Abstract
Polyamidoamine (PAMAM) dendrimer was conjugated with both carboxymethyl-β-cyclodextrin (βCD) and poly(ethylene glycol) (PEG). Cyclic RGD peptide, used as a tumor targeting ligand, was then selectively conjugated onto the distal ends of the PEG arms. The resulting βCD-PAMAM-PEG-cRGD polymer was able to form stable and uniform nanoparticles (NPs) in aqueous solution. Doxorubicin (Dox), a model hydrophobic anticancer drug, was effectively encapsulated in the NPs via an inclusion complex formed between the drug and βCD. The Dox loading level was 16.8 wt%. The cellular uptake of cRGD-conjugated Dox-loaded NPs in the U87MG cell line was much higher than that of non-targeted NPs. Furthermore, the anti-proliferative effect of the cRGD-conjugated NPs was superior to that of free drug and non-targeted NPs. These results suggest that NPs formed by βCD-PAMAM-PEG-cRGD with a high drug payload may significantly improve the anticancer efficacy by tumor-targeted delivery and enhanced cellular uptake.
Keywords: PAMAM dendrimer, β-cyclodextrin, drug loading, targeted delivery, cRGD
1. Introduction
The success of chemotherapy depends on the drug and its dosage, as well as how effectively it is delivered to the target cells. Many anticancer drugs, due to their nonspecific action, often elicit toxicity to normal tissue even under optimal conditions. To a large extent, this issue can be addressed by precise targeting of anticancer drugs to cancer cells. Over the past several decades, various nanoplatforms have been explored for tumor-targeted drug delivery utilizing the unique enhanced permeation and retention (EPR) effect exhibited by solid tumors [1-6]. In addition to tumor-targeted drug delivery, nanomedicine also provides a number of other advantages including controlled drug release at the target site [7-9].
Polyamidoamine dendrimers (PAMAM) have been extensively investigated for drug/gene delivery due to their unique nanoscale architecture and multifunctionality [10-13]. However, previous studies have found that high generation PAMAM with NH2 terminal groups are associated with cytotoxicity, hemolysis, and liver toxicity, which limit their biomedical applications [14,15]. To enhance the biocompatibility and reduce the toxicity of PAMAM-(NH2)n, different approaches have been put forward to passivate the NH2 terminal groups, including acetylation and PEGylation [16-18]. PEGylation has been more widely studied because in addition to reduced cytotoxicity, PEGylation also sterically shields the nanocarriers from opsonization thereby increasing their circulation time in the bloodstream and consequently enhancing the in vivo tumor accumulation of the therapeutic agent via the EPR effect [19-27]. However, the extent of passive tumor-targeting alone via the EPR effect is often limited [28-29]. It has been demonstrated that PEGylated nanocarriers modified using active tumor targeting ligands specific to receptors overexpressed in cancer cells can further enhance the in vivo tumor accumulation of nanocarriers [30-36]. αvβ3 integrin plays a pivotal role in the regulation of angiogenesis of many types of solid tumors including glioblastoma, melanoma, breast, prostate, and ovarian cancer [37-38]. αvβ3 integrin is also often upregulated in tumors following radiotherapy [39,40]. The high-affinity interaction between synthetic cyclic arginine–glycine–aspartic acid sequences (cRGDs) containing peptides (e.g., cRGDyK, cRGDfK, cRGDfC, etc.) and integrin αvβ3 has led to extensive interest in utilizing cRGD peptides in cancer targeting therapies [41-42]. Several studies have demonstrated that PAMAM dendrimer-cRGD conjugates could enhance the delivery of imaging agents to targeted carcinoma cells [43-44].
Another common limitation associated with dendrimers including PAMAM is its relatively small size making it difficult to encapsulate a large amount of payload. Meanwhile, cyclodextrins (CDs) have been studied extensively in the field of controlled drug delivery [45]. CDs are cyclic oligosaccharides consisting of α-D-glucose units connected through (1→4) linkages leading to the shape of truncated cones. As a consequence of this peculiar structure, CD features a conical cavity that is essentially hydrophobic in nature. The cavity provides a hydrophobic microenvironment which attracts suitably sized hydrophobic drug molecules and stabilizes them through the formation of inclusion complexes [46-47]. The formation of CD guest inclusion complexes in aqueous media has been the basis for most of the biomedical and pharmaceutical applications of CDs [48]. Reports show that CD complexed doxorubicin (Dox) exhibits anti-proliferative activities superior to those of free Dox due to higher cellular uptake [49-50]. Recently, Okamatsu et al. demonstrated folate-appended βCD as a potent nanocarrier for Dox, both in vitro and in vivo [51].
In the present study, we report a cRGD-conjugated polymer NP formed by direct chemical conjugation of PEG and βCD onto fifth generation PAMAM dendrimers with NH2 surface terminals (PAMAM-G5). The drug loading efficacy and drug release profiles of the multifunctional NP were evaluated using Dox as a model hydrophobic drug. The effect of cRGD (Cyclo(Arg-Gly-Asp-D-Phe-Cys)) cancer cell targeting on the cellular uptake of the βCD-PAMAM-PEG NPs was studied using a human primary glioblastoma cell line (U87MG). The non-toxic nature of the empty NPs and the targeted cytotoxic effect of the Dox-loaded NPs toward U87MG cells were also demonstrated.
2. Materials and Methods
2.1 Materials
Polyamidoamine dendrimer (PAMAM-G5, 128 terminal amino groups, Mn 28,826 Da) was purchased from Dentritech, (Midland, MI, USA). Carboxymethyl-β-cyclodextrin sodium salt, 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC), N-hydroxysuccinimide (NHS), triethyl-amine (TEA), and paraformaldehyde were purchased from Sigma–Aldrich (Milwaukee, WI, USA) and were used without further purification. Cyclo (Arg-Gly-Asp-D-Phe-Cys) (c(RGDfC) peptide was purchased from Peptides International (Louisville, Ky, USA). The heterobifunctional PEG derivatives, maleimide-PEG-COOH (PEG-Mal) (Mw: 5000 Da) and (OCH3)-PEG-COOH (Mw: 5000 Da), were acquired from JenKem Technology (Allen, TX, USA). Doxo-rubicin hydrochloride (Dox· HCl) was purchased from Beijing Mesochem Technology Co., Ltd. (Beijing, Beijing, China). Dulbecco's modified eagle medium (DMEM) with a high glucose content, 4’,6-diamidino-2-phenylindol, dihydrochloride (DAPI), Stempro Accutase, and anti-biotic penicillin–streptomycin were purchased from Life Technologies (Grand Island, NY, USA). Fetal bovine serum (FBS) and 3-(4,5- Dimethylthiazol-2yl)-2,5, diphenyltetrazolium bromide (MTT reagent) were purchased from Sigma–Aldrich (Milwaukee, WI, USA). U87MG human glioblastoma cells expressing high levels of integrin αvβ3 were purchased from ATCC and cultured in DMEM supplemented with 10% FBS and 1% penicillin–streptomycin. Ultrapure water (18.2 MΩ·cm resistivity) was obtained from a Milli-Q water purification system and was used for all reactions and purification processes.
2.2 Synthesis of β-Cyclodextrin-Conjugated PAMAM (βCD-PAMAM)
Carboxymethyl-β-cyclodextrin (57.31 mg, 41.62 μmol) was dissolved in 2 mL of phosphate buffered saline (PBS) with a pH of 7.4 and stirred well with equimolar amounts of EDC (41.62 μmol, 6.46 mg) and NHS (41.62 μmol, 4.79 mg). This reaction was allowed to occur for 8 h in order to activate the carboxyl group of βCD. PAMAM (50 mg, 1.734 μmol) was added to the reaction mixture and stirred well at room temperature (RT) for 24 h. The reaction mixture was then dialyzed against deionized water for 48 h and lyophilized to obtain β-cyclodextrin conjugated PAMAM (βCD-PAMAM).
2.3 PEGylation of PAMAM-βCD
NHS ester of PEG-Maleimide (PEG-Mal) (241 mg, 48.19 μmol) and methoxy PEG-succynimydyl carboxymethyl ester (572 mg, 114.46 μmol) were added to PAMAM-βCD (50 mg, 1.255 μmol) in 5 mL of PBS. The reaction mixture was stirred at RT for 24 h and dialyzed against deionized water for 48 h. It was subsequently lyophilized to obtain PEGylated PAMAM-βCD (βCD-PAMAM-PEG-Mal/OCH3).
2.4 Synthesis of cRGD Modified βCD-PAMAM-PEG-Mal/OCH3
cRGD (5.8 mg, 10.09 μmol) was added to βCD-PAMAM-PEG-Mal/OCH3 (100 mg, 0.263 μmol) in 5 mL PBS . The reaction mixture was stirred well at RT under a nitrogen atmosphere for 6 h. It was then dialyzed against deionized water for 48 h and lyophilized to obtain cRGD-modified βCD-PAMAM-PEG, abbreviated as (βCD-PAMAM-PEG-cRGD).
2.5 In Vitro Drug Loading
Doxorubicin hydrochloride (50 mg, 86.2 μmol) was stirred with 2 molar equivalents of triethylamine (17.45 mg, 172.4 μmol) in 500 μL DMSO for 4 h at RT to obtain hydrophobic Dox. Next, 100 mg of the resulting polymer was dissolved in 4.5 mL water and stirred well at RT. Hydrophobic Dox in DMSO was added to the polymer/water solution drop by drop. The resulting solution was stirred for 24 h at RT. The reaction mixture was dialyzed against water using a dialysis membrane (molecular weight cutoff (MWCO) of 10 KDa) for 16 h and kept for freeze drying.
2.6 Characterization
1H NMR spectra of all intermediate and final polymer products were recorded at 25 °C on a Bruker DPX 300 spectrometer using D2O as the solvent. The molecular weights of the polymers were determined by NMR spectra. The hydrodynamic size and size distribution of the polymer NPs were determined by dynamic light scattering (DLS) (ZetaSizer Nano ZS90, Malvern Instrument, USA) at a polymer concentration of 0.1 mg/ml. The morphology and size of the dried polymer NPs were characterized using a transmission electron microscopy (TEM) (LEO912-OMEGA, Zeiss, Germany). To prepare the TEM sample, a drop of NP solution (0.1 mg/ml) containing 1 wt.% of phosphotungstic acid was deposited onto a 200 mesh carbon-coated copper grid and dried at RT. The Dox loading level in the polymer NP was measured using a Varian Cary 300 Bio UV visible spectrophotometer based on the absorbance of Dox at 485 nm. Dox was extracted from the NP solution in methanol (10 mg/mL). After the methanol was evaporated, Dox was dissolved in 3 mL of water containing 0.01N HCl. The Dox concentration in each sample was calculated based on an established calibration curve obtained using various concentrations of Dox.
2.7 In Vitro Drug Release Kinetics
Drug release studies were performed at 37 °C in acetate buffer (pH 5.3) and phosphate buffer (pH 7.4). 50 mg of Dox-loaded NPs were dispersed in 5 ml of medium and placed in a dialysis bag (MWCO 2 KDa). The dialysis bag was immersed in 50 ml of the release medium and kept in a horizontal laboratory shaker (100 rpm) under constant temperature. Samples of 2 ml volume were periodically removed and the same volume of fresh medium was added. The amount of released Dox was analyzed with a spectrophotometer at 485 nm. The drug release studies were performed in triplicate for each sample.
2.8 Cellular Uptake Study
The cellular uptake behavior and intracellular distribution of Dox-loaded NPs were analyzed using both flow cytometry and confocal laser scanning microscopy (CLSM). For flow cytometry, U87MG cells were seeded on 6-well culture plates and cultured in DMEM (10% FBS, 1% penicillin–streptomycin) for 48 hours. Cells were then incubated with free Dox, Dox-loaded targeted (i.e., cRGD-conjugated) NPs, and Dox-loaded non-targeted NPs (all having a Dox concentration of 10 μg/mL) in DMEM (2% FBS, 1% penicillin–streptomycin) for 2 h. Subsequently, the cells were washed and suspended in PBS using Accutase. The intracellular Dox fluorescence was analyzed using a BD Accuri C6 Sampler Flow Cytometer (BD, Franklin Lakes, NJ, USA). For CLSM studies, U87MG cells were seeded onto 4-well glass-bottom chamber slides and cultured in DMEM (10% FBS, 1% penicillin–streptomycin) for 48 h. Cells were incubated with Dox, Dox-loaded targeted NPs, and Dox-loaded non-targeted NPs (all having a Dox concentration of 10 μg/mL) for 2 h. After incubation, the cells were washed with PBS and fixed using 4% PFA. The cells were then imaged with a Nikon A1R-Si confocal laser scanning microscope and images were acquired using NIS-Elements Software (Nikon, Chiyoda, Tokyo, Japan).
2.9 Cytotoxicity Assay
The in vitro cytotoxicity of pure Dox, Dox-loaded targeted NPs, and Dox-loaded non-targeted NPs to U87MG cells were characterized using the MTT assay. First, U87MG cells were seeded in 96-well culture plates and incubated for 48 h in DMEM (10% FBS, 1% penicillin–streptomycin). The cells were then incubated with Dox, Dox-loaded targeted NPs, and Dox-loaded non-targeted NPs at different Dox concentrations (5, 10, and 20 μg/mL) for 48 h. After 48 h of incubation, 0.2 mL MTT solution in DMEM (0.5mg/mL) was added and incubated for 4 h at 37 °C. After removing the media, 100 μL of DMSO was added to each well, and the plate was incubated on an orbital shaker for 30 min at RT. An absorbance at 570 nm was measured using a Promega Glo-Max Multi Detection System (Promega, Madison, WI, USA) microplate reader. The in vitro cytotoxicity of the empty targeted and non-targeted NPs (30 μg/mL, 60 μg/mL, and 120 μg/mL) were also evaluated by the same method as explained above. All cytotoxicity values were normalized to an untreated control.
3. Results and Discussion
3.1 Synthesis and Characterization of cRGD-Conjugated βCD-PAMAM-PEG NPs
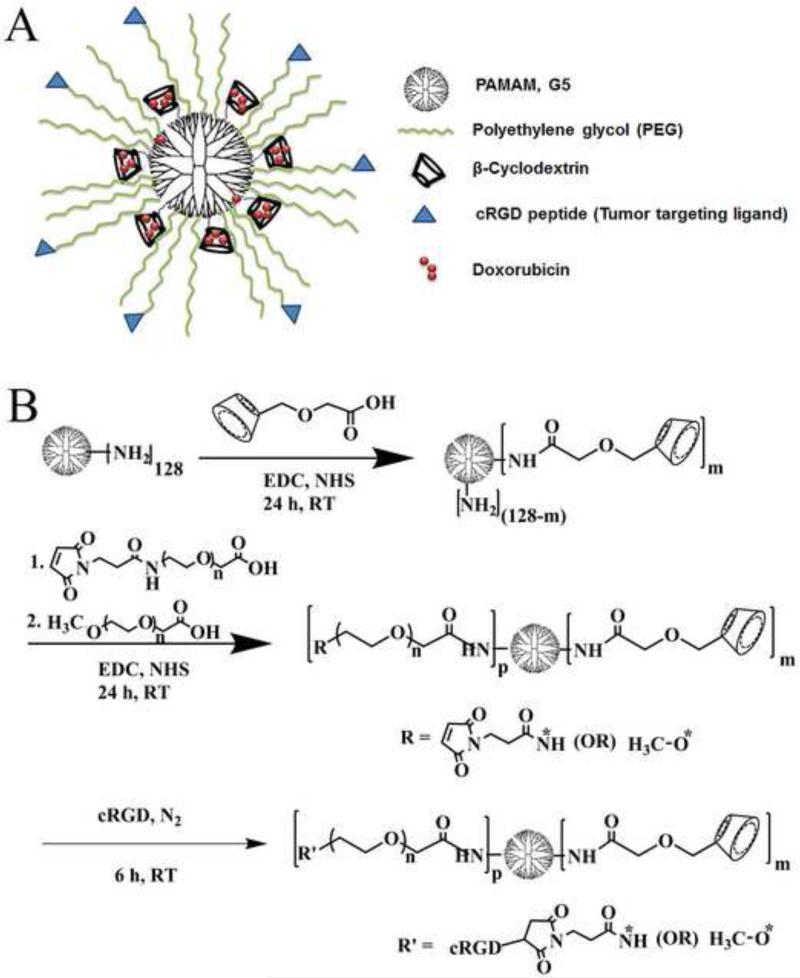
Figure 1 (A) shows a schematic of the cRGD-conjugated βCD-PAMAM-PEG NPs. A multi-arm βCD-PAMAM-PEG polymer was synthesized by covalently conjugating both β-cyclodextrin (βCD) and PEG onto the PAMAM dendrimer (G5) core having 128 NH2 terminal groups. Here βCD was used to encapsulate hydrophobic drugs such as Dox into the resulting polymer NP via the formation of inclusion complexes thereby increasing the aqueous solubility and subsequent bioavailability of Dox at tumor sites.
Figure 1.
(A) Schematic representation of multifunctional βCD-PAMAM-PEG-cRGD nanocarriers. (B) Schematic representation of the synthesis of cRGD-conjugated βCD-PAMAM-PEG NPs.
The PEG arms serve to mask the positive surface charge on PAMAM thus reducing cytotoxicity. Furthermore, the hydrophilic PEG shell can reduce the opsonization of the NPs during circulation in the bloodstream and enhance both their plasma circulation time and in vivo tumor accumulation. Finally, a tumor-targeting ligand, i.e., cRGD, was selectively conjugated onto the terminal ends of the PEG via click chemistry (reaction between the Mal group on PEG and the SH group on cRGD). A schematic representation of the preparation of cRGD-conjugated βCD-PAMAM-PEG is shown in Figure 1 (B).
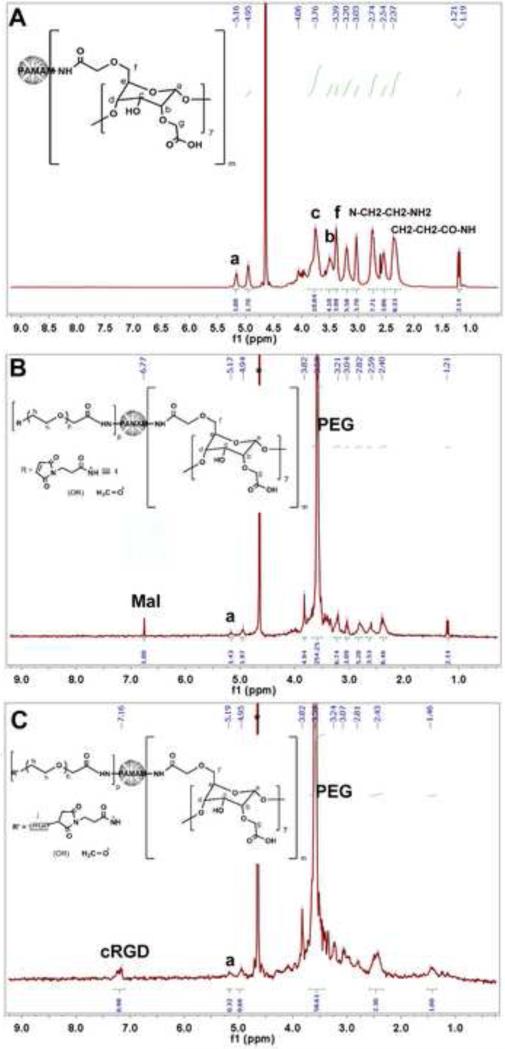
Each intermediate product during the synthesis of the βCD-PAMAM-PEG-cRGD polymer was confirmed by NMR spectra (Figure 2). Carboxymethyl β-cyclodextrin was conjugated onto the NH2 terminated PAMAM via an amidation reaction between the carboxyl group of carboxymethyl β-cyclodextrin and the terminal primary amine group of PAMAM. Conjugation of βCD onto PAMAM was confirmed by the NMR peaks located at 5.16 ppm, 3.76 ppm, and 3.51 ppm attributed to Ha, Hc, and Hb, respectively (Figure 2A) [52-53]. According to the 1H NMR spectrum, the number of βCD molecules conjugated onto PAMAM was calculated to be around 8. This calculation was based on the relative intensity ratio of the peaks at (a) 5.16 ppm and 2.74 ppm (Figure 3A) corresponding to the anomeric hydrogen in the cyclodextrin ring and the methylene group (N-CH2-CH2-NH2) of PAMAM, respectively. The remaining NH2 terminals of PAMAM (G5) were conjugated with either (Mal)-terminal PEG or (OCH3)-terminal PEG. In the NMR spectrum, the peak at 3.59 ppm was attributed to the (CH2–CH2–O) group of PEG. According to the 1H NMR spectrum (Figure 2B), the number of PEG molecules conjugated onto PAMAM was calculated to be about 43. This calculation was based on the relative intensity ratio of the peaks at 3.59 ppm and 2.4 ppm (Figure 2B) corresponding to the methylene group (CH2–CH2–O) of PEG and the methylene group (CH2–CH2–CO–NH) of PAMAM, respectively. The presence of the Mal group in the resulting βCD-PAMAM-PEG-Mal/OCH3 polymer was confirmed from the peak at 6.77 ppm (Figure 2B).[54] Peak intensities at 3.59 ppm and 6.77 ppm, corresponding to the methylene group (CH2–CH2–O) of PEG and the methylene group of (–CH=CH–) maleimide (Mal), respectively, were used for calculating the total percentage of Mal groups in the multi-arm βCD-PAMAM-PEG-Mal/OCH3 polymer. Among the 43 PEG molecules confirmed to be conjugated to PAMAM, 23 were Mal-terminated. Finally, the cRGD molecules, used as the tumor-targeting ligands, were conjugated via a nucleophillic addition reaction between the Mal group on PEG and the thiol group on the targeting ligand. cRGD peaks on the NP were confirmed based on the NMR peaks located at 7.13 to 7.32ppm (Figure 2C), corresponding to the aromatic region of the phenyl group of the cRGD peptides [55].
Figure 2.
NMR spectra of (A) βCD-PAMAM, (B) βCD-PAMAM-PEG-Mal/OCH3, and (C) βCD-PAMAM-PEG-cRGD/OCH3.
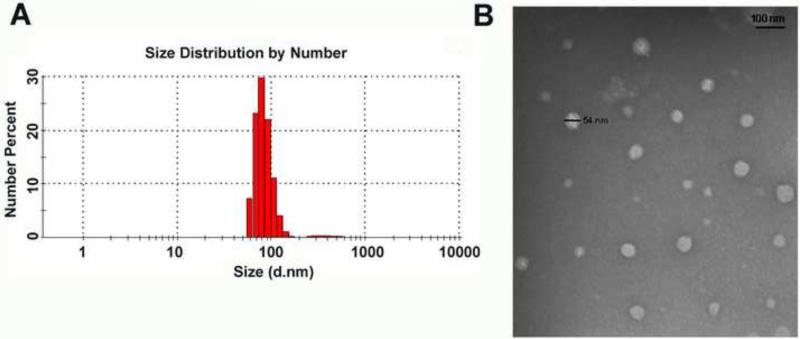
Figure 3.
(A) Size distribution of NPs as measured by DLS, and (B) morphology and size of NPs as measured by TEM.
Characterization of Dox-loaded βCD-PAMAM-PEG-cRGD NPs
Dox-loaded multi-arm βCD-PAMAM-PEG-cRGD polymer formed stable spherical NPs. The central cavity in the βCD provided a hydrophobic microenvironment capable of loading suitably sized hydrophobic drug molecules (e.g., Dox), effectively stabilizing them through the formation of inclusion complexes via hydrophobic-hydrophobic interaction, thus enhancing the aqueous solubility of Dox [56]. The amount of Dox encapsulated in the NPs was quite high at 16.8 wt %. In addition to the formation of Dox-βCD inclusion complexes, the physical entrapment of Dox in the PAMAM core via H-bonding may also have contributed to the high level of Dox loading in the NPs. A number of PAMAM-based NPs have been studied for enhanced bioavailability of Dox for targeted drug delivery [57-59]. For instance, Guo et al. demonstrated a Dox loading of 13.8 wt% for dendritic amphiphilic block copolymer PAMAM-PLA-b-PEG-TRC105 [57]. In another study Sun et al. reported a Dox loading level up to 15.6 wt% for Janus-type dendritic PAMAM amphiphiles [58]. It is well known that the morphology and stability of drug nanocarriers play an important role in the cellular internalization process as well as their in vivo performance. As the NPs were formed by individual multi-arm water-soluble βCD-PAMAM-PEG-cRGD polymers formed by covalent bonds, they are expected to have excellent in vivo stability. Unlike drug nanocarriers formed by self-assembly of multiple amphiphilic molecules such as liposomes, polymer micelles, and/or polymer vesicles these NPs are much less sensitive to various in vivo factors such as dilution, flow stress, and interaction with blood components. Particle size and size distribution studies of the βCD-PAMAM-PEG-cRGD NPs confirmed their stability in aqueous solution after one month of storage in a 4°C refrigerator. Figure 3A shows the size distribution histogram of the NPs measured at a concentration of 0.1 mg/mL with an average hydrodynamic diameter of 79 nm. Figure 3B shows a TEM image of spherically shaped NPs with diameters ranging from 35 to 54 nm.
3.2 In Vitro Drug Release Kinetics
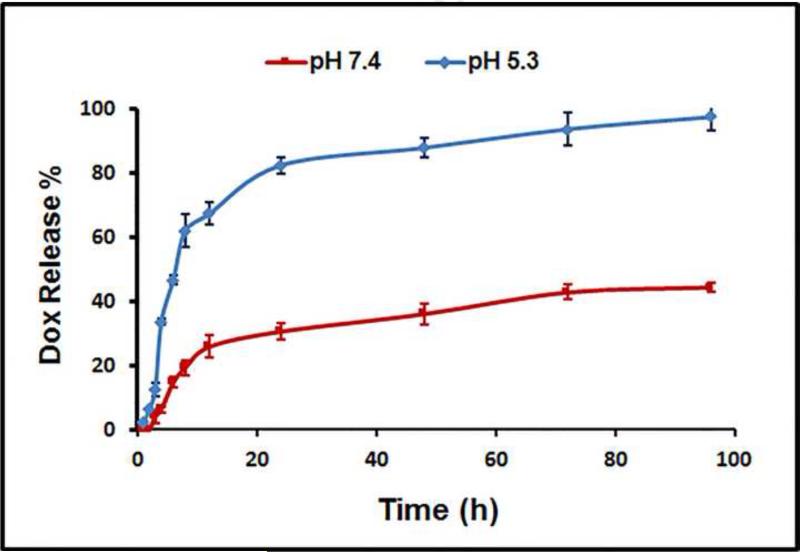
The in vitro release kinetics of Dox incorporated into NPs were studied at two pH levels at 37 °C to examine the drug release behaviour. NPs are typically taken up by cancer cells via endocytosis and it is known that the endocytic compartments (e.g., endosomes and lysosomes) are more acidic (pH, 6.5 to 4.3) than the intracellular cytosol and extracellular space. As such, phosphate buffered saline (pH 7.4) and acetate buffer (pH 5.3) were used to model the pH of blood and the endocytic compartments of the cancer cells, respectively. As shown in Figure 4, at a pH of 5.3, the NPs released Dox much faster than at a pH of 7.4. At a pH of 7.4, 19% of the total Dox was released after 8 h, and it was increased to 44% after 96 h. However, at a pH of 5.3, 62 % of the Dox was released after 8 h and 97% was released after 96 h. These results demonstrate that Dox-loaded NPs exhibited a pH-dependent in vitro release behavior, whereby a much faster release of Dox took place at an acidic pH of 5.3 than at a physiologic pH of 7.4. It may be attributed to the protonation of Dox in the acid condition since the protonation of Dox will weaken the hydrophobic-hydrophobic interaction in the inclusion complex [βCD-Dox] and release the Dox. In fact, Dox exhibited a pH-dependent release behaviour when it is encapsulated in other types of drug nanocarriers likely due to the same reason [57, 60-63]. Given the significantly higher drug release from NPs at a low pH, this nanoplatform poses less off-target threats to healthy cells and is suitable for intravenous injection [64]. Specifically, the pH-sensitive Dox-loaded NPs lose less drug payload during circulation in the blood stream (pH 7.4), but provide a desirable level of drug to effectively kill cancer cells once the NPs are internalized [65-66].
Figure 4.
In vitro drug release profiles of Dox-loaded βCD-PAMAM-PEG-cRGD NPs at pH 7.4 and pH 5.3, respectively. Error bars are standard deviation (SD) with n=3.
3.3 Effects of cRGD Targeting Ligands on the Cellular Uptake of NPs
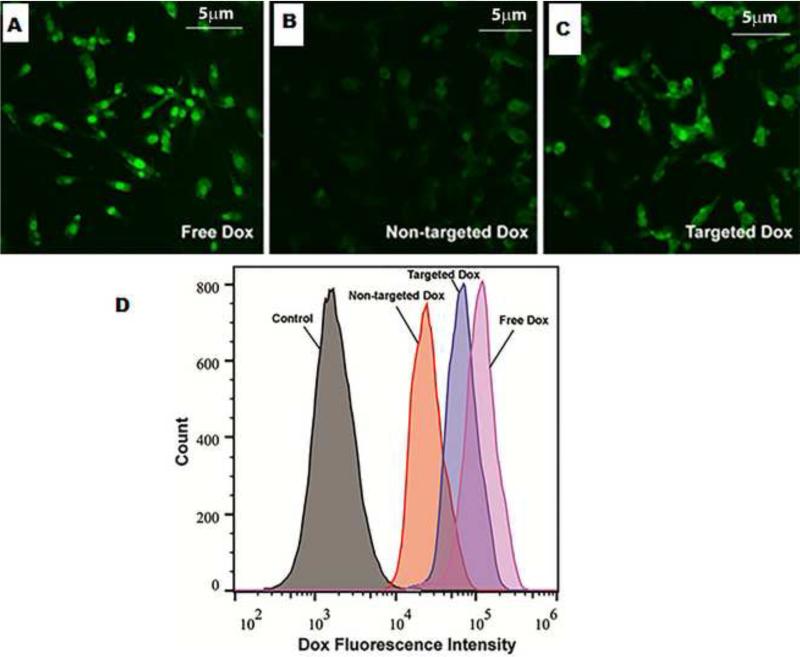
Flow cytometry and confocal laser scanning microscopy (CLSM) were used to evaluate the cellular uptake behaviour of NPs against αvβ3 receptor-positive U87MG cells. Figure 5 (A), (B) and (C) shows the CLSM images of U87MG cells incubated with free Dox, Dox-loaded targeted (cRGD-conjugated) NPs, and non-targeted NPs for 4 h. The Dox concentration used for this study was 10 μg/mL. Based on the Dox fluorescence intensity (green), the level of cellular uptake decreased in the following order: free Dox, Dox-loaded targeted NPs, and Dox-loaded non-target NPs. This was consistent with the MTT assay results and the flow cytometry findings to be discussed later. The enhanced cellular uptake exhibited by cRGD-conjugated NPs compared to non-targeted ones is attributed to receptor-mediated endocytosis [67-68]. CLSM images also show the intracellular localization of Dox in U87MG cells, clearly showing the nuclear and perinuclear localization of the targeted NPs.
Figure 5.
Cellular uptake study of the (A) free dox (B) non-targeted Dox (C) targeted Dox in U87MG cells using CLSM (D) Cellular uptake study of targeted and non-targeted NPs in U87MG cells using flow cytometry.
Figure 5 (D) shows the quantitative flow cytometry analysis data after the U87MG cells were incubated with free Dox, Dox-loaded targeted NPs, and non-targeted NPs for 4 h. Cells without any treatment were used as a negative control. Consistent with what was observed by CLSM, the flow cytometry data clearly showed that targeted NPs were taken up by the U87MG cells much more efficiently than non-targeted ones. However, free Dox showed a slightly higher cellular uptake than targeted NPs. This is likely due to the fact that small molecule drugs like Dox can efficiently diffuse into the cells through the plasma membrane.
3.4 Cytotoxicity Studies
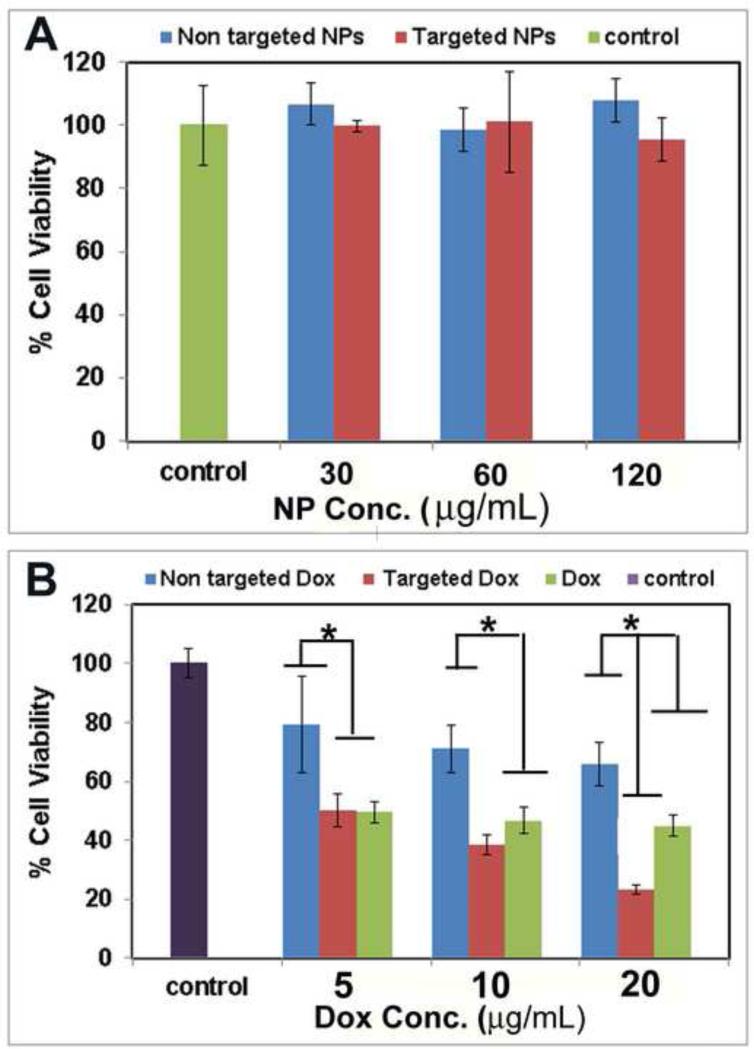
The effects of empty NPs, free Dox, Dox-loaded targeted NPs, and non-targeted NPs on the proliferative ability of U87MG cells 48 h post-treatment are shown in Figure 6. Empty (i.e., without Dox) targeted and non-targeted NPs (up to 120 μg/mL) did not induce significant changes in cell proliferation compared to the control (U87MG cells treated with media only), suggesting that these biocompatible NPs are suitable for drug delivery applications (Figure 6A). Dose-dependent cytotoxicity was observed with cells treated with free Dox and Dox-loaded targeted and non-targeted NPs (Figure 6B). At lower Dox concentrations, targeted NPs exhibited similar cytotoxicity to free Dox; however, at higher Dox concentrations, targeted NPs showed higher cytotoxicity than free Dox. For example, at 20 μg/mL, Dox-loaded targeted NPs showed ~77% cell inhibition, while free Dox had only ~55% cell inhibition. In addition, although free Dox exhibited high levels of cytotoxicity in vitro, it does not have any in vivo tumor-targeting ability. Finally, Dox-loaded non-targeted NPs exhibited the lowest cytotoxicity among the three samples tested at all concentrations, likely due to the low cellular uptake as demonstrated by both flow cytometry and CLSM.
Figure 6.
Cell viability of (A) empty (i.e., without Dox) targeted and non-targeted NPs, and (B) free Dox and Dox-loaded targeted and non-targeted NPs. Error bars are standard deviation (SD) with n=3. (*statistically significant at p<0.05).
Conclusions
cRGD-conjugated stable NPs made of multi-arm βCD-PAMAM-PEG polymers were synthesized and characterized. These NPs had a high encapsulation capacity for small hydrophobic drugs (e.g., 16.8 wt.% Dox). In vitro time-lapse diffusion analyses showed that the Dox-loaded NPs exhibited a pH-sensitive drug release behaviour desirable for targeted cancer therapy. According to flow cytometry and CLSM analyses, cRGD-conjugated NPs exhibited higher cellular uptake in U87MG cells, likely due to cRGD receptor-mediated endocytosis, resulting in higher cytotoxicity. Moreover, empty NPs were found to be minimally cytotoxic at high concentrations. Therefore, these unique NPs formed by individual multi-arm polymer molecules exhibit many desirable characteristics as promising drug nanocarriers for targeted cancer therapy.
Highlights.
Multifunctional drug nanocarriers were synthesized using poly(amido amine) (PAMAM), carboxymethyl-β-cyclodextrin (βCD), poly(ethylene glycol) (PEG) and cyclic RGD peptide.
Doxorubicin (Dox), a model hydrophobic anticancer drug, was effectively encapsulated in the nanoparticles via an inclusion complex formed between the drug and βCD.
The cellular uptake of cRGD-conjugated Dox-loaded nanopartilces in the U87MG cell line was much higher than that of non-targeted nanoparticles.
The anti-proliferative effect of the cRGD-conjugated nanoparticles was superior to that of free drug and non-targeted nanoparticles.
Acknowledgements
The authors gratefully acknowledge the financial support of the National Institutes of Health (1K25CA166178), USA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ferrari M. Nat. Rev. Cancer. 2005;5:161–171. doi: 10.1038/nrc1566. [DOI] [PubMed] [Google Scholar]
- 2.Allen TM, Cullis PR. Science. 2004;30:1818–822. doi: 10.1126/science.1095833. [DOI] [PubMed] [Google Scholar]
- 3.Maeda H, Wu J, Sawa T, Matsumura Y, Hori K. J Control Release. 2000;65:271–284. doi: 10.1016/s0168-3659(99)00248-5. [DOI] [PubMed] [Google Scholar]
- 4.Prabhakar U, Maeda H, Jain RK, Sevick-Muraca EM, Zamboni W, Farokhzad OC, Barry ST, Gabizon A, Grodzinski P, Blakey DC. Cancer Res. 2013;73:2412–2417. doi: 10.1158/0008-5472.CAN-12-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taurin S, Nehoff H, Greish K. J. Control. Release. 2012;164:265–275. doi: 10.1016/j.jconrel.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 6.Maeda H, Nakamura H, Fang J. Adv. Drug Deliver Rev. 2013;65:71–79. doi: 10.1016/j.addr.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 7.Cheng R, Meng F, Deng C, Klok HA, Zhong Z. Biomaterials. 2013;34:3647–57. doi: 10.1016/j.biomaterials.2013.01.084. [DOI] [PubMed] [Google Scholar]
- 8.Baker JR. Nanomedicine and Nanobiotechnology. 2013;5:423–9. doi: 10.1002/wnan.1226. [DOI] [PubMed] [Google Scholar]
- 9.Soppimath KS, Aminabhavi TM, Kulkarni AR, Rudzinski WE. J. Control Release. 2001;70:1–20. doi: 10.1016/s0168-3659(00)00339-4. [DOI] [PubMed] [Google Scholar]
- 10.Esfand R, Tomalia DA. Drug Discover Today. 2001;6:427–436. doi: 10.1016/s1359-6446(01)01757-3. [DOI] [PubMed] [Google Scholar]
- 11.Kesharwani P, Jain K, Jain NK. Prog Polymer Sci. 2014;39:268–307. [Google Scholar]
- 12.Zhu J, Shi X. J. Mater Chem. B. 2013;1:4199–4211. doi: 10.1039/c3tb20724b. [DOI] [PubMed] [Google Scholar]
- 13.Patri AK, Kukowska-Latallo JF, Baker JR., Jr Adv. Drug Deliver Rev. 2005;57:2203–14. doi: 10.1016/j.addr.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 14.Hong S, Bielinska AU, Mecke A, Keszler B, Beals JL, Shi X. Bioconjugate Chem. 2004;15:774–782. doi: 10.1021/bc049962b. [DOI] [PubMed] [Google Scholar]
- 15.Hong S, Leroueil PR, Janus EK, Peters JL, Kober MM, Islam MT. Bioconjugate Chem. 2006;17:728–34. doi: 10.1021/bc060077y. [DOI] [PubMed] [Google Scholar]
- 16.Jevprasesphant R, Penny J, Attwood D, McKeown NB, D'Emanuele A. Pharmaceut Res. 2003;20:1543–50. doi: 10.1023/a:1026166729873. [DOI] [PubMed] [Google Scholar]
- 17.Jevprasesphant R, Penny J, Jalal R, Attwood D, McKeown NB, D'Emanuele A. Int J Pharm. 2003;252:263–266. doi: 10.1016/s0378-5173(02)00623-3. [DOI] [PubMed] [Google Scholar]
- 18.Kolhatkar RB, Kitchens KM, Swaan PW, Ghandehari H. Bioconjugate Chem. 2007;18:2054–2060. doi: 10.1021/bc0603889. [DOI] [PubMed] [Google Scholar]
- 19.Harris JM, Chess RB. Nat Rev Drug Discov. 2003;2:214–221. doi: 10.1038/nrd1033. [DOI] [PubMed] [Google Scholar]
- 20.Guillaudeu SJ, Fox ME, Haidar YM, Dy EE, Szoka FC, Fréchet JMJ. Bioconjugate Chem. 2008;19:461–469. doi: 10.1021/bc700264g. [DOI] [PubMed] [Google Scholar]
- 21.Karthikeyan R, Kumar PM, Kumar PV. J Acute Disease. 2013;2:327–329. [Google Scholar]
- 22.Kim Y, Klutz AM, Jacobson KA. Bioconjugate Chem. 2008;19:1660–1672. doi: 10.1021/bc700483s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo D, Haverstick K, Belcheva N, Han E, Saltzman WM. Macromolecules. 2002;35:3456–3462. [Google Scholar]
- 24.Okuda T, Kawakami S, Akimoto N, Niidome T, Yamashita F, Hashida M. J Control Release. 2006;116:330–336. doi: 10.1016/j.jconrel.2006.09.012. [DOI] [PubMed] [Google Scholar]
- 25.Zhu S, Hong M, Tang G, Qian L, Lin J, Jiang Y. Biomaterials. 2010;31:1360–1371. doi: 10.1016/j.biomaterials.2009.10.044. [DOI] [PubMed] [Google Scholar]
- 26.Kaminskas LM, Boyd BJ, Karellas P, Krippner GY, Lessene R, Kelly B. Mol. Pharmaceutics. 2008;5:449–463. doi: 10.1021/mp7001208. [DOI] [PubMed] [Google Scholar]
- 27.Fang J, Nakamura H, Maeda H. Adv Drug Deliver Rev. 2011;63:136–151. doi: 10.1016/j.addr.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 28.Byrne JD, Betancourt T, Brannon-Peppas L. Adv Drug Deliver Rev. 2008;60:1615–1626. doi: 10.1016/j.addr.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 29.Okamatsu A, Motoyama K, Onodera R, Higashi T, Koshigoe T, Shimada Y. Bioconjugate Chem. 2013;24:724–733. doi: 10.1021/bc400015r. [DOI] [PubMed] [Google Scholar]
- 30.Henne WA, Kularatne SA, Hakenjos J, Carron JD, Henne KL. Bioorg Med Chem Lett. 2013;23:5810–5813. doi: 10.1016/j.bmcl.2013.08.113. [DOI] [PubMed] [Google Scholar]
- 31.Kim TH, Jo YG, Jiang HH, Lim SM, Youn YS, Lee S. J Control Release. 2012;162:422–428. doi: 10.1016/j.jconrel.2012.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sk UH, Kambhampati SP, Mishr MK, Lesniak WG, Zhang F, Kannan RM. Biomacromolecules. 2013;14:801–810. doi: 10.1021/bm3018615. [DOI] [PubMed] [Google Scholar]
- 33.Guo J, Hong H, Chen G, Gong S. Biomaterials. 2013;34:8323–8332. doi: 10.1016/j.biomaterials.2013.07.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiao YL, Hong H, Javadi A, Gong S. Biomaterials. 2012;33:3071–3082. doi: 10.1016/j.biomaterials.2011.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang X, Hong H, Grailer JJ, Gong S. Biomaterials. 2011;32:4151–4160. doi: 10.1016/j.biomaterials.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu WJ, Siddiqui LA, Nihal M, Gong S. Biomaterials. 2013;34:5244–5253. doi: 10.1016/j.biomaterials.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cai W, Chen X. Anti-Cancer Agents in Medicinal Chemistry (Formerly Current Medicinal Chemistry-Anti-Cancer Agents) 2006;6:407–428. doi: 10.2174/187152006778226530. [DOI] [PubMed] [Google Scholar]
- 38.Gasparini G, Brooks PC, Biganzoli E, Vermeulen PB, Bonoldi E, Dirix LY. Clin Cancer Res. 1998;4:2625–2634. [PubMed] [Google Scholar]
- 39.Abdollahi A, Griggs DW, Zieher H, Roth A, Lipson KE, Saffrich R. Clin Cancer Res. 2005;11:6270–6279. doi: 10.1158/1078-0432.CCR-04-1223. [DOI] [PubMed] [Google Scholar]
- 40.Danhie F, Breton AL, Preat V. Mol. Pharmaceutics. 2012;9:2961–2973. doi: 10.1021/mp3002733. [DOI] [PubMed] [Google Scholar]
- 41.Dunehoo AL, Anderson M, Majumdar S, Kobayashi N, Berkland C, Siahaan TJ. J Pharm Sci. 2006;95:1856–1872. doi: 10.1002/jps.20676. [DOI] [PubMed] [Google Scholar]
- 42.Morlieras J, Dufort S, Sancey L, Truillet C, Mignot A, Rossetti F. Bioconjugate Chem. 2013;24:1584–1597. doi: 10.1021/bc4002097. [DOI] [PubMed] [Google Scholar]
- 43.Boswell CA, Eck PK, Regino CA, Bernardo M, Wong KJ, Milenic DE. Mol. Pharmaceutics. 2008;5:527–539. doi: 10.1021/mp800022a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waite CL, Roth CM. Bioconjugate Chem. 2009;20:1908–1916. doi: 10.1021/bc900228m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang J, Ma PX. Adv Drug Deliver Rev. 2013;65:1215–1233. doi: 10.1016/j.addr.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loftsson T, Brewster ME, Másson M. Am. J Drug Deliver. 2004;2:261–275. [Google Scholar]
- 47.Del Valle E. Process Biochemistry. 2004;39:1033–1048. [Google Scholar]
- 48.Loftsson T, Brewster ME. J Pharm Sci. 1996;85:1017–1025. doi: 10.1021/js950534b. [DOI] [PubMed] [Google Scholar]
- 49.Bekers O, Beijnen J, Otagiri M, Bult A, Underberg W. J Pharmaceut Biomed. 1990;8:671–674. doi: 10.1016/0731-7085(90)80100-4. [DOI] [PubMed] [Google Scholar]
- 50.Sivasubramanian M, Thambi T, Deepagan V, Saravanakumar G, Ko H, Kang YM. J Nanosci Nanotechno. 2013;13:7271–7278. doi: 10.1166/jnn.2013.8091. [DOI] [PubMed] [Google Scholar]
- 51.Okamatsu A, Motoyama K, Onodera R, Higashi T, Koshigoe T, Shimada Y. Bioconjugate Chem. 2013;24:724–733. doi: 10.1021/bc400015r. [DOI] [PubMed] [Google Scholar]
- 52.Ayman AA, Shar SA. J Incl Phenom Macro. 2010;67:7–11. [Google Scholar]
- 53.Marcia MM, Deisy AD, Marilde TB, Roseane F, Bruno S. Bri Food J. 2001;103:281–290. [Google Scholar]
- 54.Ramesh S, Sivasamy A, Kim JH. Nanoscale Res Lett. 2012;7:350. doi: 10.1186/1556-276X-7-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li G, Hu Z, Yin H, Zhang Y, Huang X, Wang S, Li W. Int J Nanomed. 2013;8:1293–130656. doi: 10.2147/IJN.S41555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bekers O, Beijnen JH, Otagiri M, Bult A, Underberg WJM. J. Pharm. Biomed. Anal. 1990;8:671–674. doi: 10.1016/0731-7085(90)80100-4. [DOI] [PubMed] [Google Scholar]
- 57.Guo J, Hong H, Chen G, Shi S, Zheng Q, Zhang Y, Gong S. Biomaterials. 2013;34:8323–8332. doi: 10.1016/j.biomaterials.2013.07.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun L, Ma X, Dong CM, Zhu B, Zhu X. Biomacromolecules. 2012;13:3581–3591. doi: 10.1021/bm3010325. [DOI] [PubMed] [Google Scholar]
- 59.Zhang M, Guo R, Keri M, Bányai I, Zheng Y, Cao M, Shi X. J. Phys. Chem. B. 2014;118:1696–1706. doi: 10.1021/jp411669k. [DOI] [PubMed] [Google Scholar]
- 60.Zhang X, Wu Z, Gao X, Cheng C, Wang Z, Li C. Acta Biomater. 2011;7:585–592. doi: 10.1016/j.actbio.2010.08.029. [DOI] [PubMed] [Google Scholar]
- 61.Du L, Liao S, Khatib HA, Stoddart JF, Zink JI. J. Am. Chem. Soc. 2009;131:15136–15142. doi: 10.1021/ja904982j. [DOI] [PubMed] [Google Scholar]
- 62.Prabaharan M, Grailer JJ, Pilla S, Steeber DA, Gong S. Biomaterials. 2009;30:3009–3019. doi: 10.1016/j.biomaterials.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 63.Xu W, Siddiqui IA, Nihal M, Pilla S, Rosenthal K, Mukhtar H, Gong S. Biomaterials. 2013;34:5244–5253. doi: 10.1016/j.biomaterials.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schmaljohann D. Adv Drug Deliver Rev. 2006;58:1655–1670. doi: 10.1016/j.addr.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 65.Liu J, Huang Y, Kumar A, Tan A, Jin S, Mozhi A. Biotechnology Advances. 2013;32:693–710. doi: 10.1016/j.biotechadv.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 66.Prabaharan M, Grailer JJ, Pilla S, Steeber DA, Gong S. Biomaterials. 2009;30:5757–5766. doi: 10.1016/j.biomaterials.2009.07.020. [DOI] [PubMed] [Google Scholar]
- 67.Alam MR, Dixit V, Kang H, Li ZB, Chen X, Trejo J. Nucleic Acids Res. 2008;36:2764–2776. doi: 10.1093/nar/gkn115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bareford LM, Swaan PW. Adv Drug Delivery Rev. 2007;59:748–758. doi: 10.1016/j.addr.2007.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]