Abstract
Induction of antiviral immunity in vertebrates and invertebrates relies on members of the RIG-I-like receptor and Dicer families, respectively. Although these proteins have different size and domain composition, members of both families share a conserved DECH-box helicase domain. This helicase, also known as a duplex RNA activated ATPase, or DRA domain, plays an important role in viral RNA sensing. Crystallographic and electron microscopy studies of the RIG-I and Dicer DRA domains indicate a common structure and that similar conformational changes are induced by dsRNA binding. Genetic and biochemical studies on the function and regulation of DRAs reveal similarities, but also some differences, between viral RNA sensing mechanisms in nematodes, flies and mammals.
Introduction
Viral infections represent a major threat for all living organisms. Viruses consist in their most basic form of a nucleic acid encapsulated in a protein shell and their replication depends on the molecular machineries of their host cells. Both viral and host components are present in infected cells, which makes the distinction between self and non-self very challenging to the innate immune system. In addition, the error-prone viral nucleic acid polymerases enable viruses to adapt rapidly and suppress their host’s defence mechanisms. It is valuable to compare antiviral immune responses in a wide range of organisms, to understand their strategies to counter viral infections.
Although studies on antibacterial and antifungal defences revealed that important innate immunity pathways (e.g. Toll/interleukin-1 and TNF receptor pathways) have been conserved through evolution, things are more complex for antiviral immunity. In invertebrates (and in plants), RNA interference represents a major pathway of antiviral host-defence. In vertebrates, however, the response to viral infections is dominated by the interferon (IFN) system, and the induction of IFN stimulated genes (ISGs) [1]. In spite of major differences in the effectors deployed, the antiviral responses of multicellular eukaryotes are triggered by the sensing of foreign nucleic acids in the cytosol.
In invertebrates, double-stranded viral RNA generated during replication is processed into 21-23bp small interfering (si) RNA duplexes by Dicer family RNase III nucleases. These si-RNA duplexes are then loaded onto Argonaute (AGO) family nucleases within the RNA-induced silencing complex (RISC), where one of the strands will guide the RISC complex to target homologous viral RNA sequences [2]. In mice, Dicer can process viral RNA into siRNAs in some cell types [3,4]. In addition, some endogenous micro (mi)RNAs produced by Dicer can counter viral infection (e.g. [5]). However, in most tissues, viral RNA is sensed by receptors of the RIG-I-like receptor (RLR) family [6]. Upon RNA-binding, the RLRs activate a signalling cascade leading to transcription of type I and type III IFN genes (Figure 1).
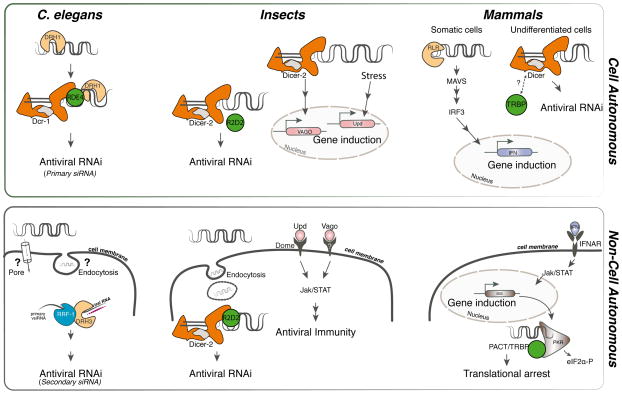
Figure 1. Antiviral innate immune pathways across species.
Schematic representation of antiviral pathways in the nematode C. elegans, insects and mammals. Signalling DRAs are shown in light orange, catalytic DRAs in dark orange (RNaseIII domains highlighted in grey) and dsRBD cofactors in green. Cell autonomous primary responses are illustrated in the top panel, and secondary responses amplifying antiviral immunity are shown in the bottom panel. In D. melanogaster, the cytokines Upd2 and Upd3 activate the Jak/STAT pathway through the receptor Domeless [49], whereas in cells from the mosquito Culex fasciculatus, Vago triggers activation of the pathway through an unknown receptor [48]. Although also associated with antiviral activity in Drosophila, the exact function of Vago in this organism is still unclear. In undifferentiated mouse cells, virus-derived siRNAs contribute to antiviral immunity [32–33].
Both Dicer nucleases and RLR receptors share an evolutionarily conserved DECH box “helicase” domain, which plays an important role in RNA sensing [7,8]. Here we review the structure and function of the DECH box proteins involved in the antiviral immune response, in vertebrates, Caenorhabditis elegans and Drosophila melanogaster.
Dicers and RLRs define a family of duplex RNA activated ATPases
Consistent with their differing functions as small RNA processing enzymes and signalling receptors, the Dicer and RLR family proteins have distinct domain organizations. Whereas Dicer enzymes contain PAZ, RNase III and dsRNA binding (dsRBD) domains, the RLRs (with the exception of LGP2) contain two amino-terminal CARD domains (Figure 2). Yet, Dicer and RLR family members share the critical DECH-box helicase, which consists of three tandem motifs, HEL1, HEL2i and HEL2 [9–12]. The HEL1 and HEL2 components of this tripartite core domain represent modified RecA domains, classically found in superfamily2 (SF2) RNA helicases, each containing a specialized motif discriminating the top (5′ to 3′) strand of the RNA duplex [13]. Furthermore, HEL1 presents an helical bridging, or pincer, domain, which is not present in SF2 helicases. This bridging domain provides an interface for interaction with the two α-helices connecting HEL2 to the C-terminal domain (CTD) [14]. The HEL1 and HEL2 domains are separated by the HEL2i domain, which sequesters the CARD2 domain in the absence of ligand (Figure 3a). Binding of dsRNA, which is facilitated by HEL2i, triggers a major conformational change of the DECH-box domain, leading to expulsion of CARD1 and CARD2 and activating signalling [7,8].
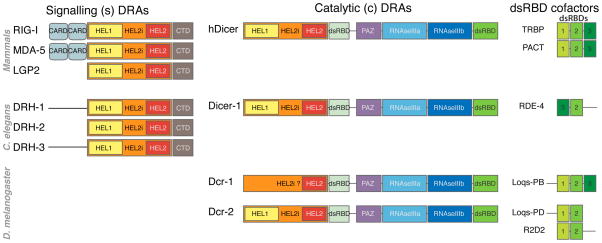
Figure 2. Conservation of s- and c-DRAs and their dsRBD cofactors.
The domain organization of DRAs from Homo sapiens, Caenorhabditis elegans and Drosophila melanogaster is shown. In RIG-I and MDA5, the CARD domains function as homotypic protein-protein interaction domains to recruit and activate the signal transducer MAVS. In Dicer enzymes, the two RNase III domains form the catalytic core of the enzyme, the PAZ domain contains a pocket anchoring the 3′OH extremity of the substrate RNA, and the dsRNA Binding Domain enhances the affinity of the enzyme for its substrate. The dsRBD cofactors contain two to three evolutionarily conserved dsRNA Binding Domains (shown with different shades of green).
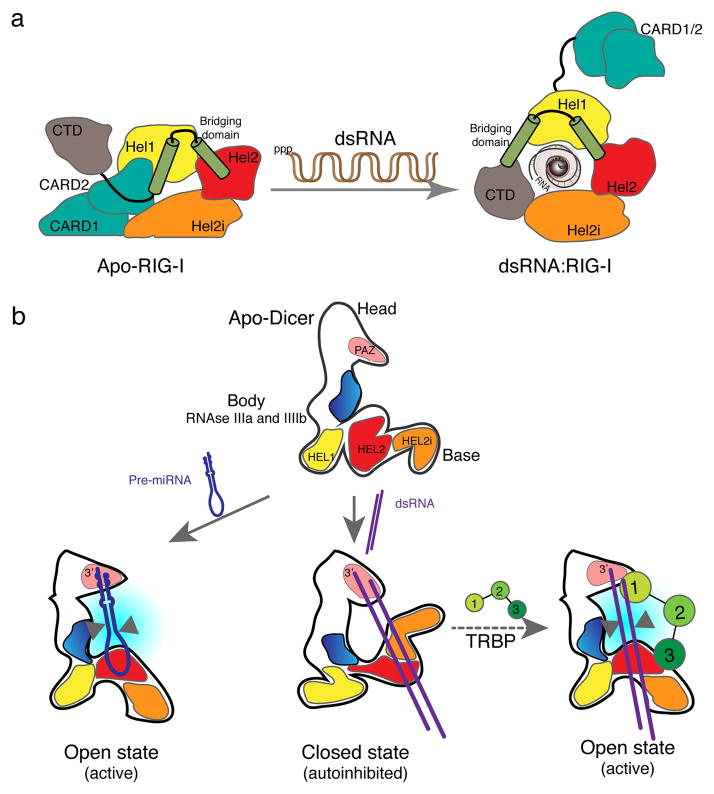
Figure 3. RNA induced conformational changes in sDRAs and cDRAs.
(a) In non-infected condition, the signalling CARD2 domain of RIG-I is sequestered by the motif HEL2i, which is not present in other SF2 helicases. Binding of dsRNA containing 5′ triphosphate extremities triggers a major conformational change, with HEL1, HEL2 and HEL2i wrapping around the dsRNA stem. The CARD domains are expelled, making them available for signalling. (b) The DRA domain of hDicer rearranges differently in the presence of pre-miRNAs or dsRNA. Binding of the pre-miR let7 triggers a bending of the base branch away from the platform, opening the conformation of the enzyme, and positioning the RNA next to the RNase III domains (blue) for cleavage. In the case of dsRNA, the binding of the free end of the duplex to the PAZ domain (pink) and of the stem to the DRA domain triggers an inward bending of the base branch. This closed conformation maintains the dsRNA at a distance from the RNase III domains, explaining the poor efficiency of the cleavage as well as the autoinhibitory function of the DRA domain. TRBP interacts with the DRA domain to trigger a different conformation, allowing processing of dsRNA. Arrowheads point to RNAse III processing. Redrawn with modifications from references [8] and [31].
Metazoan Dicer enzymes are complex multi-domain proteins, typically larger than 200kDa, and difficult to crystallize. However, the characteristic HEL1, HEL2i and HEL2 motifs are conserved in most Dicers [7]. Furthermore, electron microscopy (EM)-based 3D reconstructions of human Dicer and Drosophila Dicer-2 reveal “L”-shaped particles composed of three distinct regions [15] (Figure 3b). The PAZ domain, which binds the extremity of the dsRNA helix, is located at the head of the structure. The RNase III domains are in the long arm body of the L. Finally, the tripartite “helicase” domain extends along the base of the L (Figure 3b). The crystal structure of the RIG-I DECH-box helicase can be mapped to fit into the homologous region of Dicer [15]. The RIG-I helicase domain binds dsRNA which then appears to be clamped by the ligand-induced conformational change [15]. Similar conformational changes following dsRNA binding may occur in both protein families (Figure 3), although this remains to be determined directly for Dicer.
Importantly, neither Dicer nor RIG-I has been shown to function as a bona fide helicase. Thus, the generic acronym DRA has been proposed to include both these families of proteins that sense and respond to viral RNA [13]: DRA corresponds to Duplex RNA activated ATPases, (or alternatively, Dicer/RIG-I like ATPases). In metazoa, two groups of DRAs participate in antiviral immunity: the signalling sDRAs and the catalytic (RNase III) cDRAs. While flies and other insects, lack sDRAs, they have two cDRAs, one of which (Dicer-2) is dedicated to antiviral immunity. C. elegans and mammals, on the other hand, have a single cDRA and multiple sDRAs (Figure 2). Interestingly, sDRAs participate in different antiviral pathways in C. elegans and mammals.
An ancient role of sDRAs in sensing viral RNA
In mammals, differences in the CTD domain account for the different binding specificities of RIG-I and MDA5. The RIG-I CTD domain accommodates the terminal 5′ tri- or di- phosphates of dsRNA [6,16]. By contrast, the MDA5 CTD binds to the internal segments of long dsRNAs, rather than at their extremities [17] (Table I). This is consistent with critical role of MDA5 in sensing of picornaviruses, which produce long dsRNAs that lack terminal 5′-di or tri- phosphates. [18]. In addition, the LGP2 sDRA lacks terminal CARD domains (Figure 2) and functions as a coreceptor together with MDA5 [19,20].
Table I.
General RNA structure of DRA ligands and products
| Species | cDRA | sDRA | RNA structure |
|---|---|---|---|
| C. elegans | Dicer-1 | 5′p-23 nt siRNA duplexes with 2 nt 3′ overhang (product) | |
|
| |||
| DRH1 | viral dsRNA, uncharacterized features (ligand) | ||
| DRH3 | 5′ppp-22nt secondary siRNAs with 5′ G (product) | ||
|
| |||
| D. melanogaster | Dcr-2 | viral dsRNA, uncharacterized features (ligand) | |
|
| |||
| Dcr-2 | 5′p-21 nt siRNA duplexes with 2 nt 3′ overhang (product) | ||
|
| |||
| M. musculus | Dicer | 5′p-21 to 23 nt siRNA duplexes with 2 nt 3′ overhang (product) | |
|
| |||
| RIG-I | 5′ppp-or 5′pp- short viral dsRNA (ligand) | ||
|
| |||
| MDA5 | long viral dsRNA (ligand) | ||
In C. elegans, Dicer-1 functions in competing RNAi pathways, while specificity is determined by associated RNA binding proteins. The alternative co-factors recruit Dicer-1 to form complexes specific for viral replication intermediates, aberrant endogenous RNA (endoRNAi), or exogenous dsRNA (exoRNAi). The identification of a natural viral pathogen infecting C. elegans has opened the way for the genetic characterization of antiviral resistance [21]. One major locus associated with sensitivity to virus infection is the sDRA Dicer-related helicase-1 (DRH-1), which senses viral RNAs and recruits Dicer-1 to the antiviral RNAi pathway [22,23]. Interestingly, DRH-1 is not required for processing exogenous dsRNAs into siRNAs, indicating that it discriminates something more than the double-strandedness of viral RNAs. The requirement of the CTD domain for the antiviral activity of DRH-1 suggests, by analogy with RIG-I, that DRH-1 may recognize 5′-di or triphosphates to improve viral RNA recognition and processing by Dicer-1.
The antiviral function of DRH-1 requires not only the DRA homology domain and the CTD, but also the N-terminal domain (NTD), which is not conserved in RIG-I. This NTD domain may function to recruit effector molecules, as do the CARD domains of RLRs [22,23]. A second member of the DRH family, DRH-3, interacts with Dicer-1 in silencing aberrant endogenous RNAs during spermatogenesis and embryogenesis (the ERI pathway) [24]. DRH-3 also potentiates the antiviral response, but in a Dicer-1 independent manner (see below) [23].
In summary, in both worm and vertebrate sDRAs, homologous DRA and CTD domains interact with viral RNAs while species-specific NTD domains activate different responses. In C. elegans a sDRA mediates recruitment of Dicer-1 to viral RNA substrates, raising the question of the substrate specificity of the human and fly Dicer enzymes.
Sensing RNAs by cDRAs
In Drosophila, two Dicer orthologues produce miRNAs (Dcr-1) and siRNAs (Dcr-2) [25]. Dcr-1 lacks conserved residues in the modified HEL1 domain (Figure 2), which may be critical to process dsRNAs. Although purified Dcr-2 can cleave pre-miRNAs in vitro, this activity is blocked by the R2D2 protein and inorganic phosphate [26]. R2D2 contains a dsRNA binding domain (dsRBD) and associates with Dcr-2 (see below), while inorganic phosphate occupies a pocket in the PAZ domain of Dcr-2, precluding binding of the 5′ phosphate extremity of pre-miRNAs [27,28].
The DRA domain of Dcr-2 appears to play at least two roles. Firstly, it initiates dicing of blunt-ended dsRNAs, perhaps by unwinding the first dsRNA bases and allowing positioning of a 3′-OH extremity in the PAZ domain [29]. Secondly, the DRA domain is required for translocation of Dcr-2 along the dsRNA molecule. Consequently, the DRA domain allows Dcr-2 to generate many siRNAs from a single long dsRNA substrate [26,29]. Thus, the DRA domain converts a distributive double–strand nuclease into a processive enzyme.
The single mammalian Dicer orthologue has the same domain organization as Drosophila Dcr-2 (Figure 2), but processes pre-miRNAs rather than dsRNAs. Human Dicer binds to both substrates with the same affinity [30], but pre-miRNAs trigger specific repositioning of the DRA domain, which confers substrate specificity [31] (Figure 3b). Accordingly, DRA domain deleted hDicer can process dsRNA in vitro [32]. DRA truncated isoforms of Dicer, with enhanced dsRNA activity, are found in rodent oocytes and nematodes [33,34]. These truncated isoforms may participate in the control of viral infections (e.g. [3,4]). Importantly, full-length hDicer can also process dsRNA, when associated with TRBP.
dsRBD cofactors of cDRAs
In mammals and invertebrates, cDRAs function in protein complexes with dsRBD cofactors (Figure 2). The human TRBP and PACT cofactors, are both composed of three contiguous dsRBDs, which associate with the DRA domain of hDicer. TRBP binding triggers repositioning of the DRA domain, allowing efficient processing of dsRNA [31] (Figure 3b). C. elegans has a unique TRBP/PACT orthologue, RDE4 [35]. The nuclease activity of Dicer-1 requires RDE4 for viral RNA and exogenous dsRNA substrates, but not for endogenous dsRNAs or pre-miRNAs [23,36].
The situation is more complex in flies, possibly reflecting the absence of sDRAs. The fly orthologue of the genes TRBP and RDE4 is loquacious (loqs) [37]. Two isoforms, Loqs-PB and Loqs-PD, have been characterized. Loqs-PB binds specifically to Dcr-1 through its dsRBD3 domain, and is important for the production of miRNAs. The Loqs-PD isoform, lacks the dsRBD3 domain and acts as a cofactor of Dcr-2 in the production of exogenous and endogenous siRNAs [38,39]. Surprisingly, however, Loqs is not required for the resistance to viral infections, or the production of virus-derived siRNAs by Dicer-2 [40]. This implies that Dcr-2 may have an intrinsic capacity to sense and engage viral RNAs. In insects, a third dsRBD-containing co-factor, R2D2, participates in antiviral RNAi. However, R2D2 acts downstream of Dcr-2 processing, playing a critical role in the loading of siRNAs onto AGO2 [39].
From cell intrinsic to systemic antiviral immunity
Given the rapid propagation of most viruses, an essential aspect of antiviral immunity is the capacity to trigger paracrine signalling, leading to antiviral immunity in non-infected cells. This task is achieved by IFNs in vertebrates, which induce the production of antiviral molecules such as PKR. Upon infection, this dsRNA-dependent protein kinase phosphorylates eIF2α to shut down translation, thus ensuring an immediate response of the cell. Interestingly, TRBP and PACT interact with PKR and regulate its activity [41,42] (Figure 1).
In invertebrates, the systemic antiviral response utilises different mechanisms [43]. In C. elegans, the RNAi response is divided into primary and secondary steps and two types of siRNAs are produced. Primary siRNAs are produced by Dicer-1 and have a size of 23nt and a monophosphate at their 5′ extremities. These siRNAs are transferred to non-infected cells by poorly characterized mechanisms, where they trigger the synthesis of 5′ triphosphorylated 22nt-long secondary siRNAs by an endogenous RNA-dependent RNA polymerase (RdRP). Secondary siRNAs, which have a strong bias for a G at the 5′ position, appear to be loaded directly onto members of an expanded clade of worm specific AGOs, without Dicer-1 processing [44] (Table I). Importantly, the sDRA, DRH-3, is essential for the biogenesis of RdRP-derived small RNAs in C. elegans, and plays a critical role in antiviral immunity [22,23]. This indicates first that secondary siRNAs contribute to host defence by amplifying the antiviral response and, second, that a sDRA can function in antiviral immunity in a Dicer-independent manner in nematodes.
In insects, two mechanisms appear to contribute to the amplification of the antiviral response. The first is the dsRNA uptake pathway [45], which allows uninfected cells to sample their environment for the presence of dsRNA and trigger a preventive RNAi response [46]. The second mechanism involves the inducible expression of cytokines, such as Unpaired or Vago, following cellular stress or the detection of dsRNA. Such cytokines contribute to host-defence by ill-defined mechanisms [47–49]. The Drosophila genome does not encode RdRPs that could mediate signal amplification. However, the recent discovery that viral RNAs can be reverse transcribed into DNA in persistent viral infections raises the possibility that spreading of antiviral silencing might involve DNA- rather than RNA-dependent polymerases [50].
Conclusion and perspectives
Important knowledge has been gained in recent years on the function and regulation of DRAs in antiviral immunity across species. Furthermore, structural studies on DRAs suggest common mechanisms for binding viral dsRNA and in substrate-induced conformational changes in activity. These data pave the way for more detailed structure-function studies on the discrimination between viral RNAs of different origins by DRAs. In this regard, the available collection of Dcr-2 alleles with point mutations in the DRA domain represents a powerful resource for studies in Drosophila [25].
The conserved set of s-cDRAs and dsRBD cofactors forms a core viral dsRNA sensing and response mechanism in Metazoans, although these proteins intervene at different levels in the antiviral responses across species. Such modifications probably reflect the constant arms race between viruses and their hosts, requiring rapid and constant evolution of antiviral innate immunity pathways (e.g. [51,52]). Thus, the nature of the ancestral response to viral infection remains uncertain. The importance of RNAi in antiviral immunity in worms and insects points to the ancient origin of this antiviral pathway. However, sDRAs with CARD domains are present in the genomes of oysters and sponges, suggesting that bona fide RLRs are present in invertebrates, but were lost in nematodes and insects [53]. Functional characterization of antiviral innate immunity in other groups of invertebrates will shed light on the ancestral function of DRAs and the complex interrelationship between signalling and catalytic members of the family.
Highlights.
Dicers and RLRs are important for antiviral immunity in Metazoans.
Dicers and RLRs share a conserved Duplex RNA activated ATPase (DRA) domain.
DRAs from Dicers and RLRs undergo similar conformational changes upon dsRNA binding.
Signaling DRAs trigger different pathways upon sensing viral RNA in worms and mammals.
dsRNA binding cofactors regulate activity of catalytic DRAs.
Acknowledgments
We thank Pr. Joao T. Marques and Dr. David Gubb for critical reading of the manuscript, and NIH (PO1 AI070167), ANR (ANR-10-LABX-36; ANR-11-ASV3-002), USIAS and CNRS for financial support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
- 1.Beutler B, Eidenschenk C, Crozat K, Imler JL, Takeuchi O, Hoffmann JA, Akira S. Genetic analysis of resistance to viral infection. Nature reviews in Immunology. 2007;7:753–766. doi: 10.1038/nri2174. [DOI] [PubMed] [Google Scholar]
- 2.Ding SW. RNA-based antiviral immunity. Nature reviews. 2010;10:632–644. doi: 10.1038/nri2824. [DOI] [PubMed] [Google Scholar]
- 3.Maillard PV, Ciaudo C, Marchais A, Li Y, Jay F. Antiviral RNA interference in mammalian cells. Science. 2013;342:235–238. doi: 10.1126/science.1241930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Y, Lu J, Han Y, Fan X, Ding SW. RNA interference functions as an antiviral immunity mechanism in mammals. Science. 2013;342:231–234. doi: 10.1126/science.1241911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Otsuka M, Jing Q, Georgel P, New L, Chen J, Mols J, Kang YJ, Jiang Z, Du X, Cook R, et al. Hypersusceptibility to Vesicular Stomatitis Virus Infection in Dicer1-Deficient Mice Is Due to Impaired miR24 and miR93 Expression. Immunity. 2007;27:123–134. doi: 10.1016/j.immuni.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 6.Goubau D, Deddouche S, Sousa CRE. Cytosolic sensing of viruses. Immunity. 2013;38:855–869. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rawling DC, Pyle AM. Parts, assembly and operation of the RIG-I family of motors. Curr Opin Struct Biol. 2014;25:25–33. doi: 10.1016/j.sbi.2013.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •8.Kolakofsky D, Kowalinski E, Cusack S. A structure-based model of RIG-I activation. RNA. 2012;18:2118–2127. doi: 10.1261/rna.035949.112. A thorough review on the molecular events leading to RIG-I activation by dsRNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo D, Ding SC, Vela A, Kohlway A, Lindenbach BD, Pyle AM. Structural Insights into RNA Recognition by RIG-I. Cell. 2011;147:409–422. doi: 10.1016/j.cell.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kowalinski E, Lunardi T, McCarthy AA, Louber J, Brunel J, Grigorov B, Gerlier D, Cusack S. Structural Basis for the Activation of Innate Immune Pattern-Recognition Receptor RIG-I by Viral RNA. Cell. 2011;147:423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- 11.Jiang F, Ramanathan A, Miller MT, Tang G-Q, Gale MJ, Patel SS, Marcotrigiano J. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature. 2011;479:423–U184. doi: 10.1038/nature10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Civril F, Bennett M, Moldt M, Deimling T, Witte G, Schiesser S, Carell T, Hopfner K-P. The RIG-I ATPase domain structure reveals insights into ATP-dependent antiviral signalling. EMBO Rep. 2011;12:1127–1134. doi: 10.1038/embor.2011.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••13.Luo D, Kohlway A, Pyle AM. Duplex RNA Activated ATPases (DRAs) Platforms for RNA sensing, signaling and processing. RNA Biol. 2013;10:111–120. doi: 10.4161/rna.22706. An insightful comprehensive analysis on Dicer and RIG-I-like ATPases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rawling DC, Kohlway AS, Luo D, Ding SC, Pyle AM. The RIG-I ATPase core has evolved a functional requirement for allosteric stabilization by the Pincer domain. Nucleic acids research. 2015;42:11601–11611. doi: 10.1093/nar/gku817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••15.Lau P-W, Guiley KZ, De N, Potter CS, Carragher B, MacRae IJ. The molecular architecture of human Dicer. Nature structural & molecular biology. 2012;19:436–440. doi: 10.1038/nsmb.2268. An elegant microscopy analysis defining the molecular architecture of Dicer enzymes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goubau D, Schlee M, Deddouche S, Pruijssers AJ, Zillinger T, Goldeck M, Schuberth C, Van der Veen AG, Fujimura T, Rehwinkel J, et al. Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5′-diphosphates. Nature. 2014;514:372–375. doi: 10.1038/nature13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu B, Peisley A, Richards C, Yao H, Zeng X, Lin C, Chu F, Walz T, Hur S. Structural Basis for dsRNA Recognition, Filament Formation, and Antiviral Signal Activation by MDA5. Cell. 2013;152:276–289. doi: 10.1016/j.cell.2012.11.048. [DOI] [PubMed] [Google Scholar]
- 18.Feng Q, Hato SV, Langereis MA, Zoll J, Virgen-Slane R, Peisley A, Hur S, Semler BL, van Rij RP, van Kuppeveld FJM. MDA5 Detects the Double-Stranded RNA Replicative Form in Picornavirus-Infected Cells. Cell Rep. 2012;2:1187–1196. doi: 10.1016/j.celrep.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bruns AM, Leser GP, Lamb RA, Horvath CM. The innate immune sensor LGP2 activates antiviral signaling by regulating MDA5-RNA interaction and filament assembly. Molecular cell. 2014;55:771–781. doi: 10.1016/j.molcel.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deddouche S, Goubau D, Rehwinkel J, Chakravarty P, Begum S, Maillard PV, Borg A, Matthews N, Feng Q, van Kuppeveld FJM, et al. Identification of an LGP2-associated MDA5 agonist in picornavirus-infected cells. Elife. 2014;3:e01535. doi: 10.7554/eLife.01535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Felix M-A, Ashe A, Piffaretti J, Wu G, Nuez I, Belicard T, Jiang Y, Zhao G, Franz CJ, Goldstein LD, et al. Natural and Experimental Infection of Caenorhabditis Nematodes by Novel Viruses Related to Nodaviruses. PLoS biology. 2011;9:e10000586. doi: 10.1371/journal.pbio.1000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••22.Guo X, Zhang R, Wang J, Ding S-W, Lu R. Homologous RIG-I-like helicase proteins direct RNAi-mediated antiviral immunity in C. elegans by distinct mechanisms. Proc Natl Acad Sci US A. 2013;110:16085–16090. doi: 10.1073/pnas.1307453110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••23.Ashe A, Belicard T, Le Pen J, Sarkies P, Frezal L, Lehrbach NJ, Felix M-A, Miska EA. A deletion polymorphism in the Caenorhabditis elegans RIG-I homolog disables viral RNA dicing and antiviral immunity. Elife. 2013;2:e00994. doi: 10.7554/eLife.00994. Present elegant genetic and molecular analysis establishing the importance of sDRAs in antiviral immunity in C. elegans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thivierge C, Makil N, Flamand M, Vasale JJ, Mello CC, Wohlschlegel J, Conte DJ, Duchaine TF. Tudor domain ERI-5 tethers an RNA-dependent RNA polymerase to DCR-1 to potentiate endo-RNAi. Nature structural & molecular biology. 2012;19:90–U114. doi: 10.1038/nsmb.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 26.Cenik ES, Fukunaga R, Lu G, Dutcher R, Wang Y, Hall TM, Zamore PD. Phosphate and R2D2 Restrict the Substrate Specificity of Dicer-2, an ATP-Driven Ribonuclease. Molecular cell. 2011;42:172–184. doi: 10.1016/j.molcel.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fukunaga R, Colpan C, Han BW, Zamore PD. Inorganic phosphate blocks binding of pre-miRNA to Dicer-2 via its PAZ domain. The EMBO journal. 2014;33:371–384. doi: 10.1002/embj.201387176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tian Y, Simanshu DK, Ma J-B, Park J-E, Heo I, Kim VN, Patel DJ. A Phosphate-Binding Pocket within the Platform-PAZ-Connector Helix Cassette of Human Dicer. Molecular cell. 2014;53:606–616. doi: 10.1016/j.molcel.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Welker NC, Maity TS, Ye X, Aruscavage PJ, Krauchuk AA, Liu Q, Bass BL. Dicer’s helicase domain discriminates dsRNA termini to promote an altered reaction mode. Molecular cell. 2011;41:589–599. doi: 10.1016/j.molcel.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chakravarthy S, Sternberg SH, Kellenberger CA, Doudna JA. Substrate-Specific Kinetics of Dicer-Catalyzed RNA Processing. Journal of molecular biology. 2010;404:392–402. doi: 10.1016/j.jmb.2010.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •31.Taylor DW, Ma E, Shigematsu H, Cianfrocco MA, Noland CL, Nagayama K, Nogales E, Doudna JA, Wang H-W. Substrate-specific structural rearrangements of human Dicer. Nature structural & molecular biology. 2013;20:662–670. doi: 10.1038/nsmb.2564. A comprehensive in vitro analysis of the conformations of hDicer in the absence or presence of RNA substrates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma E, MacRae IJ, Kirsch JF, Doudna JA. Autoinhibition of human dicer by its internal helicase domain. Journal of molecular biology. 2008;380:237–243. doi: 10.1016/j.jmb.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •33.Flemr M, Malík R, Franke V, Nejepinska J, Sedlacek R, Vlahovicek K, Svoboda P. A Retrotransposon-Driven Dicer Isoform Directs Endogenous Small Interfering RNA Production in Mouse Oocytes. Cell. 2013;155:807–816. doi: 10.1016/j.cell.2013.10.001. In vivo demonstration that a Dicer isoform with a truncated DRA domain can process dsRNA. [DOI] [PubMed] [Google Scholar]
- 34.Sawh AN, Duchaine TF. A Truncated Form of Dicer Tilts the Balance of RNA Interference Pathways. Cell Rep. 2013;4:454–463. doi: 10.1016/j.celrep.2013.07.013. [DOI] [PubMed] [Google Scholar]
- 35.Tabara H, Yigit E, Siomi H, Mello CC. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell. 2002;109:861–871. doi: 10.1016/s0092-8674(02)00793-6. [DOI] [PubMed] [Google Scholar]
- 36.Lu R, Yigit E, Li WX, Ding SW. An RIG-I-Like RNA helicase mediates antiviral RNAi downstream of viral siRNA biogenesis in Caenorhabditis elegans. PLoS pathogens. 2009;5:e1000286. doi: 10.1371/journal.ppat.1000286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Förstemann K, Tomari Y, Du TT, Vagin VV, Denli AM, Bratu DP, Klattenhoff C, Theurkauf WE, Zamore PD. Normal microRNA maturation and germ-line stem cell maintenance requires loquacious, a double-stranded RNA-binding domain protein. PLoS biology. 2005;3:1187–1201. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hartig JV, Esslinger S, Boettcher R, Saito K, Foerstemann K. Endo-siRNAs depend on a new isoform of loquacious and target artificially introduced, high-copy sequences. The EMBO journal. 2009;28:2932–2944. doi: 10.1038/emboj.2009.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marques JT, Kim K, Wu P-H, Alleyne TM, Jafari N, Carthew RW. Loqs and R2D2 act sequentially in the siRNA pathway in Drosophila. Nature structural & molecular biology. 2010;17:24–30. doi: 10.1038/nsmb.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •40.Marques JT, Wang J-P, Wang X, de Oliveira KPV, Gao C, Aguiar ERGR, Jafari N, Carthew RW. Functional specialization of the small interfering RNA pathway in response to virus infection. PLoS pathogens. 2013;9:e1003579–e1003579. doi: 10.1371/journal.ppat.1003579. In vivo demonstration that the dsRBD cofactor Loqs-PD, important for exoRNAi, is not required for antiviral immunity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sadler AJ, Williams BRG. Structure and function of the protein kinase R. Current topics in microbiology and immunology. 2007;316:253–292. doi: 10.1007/978-3-540-71329-6_13. [DOI] [PubMed] [Google Scholar]
- 42.Kim Y, Yeo J, Lee JH, Cho J, Seo D, Kim J-S, Kim VN. Deletion of Human tarbp2 Reveals Cellular MicroRNA Targets and Cell-Cycle Function of TRBP. Cell Rep. 2014;9:1061–1074. doi: 10.1016/j.celrep.2014.09.039. [DOI] [PubMed] [Google Scholar]
- 43.Guo X, Zhang R, Wang J, Lu R. Antiviral RNA Silencing Initiated in the Absence of RDE-4, a Double-Stranded RNA Binding Protein, in Caenorhabditis elegans. Journal of virology. 2013;87:10721–10729. doi: 10.1128/JVI.01305-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gu W, Shirayama M, Conte DJ, Vasale J, Batista PJ, Claycomb JM, Moresco JJ, Youngman EM, Keys J, Stoltz MJ, et al. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Molecular cell. 2009;36:231–244. doi: 10.1016/j.molcel.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saleh MC, van Rij RP, Hekele A, Gillis A, Foley E, O’Farrell PH, Andino R. The endocytic pathway mediates cell entry of dsRNA to induce RNAi silencing. Nature cell biology. 2006;8:793–802. doi: 10.1038/ncbl439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saleh MC, Tassetto M, van Rij RP, Goic B, Gausson V, Berry B, Jacquier C, Antoniewski C, Andino R. Antiviral immunity in Drosophila requires systemic RNA interference spread. Nature. 2009;458:346–350. doi: 10.1038/nature07712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deddouche S, Matt N, Budd A, Mueller S, Kemp C, Galiana-Arnoux D, Dostert C, Antoniewski C, Hoffmann JA, Imler JL. The DExD/H-box helicase Dicer-2 mediates the induction of antiviral activity in drosophila. Nature immunology. 2008;9:1425–1432. doi: 10.1038/ni.1664. [DOI] [PubMed] [Google Scholar]
- 48.Paradkar PN, Trinidad L, Voysey R, Duchemin J-B, Walker PJ. Secreted Vago restricts West Nile virus infection in Culex mosquito cells by activating the Jak-STAT pathway. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:18915–18920. doi: 10.1073/pnas.1205231109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kemp C, Mueller S, Goto A, Barbier V, Paro S, Bonnay F, Dostert C, Troxler L, Hetru C, Meignin C, et al. Broad RNA interference-mediated antiviral immunity and virus-specific inducible responses in Drosophila. J Immunol. 2013;190:650–658. doi: 10.4049/jimmunol.1102486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goic B, Vodovar N, Mondotte JA, Monot C, Frangeul L, Blanc H, Gausson V, Vera-Otarola J, Cristofari G, Saleh M-C. RNA-mediated interference and reverse transcription control the persistence of RNA viruses in the insect model Drosophila. Nature immunology. 2013;14:396–403. doi: 10.1038/ni.2542. [DOI] [PubMed] [Google Scholar]
- 51.Obbard DJ, Jiggins FM, Halligan DL, Little TJ. Natural selection drives extremely rapid evolution in antiviral RNAi genes. Curr Biol. 2006;16:580–585. doi: 10.1016/j.cub.2006.01.065. [DOI] [PubMed] [Google Scholar]
- 52.van Mierlo JT, Overheul GJ, Obadia B, van Cleef KWR, Webster CL, Saleh M-C, Obbard DJ, van Rij RP. Novel Drosophila viruses encode host-specific suppressors of RNAi. PLoS pathogens. 2014;10:e1004256. doi: 10.1371/journal.ppat.1004256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Y, Yu F, Li J, Tong Y, Zhang Y, Yu Z. The first invertebrate RIG-I-like receptor (RLR) homolog gene in the pacific oyster Crassostrea gigas. Fish Shellfish Immunol. 2014;40:466–471. doi: 10.1016/j.fsi.2014.07.029. [DOI] [PubMed] [Google Scholar]