported by NIH grants (GM067113 and CA1638311 to Y.X.; CA132809 and CA108941 to K.L.G.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Cancer Genome Atlas Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Chen Y, Bian C, Fujiki R, Yu X. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature. 2013;493:561–564. doi: 10.1038/nature11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury R, Yeoh KK, Tian YM, Hillringhaus L, Bagg EA, Rose NR, Leung IK, Li XS, Woon EC, Yang M, et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO reports. 2011;12:463–469. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa Y, Ding J, Theunissen TW, Faiola F, Hore TA, Shliaha PV, Fidalgo M, Saunders A, Lawrence M, Dietmann S, et al. NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature. 2013;495:370–374. doi: 10.1038/nature11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty MM, Breiling A, Le T, Raddatz G, Barrasa MI, Cheng AW, Gao Q, Powell BE, Li Z, Xu M, et al. Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev Cell. 2013;24:310–323. doi: 10.1016/j.devcel.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Masse A, Kosmider O, Le Couedic JP, Robert F, Alberdi A, et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360:2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- Deplus R, Delatte B, Schwinn MK, Defrance M, Mendez J, Murphy N, Dawson MA, Volkmar M, Putmans P, Calonne E, et al. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. The EMBO journal. 2013;32:645–655. doi: 10.1038/emboj.2012.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doege CA, Inoue K, Yamashita T, Rhee DB, Travis S, Fujita R, Guarnieri P, Bhagat G, Vanti WB, Shih A, et al. Early-stage epigenetic modification during somatic cell reprogramming by Parp1 and Tet2. Nature. 2012;488:652–655. doi: 10.1038/nature11333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Mercado M, Yip BH, Pellagatti A, Davies C, Larrayoz MJ, Kondo T, Perez C, Killick S, McDonald EJ, Odero MD, et al. Mutation patterns of 16 genes in primary and secondary acute myeloid leukemia (AML) with normal cytogenetics. PLoS One. 2012;7:e42334. doi: 10.1371/journal.pone.0042334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huff V. Wilms' tumours: about tumour suppressor genes, an oncogene and a chameleon gene. Nat Rev Cancer. 2011;11:111–121. doi: 10.1038/nrc3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MK, McGarry TJ, P OB, Flatow JM, Golden AA, Licht JD. An integrated genome screen identifies the Wnt signaling pathway as a major target of WT1. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:11154–11159. doi: 10.1073/pnas.0901591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney SM, Chin HG, Vaisvila R, Bitinaite J, Zheng Y, Esteve PO, Feng S, Stroud H, Jacobsen SE, Pradhan S. Tissue-specific distribution and dynamic changes of 5-hydroxymethylcytosine in mammalian genomes. The Journal of biological chemistry. 2011;286:24685–24693. doi: 10.1074/jbc.M110.217083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, Bandukwala HS, An J, Lamperti ED, Thompson EC, Hastie R, Tsangaratou A, Rajewsky K, Koralov SB, Rao A. Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc Natl Acad Sci U S A. 2011;108:14566–14571. doi: 10.1073/pnas.1112317108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502:472–479. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F, Collins RE, De Cegli R, Alpatov R, Horton JR, Shi X, Gozani O, Cheng X, Shi Y. Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature. 2007;448:718–722. doi: 10.1038/nature06034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Cai X, Cai CL, Wang J, Zhang W, Petersen BE, Yang FC, Xu M. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood. 2011;118:4509–4518. doi: 10.1182/blood-2010-12-325241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang DC, Liu HC, Yang CP, Jaing TH, Hung IJ, Yeh TC, Chen SH, Hou JY, Huang YJ, Shih YS, et al. Cooperating gene mutations in childhood acute myeloid leukemia with special reference on mutations of ASXL1, TET2, IDH1, IDH2, and DNMT3A. Blood. 2013;121:2988–2995. doi: 10.1182/blood-2012-06-436782. [DOI] [PubMed] [Google Scholar]
- Maheswaran S, Englert C, Bennett P, Heinrich G, Haber DA. The WT1 gene product stabilizes p53 and inhibits p53-mediated apoptosis. Genes Dev. 1995;9:2143–2156. doi: 10.1101/gad.9.17.2143. [DOI] [PubMed] [Google Scholar]
- Maheswaran S, Park S, Bernard A, Morris JF, Rauscher FJ, 3rd, Hill DE, Haber DA. Physical and functional interaction between WT1 and p53 proteins. Proc Natl Acad Sci U S A. 1993;90:5100–5104. doi: 10.1073/pnas.90.11.5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F, Weber M, Schubeler D, Roloff TC. Methylated DNA immunoprecipitation (MeDIP) Methods Mol Biol. 2009;507:55–64. doi: 10.1007/978-1-59745-522-0_5. [DOI] [PubMed] [Google Scholar]
- Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, Figueroa ME, Vasanthakumar A, Patel J, Zhao X, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20:11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel JP, Gonen M, Figueroa ME, Fernandez H, Sun Z, Racevskis J, Van Vlierberghe P, Dolgalev I, Thomas S, Aminova O, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366:1079–1089. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quivoron C, Couronne L, Della Valle V, Lopez CK, Plo I, Wagner-Ballon O, Do Cruzeiro M, Delhommeau F, Arnulf B, Stern MH, et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell. 2011;20:25–38. doi: 10.1016/j.ccr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- Rivera MN, Haber DA. Wilms' tumour: connecting tumorigenesis and organ development in the kidney. Nat Rev Cancer. 2005;5:699–712. doi: 10.1038/nrc1696. [DOI] [PubMed] [Google Scholar]
- Rocquain J, Carbuccia N, Trouplin V, Raynaud S, Murati A, Nezri M, Tadrist Z, Olschwang S, Vey N, Birnbaum D, et al. Combined mutations of ASXL1, CBL, FLT3, IDH1, IDH2, JAK2, KRAS, NPM1, NRAS, RUNX1, TET2 and WT1 genes in myelodysplastic syndromes and acute myeloid leukemias. BMC Cancer. 2010;10:401. doi: 10.1186/1471-2407-10-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Wu H, Diep D, Yamaguchi S, D'Alessio AC, Fung HL, Zhang K, Zhang Y. Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell. 2013;153:692–706. doi: 10.1016/j.cell.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E, Shilatifard A. The chromatin signaling pathway: diverse mechanisms of recruitment of histone-modifying enzymes and varied biological outcomes. Molecular cell. 2010;40:689–701. doi: 10.1016/j.molcel.2010.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CX, Szulwach KE, Dai Q, Fu Y, Mao SQ, Lin L, Street C, Li Y, Poidevin M, Wu H, et al. Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell. 2013;153:678–691. doi: 10.1016/j.cell.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tefferi A, Pardanani A, Lim KH, Abdel-Wahab O, Lasho TL, Patel J, Gangat N, Finke CM, Schwager S, Mullally A, et al. TET2 mutations and their clinical correlates in polycythemia vera, essential thrombocythemia and myelofibrosis. Leukemia. 2009;23:905–911. doi: 10.1038/leu.2009.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC, Wartman LD, Lamprecht TL, Liu F, Xia J, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150:264–278. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, D'Alessio AC, Ito S, Xia K, Wang Z, Cui K, Zhao K, Sun YE, Zhang Y. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011;473:389–393. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Wang P, Xiao MT, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Xu C, Kato A, Tempel W, Abreu JG, Bian C, Hu Y, Hu D, Zhao B, Cerovina T, et al. Tet3 CXXC domain and dioxygenase activity cooperatively regulate key genes for Xenopus eye and neural development. Cell. 2012;151:1200–1213. doi: 10.1016/j.cell.2012.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Chen W, Iyer LM, Hu J, Wang G, Fu Y, Yu M, Dai Q, Aravind L, He C. A TET homologue protein from Coprinopsis cinerea (CcTET) that biochemically converts 5-methylcytosine to 5-hydroxymethylcytosine, 5-formylcytosine, and 5-carboxylcytosine. Journal of the American Chemical Society. 2014;136:4801–4804. doi: 10.1021/ja500979k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Szulwach KE, Hon GC, Song CX, Park B, Yu M, Lu X, Dai Q, Wang X, Street CR, et al. Tet-mediated covalent labelling of 5-methylcytosine for its genome-wide detection and sequencing. Nature communications. 2013;4:1517. doi: 10.1038/ncomms2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.