Abstract
JC Virus (JCV) infection of the brain can cause Progressive Multifocal Leukoencephalopathy, JCV Granule Cell Neuronopathy and JCV Encephalopathy (JCVE). JCVCPN, isolated from the brain of a patient with JCVE, is a naturally occurring strain of JCV with a 143 base pair deletion in the agnogene. Cell culture studies of JCVCPN have shown that the loss of these nucleotides in the agnogene results in impaired expression of VP1 and infectious virion production. To better understand the role of this DNA sequence in JCV replication, we generated a series of deletions in the agnogene on the backbone of a virus which has a mutated agnoprotein start codon preventing agnoprotein expression. We found that deletion of nucleotides 376-396 results in decreased levels of viral DNA replication and a lack of VP1 expression. These results indicate that these nucleotides play a crucial role in JCV replication.
Keywords: JC virus, agnogene, agnoprotein, VP1 expression
INTRODUCTION
JC Virus (JCV) is the etiological agent of the fatal demyelinating disease Progressive Multifocal Leukoencephalopathy (PML), as well as the more recently described JCV Granule Cell Neuronopathy (JCV GCN) and JCV Encephalopathy (JCVE) (Koralnik et al., 2005; Tan and Koralnik, 2010; Wuthrich et al., 2009). JCV infection is widespread and asymptomatic in healthy individuals (Knowles et al., 2003; Weber et al., 1997). Individuals with immune suppression, such as patients with AIDS (Berger et al., 1987), organ transplants (Molloy and Calabrese, 2009) and hematological malignancies (Shimizu et al., 1999), are at risk for developing PML.
JCV is a polyomavirus with a circlular double stranded DNA genome. The genome has 3 regions, the highly conserved early and late coding regions and the hypervariable regulatory region (RR) (Gheuens et al., 2013). The regulatory proteins small t antigen and large T antigen (T Ag) are encoded by the early coding region while the structural proteins VP1, VP2 and VP3 and the agnoprotein are encoded by the late coding region (Gheuens et al., 2013). The origin of replication and the early and late promoters are contained by the RR (Jensen and Major, 2001).
The agnoprotein of JCV is a 71 amino acid, highly basic protein that is non-essential for viral infection (Khalili et al., 2005). It is expressed late in infection, primarily in the cytoplasm, and is not incorporated into virions (Khalili et al., 2005; Okada et al., 2001). Recent research into agnoprotein function has begun to shed light on its roles in JCV replication, but there is still much to be learned. The agnoprotein can be phosphorylated (Sariyer et al., 2006; Sariyer et al., 2008) and form both homodimers and oligomers, an interaction which is mediated by the Leu/Ile/Phe rich domain of the protein (Saribas et al., 2013; Saribas et al., 2012). The agnoprotein has been implicated in a number of different viral processes, including DNA replication (Safak et al., 2001; Saribas et al., 2012), expression and splicing of viral transcripts (Okada et al., 2001; Safak et al., 2001; Safak et al., 2002; Saribas et al., 2013), early and late protein expression (Sariyer et al., 2011) and virion formation and release (Sariyer et al., 2011; Suzuki et al., 2012). It has also been reported that agnoprotein may act as a viroporin (Suzuki et al., 2010). While these studies have given insights into agnoprotein function, the exact mechanisms of action are still unclear.
Studies have shown that deleting the agnogene DNA is more detrimental to JCV replication than loss of agnoprotein expression without deletion of the DNA sequence, indicating that the agnogene may contain a cis-acting regulatory element (Akan et al., 2006; Ellis et al., 2013). Additionally, 3 potential host-factor binding sites have been identified in the agnogene (Akan et al., 2006).
JCVCPN was isolated from the brain of a patient with JCVE, and has an archetype-like RR and a 143 base pair deletion from nucleotide 300-442 in the agnogene (Dang et al., 2012). It was named JCVCPN since it causes productive infection in Cortical Pyramidal Neurons. This was the first time a large deletion in the agnogene was isolated in a naturally occurring viral isolate. We have recently studied the replication of JCVCPN in cell culture to better understand both the pathogenesis of JCVE and the role of the RR and agnogene in JCV replication (Ellis et al., 2013). Our results indicated that, in cell culture, JCVCPN was incapable of establishing a high level persistent infection compared to the prototype strain JCVMad-1 (Frisque et al., 1984) and that this was due to a lack of VP1 expression and infectious virion release (Ellis et al., 2013). Further experiments identified the deletion in the agnogene, specifically the loss of the agnogene DNA more so than the loss of a full length agnoprotein, as the primary cause of this phenotype (Ellis et al., 2013).
To further characterize the role of the agnogene in JCV replication, we generated a series of deletions in the agnogene. All of the deletions were introduced into JCVMad-1 with a mutated agnoprotein start codon (Mad-1 Pt), which prevents protein expression. This allowed us to study the function of the deleted regions in isolation from the functions of the agnoprotein. We studied the replication of these viruses in Cos-7 cells, an African green monkey kidney cell line that expresses SV40 T Ag, which allows JCV to replicate efficiently in these cells. We compared the replication of these deletion mutants to JCVMad-1 in cell culture by measuring DNA replication, early and late mRNA expression and VP1 protein production to identify smaller regions of the agnogene which are crucial for viral replication.
RESULTS
Generation of agnogene deletion mutants
Our previous studies have indicated that the deletion of nucleotides 300-442 in the agnogene in JCVCPN leads to decreased viral replication and a lack of detectable VP1 protein expression, and that this phenotype is due primarily to the loss of the DNA sequence in the agnogene and not the loss of full length agnoprotein expression (Ellis et al., 2013). To further narrow down the regions of the agnogene which are critical for viral replication and VP1 protein expression we generated a series of additional agnogene deletion mutants. All additional deletions were introduced onto the Mad-1 Pt backbone, which has a mutated start codon to prevent expression of the agnoprotein, in order to investigate only the effects of deletions in the agnogene. We chose the Mad-1 Pt virus with a rearranged regulatory region rather than the JCVCPN regulatory region in order to focus on the effects of the agnogene deletion without having any regulatory region effects involved.
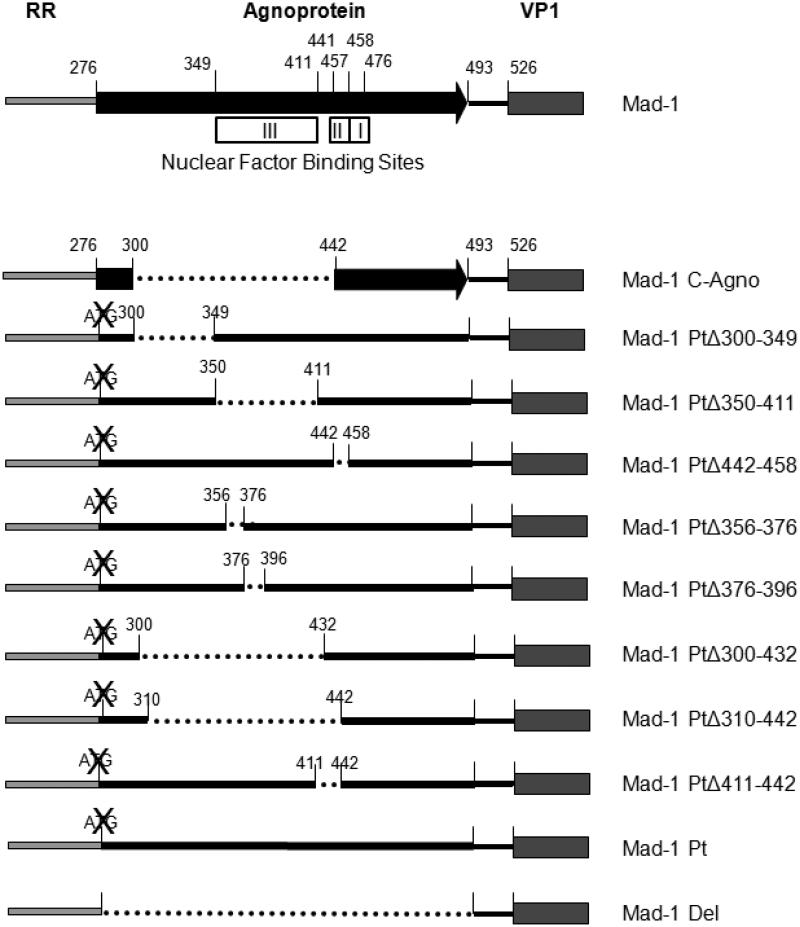
Figure 1 shows the locations of the additional mutants that were generated, as well as the locations of the predicted host cell protein binding sites (Akan et al., 2006). Mad-1 PtΔ300-349 encompasses the region from the beginning of the JCVCPN deletion to the start of binding site III. Mad-1 PtΔ350-411 and Mad-1 PtΔ442-458 delete binding sites III and II, respectively. Both Mad-1 PtΔ356-376 and Mad-1 PtΔ376-396 are located within binding site III. Mad-1 PtΔ356-376 deletes a region predicted to be a possible transcription factor binding site by AliBaba 2 software (http://www.generegulation.com/pub/programs.html#alibaba2) and Mad-1 PtΔ376-396 deletes a region of high homology between JCV, BKV and SV40. Mad-1 PtΔ300-432 and Mad-1 PtΔ310-442 encompass the region deleted in JCVCPN (300-442) with the last 10 or first 10 nucleotides of the deletion added back, respectively. Finally, Mad-1 PtΔ411-442 is the region between binding sites III and II. As controls, we used the previously characterized viruses Mad-1 Pt, a start codon deletion mutant, Mad-1 Del, a full agnogene deletion mutant (Akan et al., 2006), and Mad-1 C-Agno, JCVMad-1 with the JCVCPN agnogene, which has a deletion of nucleotides 300-442 (Ellis et al., 2013).
Fig. 1. Agnogene deletion mutants.
This diagram shows all of the viruses used in this study. Boxes represent open reading frames. Lines represent DNA sequences. Dotted lines represent deletions. ATG with an X through it indicates a mutated start codon preventing agnoprotein expression. Nuclear factor binding sites indicate the regions identified by Akan et al (Akan et al., 2006).
Deletion of nucleotides 376-396 decreases levels of viral DNA replication at late time points post-transfection
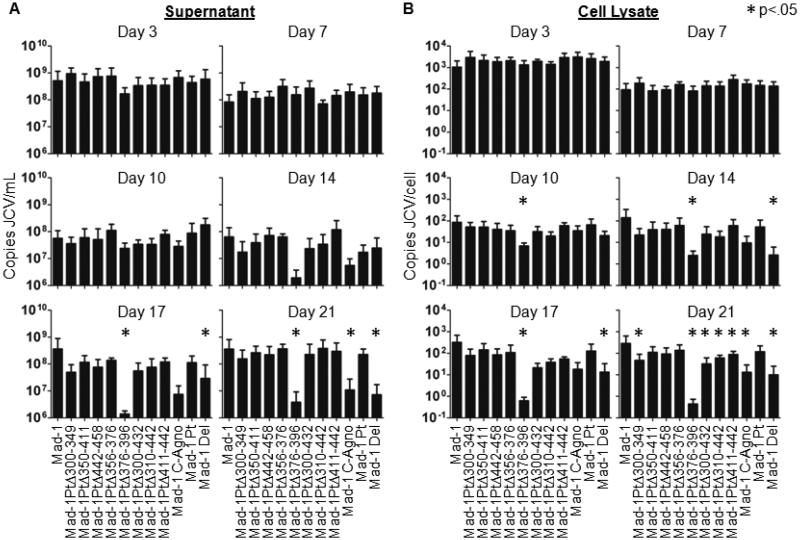
Linearized JCV genomes were transfected into Cos-7 cells and viral DNA was quantified in the supernatant (Figure 2A) and cell lysate (Figure 2B) by qPCR 3, 7, 10, 14, 17 and 21 days post-transfection compared to JCVMad-1. As previously reported (Ellis et al., 2013), Mad-1 C-Agno and Mad-1 Del show lower levels of viral DNA in the supernatant and cell lysate from 14-21 days post-transfection. Most of the new agnogene deletion mutants have replication levels similar to Mad-1 and Mad-1 Pt. Interestingly, Mad-1 PtΔ376-396 has drastically decreased levels of JCV DNA in the supernatant and cell lysates at later times post-transfection, similar to Mad-1 C-Agno and Mad-1 Del. Levels of Mad-1 PtΔ376-396 DNA are significantly lower than JCVMad-1 starting 10 days post-transfection in cell lysate and 17 days post-transfection in supernatant. This indicates that nucleotides 376-396 are important for JCV replication.
Fig. 2. Deletion of nucleotides 376-396 reduces viral DNA replication.
Cos-7 cells were transfected with linearized JCV genomes and cells were subcultured every 3-4 days. DNA was extracted from the supernatant and cell samples, digested with DpnI to remove input plasmid DNA, and analyzed by qPCR. Data represents the average of 3-5 independent experiments. Mad-1 PtΔ376-396 has decreased levels of viral DNA compared to JCVMad-1 at 17-21 days post-transfection in supernatant (A) and 10-21 in cell lysate (B). Mock transfected cells remained JCV negative. Error bars represent standard deviation. P-values were calculated for all viruses compared to JCVMad-1 using the Wilcoxon Rank Test. * indicates p<.05
Deletion of nucleotides 376-396 result in decreased levels of late mRNA expression
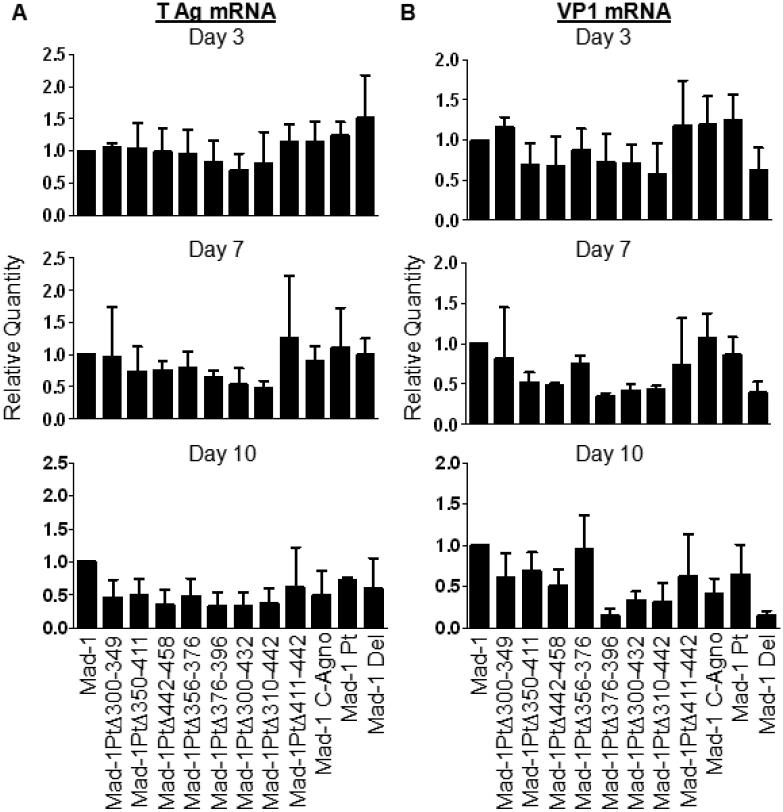
RNA was extracted from JCV transfected Cos-7 cells 3, 7 and 10 days post-transfection. RNA was reverse transcribed to cDNA and levels of early (T Ag) and late (VP1) mRNA were determined using qPCR. While there was some decrease in T Ag mRNA expression 10 days post-transfection with most of the deletion mutants, there were no significant decreases observed (Figure 3A). Greater decreases in expression levels were observed for VP1 mRNA, although none were statistically significant (Figure 3B). The greatest decrease in late mRNA expression was seen with Mad-1 PtΔ376-396, which has levels equivalent to Mad-1 Del 7 and 10 days post-transfection. Additionally, Mad-1 PtΔ300-432 and Mad-1 PtΔ310-442 also have decreased levels of VP1 mRNA expression 7 and 10 days post-transfection. These results indicate that nucleotides 376-396 are involved in the expression of late mRNAs.
Fig. 3. Deletion of nucleotides 376-396 results in a reduction of late mRNA expression.
Cos-7 cells were transfected with linearized JCV genomes. Cell pellets were collect and RNA was extracted and reverse transcribed into cDNA. qPCR was used to measure the levels of early (T Ag) and late (VP1) transcripts as well as the endogenous control TATA-Box Binding Protein (TBP). Relative Quantity (RQ) was calculated using the ΔΔCt method, with TBP as the endogenous control and JCVMad-1 as the calibrator sample. Data represents the average of 3 independent experiments. (A) While some decrease in T Ag mRNA is observed 10 days post-transfection, no significant differences were observed for any of the viruses tested. (B) Mad-1 PtΔ376-396 expresses decreased levels of VP1 mRNA 7 and 10 days post-transfection, with levels similar to Mad-1 Del. Mad-1 PtΔ300-432 and Mad-1 PtΔ310-442 also show some decrease in VP1 mRNA expression. None of these differences reached statistical significance. Mock transfected samples remained negative for JCV mRNAs. Error Bars represent standard deviation. P values were calculated for all viruses compared to JCVMad-1 with Student’s t test using univariate analysis.
Nucleotides 376-396 are critical for VP1 expression
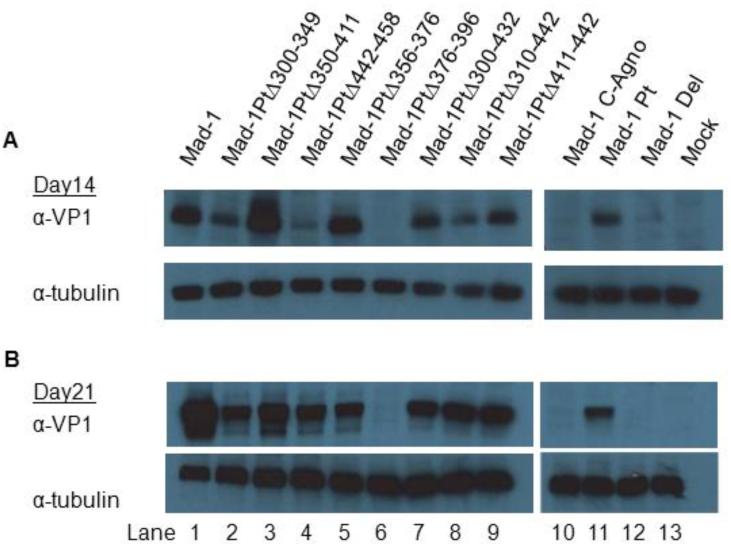
Western blotting was done for VP1 expression in cell lysate collected from Cos-7 cells 14 and 21 days post-transfection (Figure 4). As previously published, JCVMad-1 (Lane 1) and Mad-1 Pt (Lane 11) express detectable levels of VP1 protein, while Mad-1 C-Agno (Lane 10) and Mad-1 Del (Lane 12) do not (Ellis et al., 2013). Interestingly, all of the additional agnogene deletion mutants except Mad-1 PtΔ376-396 (Lane 6) express VP1, although at somewhat decreased levels. Mad-1 PtΔ376-396 does not express detectable levels of VP1 14 or 21 days post-transfection. This indicates that nucleotides 376-396 are crucial for VP1 protein expression.
Fig. 4. Deletion of nucleotides 376-396 prevents VP1 expression.
Western blots were done with PAB597 (anti-VP1) (upper panels) using cell lysate collected from Cos-7 cells (A) 14 and (B) 21 days post-transfected. VP1 is detectable with most of the viruses tested, except Mad-1 PtΔ376-396 (Lane 6), Mad-1 C-Agno (Lane 10) and Mad-1 Del (Lane 12). Anti-tubulin is used as a loading control (lower panels). Blots are representative of results observed in 3 independent experiments.
DISCUSSION
Recent studies of the agnoprotein have identified a number of roles for the protein in JCV replication, but there is still a lot to be learned. Additionally, a cis-acting regulatory DNA element and host cell binding sites in the agnogene have been identified (Akan 2006), although the mechanism of action is unclear. We have previously studied JCVCPN, a naturally occurring isolate which contains a 143-base pair deletion in the agnogene, the first large agnogene deletion found in nature (Dang et al., 2012, Ellis et al., 2013). The agnogene deletion in JCVCPN was found to impair VP1 expression and virion production, and this phenotype was primarily due to the loss of the agnogene DNA, rather than lack of a full length agnoprotein.
In this study, we generated a series of agnogene deletion mutants. In order to rule out any effect due to mutation of the agnoprotein, these deletions were introduced into Mad-1 Pt, which has as start codon mutation preventing agnoprotein expression. We transfected these deletion mutants, along with a number of previously characterized control viruses, into Cos-7 cells and studied their replication by measuring levels of DNA replication, mRNA expression and VP1 protein expression.
We have previously found that, compared to JCVMad-1, both Mad-1 Del and Mad-1 C-Agno had decreased levels of DNA replication at later times post-transfection and lacked expression of VP1 protein. Most of the new deletion mutants had similar phenotypes to Mad-1 Pt and JCVMad-1. The most striking phenotype was observed with Mad-1 PtΔ376-396, which had decreased levels of DNA replication at later time points post-transfection, and was the only new deletion mutant which did not express VP1.
This suggests that DNA replication is not affected by this deletion, and the decrease in viral DNA at late time points is due to the lack of VP1 protein expression. All of the viruses have approximately the same levels of DNA at early time points, because equal amounts of DNA are transfected into the cells. Without the expression of VP1 protein, the DNA replicated in these cells cannot be packaged into viral particles at early time points, which prevents the production of new virions and spread of the infection within the culture. This, in turn, leads to a decrease in viral load as the cells are subcultured and new cells are not being infected. This is consistent with the results reported by Saribas et al (Saribas et al., 2013). Interestingly, some of the larger deletions that include these 20 nucleotides do express VP1. There are a number of possible mechanisms which may explain this phenotype. Nucleotides 376-396 fall within the predicted binding site III for host cell factors. If there are multiple host cell proteins binding to the agnogene, Mad-1 PtΔ376-396 may have lost only one of them. This could change the interactions and effects of other host cell proteins binding to the agnogene. Alternatively, this deletion may have created a new binding site. It is also possible that these nucleotides are involved in proper splicing of the late mRNA, or that this deletion has created a new RNA secondary structure which affects translation of the VP1 protein. In this study, we cannot distinguish between effects on transcription or translation, and either or both may be responsible for the phenotypes observed.
Recently, Saribas et al published a study where they included an agnoprotein mutant with amino acids 34-36 deleted, named Mad-1 Agno Δ(34-36) (Saribas et al., 2013). This region is within the deleted region of Mad-1 PtΔ376-396. In this study, they found that these residues are crucial for agnoprotein oligomerization and dimerization. They also see reduced viral replication 15 days post-transfection, and decreased levels of late transcript and altered relative levels of the M1 and M2 late transcript splice variants with Mad-1 Agno Δ(34-36). Our results indicate that some of these phenotypes may be due to the loss of the DNA in the agnogene rather than expressing an agnoprotein which cannot form dimers and oligomers.
Our study adds to the body of evidence that there is a cis-acting DNA element in the agnogene. Further studies examining the mechanisms of this phenotype are warranted. Identification of the host cell proteins which bind to the agnogene would be valuable in understanding the role of the agnogene in JCV replication, and in the future development of therapeutics for treatment of PML and other brain diseases caused by JCV infection
MATERIALS AND METHODS
Cell Lines
Cos-7 cells were purchased from the ATCC (Gluzman, 1981). Cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 500 units/mL Penicillin and 500 μg/mL Streptomycin.
Plasmids
Construction of JCVMad-1 plasmid was previously described by Dang et al. (Dang et al., 2012). Construction of Mad-1 Pt, Mad-1 Del and Mad-1 C-Agno was previously described by Ellis et al. (Ellis et al., 2013). Additional agnogene deletion mutants were introduced using the QuikChange Lightening Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer’s protocol. All deletions were made on the Mad-1 Pt backbone. The primer sequences used were: Mad-1 PtDel300-349 (del300-349F 5’-GGTTCTTCGCCAGCTGTCACGCTCAAAGGATTTTAATTTTTTT-3’ and del300-349R 5’- AAAAAAATTAAAATCCTTTGAGCGTGACAGCTGGCGAAGAACC-3’), Mad-1 PtDel350-411 (del350-411F 5’- CTGGAGTGGAACTAAAAAAAGAGGTAGACGGGAAAAAAAGAC-3’ and del350-411R 5’- GTCTTTTTTTCCCGTCTACCTCTTTTTTTAGTTCCACTCCAG-3’), Mad-1 PtDel442-458 (del442-458F 5’-AAAAAAAGACAGAGACACAGTATACAGTGCTTTGCCTGAACC-3’ and del442-458R 5’-GGTTCAGGCAAAGCACTGTATACTGTGTCTCTGTCTTTTTTT-3’), Mad-1 PtDel356-376 (del356-376F 5’- GTGGAACTAAAAAAAGAGCTCAAAAATTTTTGCTGGACTTTTGCACAG-3’ and del356-376R 5’- CTGTGCAAAAGTCCAGCAAAAATTTTTGAGCTCTTTTTTTAGTTCCAC-3’), Mad-1 PtDel376-396 (del376-396F 5’- TAAAAAAAGAGCTCAAAGGATTTTAATTTTTTTGTTAACAGGTGAAGACAGTGTAG-3’ and del376-396R 5’- CTACACTGTCTTCACCTGTTAACAAAAAAATTAAAATCCTTTGAGCTCTTTTTTTA-3’), Mad-1 PtDel300-432 (del300-432F 5'- TTCGCCAGCTGTCACGTAGACACAGTGGTTTG-3' and del300-432R 5'- CAAACCACTGTGTCTACGTGACAGCTGGCGAA-3'), Mad-1 PtDel310-442 (del310-442F 5'-GTCACGTAAGGCTTCTGGTTTGACTGAGCAGACA-3' and del310-442R 5'- TGTCTGCTCAGTCAAACCAGAAGCCTTACGTGAC-3') and Mad-1 PtDel411-442 (del411-442F 5'-GCACAGGTGAAGACAGGTTTGACTGAGCAGAC-3' and del411-442R 5'-GTCTGCTCAGTCAAACCTGTCTTCACCTGTGC-3'). Plasmids were transformed into TOP10 (Invitrogen) or XL1-Blue cells (Agilent) and plasmid DNA was maxi prepped (QIAGEN). All plasmids were fully sequenced to confirm the presence of the desired deletions.
Transfection
Full-length JCV genomes were digested out of the plasmid backbone using EcoRI (NEB). Digested DNA was run on a 0.8% agarose gel and the virus band (about 5kb) was purified using the QIAquick Gel Extraction Kit (QIAGEN). Cells were transfected with 2 μg of purified JCV DNA using FuGENE6 transfection reagent (Promega) in 6 well plates. 3 days post-transfection, cells were passaged 1:3 into T25 flasks, and subsequently every 3-4 days 1:4 in T25 flasks. Supernatant and cell pellets were collected at each passage and stored at −80°C until further analysis.
DNA Extraction and Quantitative PCR (qPCR)
DNA was extracted from cell pellets and supernatant samples using the QIAamp DNA Blood Mini Kit (QIAGEN) according to the manufacturer’s protocol. QPCR was performed as previously described (Ellis et al., 2013). An RNAase P primer/probe set (Applied Biosystems) was used to determine the number of input cells for each reaction and the copies JCV/cell as previously described (Ellis et al., 2013). All samples were run in triplicate.
RNA Extraction and qRT-PCR
RNA was extracted and reverse transcribed as previously described (Ellis et al., 2013). QPCR was performed on a 7300 Real-time PCR System using Gene Expression Master Mix (Applied Biosystems). Primer/probe sets spanning the Large T Ag splice site and located in the VP1 gene were used as previously described (Ellis et al., 2013). TATA-box binding protein (TBP) (Invitrogen) was used as the endogenous control to determine the relative quantity by the comparative CT method (also known as the ΔΔCt method) (Schmittgen and Livak, 2008). All samples were run in triplicate.
Western Blotting
Cells were lysed for 30 minutes in TNN lysis buffer (50 mM TrisHCl pH 7.5, 150 mM NaCl, 0.1% NP40) with 0.2 mM Na-Orthovanadate and 1% protease inhibitor cocktail on ice. Cell lysate was centrifuged at 8000 rpm for 4 min to remove cell debris. Laemmli Buffer (Bio-Rad) was added and cell lysates were boiled for 10 min. Samples were run on a 10% SDS-PAGE gel in Tris/Glycine/SDS running buffer (Bio-Rad) and transferred to a nitrocellulose membrane using the iBlot system (Invitrogen). Membranes were blocked with 5% milk in PBST and then incubated overnight at 4°C in either VP1 antibody pAB597 (1 mg/mL) diluted 1:1000 or loading control anti-alpha tubulin [DM1A] (abcam) in 2% milk in PBST.. Membranes were washed and incubated with HRP conjugated goat-anti mouse IgG secondary antibody (Bio-Rad) for 1 hour at room temperature, followed by detection with ECL Plus reagent (Thermo Scientific). Signal was detected on film.
Statistical analysis
P values were calculated using SAS9.3 software. Before analysis, data was tested for normality. The non-parametric Wilcoxon Rank Test was used for DNA levels and univariate analysis to determine student’s t p values was used for mRNA levels.
Acknowledgments
This work was supported in part by NIH R01 NS 074995 to IJK
Footnotes
Conflict of Interest
The authors declare that they have no conflict of interest.
REFERENCES
- Akan I, Sariyer IK, Biffi R, Palermo V, Woolridge S, White MK, Amini S, Khalili K, Safak M. Human polyomavirus JCV late leader peptide region contains important regulatory elements. Virology. 2006;349:66–78. doi: 10.1016/j.virol.2006.01.025. [DOI] [PubMed] [Google Scholar]
- Berger JR, Kaszovitz B, Post MJ, Dickinson G. Progressive multifocal leukoencephalopathy associated with human immunodeficiency virus infection. A review of the literature with a report of sixteen cases. Annals of internal medicine. 1987;107:78–87. doi: 10.7326/0003-4819-107-1-78. [DOI] [PubMed] [Google Scholar]
- Dang X, Wuthrich C, Gordon J, Sawa H, Koralnik IJ. JC virus encephalopathy is associated with a novel agnoprotein-deletion JCV variant. PloS one. 2012;7:e35793. doi: 10.1371/journal.pone.0035793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis LC, Norton E, Dang X, Koralnik IJ. Agnogene deletion in a novel pathogenic JC virus isolate impairs VP1 expression and virion production. PloS one. 2013;8:e80840. doi: 10.1371/journal.pone.0080840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisque RJ, Bream GL, Cannella MT. Human polyomavirus JC virus genome. Journal of virology. 1984;51:458–469. doi: 10.1128/jvi.51.2.458-469.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheuens S, Wuthrich C, Koralnik IJ. Progressive multifocal leukoencephalopathy: why gray and white matter. Annual review of pathology. 2013;8:189–215. doi: 10.1146/annurev-pathol-020712-164018. [DOI] [PubMed] [Google Scholar]
- Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981;23:175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- Jensen PN, Major EO. A classification scheme for human polyomavirus JCV variants based on the nucleotide sequence of the noncoding regulatory region. Journal of neurovirology. 2001;7:280–287. doi: 10.1080/13550280152537102. [DOI] [PubMed] [Google Scholar]
- Khalili K, White MK, Sawa H, Nagashima K, Safak M. The agnoprotein of polyomaviruses: a multifunctional auxiliary protein. Journal of cellular physiology. 2005;204:1–7. doi: 10.1002/jcp.20266. [DOI] [PubMed] [Google Scholar]
- Knowles WA, Pipkin P, Andrews N, Vyse A, Minor P, Brown DW, Miller E. Population-based study of antibody to the human polyomaviruses BKV and JCV and the simian polyomavirus SV40. Journal of medical virology. 2003;71:115–123. doi: 10.1002/jmv.10450. [DOI] [PubMed] [Google Scholar]
- Koralnik IJ, Wuthrich C, Dang X, Rottnek M, Gurtman A, Simpson D, Morgello S. JC virus granule cell neuronopathy: A novel clinical syndrome distinct from progressive multifocal leukoencephalopathy. Annals of neurology. 2005;57:576–580. doi: 10.1002/ana.20431. [DOI] [PubMed] [Google Scholar]
- Molloy ES, Calabrese LH. Progressive multifocal leukoencephalopathy: a national estimate of frequency in systemic lupus erythematosus and other rheumatic diseases. Arthritis and rheumatism. 2009;60:3761–3765. doi: 10.1002/art.24966. [DOI] [PubMed] [Google Scholar]
- Okada Y, Endo S, Takahashi H, Sawa H, Umemura T, Nagashima K. Distribution and function of JCV agnoprotein. Journal of neurovirology. 2001;7:302–306. doi: 10.1080/13550280152537148. [DOI] [PubMed] [Google Scholar]
- Safak M, Barrucco R, Darbinyan A, Okada Y, Nagashima K, Khalili K. Interaction of JC virus agno protein with T antigen modulates transcription and replication of the viral genome in glial cells. Journal of virology. 2001;75:1476–1486. doi: 10.1128/JVI.75.3.1476-1486.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safak M, Sadowska B, Barrucco R, Khalili K. Functional interaction between JC virus late regulatory agnoprotein and cellular Y-box binding transcription factor, YB-1. Journal of virology. 2002;76:3828–3838. doi: 10.1128/JVI.76.8.3828-3838.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saribas A, Abou-Gharbia M, Childers W, Sariyer IK, White MK, Safak M. Essential roles of Leu/Ile/Phe-rich domain of JC virus agnoprotein in dimer/oligomer formation, protein stability and splicing of viral transcripts. Virology. 2013;443:161–176. doi: 10.1016/j.virol.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saribas AS, White MK, Safak M. JC virus agnoprotein enhances large T antigen binding to the origin of viral DNA replication: evidence for its involvement in viral DNA replication. Virology. 2012;433:12–26. doi: 10.1016/j.virol.2012.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sariyer IK, Akan I, Palermo V, Gordon J, Khalili K, Safak M. Phosphorylation mutants of JC virus agnoprotein are unable to sustain the viral infection cycle. Journal of virology. 2006;80:3893–3903. doi: 10.1128/JVI.80.8.3893-3903.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sariyer IK, Khalili K, Safak M. Dephosphorylation of JC virus agnoprotein by protein phosphatase 2A: inhibition by small t antigen. Virology. 2008;375:464–479. doi: 10.1016/j.virol.2008.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sariyer IK, Saribas AS, White MK, Safak M. Infection by agnoprotein-negative mutants of polyomavirus JC and SV40 results in the release of virions that are mostly deficient in DNA content. Virology journal. 2011;8:255. doi: 10.1186/1743-422X-8-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nature protocols. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Shimizu N, Imamura A, Daimaru O, Mihara H, Kato Y, Kato R, Oguri T, Fukada M, Yokochi T, Yoshikawa K, et al. Distribution of JC virus DNA in peripheral blood lymphocytes of hematological disease cases. Intern Med. 1999;38:932–937. doi: 10.2169/internalmedicine.38.932. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Orba Y, Okada Y, Sunden Y, Kimura T, Tanaka S, Nagashima K, Hall WW, Sawa H. The human polyoma JC virus agnoprotein acts as a viroporin. PLoS pathogens. 2010;6:e1000801. doi: 10.1371/journal.ppat.1000801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Semba S, Sunden Y, Orba Y, Kobayashi S, Nagashima K, Kimura T, Hasegawa H, Sawa H. Role of JC virus agnoprotein in virion formation. Microbiology and immunology. 2012;56:639–646. doi: 10.1111/j.1348-0421.2012.00484.x. [DOI] [PubMed] [Google Scholar]
- Tan C, Koralnik I. Beyond progressive multifocal leukoencephalopathy: expanded pathogenesis of JC virus infection in the central nervous system. Lancet Neurology. 2010;9:425–437. doi: 10.1016/S1474-4422(10)70040-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber T, Trebst C, Frye S, Cinque P, Vago L, Sindic CJ, Schulz-Schaeffer WJ, Kretzschmar HA, Enzensberger W, Hunsmann G, Luke W. Analysis of the systemic and intrathecal humoral immune response in progressive multifocal leukoencephalopathy. The Journal of infectious diseases. 1997;176:250–254. doi: 10.1086/514032. [DOI] [PubMed] [Google Scholar]
- Wuthrich C, Dang X, Westmoreland S, McKay J, Maheshwari A, Anderson MP, Ropper AH, Viscidi RP, Koralnik IJ. Fulminant JC virus encephalopathy with productive infection of cortical pyramidal neurons. Annals of neurology. 2009;65:742–748. doi: 10.1002/ana.21619. [DOI] [PMC free article] [PubMed] [Google Scholar]