Abstract
FadR is a master regulator of fatty acid metabolism and influences virulence in certain members of Vibrionaceae. Among FadR homologs of the GntR family, the Vibrionaceae protein is unusual in that it contains a C-terminal 40-residue insertion. Here we report the structure of Vibrio cholerae FadR (VcFadR) alone, bound to DNA, and in the presence of a ligand, oleoyl-CoA. Whereas Escherichia coli FadR (EcFadR) contains only one acyl-CoA binding site in each monomer, crystallographic and calorimetric data indicate that VcFadR has two. One of the binding sites resembles that of E. coli FadR, whereas the other, comprised of residues from the insertion, has not previously been observed. Upon ligand binding, VcFadR undergoes a dramatic conformational change that would more fully disrupt DNA binding than EcFadR. These findings suggest that the ability to bind and respond to an additional ligand allows FadR from Vibrionaceae to function as a more efficient regulator.
Introduction
In all organisms, fatty acids (FAs) are essential components of membranes and important sources of metabolic energy. FadR is a member of the GntR family of transcription factors that coordinately controls the pathways of FA degradation and unsaturated fatty acid (UFA) biosynthesis in enteric bacteria1. In the absence of exogenous long chain fatty acids (LCFAs), FadR functions as a repressor of FA degradation by binding to a site in the promoters of the fad genes and interfering with RNA polymerase2 (Fig. 1a). These genes include fadL, fadD, fadBA, fadE, and fadH that encode proteins required for the transport, activation and β-oxidation of LCFAs3. In V. cholerae, FadR also represses the expression of the plsB gene involved in membrane phospholipid biosynthesis4. In the absence of exogenous LCFAs, FadR simultaneously activates the expression of the fabA and fabB genes that encode proteins required for the biosynthesis of UFAs5,6. When exogenous LCFAs are present (Fig. 1b), they are transported across the outer membrane by FadL and activated by the inner membrane-associated acyl-coenzyme A (CoA) ligase FadD7 to produce long chain fatty acyl-CoAs (LCFA-CoAs). These LCFA-CoAs bind directly to FadR and induce a conformational change that releases FadR from its binding sites8. This derepresses fad gene expression to utilize the LCFAs and decreases the expression of fabA and fabB since UFA biosynthesis is no longer necessary.
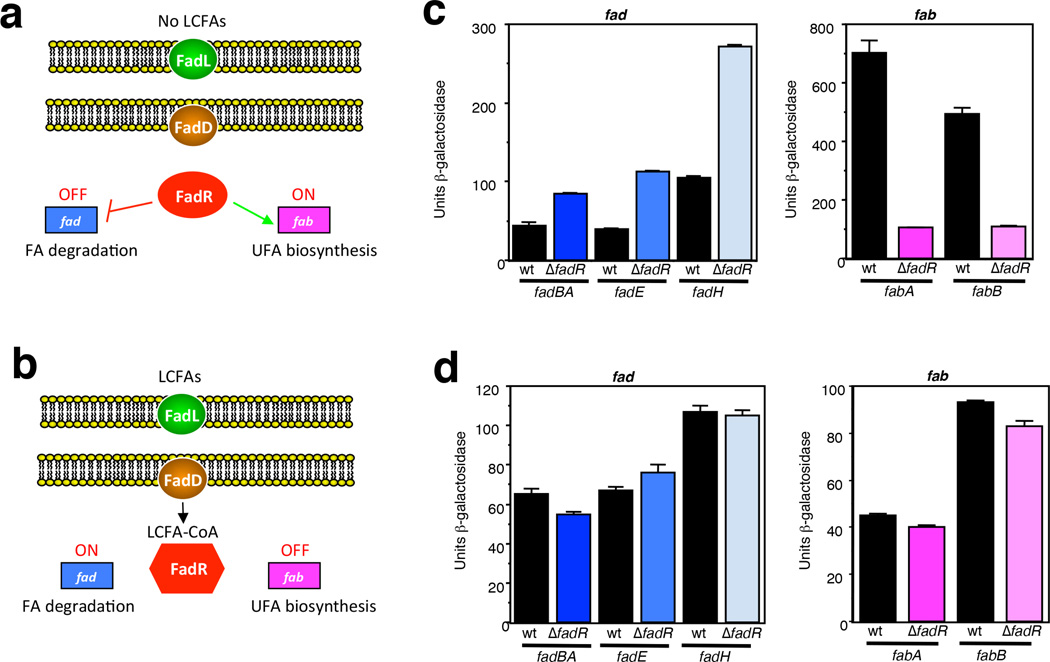
Figure 1. Roles of FadR in V. cholerae.
(a, b) Schematic showing FadR-mediated regulation in V. cholerae. In the absence of LCFAs, (a), FadR represses FA degradation by binding to a site in the promoters of the fad genes and activates UFA biosynthesis by binding to a site in the promoters of the fabA and fabB genes. In the presence of LCFAs, (b), they are transported into the cell by FadL and activated to LCFA-CoAs by FadD. The resulting activated LCFA-CoAs bind to FadR causing a conformational change that releases it from DNA and results in derepression and failure to activate its regulated promoters. (c, d) Influence of a ΔfadR mutation on the expression of fadBA, fadE, fadH, fabA and fabB promoter-lacZ fusions in V. cholerae. Strains were grown in Tryptone medium with aeration for 5 h at 37°C in the absence (c) and presence (d) of 2.5 mM oleate. From left to right: WL1031 (fadBA-lacZ), WL1035 (ΔfadR), WL1027 (fadE-lacZ), WL1029 (ΔfadR), WL1040 (fadH-lacZ), WL1042 (ΔfadR), WL1005 (fabA-lacZ), WL1007 (ΔfadR), GK1609 (fabB-lacZ), GK1610 (ΔfadR). Error bars indicate the standard deviation of at least two replicates. Results are representative of at least three independent experiments.
In addition to controlling the activity of FadR, UFAs, which are components of bile in the intestinal lumen, are important signals present in the host environment that influence the expression of virulence genes in V. cholerae9. Members of Vibrionaceae are ubiquitous in marine and fresh water environments, with species found in open water, estuaries and marine sediments as either free-living or in association with phyto- and zooplankton10,11. Vibrio cholerae is the causative agent of the acute intestinal infection cholera. Upon entry into the host intestine, V. cholerae induces a transcriptional cascade resulting in the expression of the AraC-type master virulence regulator, ToxT. ToxT directly activates the expression of the two primary virulence factors of V. cholerae, the toxin-coregulated pilus (TCP)12 and cholera toxin (CT)13. UFAs have been found to directly bind into the pocket of ToxT and influence its activity by impairing the ability of the protein to dimerize and bind to DNA14. The location of the UFA, buried in the ligand pocket at the interface between the N- and C- terminal domains is thought to promote a “closed” conformation of ToxT that is not capable of binding to DNA. Once the bacteria have penetrated the mucus of the intestine, and the concentrations of UFAs are reduced, the closed conformation is presumably destabilized, and the protein becomes competent for DNA binding and activation of gene expression14.
The majority of FadR proteins that have been identified are members of the GntR family, as typified by E. coli FadR (EcFadR), which has been crystallized in its apo, ligand-bound, and DNA-bound forms15–17. EcFadR has an N-terminal DNA binding domain containing a winged helix-turn-helix (wHTH) motif, and a C-terminal domain consisting of a seven-helical bundle containing a large cavity that is involved in acyl-CoA binding15. Upon ligand binding, effector induced conformational changes occur in the protein that lead to displacement of the entire DNA binding domain with respect to the effector binding domain, leading to disruption of the protein-DNA complex16.
FadR from Vibrionaceae is unusual among FadRs of the GntR family in that it contains a 40 amino acid insertion in its C-terminal domain18. Although FadR from V. cholerae appears to repress gene expression as well as other FadR homologs that have been examined, it has a higher binding affinity for acyl-CoAs than the other homologs and it induces the expression of genes involved in FA utilization (i.e. it is derepressed) more efficiently in the presence of ligand18. It has been suggested that these properties may be related to the 40 amino acid insertion confined to the family. To gain insights into the role of this 40 amino acid insertion for V. cholerae FadR (VcFadR), we have crystallized the VcFadR protein in three forms: apo, bound to its cognate DNA, and in the presence of a ligand, oleoyl-CoA. Our results indicate the insertion region in VcFadR potentially stabilizes the wing of the wHTH DNA binding domain in the absence of DNA and, strikingly, facilitates the binding of a second oleoyl-CoA ligand in each monomer. The large conformational changes that occur in the protein upon binding of this second ligand are predicted to more fully disrupt DNA binding than what has been observed in the E. coli protein, possibly explaining why VcFadR is a more efficient regulatory protein.
Results
VcFadR is unusual but still regulates FA metabolism
FadR is present in a variety of bacterial species where it plays a role in the transcriptional regulation of genes involved in FA metabolism19. The regulation of genes involved in FA degradation and UFA biosynthesis by FadR in V. cholerae has been inferred from bioinformatics analyses19,20. To confirm that VcFadR functions as a regulator of these processes in V. cholerae, fusions of a promoterless lacZ gene from E. coli were made to the upstream regions of the V. cholerae fadBA, fadE, and fadH genes involved in FA degradation and to the fabA and fabB genes involved in UFA biosynthesis. The fusions were introduced into the V. cholerae chromosome at the lacZ locus, and examined in the presence and absence of FadR and LCFAs. As shown in Fig. 1c, the loss of FadR increased the expression of the fadBA, fadE and fadH promoters 2, 2.8 and 2.5 fold, respectively, whereas it decreased the expression of the fabA and fabB promoters 6.7 and 4.5 fold, respectively, in the absence of LCFAs. In the presence of LCFAs (Fig. 1d), no significant difference in the expression of the fusions was observed between the wild-type and ΔfadR mutant strains. These results show that FadR functions as a regulator of FA metabolism in V. cholerae.
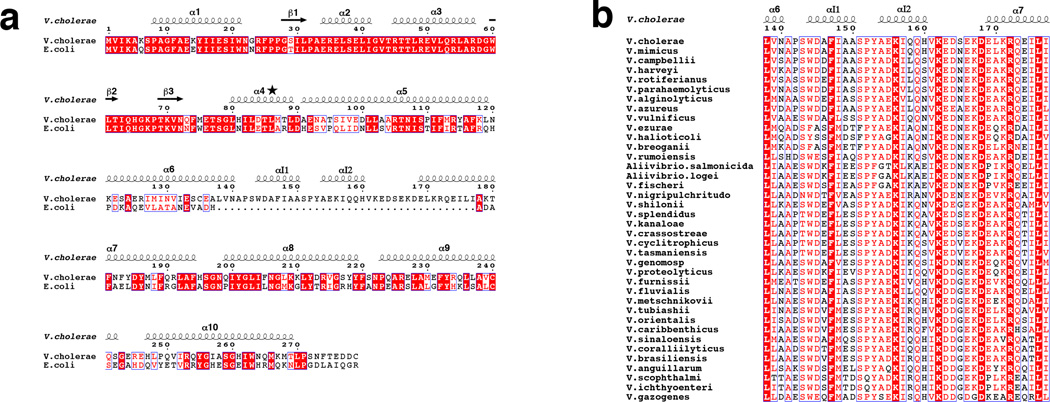
VcFadR shares 52% sequence identity with EcFadR (Fig. 2a). However, the protein from V. cholerae, as well as from other members of Vibrionaceae, shows a major difference from all other GntR family FadRs that have been sequenced in that it contains 40 additional residues in the C-terminal domain (Figs 2a, b and Supplementary Fig.1). A Blast search21 with V. cholerae FadR also reveals several related proteins with a 44-residue insertion (Supplementary Fig. 1). Thus, it appears that Vibrionaceae FadRs acquired an insertion at some point in their evolution; retention of this insertion suggests it provides a biological advantage for the organism.
Figure 2. Sequence alignment of FadR.
(a) Alignment of FadR between V. cholerae and E. coli. Identical residues are indicated by red background. Incomplete identities among aligned sequences are in red text. Secondary structure is shown above the sequence and the numbering is based on EcFadR (PDB 1E2X)15. Helix α4, marked with the star, undergoes a helix to loop transition upon ligand binding. (b) Alignment of 40-residue insertions from Vibrionaceae. Colors are the same as in (a). The alignments were prepared with Clustal W41 and ESPript 3.042 (http://espript.ibcp.fr/).
Structure of apo-VcFadR and comparison with EcFadR
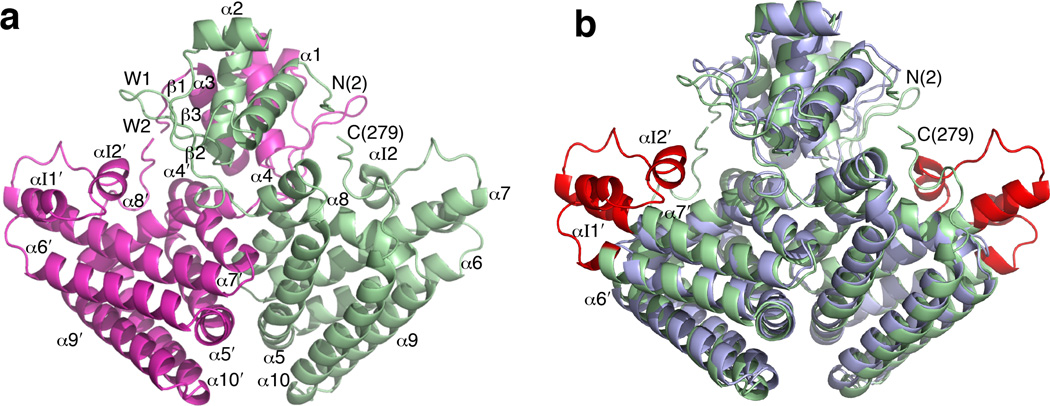
To shed light on the role of the insertion for Vibrionaceae, the structure of V. cholerae apo-FadR was determined (Fig. 3a and Table 1). The asymmetric unit contains a dimer formed in a manner similar to that of EcFadR (PDB 1E2X and 1HW1)15,17. Each monomer consists of an α/β N-terminal domain (residues 2–72, α1-β1-α2-α3-β2-W1-β3-W2, ‘W’ denotes a ‘wing’), a helical C-terminal domain (residues 94–279, α5–α6–αI1-αI2-α7-α8–α9–α10, ‘I’ denotes that the helix is derived from the insertion), and a short helical linker (residues 81–89, α4) (Fig. 3a). The structure of the N-terminal domain conforms to the so-called wHTH motif22, consistent with its role in binding DNA, and contains the most flexible loop in the structure as suggested by its B-factors (Supplementary Fig. 2a).
Figure 3. Structure of VcFadR.
(a) The VcFadR dimer. One monomer is magenta and the other is green. The N and C termini are labeled and the amino acid positions are shown in parentheses. αI1 and αI2 denote the helices from the insertion and W1 and W2 denote wings. Helices from chain B are distinguished by a prime. (b) Superposition of the structures of the VcFadR and EcFadR dimers (PDB 1E2X). The 40-residue insertion in VcFadR is red and the rest of the protein is green. EcFadR is shown in blue.
Table 1.
Data collection and refinement statistics
| VcFadR | VcFadR-DNA |
VcFadR- oleoyl-CoA |
|
|---|---|---|---|
| Data collection | |||
| Space group | P41 | P1 | P212121 |
| Cell dimensions | |||
| a, b, c (Å) | 87.48 87.48 81.43 | 84.6 94.7 101.7 | 116.92 88.69 62.86 |
| α, β, γ, (°) | 90 90 90 | 89.8 114.6 116.5 | 90 90 90 |
| Resolution (Å) | 19.56–2.20 (2.28–2.20)a | 19.89–3.21 (3.32–3.21) | 19.73–2.80 (2.90–2.80) |
| VM(Å3//Da)/solvent content (%) | 2.4/49.7 | 3.9/71.0 | 2.4/48.4 |
| No. of dimers in asymmetric unit | 1 | 2 | 1 |
| Rsym | 7.9 (49.3) | 8.9 (36.6) | 13.1(48.3) |
| I / σI | 33.8 (6.6) | 13.0 (3.0) | 11.5 (3.2) |
| Completeness (%) | 99.8 (100.0) | 97.8 (97.1) | 99.7(99.6) |
| Redundancy | 15.3 (15.2) | 3.5 (3.5) | 3.6 (3.7) |
| Refinement | |||
| Resolution (Å) | 19.56–2.2 | 19.89–3.21 | 19.73–2.8 |
| No. reflections | 31200 | 40186 | 16577 |
| Rwork / Rfree | 0.174/0.205 | 0.218/0.252 | 0.259/0.298 |
| No. atoms | |||
| Protein | 4488 | 4402 | 4240 |
| Ligand/DNA | - | 1271 | 268 |
| Water | 387 | - | - |
| B-factors (Å2) | |||
| Wilson B-factor | 25.9 | 73.3 | 35.7 |
| Protein | 27.0 | 83.7 | 74.1 |
| Ligand/DNA | - | 98.5 | 59.5 |
| Water | 30.30 | - | - |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.004 | 0.007 | 0.010 |
| Bond angles (°) | 0.9 | 1.4 | 2.2 |
Each dataset was collected from a single crystal.
Values in parentheses are for highest-resolution shell.
Superposition of the VcFadR dimer onto the EcFadR dimer (PDB 1E2X)15 (Fig. 3b) reveals that overall the two proteins are very similar [root mean square deviation (RMSD) over 424 alpha carbon atoms is 1.64 Å] except for the insertion (residues 138–177), which lies between helix α6 and helix α7. The insertion region elongates helices α6 and α7, as previously predicted18, and forms two additional short α-helices, αI1 and αI2, along with several connecting loops.
Structure of VcFadR with DNA and comparison with EcFadR
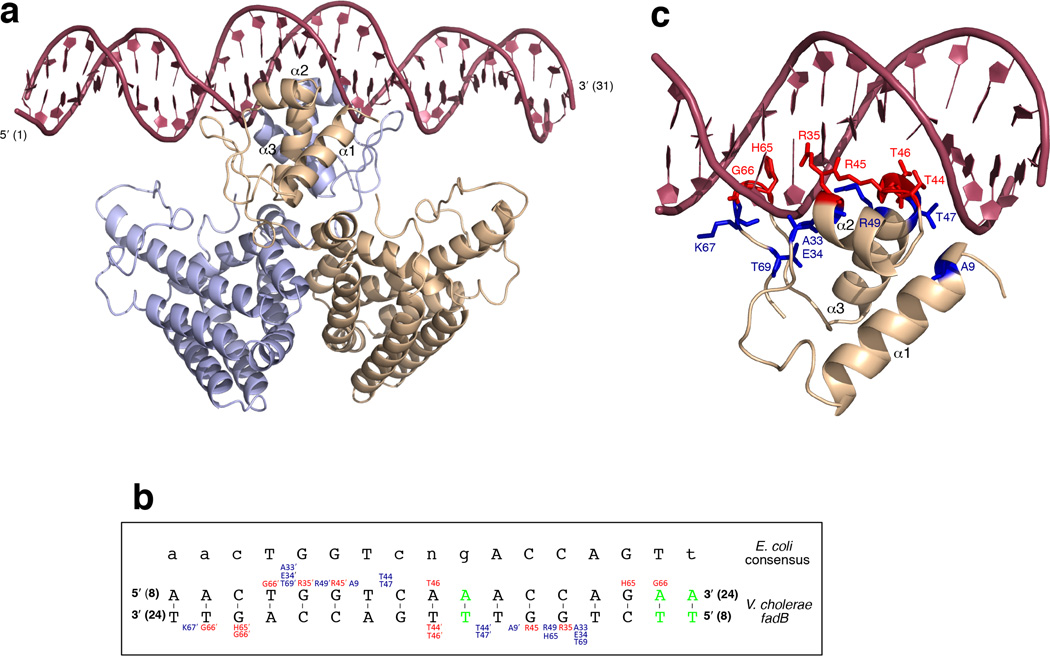
VcFadR was crystallized in complex with a 31-bp oligonucleotide derived from the V. cholerae fadBA promoter (Figs 4a, b, and Table 1). As in the EcFadR-DNA complex (PDB 1H9T and 1HW2)16,17, the DNA binding site is recognized by residues from helices α1, α2, and α3, which interact with the major groove, as well as the two wings (W1 and W2) that bind to the adjacent minor grooves (Figs 4a, c), stabilizing these loops significantly (Supplementary Fig. 2b). As in the EcFadR-DNA complex, the DNA in the V. cholerae FadR-DNA complex has a B-form conformation with a curvature of ~20° toward the protein, resulting in a contraction of the central major groove and an expansion of the opposite minor groove. As the amino acid sequences of the DNA binding domains of the V. cholerae and E. coli proteins are nearly identical (Fig. 2a), the protein-DNA contacts are also very similar. For example, there are five residues in both proteins that specifically recognize DNA base pairs (R35 in α2; R45 and T46 in α3; and H65 and G66 in the tip of the wing) (Figs 4b, c and Supplementary Fig. 3). There are also similarities in a number of nonspecific interactions with the DNA sugar-phosphate backbone, including A9, E34, T44, T47, R49 and T69 (Figs 4b, c and Supplementary Fig. 3). All contacts are symmetrical, except for T44, H65 and G66 (which have both symmetrical and non-symmetrical contacts) and K67 (which only has a non-symmetrical contact) (Fig. 4b).
Figure 4. Structure of the VcFadR-DNA complex.
(a) The VcFadR dimer bound to DNA. One monomer is slate blue and the other is wheat. (b) Schematic overview of the VcFadR-DNA contacts. Residues that form specific contacts with the DNA are red and those that form nonspecific interactions with the DNA sugar-phosphate are dark blue. The base pairs in the V. cholerae fadB recognition sequence that are different from the E. coli consensus sequence43 are green. Within the consensus sequence, upper and lowercase letters represent nucleotides found more and less frequently, respectively. Residues from chain B are distinguished by a prime following the amino acid number. (c) Stereoview showing a close up of the VcFadR-DNA interface within one monomer. Colors are the same as in (b).
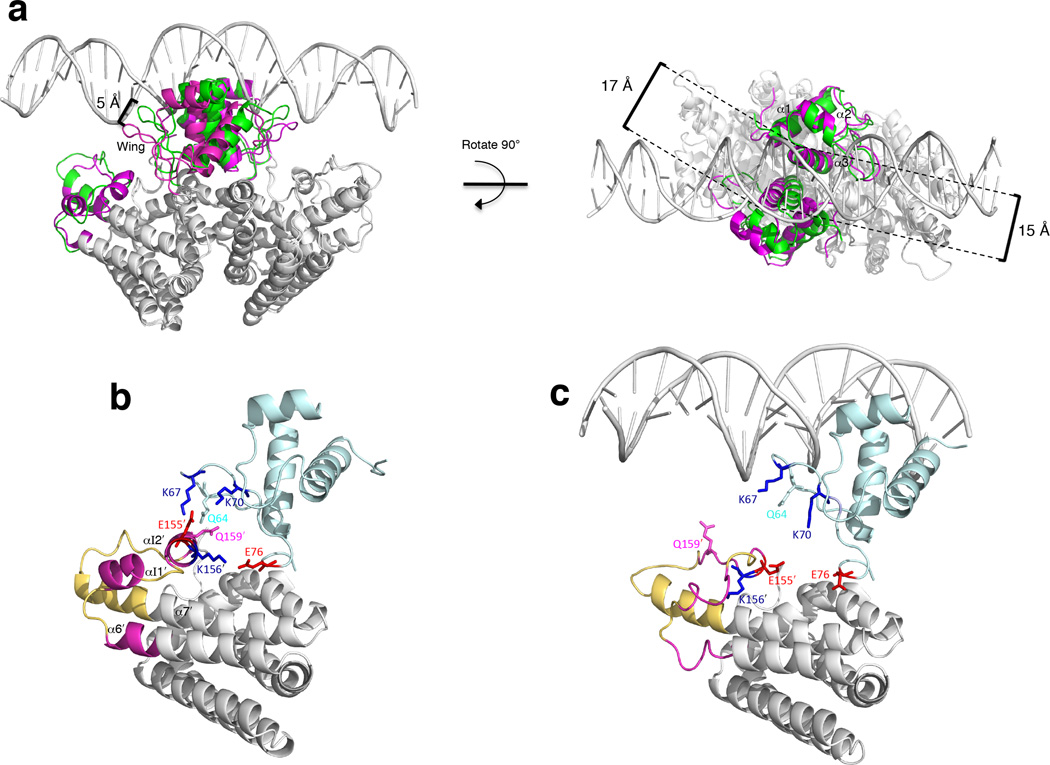
Superposition of the structures of the dimeric VcFadR-DNA complex and the apo-VcFadR dimer (Fig. 5a) shows that the DNA binding domain and the insertion region are altered upon DNA binding. Within the DNA binding domain, the distance between the two DNA recognition helices (α3) narrows from 17 Å in the apo-VcFadR structure to 15 Å in the VcFadR-DNA complex such that it is better able to interact with the DNA (Fig. 5a). In addition, the tip of the wing in each wHTH DNA binding domain is shifted by 5 Å, forming interactions with DNA (Fig. 5a).
Figure 5. Conformational changes in VcFadR upon DNA binding.
(a) Superposition of the VcFadR-DNA complex and apo-VcFadR shown in two orientations (along and perpendicular to the pseudo 2-fold axis). The two primary regions that are altered upon DNA binding are shown in color (green, VcFadR-DNA complex; magenta, apo-VcFadR) and the rest of the protein is white. The tip of the wing (H65) in each DNA wHTH binding domain is shifted by up to 5 Å between the two structures and the distance between the two DNA recognition helices (R45 at the beginning of helix α3) is wider in apo-VcFadR structure (17 Å) compared to that in the VcFadR-DNA complex (15 Å). (b) Detailed view of the interaction between the insertion region and the wing of wHTH in the absence of the DNA. The N-terminal DNA binding domain is pale cyan. The region that undergoes a helix to loop transition upon DNA binding (helices αI1, αI2 and the C-terminal end of α6 in apo-VcFadR) is magenta and the rest of the 40-residue insertion region is yellow. Residues that are positively charged are blue and those that are negatively charged are red. Helices and residues from chain B are distinguished by a prime. (c) Same view as in (b) but in the presence of DNA. The colors are the same as in (b).
Although the insertion region (αI1, αI2 and several connecting loops) does not directly contact DNA (Fig. 5a), it undergoes a conformational change upon DNA binding. In the absence of DNA, the wing of the wHTH of chain A interacts with αI2 from the insertion region of chain B (Fig. 5b) via hydrogen bonds between the side chains of Q64 and K67 and the carbonyl oxygen of Q1592 (residues from chain B are distinguished by a prime) and two salt bridges (K70-E1552 and E76-K1562), suggesting the insertion stabilizes the wing. These interactions are also symmetric, although the distances are slightly longer between the insertion of chain A and the wing of chain B. In the presence of DNA (Fig. 5c), the wing of the wHTH moves away from the insertion to interact with DNA and helices αI1, αI2 and the C-terminal end of α6 undergo a helix to loop transition. This conformational change in the insertion region when VcFadR is bound to DNA may be due to the loss of interactions with the wing.
Structure of VcFadR with ligand and comparison with EcFadR
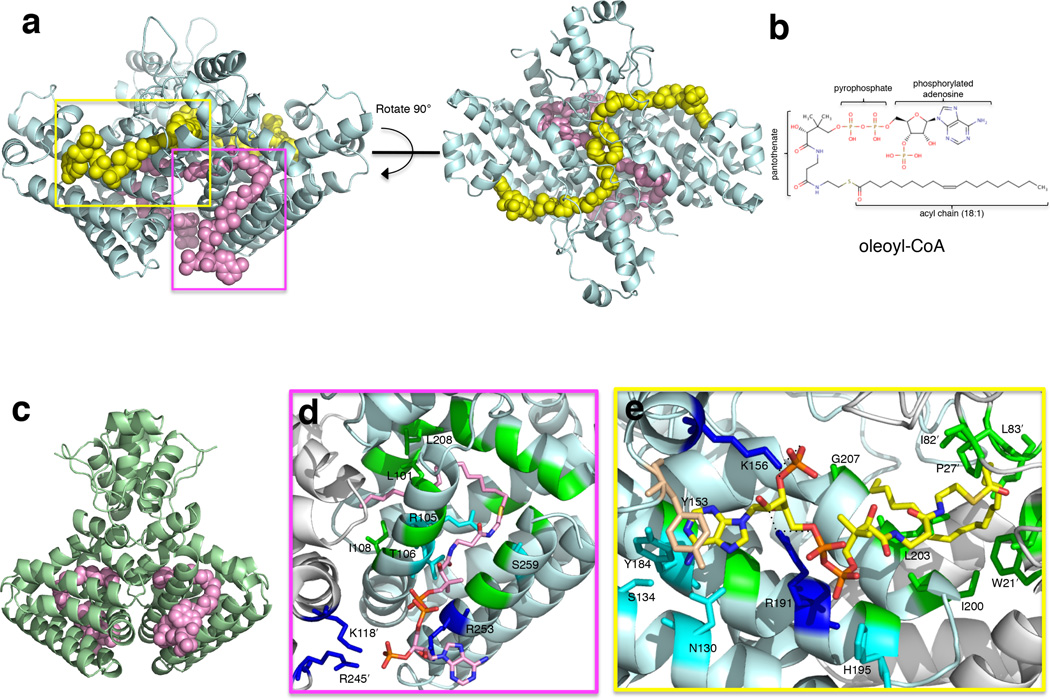
In order to visualize the effects of ligand binding on VcFadR, the structure was determined in the presence of the high binding affinity ligand, oleoyl-CoA18 (18:1) (Figs 6a, b, and Table 1). Interestingly, in contrast to EcFadR, which binds one molecule of ligand [myristoyl-CoA (14:0) (PDB 1H9G)16] per monomer (Fig. 6c), VcFadR binds two ligand molecules, with one (colored pink in Fig. 6a) located in the site corresponding to the EcFadR ligand binding site (designated site #1) and the other (colored yellow in Fig. 6a) in a binding site derived in part from the 40-residue insertion region (designated site #2) that is structurally distinct from site #1 (electron density is shown in Supplementary Fig. 4). Thus, the insertion in FadR from Vibrionaceae facilitates the binding of a second molecule of ligand into each monomer; this has not been observed in any FadR structure that has been solved to date.
Figure 6. Structures of VcFadR and EcFadR with ligand.
(a) The VcFadR-oleoyl-CoA complex in two orientations. The ligand in the site corresponding to EcFadR (see c) is shown in pink spheres whereas the ligand in the site that involves the insertion is in yellow spheres. (b) The chemical structure of oleoyl-CoA. (c) Structure of EcFadR bound to myristoyl-CoA (PDB 1H9G)16. (d–e) Regions boxed in (a) highlighting critical residues interacting with the ligand in the pocket of VcFadR corresponding to that of E. coli (d, site #1, monomer A) and in the pocket derived from the insertion (e, site #2, monomer A). Positively charged residues that interact with ligand phosphates are in blue. Residues forming hydrogen bonds are cyan, and those forming hydrophobic interactions are in green. Y153, which forms π-stacking with adenosine of the ligand, is wheat. Residues from chain B are distinguished by a prime following the amino acid number. The ligand is shown as sticks in (d) and (e). The carbon atoms are colored pink in (d) and yellow in (e). Other atoms of the ligand are colored as follows: nitrogen, blue; oxygen, red; sulfur, dark yellow; phosphorous, orange.
For the ligand bound to site #1 in monomer A, salt bridges are formed between ligand phosphates and residues R253, K1182 and R2452 (Figs 6d, Supplementary Fig. 5a). In monomer B, the phosphorylated adenosine head group of the site #1 ligand has a different and more disordered conformation than that in monomer A (Supplementary Fig. 6), and electron density can be observed for multiple positions of the head group. The arrangements of salt bridges in VcFadR differ significantly from the EcFadR-myristoyl-CoA structure (1H9G)16 where only one salt bridge forms between the ligand phosphate and EcFadR R213 (corresponding to VcFadR R253). In VcFadR, additional hydrogen bonds are formed between carbonyl oxygens in the pantothenic acid moiety and the polar side chains T106 and S259 (Fig. 6d and Supplementary Fig. 5a), homologous to hydrogen bonds involving EcFadR T106 and S219). Finally, as in the EcFadR-myristoyl-CoA structure (1H9G)16, the C18 acyl chain adopts a bent conformation and terminates in a hydrophobic pocket formed by residues L101, I108 and L208 from helices α5 and α8 (Figs 6d and Supplementary Fig. 5a). The FadR ligand pocket thus can clearly accommodate ligands of different lengths, as the longer acyl chain in VcFadR structure extends ~4 Å farther into the pocket compared with the C14 acyl chain of myristoyl-CoA in EcFadR (1H9G)16. It is unclear from the structure if the unsaturated bond between C9 and C10 of oleoyl-CoA plays an important role in binding, however comparison of the VcFadR and EcFadR pockets in this region does show one change that might be significant; M168 in EcFadR is replaced by L208 in VcFadR, which is positioned relatively further away from the ligand, perhaps to accommodate the kink in the fatty acyl chain introduced by the double bond (Supplementary Fig. 7)
For ligand binding site #2, residues involved in specific interactions with the ligand are shown in Fig. 6e and Supplementary Fig. 5b. Both K156 from αI2 and R191 from α7 form salt bridges with ligand phosphate groups, and Y153 from αI2 forms a π-stacking interaction with the adenine ring. N130, S134, and Y184 form hydrogen bonds with the adenine ring (Fig. 6e). The C18 acyl chain is buried in a pocket surrounded by hydrophobic residues and the tails of the acyl chains from each monomer pack closely together (Fig. 6a, right-center). The dominantly hydrophobic pocket is mainly formed by residues from helix α8 and the loop region connecting the DNA-binding and the ligand binding domains (e.g. I822, I832, I200, L203, G207) (Fig. 6e and Supplementary Fig. 5b). In addition, upon ligand binding, residues in the DNA binding domain (e.g. W212, P272) move closer to the C-terminal domain and interact with the ligand (see following section). Interestingly, the bent helix α8 (residues 199–221, notably G207) forms part of both binding sites #1 and #2. The above-mentioned residues that are interacting with ligand in binding site #2 are highly conserved among the other FadR proteins with the insertion (Fig. 2b and Supplementary Fig. 1), suggesting that they will also bind the second ligand.
To determine the thermodynamic parameters for binding of the oleoyl-CoA ligand to VcFadR, isothermal titration calorimetry (ITC) was used to measure the dissociation constant (Kd), enthalpy (ΔH), and the stoichiometry of binding (n). Typical ITC profiles of the binding of oleoyl-CoA to VcFadR are shown in Supplementary Fig. 8. Consistent with the crystal structure, the stoichiometry of binding indicates that every monomer of VcFadR protein binds two molecules of oleoyl-CoA. The biphasic nature of the isotherm indicates the presence of nonequivalent binding sites with different affinities for oleoyl-CoA (6 nM and 88 nM). Although it is not possible to determine which binding site has the higher affinity from the ITC data, computed scoring functions obtained from X-SCORE23 and DSX24 predict the higher affinity binding pocket to be site #1.
Comparison of effector mediated conformational changes
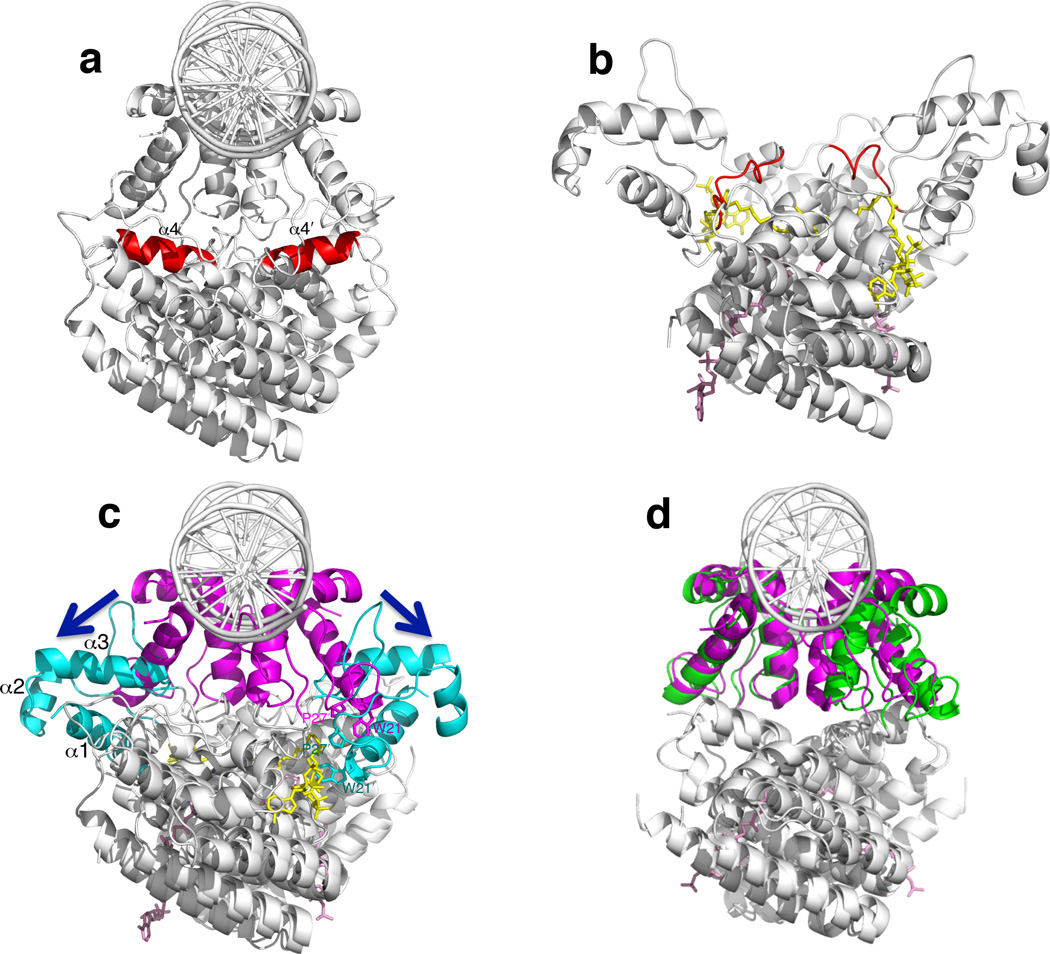
Having defined the structures of V. cholerae apo-FadR, FadR-DNA and FadR-oleoyl-CoA ligand complexes, a comparison can now be made between the effects of ligand binding on VcFadR and EcFadR. In VcFadR, the linker helix α4, which is observed in the VcFadR-DNA and apo-VcFadR structures (Figs 7a, b and Supplementary Fig. 9), unwinds upon ligand binding to site #2 to avoid potential clashes with the ligand (Supplementary Fig. 2c). This transition also appears to destabilize the extensive hydrophobic contacts of α4 with the adjacent DNA binding and the ligand binding domains (Supplementary Fig. 9c), enabling the DNA binding domain to become uncoupled from the ligand binding domain.
Figure 7. Comparison of effector mediated conformational changes between V. cholerae and E. coli.
(a) The VcFadR-DNA complex. The linker helix α4 is red and changes to a loop in the VcFadR-ligand complex (b). α4 from chain B is distinguished by a prime. (c) Superposition of the structures of VcFadR-DNA complex and VcFadR-ligand complex. The DNA binding domain is in magenta (VcFadR-DNA complex) or cyan (VcFadR-ligand complex) and the rest of the protein is white. The arrows show that, upon ligand binding, the DNA binding domain is widened to almost 180°. Residues from chain B are distinguished by a prime following the amino acid number. (d) Superposition of the structures of EcFadR-DNA complex (PDB 1H9T)16 and EcFadR-ligand complex (PDB 1H9G)16. The DNA binding domain is magenta (EcFadR-DNA complex) or green (EcFadR-ligand complex) and the rest of the protein is white.
Comparison of the VcFadR-oleoyl-CoA and VcFadR-DNA complexes shows that ligand binding induces the DNA binding domain to move as a rigid body towards the C-terminal domain (Fig. 7c). This allows residues W212 and P272 from the DNA binding domain to interact with the ligand (Figs 6e, 7c); in turn, this could stabilize the DNA binding domain in this conformation. This movement leads to a large change in the distance between the two recognition helices (α3), from 15 Å in the VcFadR-DNA complex (17 Å in the apo-VcFadR structure) to 65 Å in VcFadR-oleoyl-CoA complex (Fig. 7c and Supplementary Fig. 10a). Based on these conformational changes, we propose that binding of the second ligand to VcFadR induces a dramatic separation of the two recognition helices, strongly inhibiting the ability of VcFadR to interact with DNA.
In contrast to VcFadR, ligand binding to EcFadR does not alter the conformation of linker helix α4 as significantly. Instead of the unwinding of α4 described above, ligand binding to EcFadR pushes α4 towards the N-terminal DNA binding domain, where it forms additional contacts with helix α1. The resulting tilt in α1 moves the DNA binding domain ~13° away from the DNA16. This results in smaller separation of the two recognition helices of EcFadR than observed in VcFadR; from 15Å in the EcFadR-DNA complex (PDB 1H9T; 15Å in the apo-EcFadR structure, PDB 1E2X)15,16 to 23Å in the EcFadR-myristoyl-CoA structure (PDB 1H9G)16 (Fig. 7d and Supplementary Fig. 10b).
Discussion
FadR is the master regulator of FA metabolism in bacteria and coordinately controls the pathways of FA degradation and UFA biosynthesis. In V. cholerae, FadR also regulates these processes, but the protein is different from other GntR family homologs in that it has a 40-residue insertion in its C-terminal domain. In all of the structures of FadR that have been solved to date, each monomer binds only one molecule of the acyl-CoA ligand. Although the overall structure of VcFadR and the manner in which it binds to DNA described here is similar to EcFadR, the insertion in the V. cholerae protein elongates the acyl-CoA binding domain and accommodates a second ligand of oleoyl-CoA. One of these sites (site #1) is structurally similar to the ligand binding site in EcFadR whereas the other (site #2) is comprised of residues from the insertion as well as from the N- and C- terminal domains of the protein. Although the two sites share some structural features, such as the involvement of residues within α8, VcFadR appears to have evolved a new way to bind an additional molecule of the same ligand.
We are aware of only two other transcription factors that have been shown to bind more than one molecule of the same ligand in distinct sites within a monomer. The E. coli catabolite gene activator protein (CAP) was crystallized in the presence of DNA with two molecules of cAMP bound to each monomer (PDB 2CGP); however it is not clear if binding of the second ligand is biologically relevant25. BenM, a LysR family transcription factor, was also crystallized with one molecule of benzoate bound at a primary site and another molecule of benzoate (PDB 2F78) bound at a secondary binding site within the same monomer; however, a different ligand, muconate, is also capable of binding at the primary site26. In the presence of both effectors, a unique conformation capable of high level transcriptional activation is achieved26.
The two ligand binding sites in VcFadR appear to have significantly different affinities for oleoyl-CoA. Site #1, the site structurally similar to EcFadR, is predicted to be the higher affinity site and appears to bind ligand with a higher affinity (6 nM) than the E. coli protein, determined for oleoyl-CoA to be 70 nM. In contrast, site #2, derived from the insertion, is predicted to be the lower affinity site and binds ligand with a reduced affinity (88 nM) compared to the E. coli protein. Both site #1 and #2 of VcFadR bind ligand with a higher affinity compared to S. enterica (143 nM), P. multocida (2231 nM) and H. influenzae protein (2636 nM)18. Analysis of VcFadR binding site #1 shows a greater number of salt bridges are formed with ligand phosphates than for EcFadR, and these additional interactions could be responsible for the increased affinity for ligand at this site. It is also possible that the presence of the insertion in VcFadR alters the dynamics of the entire protein so as to increase the affinity for ligand at site #127. For example, in addition to providing a second binding site for the ligand, the insertion also appears to stabilize the wing of the wHTH binding domain in the absence of DNA. This interaction could change the protein such that its affinity for ligand increases.
It has previously been shown that addition of oleate to V. cholerae FadR allows for nearly complete derepression of genes involved in FA utilization, suggesting that in the presence of ligand, VcFadR decreases its affinity for DNA to a greater extent than other regulators18. These findings are consistent with our observation that the V. cholerae protein undergoes dramatic conformational changes upon ligand binding that would more fully disrupt DNA binding than the ligand induced movements in EcFadR and suggest that the binding of ligand to site #2 is physiologically relevant. In VcFadR, ligand binding to site #2 induces a helix to loop transition in α4 that enables the DNA binding domain to swing towards the C-terminal domain, forming stabilizing interactions with the site #2 ligand. We propose this conformational change allows for a more complete release of the protein from the DNA, leading to fuller expression of the FA utilization genes. In addition, the presence of two binding sites with different affinities likely gives VcFadR a broader response range to different concentrations of ligand than EcFadR. For example, the model predicts that, at very low concentrations, VcFadR binds only one ligand per monomer (likely at site #1) causing VcFadR to have a reduced affinity for DNA, similar to that of EcFadR. At higher concentrations of ligand, VcFadR binds two ligands per monomer (at both sites #1 and #2) further reducing the affinity of the protein for DNA in comparison to EcFadR. Unlike other enteric bacteria such as E. coli that colonize the digestive tract, Vibrionaceae are frequently found in aquatic environments where they acquire FAs from the sediment28,29. The need for Vibrionaceae to have a more efficient mechanism for utilizing FAs in this environment may have selected for an additional ligand binding site in FadR that allows the protein to function as a more dynamic regulatory switch.
Methods
Expression and purification of FadR
V. cholerae FadR was purified using the IMPACT-CN fusion protein system (New England Biolabs). The full-length fadR gene was PCR amplified from V. cholerae C6706 str2 using primers FadR5 and FadR6 (Supplementary Table 1). The resulting fragment was inserted into pTXB1 (New England Biolabs) to produce the FadR-intein/CBD (chitin binding domain) fusion construct pWEL225. FadR was expressed in BL21 (DE3) by autoinduction30 in ZYM-5052 media. Cells were harvested by centrifugation, resuspended in column buffer (20 mM HEPES pH 7.8, 1 mM EDTA, and 500 mM NaCl), lysed by sonication, clarified by centrifugation, and filtered through a 0.45 µm filter. Chitin beads (New England Biolabs) were equilibrated with cold column buffer, mixed with the clarified supernatant, and incubated at 4°C with gentle rocking. The chitin bead slurry was then loaded onto a gravity flow column, washed with 50 column volumes of high salt wash buffer (20 mM HEPES pH 7.8, 1 mM EDTA, and 1 M NaCl), and equilibrated with five column volumes of cleavage buffer (20 mM HEPES pH 7.8, 1 mM EDTA, and 50 mM NaCl). The intein with the CBD was cleaved from FadR using cleavage buffer with 100 mM β-mercaptoethanol and left at 16 °C for 16 h. Eluant from the chitin column in cleavage buffer was then loaded onto a new chitin column and followed by a HiTrap Sepharose prepacked (Q) fast flow (FF) strong anionic exchange column (GE) using a NaCl gradient. This separates the FadR-intein/CBD fusion protein and the intein tag that coeluted with the native FadR. The fractions containing FadR were identified by SDS-polyacrylamide gel electrophoresis and pooled. The pooled fractions were concentrated with an Amicon Ultra Centrifugal filter unit (Millipore) and applied to a Superdex 75 Prep Grade column (GE) equilibrated in crystallization buffer (20 mM HEPES pH 7.8, 1 mM EDTA, and 100 mM NaCl). FadR eluted as a single peak at a position consistent with a dimer and was essentially pure as judged by SDS-polyacrylamide gel electrophoresis.
To prepare the VcFadR-DNA complex for crystallization, the oligonucleotides WS1 and WS2 (Supplementary Table 1) were dissolved in low salt buffer (100 mM NaCl and 10 mM NaOH) and purified by MonoBeads prepacked Mono Q strong anionic exchange column (GE) with a NaCl gradient in 10 mM NaOH. The fractions containing pure oligonucleotides were pooled and concentrated as described above and the buffer was exchanged into crystallization buffer by a desalting column (GE). The purified oligonucleotides were annealed, concentrated and applied to a Superdex 75 Prep Grade column (GE) equilibrated in crystallization buffer to eliminate unannealed oligonucleotides. The purified dsDNA was mixed with purified VcFadR at a protein/DNA ratio of 1:1.5. The VcFadR-DNA complexes were concentrated as described above and applied to a Superdex 200 Prep Grade column (GE) equilibrated in crystallization buffer. The fractions containing the VcFadR-DNA complexes were collected and concentrated as described above.
Crystallization
Crystals of apo-VcFadR were obtained using the sitting drop procedure by mixing equal volumes of the protein solution at 6 mg/ml with a reservoir solution containing 0.2 M sodium chloride, 0.1 M MES pH 6.5, 10% (w/v) PEG 4000. Crystals typically grew within 1 week. VcFadR crystals were cryoprotected in a 30% v/v solution of glycerol/mother liquor and flash frozen in liquid nitrogen for X-ray data collection. Crystals of the VcFadR-DNA complex were also obtained by the sitting drop method by mixing equal volumes of 8 mg/ml complex solution with a reservoir containing 0.2 M lithium sulfate, 0.1 M Tris pH 8.5, 1.26 M ammonium sulfate. Crystals grew in 2 weeks. VcFadR-DNA complex crystals were cryoprotected in a 30% v/v solution of glycerol/mother liquor and flash frozen in liquid nitrogen for X-ray data collection. Crystals of the VcFadR-oleoyl-CoA complex, also obtained by the sitting drop method, were produced by mixing equal volumes of VcFadR-ligand complex (VcFadR=4 mg/ml; ligand (oleoyl coenzyme A lithium salt (Sigma))/VcFadR monomer molar ratio=3.6:1) solution with a reservoir containing 17% PEG 3350, 0.23 M magnesium formate. Crystals grew in about 1 week. VcFadR-oleoyl-CoA complex crystals were cryoprotected in a 30% v/v solution of glycerol/mother liquor and flash frozen in liquid nitrogen for X-ray data collection.
Data collection and structure solution
Crystals of apo-VcFadR diffracted to 2.2 Å at NSLS beamline X6A at Brookhaven National Laboratory. Crystals belonged to the tetragonal space group P41 with unit cell dimensions a=87.48 Å, b=87.48 Å, c=81.43 Å, α=90.0°,β=90.0°, γ=90.0°. The asymmetric unit contained one dimer of VcFadR, giving a crystal volume per protein mass (VM) of 2.4 Å3/Da and a solvent content of 49.7%. Data were processed in XDS31 and structure determination was carried out using AutoMR in Phenix32. Two copies of the E. coli FadR monomer divided into the N-terminal (residues 6 to 137) and C-terminal domain (residues 138 to 226) were used as search models for molecular replacement. The insertion region was built back manually in Coot33. Multiple rounds of refinement and rebuilding were carried out in Phenix.refine34 and Coot33, respectively, using data to 2.2 Å. The resulting final structure has a R-work 17.44% of and R-free of 20.46% with good stereochemistry (Table 1) and continuous density for the backbone from residue 2 to 279 out of 279 possible residues (Fig. 3a). Ramachandran statistics show that 97% and 2.82% of the residues are in the preferred and allowed regions, respectively. In MOLPROBITY35 validation results, this structure was assigned a MOLPROBITY score of 1.44, which is the 99th percentile among structures of comparable resolution.
Diffraction data for the VcFadR-DNA complex were collected to 3.2 Å at NSLS beamline X6A at Brookhaven National Laboratory. Crystals belonged to the triclinic space group P1 with unit cell dimensions a=84.6 Å, b=94.7 Å, c=101.7 Å, α=89.8°, β=114.6°, γ=116.5°. The asymmetric unit contained two molecules of VcFadR-DNA complex, giving a crystal volume per protein mass (VM) of 3.9 Å3/Da and a solvent content of 71.0%. Data were processed in XDS31 and structure determination was carried out using AutoMR in Phenix32. Four copies of the V. cholerae FadR monomer without the insertion region and two copies of DNA from E. coli FadR-DNA complex were used as search models for molecular replacement. The resulting maps showed density for the insertion region and density for additional DNA duplex. The insertion region was built back manually in Coot33; the DNA duplex was mutated to the V. cholerae FadR DNA sequence and additional bases were built back manually in Coot33. Multiple rounds of refinement and rebuilding were carried out in Phenix.refine34 and Coot33, respectively, using data to 3.2 Å. The resulting final structure has a R-work of 21.76% and R-free of 25.23%, with good stereochemistry (Table 1) and continuous density that included all atoms in DNA and residues from residue 6 to 277 out of 279 possible residues (Fig. 4a). Ramachandran statistics show that 91% and 7.6% of the residues are in the preferred and allowed regions, respectively. In MOLPROBITY35 validation results, this structure was assigned a MOLPROBITY score of 2.55, which is the 96th percentile among structures of comparable resolution.
Diffraction data for VcFadR with ligand were collected to 2.8 Å at NSLS beamline X6A at Brookhaven National Laboratory. Crystals belonged to the orthorhombic space group P212121 with unit cell dimensions a=116.92 Å, b=88.69 Å, c=62.86 Å, α=90°, β=90°, γ=90°. The asymmetric unit contained one dimer of VcFadR-oleoyl-CoA complex, giving a crystal volume per protein mass (VM) of 2.4 Å3/Da and a solvent content of 48.4%. Data were processed in XDS31 and structure determination was carried out using AutoMR in Phenix32. Initial molecular replacement with V. cholerae apo-FadR as a search model, was unsuccessful, suggesting that significant structural rearrangement might have occurred upon binding of oleoyl-CoA. Subsequent molecular replacement with only the C-terminal acyl-CoA binding domain gave a single solution. Refinement in Phenix34 resulted in maps in which the ligands and the N-terminal DNA binding domain could be positioned manually in Coot33. Multiple rounds of refinement and rebuilding were carried out in Phenix.refine34 and Coot33, respectively, using data to 2.8 Å. The resulting final structure has a R-work of 25.57% and R-free of 30.35%, with good stereochemistry (Table 1) and continuous density that included all atoms in the ligand and residues from residue 8 to 266 out of 279 possible residues (Fig. 5b). Ramachandran statistics show that 87% and 6.8% of the residues are in the preferred and allowed regions, respectively. In MOLPROBITY35 validation results, this structure was assigned a MOLPROBITY score of 3.15, which is the 53rd percentile among structures of comparable resolution.
Construction of V. cholerae strains and mutations
The fabA-lacZ fusion was constructed by amplifying a DNA fragment from C6706 str2 using primers FabA1 and FabA3 (Supplementary Table 1). The fragment, together with a promoterless lacZ fragment from pVC20036, were inserted into pKAS18037 generating pWEL234. Plasmid pWEL234 was used for allelic exchange37 into the lacZ locus of V. cholerae strain KSK26238 generating WL1005. Plasmid pWEL236 was constructed by amplifying a DNA fragment from C6706 str2 using primers Chr1 and Chr2 and a DNA fragment from pVC20036 using primers LacNot and LacBgl. The resulting fragments were inserted into pKAS15439. The fadE, fadBA, fadH and fabB-lacZ fusions were constructed by amplifying DNA fragments from C6706 str2 using primers FadE1 and FadE3, FadB1 and FadB4, and FadH1 and FadH3, and FabB3 and FabB4. The resulting fragments were inserted into pWEL236, generating plasmids pWEL239, pWEL240, pWEL242, and pGKK469, respectively. The fusions in the resulting plasmids were introduced into the lacZ locus of KSK262 by allelic exchange generating strains WL1027, WL1031, WL1040, and GK1609. The ΔfadR mutation was constructed by amplifying two DNA fragments from C6706 str2 using primers FadR1 with FadR2 and FadR3 with FadR4. The fragments were inserted into pKAS15439, generating plasmid pGKK367, and the deletion was introduced into the chromosome of V. cholerae by allelic exchange.
β-Galactosidase assays
β-Galactosidase assays40 were carried out by growing cultures in Tryptone medium with aeration for 5 h at 37°C.
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Health Grant AI072661 (to F.J.K) and Grant AI039654 (to R.K.T). Research carried out at the X6A beam line was funded by the NIH (NIGMS) under agreement GM-0080. We thank the staff of National Synchrotron Light Source (NSLS) beamline X6A, particularly V. Stojanoff and E. Lazo, for beam time and help with data collection; D. Wilcox and M. Carpenter for training, guidance, and usage of the VP-ITC; S. Almagro-Moreno for helpful discussions and critical reading of the manuscript and our colleagues in the Kull laboratory for intellectual discussions. The authors also thank John Cronan for strains and immunoreagents for the detection of FadR, as well as insightful discussions.
Footnotes
Author Contributions
W.S. purified, crystalized the VcFadR, VcFadR-DNA complex and VcFadR-ligand complex and also collected data on the crystals and determined the structures. W.S. performed ITC experiments and analyzed the data. G.K and W.L. constructed the V. cholerae plasmids and strains and carried out the β-galactosidase assays. K.S., R.K.T. and F.J.K. were involved in study conception and study design. The manuscript was prepared by W.S., K.S., and F.J.K.
Accession codes: Atomic coordinates and structure factor files have been deposited in the Protein Data Bank (ID codes: apo-VcFadR, 4P96; VcFadR-DNA complex, 4P9U; VcFadR-ligand complex, 4PDK).
Competing financial interests: The authors declare no competing financial interests.
References
- 1.Zhang YM, Rock CO. Transcriptional regulation in bacterial membrane lipid synthesis. J. Lipid Res. 2009;50(Suppl):S115–S119. doi: 10.1194/jlr.R800046-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cronan JE, Jr, Subrahmanyam S. FadR, transcriptional co-ordination of metabolic expediency. Mol. Microbiol. 1998;29:937–943. doi: 10.1046/j.1365-2958.1998.00917.x. [DOI] [PubMed] [Google Scholar]
- 3.Clark DP, Cronan JE., Jr . In: Escherichia coli and Salmonella : cellular and molecular biology. Neidhardt Frederick C., editor. ASM Press; 1996. p. 343.p. 357. [Google Scholar]
- 4.Feng Y, Cronan JE. The Vibrio cholerae fatty acid regulatory protein, FadR, represses transcription of plsB, the gene encoding the first enzyme of membrane phospholipid biosynthesis. Mol. Microbiol. 2011;81:1020–1033. doi: 10.1111/j.1365-2958.2011.07748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henry MF, Cronan JE., Jr Escherichia coli transcription factor that both activates fatty acid synthesis and represses fatty acid degradation. J. Mol. Biol. 1991;222:843–849. doi: 10.1016/0022-2836(91)90574-p. [DOI] [PubMed] [Google Scholar]
- 6.Campbell JW, Cronan JE., Jr Escherichia coli FadR positively regulates transcription of the fabB fatty acid biosynthetic gene. J. Bacteriol. 2001;183:5982–5990. doi: 10.1128/JB.183.20.5982-5990.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DiRusso CC, Black PN. Bacterial long chain fatty acid transport: gateway to a fatty acid-responsive signaling system. J. Biol. Chem. 2004;279:49563–49566. doi: 10.1074/jbc.R400026200. [DOI] [PubMed] [Google Scholar]
- 8.DiRusso CC, Heimert TL, Metzger AK. Characterization of FadR, a global transcriptional regulator of fatty acid metabolism in Escherichia coli. Interaction with the fadB promoter is prevented by long chain fatty acyl coenzyme A. J. Biol. Chem. 1992;267:8685–8691. [PubMed] [Google Scholar]
- 9.Chatterjee A, Dutta PK, Chowdhury R. Effect of fatty acids and cholesterol present in bile on expression of virulence factors and motility of Vibrio cholerae. Infect. Immun. 2007;75:1946–1953. doi: 10.1128/IAI.01435-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reen FJ, Almagro-Moreno S, Ussery D, Boyd EF. The genomic code: inferring Vibrionaceae niche specialization. Nat. Rev. Microbiol. 2006;4:697–704. doi: 10.1038/nrmicro1476. [DOI] [PubMed] [Google Scholar]
- 11.Takemura AF, Chien DM, Polz MF. Associations and dynamics of Vibrionaceae in the environment, from the genus to the population level. Front. Microbiol. 2014;5:38. doi: 10.3389/fmicb.2014.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taylor RK, Miller VL, Furlong DB, Mekalanos JJ. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc. Natl. Acad. Sci. U.S.A. 1987;84:2833–2837. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waldor MK, Mekalanos JJ. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 14.Lowden MJ, et al. Structure of Vibrio cholerae ToxT reveals a mechanism for fatty acid regulation of virulence genes. Proc. Natl. Acad. Sci. U.S.A. 2010;107:2860–2865. doi: 10.1073/pnas.0915021107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Aalten DM, DiRusso CC, Knudsen J, Wierenga RK. Crystal structure of FadR, a fatty acid-responsive transcription factor with a novel acyl coenzyme A-binding fold. EMBO J. 2000;19:5167–5177. doi: 10.1093/emboj/19.19.5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Aalten DM, DiRusso CC, Knudsen J. The structural basis of acyl coenzyme A-dependent regulation of the transcription factor FadR. EMBO J. 2001;20:2041–2050. doi: 10.1093/emboj/20.8.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu Y, Heath RJ, Li Z, Rock CO, White SW. The FadR.DNA complex. Transcriptional control of fatty acid metabolism in Escherichia coli. J. Biol. Chem. 2001;276:17373–17379. doi: 10.1074/jbc.M100195200. [DOI] [PubMed] [Google Scholar]
- 18.Iram SH, Cronan JE. Unexpected functional diversity among FadR fatty acid transcriptional regulatory proteins. J. Biol. Chem. 2005;280:32148–32156. doi: 10.1074/jbc.M504054200. [DOI] [PubMed] [Google Scholar]
- 19.Kazakov AE, et al. Comparative genomics of regulation of fatty acid and branched-chain amino acid utilization in proteobacteria. J. Bacteriol. 2009;191:52–64. doi: 10.1128/JB.01175-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sadovskaya NS, Laikova ON, Mironov AA, Gelfand MS. Study of regulation of long-chain fatty acid metabolism using computer analysis of complete bacterial genomes. Mol. Biol. 2001;35:1010–1014. [PubMed] [Google Scholar]
- 21.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 22.Gajiwala KS, Burley SK. Winged helix proteins. Curr. Opin. Struct. Biol. 2000;10:110–116. doi: 10.1016/s0959-440x(99)00057-3. [DOI] [PubMed] [Google Scholar]
- 23.Wang R, Lai L, Wang S. Further development and validation of empirical scoring functions for structure-based binding affinity prediction. J. Comput. Aided Mol. Des. 2002;16:11–26. doi: 10.1023/a:1016357811882. [DOI] [PubMed] [Google Scholar]
- 24.Neudert G, Klebe G. DSX: a knowledge-based scoring function for the assessment of protein-ligand complexes. J. Chem. Inf. Model. 2011;51:2731–2745. doi: 10.1021/ci200274q. [DOI] [PubMed] [Google Scholar]
- 25.Passner JM, Steitz TA. The structure of a CAP-DNA complex having two cAMP molecules bound to each monomer. Proc. Natl. Acad. Sci. U.S.A. 1997;94:2843–2847. doi: 10.1073/pnas.94.7.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ezezika OC, Haddad S, Clark TJ, Neidle EL, Momany C. Distinct effector-binding sites enable synergistic transcriptional activation by BenM, a LysR-type regulator. J. Mol. Biol. 2007;367:616–629. doi: 10.1016/j.jmb.2006.09.090. [DOI] [PubMed] [Google Scholar]
- 27.Motlagh HN, Wrabl JO, Li J, Hilser VJ. The ensemble nature of allostery. Nature. 2014;508:331–339. doi: 10.1038/nature13001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giles DK, Hankins JV, Guan Z, Trent MS. Remodelling of the Vibrio cholerae membrane by incorporation of exogenous fatty acids from host and aquatic environments. Mol. Microbiol. 2011;79:716–728. doi: 10.1111/j.1365-2958.2010.07476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Urakawa H, Yoshida T, Nishimura M, Ohwada K. Characterization of depth-related population variation in microbial communities of a coastal marine sediment using 16S rDNA-based approaches and quinone profiling. Environ. Microbiol. 2000;2:542–554. doi: 10.1046/j.1462-2920.2000.00137.x. [DOI] [PubMed] [Google Scholar]
- 30.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 31.Kabsch W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCoy AJ, et al. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 34.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parsot C, Mekalanos JJ. Expression of ToxR, the transcriptional activator of the virulence factors in Vibrio cholerae, is modulated by the heat shock response. Proc. Natl. Acad. Sci. U.S.A. 1990;87:9898–9902. doi: 10.1073/pnas.87.24.9898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skorupski K, Taylor RK. Positive selection vectors for allelic exchange. Gene. 1996;169:47–52. doi: 10.1016/0378-1119(95)00793-8. [DOI] [PubMed] [Google Scholar]
- 38.Kovacikova G, Skorupski K. A Vibrio cholerae LysR homolog, AphB, cooperates with AphA at the tcpPH promoter to activate expression of the ToxR virulence cascade. J. Bacteriol. 1999;181:4250–4256. doi: 10.1128/jb.181.14.4250-4256.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kovacikova G, Skorupski K. Regulation of virulence gene expression in Vibrio cholerae by quorum sensing: HapR functions at the aphA promoter. Mol. Microbiol. 2002;46:1135–1147. doi: 10.1046/j.1365-2958.2002.03229.x. [DOI] [PubMed] [Google Scholar]
- 40.Miller JH. Experiments in molecular genetics. Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 41.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 42.Gouet P, Robert X, Courcelle E. ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003;31:3320–3323. doi: 10.1093/nar/gkg556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gui L, Sunnarborg A, LaPorte DC. Regulated expression of a repressor protein: FadR activates iclR. J. Bacteriol. 1996;178:4704–4709. doi: 10.1128/jb.178.15.4704-4709.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.