Abstract
The ongoing threat of influenza epidemics and pandemics has emphasized the importance of developing safe and effective vaccines against infections from divergent influenza viruses. In this review, we first introduce the structure and life cycle of influenza A viruses, describing major influenza A virus-caused pandemics. We then compare different types of influenza vaccines and discuss current advancements in the development of subunit influenza vaccines, particularly those based on nucleoprotein (NP), extracellular domain of matrix protein 2 (M2e) and hemagglutinin (HA) proteins. We also illustrate potential strategies for improving the efficacy of subunit influenza vaccines.
Keywords: Influenza A virus, subunit vaccines, nucleoprotein, matrix protein, hemagglutinin
1. Introduction
Influenza viruses belong to the family Orthomyxoviridae, consisting of five genera that include influenza A virus, influenza B virus, influenza C virus, Thogotovirus and Isavirus, as defined by the International Committee on Taxonomy of Viruses [1]. On the basis of their nucleoprotein (NP) and matrix (M) protein antigens, influenza A and B viruses contain hemagglutinin (HA) and neuraminidase (NA) activities, whereas influenza C virus has no NA, but does have a hemagglutinin–esterase fusion (HEF) protein [1]. Only influenza A and B viruses can cause clinical diseases. Influenza B viral infections are often limited to localized outbreaks, while influenza A virus is the primary pathogen for humans and is thus the principal cause of larger epidemics and pandemics.
Influenza A virus is further divided into different subtypes based on the antigenicity of the two surface glycoproteins, HA and NA. Currently, influenza A virus has 18 HA subtypes (H1-H18) and 11 NA subtypes (N1-N11) [2], theoretically leading to 198 potential combinations. In fact, not all of these subtypes cause human diseases. Seroarchaeology data from the late 19th and early 20th centuries indicated that the H1, H2 and H3 influenza virus subtypes were successfully transmitted among humans [3]. Particularly, the H5 subtype has threatened to emerge as a human pandemic pathogen since 1997, when it killed 6 out of 18 infected humans [3]. The H7 subtype is also worthy of concern because of the newly emerged 2013 avian influenza A/H7N9 pandemic in China [4]. The highly pathogenic H5 and H7 subtypes possess a unique ability to accumulate multiple basic amino acids at the HA cleavage site, increasing the ability of the viruses to spread systemically in an infected host and cause significant disease [5].
2. Influenza A virus structure and life cycle
The influenza A virus genome consists of eight negative sense, single-stranded RNA segments encoding eleven viral proteins which are essential for viral replication and packaging of progeny virus. Major viral proteins include structure proteins HA, NA, M1, M2 and NP; three RNA polymerases, such as polymerase basic protein 1 (PB1), PB2 and polymerase acidic protein (PA); as well as three non-structural proteins (NS) named NS1, NS2 (nuclear export protein, NEP) and PB1-F2 (Fig. 1) [6]. These proteins play significant roles in influenza virus replication, including their assistance in cell membrane recognition and endosomal fusion and acidification, inducing ribonucleoprotein (RNP) delivery into the nucleus, catalyzing polymerase holoenzyme of transcription and replication, and promoting protein and RNA binding and sialidase activity. Specific functions of these proteins are listed below.
Fig. 1. Schematic diagram of the influenza A virus structure.
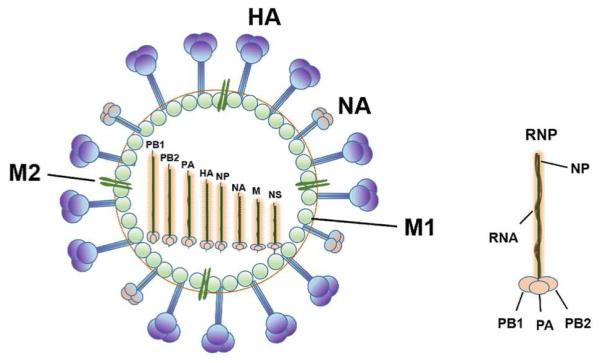
The virus contains three trans-membrane proteins, including HA, NA, and M2 ion channel. The matrix protein M1 forms the protein layer beneath the lipid bilayer. Within the viral envelope is ribonucleoprotein (RNP) consisting of RNA segments associated with NP and the PA, PB1, and PB2 polymerase proteins.
Influenza virus M1 protein is important in RNP coating during viral assembly [7], while M2 protein is a transmembrane protein that forms an ion channel tetramer, exhibiting pH-inducible proton transport activity. During initial virus infection, M2 regulates the pH of viral core after virus uptake into the host cell’s endosomal compartment, and at the late stage of infection, it transports viral transmembrane proteins to the cell surface [8]. HA glycoprotein forms spikes on the surface of virions, mediating attachment of the virus to host cell receptors, thereby enabling the virus to gain entry through membrane fusion. NA glycoprotein forms knob-like structures on the viral surface to catalyze progeny virus from infected cells, thereby allowing the virus to spread with resulting infection of new host cells. NP protein is a core antigen that plays an essential role in viral replication and transcription. NS2 protein may help catalyze the nuclear export of newly synthesized viral RNPs from nucleus to cytoplasm, where assembly of the progeny virions occurs [9].
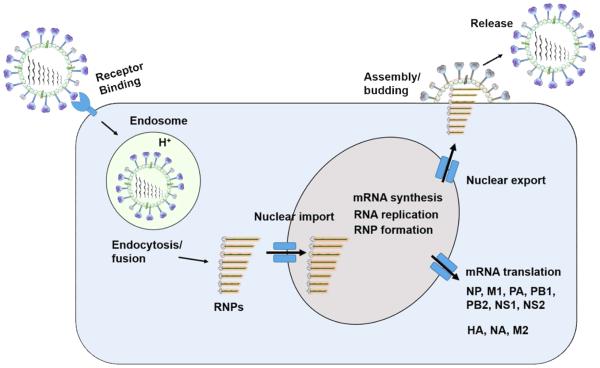
The life cycle of influenza A virus involves the participation of major viral proteins in several important steps (Fig. 2). First, influenza virus infects host cells via HA-mediated binding to cell surface sialic acids, internalizing via receptor-mediated endocytosis. Then, the virus with proteolytically activated HA fuses with endosomal membrane through an acidic pH-promoted conformational change, followed by acidification of the viral core via M2 proton channel, leading to dissociation of the M1 coat protein and release of viral RNPs into the cytoplasm. Nuclear-localized signal on NP facilitates the transport of RNPs to the nucleus, where viral mRNA transcription and genomic replication occur. Once translated, NS1 protein binds to double-stranded RNA and host mRNA processing factors to inhibit cellular interferon-induced antiviral responses [10]. Subsequently, the polymerase performs host mRNA cap-recognition, providing capped mRNA primers for initiation of viral transcription [11]. Replicated RNPs, assisted by NEP, are then exported from the nucleus and transported to plasma membrane for assembly with envelope proteins (HA, NA, M1 and M2). Finally, NA protein plays a crucial role in removing sialic acid from sialyl oligosaccharides, thereby releasing newly assembled virions from the cell surface and preventing self-aggregation of the viral particles [6].
Fig. 2. Schematic diagram of influenza A virus life cycle.
The following five steps describe the life cycle of infectious influenza A virus. 1) HA protein binds target cells containing sialic acids, followed by endocytosis and fusion with an endosome, leading to the fusion of viral membrane and endosomal membrane. 2) Viral RNPs are released into the cytoplasm and then transported into the nucleus. 3) Viral genome is either replicated or transcribed into viral mRNA and then transported into the cytoplasm for translation of viral proteins. 4) Viral RNPs are exported from the nucleus. 5) Mature virus is assembled and budded at the cytoplasmic membrane of the host cell, acquiring viral surface proteins.
3. Influenza A virus-caused pandemics and the importance of developing effective influenza vaccines
Antigenic shift causes emergence of new influenza viruses, resulting in efficient human-to-human transmission and influenza pandemics. Influenza pandemics occur frequently, with three major outbreaks in the 20th century and two outbreaks in the 21st century, including the 2009 pandemic H1N1 and the newly emerged 2013 avian influenza A/H7N9 pandemic. In addition, the highly pathogenic avian influenza (HPAI) A/H5N1 with pandemic potential has caused serious diseases. This section introduces major pandemics caused by influenza A viruses, pointing out the urgency for developing influenza vaccines.
3.1. Three major influenza pandemics in the 20th century
The three major influenza pandemics in the 20th century are 1918 Spanish flu, 1957-1958 Asian influenza pandemic, and 1968 Hong Kong influenza pandemic, which were caused by A/H1N1, A/H2N2 and A/H3N2 influenza viruses, respectively [12]. The 1918 Spanish flu was the most severe influenza pandemic in history, with an estimated 50 million deaths globally [13], while the pandemics caused by 1957-1958 Asian and 1968 Hong Kong influenza pandemics were less severe, with more than one million and around 700,000 deaths, respectively (http://www.flu.gov/pandemic/history/).
The gene sequences and phylogenetic analyses of the virus causing the 1918 pandemic suggest that the genes encoding HA and NA proteins were derived from an avian-like influenza virus strain which had not circulated widely in humans and swine before the pandemic [14-16]. In the 1957-1958 Asian flu, three new gene segments, including HA, NA and PB1, were introduced from an avian-like H2 and N2 species [17, 18], while the other 5 gene segments were derived from the 1918-derived H1N1 lineage. In the case of the 1968 Hong Kong influenza pandemic, two new gene segments, including HA and PB1, were introduced from avian-like H3 and PB1 species, and the other 6 gene segments were derived from the 1957 H2N2 virus [17-19]. The relative mildness of this pandemic might have resulted from the retention of NA protein [20].
3.2. The 2009 influenza A/H1N1 pandemic
In April 2009, a novel swine-origin influenza A/H1N1 virus emerged in Mexico and then quickly spread around the world. As of September 13, 2009, WHO reported over 296,000 confirmed cases in more than 100 countries with at least 3400 deaths (http://www.who.int/csr/don/2009_09_18/en/). The 2009 pandemic H1N1 influenza virus was derived from the reassortment of North American avian H3N2 virus, H1N2 swine virus and Eurasian H1N1 swine lineage virus [21-23]. Like the 1957 pandemic H2N2 and 1968 pandemic H3N2 viruses, the 2009 pandemic H1N1 virus was also a descendant of the 1918 pandemic H1N1 virus [24]. The PB2 and PA gene segments were in the swine triple reassortant lineage which was avian in origin and entered pigs in North America around 1998, while the avian-originated PB1 entered humans and then pigs around 1968 and 1998, respectively [25]. In addition, the avian-originated HA, NP and NS gene segments entered pigs around 1918 and subsequently circulated in classical swine viruses and triple reassortment swine viruses [26], but the NA and M gene segments, which were in the Eurasian swine lineage, originally derived from an avian influenza virus and entered the Eurasian swine population in 1979 [27].
3.3. The newly emerged 2013 avian influenza A/H7N9 pandemic
In February and March of 2013, three patients were hospitalized with severe lower respiratory tract disease associated with a novel avian-origin reassortant influenza A (H7N9) virus infection [4]. A total of 450 laboratory-confirmed cases of human infection with avian influenza A (H7N9) virus, including 165 deaths, were reported to WHO as of June 27, 2014 (http://www.who.int/influenza/human_animal_interface/influenza_h7n9/riskassessment_h7n9_2 7june14.pdf?ua=1). Although this virus type does not appear to transmit easily from person to person and sustained human-to-human transmission has not been reported, understanding the source and mode of transmission of viral infections and taking further surveillance and appropriate countermeasures, such as vaccine development, are urgently required.
3.4. HPAI A/H5N1-caused diseases
HPAI is an extremely contagious, multi-organ systemic disease of poultry, leading to high mortality [28]. The HPAI A/H5N1 viruses were first isolated from sick geese in Guangdong Province in China in 1996 [29]. H5N1 viruses have spread in poultry and wild birds in Asia, Europe, and Africa since 2003, posing a potential risk of outbreaks in poultry and maintaining a high frequency of annual endemics [2]. The transmission of H5N1 virus to humans in Hong Kong in 1997 was the first confirmed avian-to-human transmission of this influenza virus type. As a result, 18 people were hospitalized and six died [30]. Since then, sporadic cases of H5N1 virus infection in humans have been increasing every year. As of July 27, 2014, a total of 667 infected people, including 393 deaths (mortality rate approaching 60%), were reported to WHO (http://www.who.int/influenza/human_animal_interface/EN_GIP_20140727CumulativeNumber H5N1cases.pdf?ua=1) for the period 2003-2014. H5N1 virus is a continuing threat by its capacity to cross species barriers and generate severe infection and high mortality in humans.
The continuous pandemic potential of HPAI A/H5N1 and other emerging or reemerging influenza virus types reinforces the importance of developing effective vaccines and antiviral agents against influenza virus infection. Although antiviral drugs, such as NA and M2 inhibitors, are, to some degree, effective for pandemic control, a number of drug-resistant virus strains have been reported [31-35]. Accordingly, vaccination is currently considered as one of the most effective measures to fight against influenza pandemics [36].
4. Influenza vaccines
Currently developed influenza vaccines include inactivated virus-based, live-attenuated virus-based, DNA-based, viral vector-based, virus-like particle (VLP)-based, and subunit vaccines. The remainder of this review will briefly discuss these vaccine types, pointing out the importance of developing subunit vaccines based on specific viral antigens. The advantages and disadvantages of these vaccine types, as well as the specific adjuvants used for each vaccine type, are summarized in Table 1. While adjuvants are required for inactivated and live-attenuated virus-based, particularly subunit-based vaccines, to promote their efficacy against influenza virus infection, they are not necessary for influenza vaccines based on DNAs, viral vectors and VLPs (Table 1).
Table 1.
Potential advantages and disadvantages of different types of influenza vaccines
| Vaccine types | Advantages | Disadvantages | Adjuvants used | References |
|---|---|---|---|---|
| Inactivated virus and live-attenuated virus-based | Induces robust humoral and cellular immune responses and protection. | Safety problems; potential allergic reactions to eggs. | Rintatolimod, MF59, AS03B | [40, 41, 43, 44] |
| DNA-based | Induces humoral and cellular immune responses and protection; safe and easy to scale up. | Low immunogenicity; potential host autoimmunity; risk of malignancy. | NA | [50-57] |
| Viral vector-based | Induces high humoral and cellular immune responses and cross-protection. | Safety problems; pre-existing immunity. | NA | [60, 61, 106, 107] |
| VLP-based | Genome-free particles containing only viral envelope proteins; can be prepared in a variety of expression systems; induces immune responses and protection. | Relatively lower immunogenicity. | NA | [62-66] |
| Subunit vaccines | High safety profile without involving infectious viruses; rapid, stable, consistent and scalable production; induces humoral and cellular immune responses, neutralizing antibodies and cross-protection. | Require appropriate adjuvants, suitable administration route, fusion with several proteins, or combination with different vaccination strategies to improve immunogenicity. | Adjuvants can be fused with influenza protein vaccines or mixed together with these vaccines | [67] |
| M2e-based | Induces M2e-specific humoral and cellular immune responses; elicits broad cross-protection against divergent virus strains; can be developed as a universal influenza vaccine | Single M2e molecule induces lower immune responses, which may be significantly improved by linking several M2e molecules, or fusing M2e to an appropriate carrier, or other immunogenic protein components. | Iscomatrix, alum, and Freund's adjuvants, or use of fused protein carriers as adjuvants | [70-76, 103-105] |
| NP-based | Contains conserved CD8 T-cell epitopes that induces cross-reactive T cell response and cross-protective immunity, thus having potential for developing universal influenza vaccines. | It is less effective to induce specific antibody responses, especially the neutralizing antibodies. | Alum, Ribi Adjuvant System | [59, 77-79, 81] |
| HA-based | Induces strong HA-specific antibodies, HI and neutralizing antibodies; elicits potent protection. Highly conserved HA2 stem region can be used as a universal vaccine target to develop broad-spectrum influenza vaccines. | HA1 usually induces strain-specific protection resulting from sequence variation. | PELC/CpG, Freund's, Sigma Adjuvant System, Montanide ISA51, CpG7909, GPI-0100, rCTB | [84-89, 95, 96] |
| NA-based | Induces antibodies and protection against homologous virus strains | Induces less cross-reactivity and cross-protection against divergent virus strains. | Iscomatrix | [91-94] |
4.1. Inactivated and live-attenuated influenza vaccines
Influenza vaccines based on inactivated and live-attenuated virus are widely developed, and some of them are in clinical trials [37-39]. Intranasal inoculation of mice, pigs, horses and macaques with NS1-modified influenza virus strains was shown to induce robust humoral and cellular immune responses and generate protective immunity against challenge with wild-type virus [40]. Research has also found that intranasal immunization of humans with a trivalent seasonal influenza vaccine and a TLR-3 agonist (rintatolimod) elicits the production of cross-reactive IgA antibody, protecting against avian influenza virus strains H5N1 and H7N9 [41], in which rintatolimod is served as a potential mucosal adjuvant to boost this protection. However, the use of inactivated and live-attenuated influenza vaccines has raised safety concerns arising from the possible reassortment between specific gene segments and circulating influenza viruses [42], which could, in turn, result in a new pandemic potential. In addition, traditional egg-based vaccines have the risk of allergic reaction. Furthermore, the presence of retroviruses in eggs has raised concerns for their possible effects on the production of live-attenuated influenza virus vaccines [43, 44].
4.2. Plasmid DNA-based influenza vaccines
Plasmid DNA vaccines are considered alternatives to inactivated influenza virus vaccines [45]. Influenza DNA vaccines based on viral HA, M2e, NA and NP genes have shown promising results [46-49]. Vaccination with HA-, HA ectodomain (HAe)- or HA1-based DNA vaccines was reported to stimulate protective immunity in animals. Immune responses induced by HA1 were lower when compared to those induced by either HA or HAe [45]. In addition, immunization of mice with DNA vaccine expressing the full-length consensus-sequence M2 induced specific antibody responses, protecting mice against challenge of lethal influenza virus [50]. Reports have also indicated that a polyvalent DNA vaccine containing HA and NA genes triggered effective immune responses against homologous and heterologous virus strains, protecting pigs against 2009 H1N1 influenza virus infection [51].
DNA-based influenza vaccines have several advantages. Safety can be attributed to their non-replicating and noninfectious features. Also, such vaccines allow for flexible optimization of viral genetic sequences of the encoding genes and other related components, as well as the ease of scaling up by fermentation techniques [52-55]. However, vaccines based on DNAs suffer from low transduction and expression of multiple genes, the potential of inducing host autoimmunity, and the possible risk of malignancy arising from integration of exogenous DNA into the host genome [56]. Additionally, DNA-based vaccines are less effective in the elderly and newborn [57].
4.3. Viral vector-based influenza vaccines
Influenza vaccines based on viral vectors have been studied extensively with some success [58, 59]. For example, a replication-defective adenovirus vector expressing HA from different subtypes and NP from one subtype induced high humoral and cellular immune responses and conferred cross-protection against challenge from H5, H7 and H9 influenza virus [60]. In addition, intranasal administration of a recombinant adenovirus encoding HA2 of influenza A/California/7/2009(H1N1) virus fused to a trimerized form of murine CD40L enhanced specific mucosal IgA and serum IgG antibodies, induced HA2-specific, CD40L-dependent T-cell responses, and completely protected against lethal challenge of divergent influenza A viruses, including H1N1, H3N2, and H9N2, highlighting the possibility of developing HA2-based universal influenza vaccines [61]. Although adenovirus vector vaccines have a number of merits, potential safety risks arising from the prevalence of pre-existing immunity significantly prevent its use as a promising vaccine candidate.
4.4. VLP-based influenza vaccines
VLPs have been applied as influenza vaccines [62]. VLPs are genome-free particles containing only viral envelope proteins. VLPs can be produced in mammalian cells, yeast and baculovirus expression systems, as well as recombinant vaccinia virus systems [63-65]. They can display viral surface proteins in high density, and present adjuvanticity, resulting in the induction of effective immune responses [62]. Reports have shown that an encapsulated recombinant H5 HA expressed on baculovirus surface induced protective immune response against H5N1 infection in mice and that a VLP containing two M2e peptides fused with HBcAg and B subunit of E.coli heat labile enterotoxin (LTB) significantly enhanced mucosal immune responses in mice [66]. However, since immune responses induced by VLPs might be relatively lower, further application of this vaccine type requires more study.
5. Advancements in the development of subunit influenza vaccines
Based on their rapid, stable, consistent and scalable production, recombinant subunit vaccines have been proven an effective strategy for meeting the demands of a possible influenza pandemic [67]. Compared with other vaccine types, subunit vaccines maintain the highest safety profile by the absence of infectious viruses. Subunit vaccines against influenza viruses with pandemic potential, including H5N1 and H7N9, are under development, among which viral structural proteins, such as M2e, HA and NP, are attractive vaccine targets. Other proteins, such as M1 and NA, also have potential for development as influenza subunit vaccines.
5.1. Subunit vaccines based on conserved M2e proteins
Influenza virus M2 tetramers are expressed at high density in the plasma membrane of infected cells [68]. The extracellular domain of the M2 protein, M2e, which contains 24 residues at the N-terminus, is highly conserved among influenza A viruses [68], in which the N-terminal epitope SLLTEVET (residues 2-9) shows nearly 100% homology in all subtypes of influenza A viruses [69]. Therefore, M2e serves as an attractive target for development of universal influenza subunit vaccines.
It should be noted that immunogenicity induced by a single M2e molecule is relatively low, requiring some modifications to produce effective immune responses. Therefore, construction of protein vaccines containing several M2e molecules is an option to improve the efficacy of M2e vaccines. An E.coli-expressed M2e containing tandem copies of different M2e molecules has been shown to yield better protection against highly pathogenic A/chicken/Guangdong/04 (H5N1) in chickens [70]. Fusing M2e to an appropriate carrier is another approach to improve immunogenicity of M2e-based subunit vaccines. For example, M2e vaccines linked with a multiple antigenic peptide (MAP) induced strong M2e-specific IgG antibodies, protecting immunized mice against lethal challenge with PR8 A/H1N1, 2009 pandemic H1N1, or divergent H5N1 influenza viruses [71-73].
Other strategies, such as fusing M2e with immunogenic protein components, are also under development to improve efficacy induced by M2e-based influenza subunit vaccines. With this vaccination strategy, protein carriers themselves have potential adjuvanticity, thus avoiding the use of strong adjuvants or additional adjuvants during immunization [74, 75], a step which simplifies the immunization process, but achieves similar immune effects. Therefore, this vaccination strategy is more practical, representing a future direction for the development of M2e-based universal subunit vaccines. Studies have also demonstrated that fusion of four tandem copies of M2e molecules with the immunogenic decameric protein Brucella abortus lumazine synthase, BLS-4M2e, resulted in survival rates of 100% and 80% for mice challenged with influenza virus in the presence of Iscomatrix or alum adjuvant, respectively, while 60% of these mice still survived in the absence of such adjuvants [75]. We have also found that a recombinant fusion protein linking three tandem copies of the H5N1 M2e consensus sequence to activation associated protein-1 (ASP-1) adjuvant (M2e3-ASP-1) was able to provide significant M2e-specific immune responses and cross-clade protective immunity against divergent H5N1 viruses without the requirement of additional adjuvants [76].
5.2. Subunit vaccines based on conserved NP protein
The highly conserved influenza virus NP is an internal protein capable of inducing cross-protective immunity against different influenza A viruses, making it an ideal target for developing universal influenza vaccines [59, 77]. A robust CD4+ T cell response was elicited against peptides of two conserved epitopes (NP265-274 and NP174-184) [78], suggesting that these two epitopes may be candidates able to provide partial immunity to pandemic H5N1 virus. NP may also induce specific CD8+ T cell response which correlates with protection [79]. At least 14 human NP peptides have been identified as epitopes of cytotoxic T lymphocytes (CTL) [80]. In addition, immunization with NP plus Ribi Adjuvant System (RAS) could increase humoral and cellular immune responses compared to unadjuvanted NP [81], suggesting that suitable adjuvants will be needed in subunit vaccines based on NP. However, immunity induced by NP might be low, and with the absence of neutralizing activity, it would be unable to induce highly potent protection against virus infection.
5.3. Subunit vaccines based on HA proteins
In addition to highly conserved M2e and NP, which serve as important targets for subunit influenza vaccines, viral surface HA glycoprotein is also an ideal antigen for the induction of protective immune responses against influenza virus infection. Subunit vaccines based on the full-length HA protein have shown their ability to induce protective immunity in preclinical tests and clinical trials [82]. For example, immunization with a baculovirus system-expressed recombinant protein (rH5HA) against the HA of a highly pathogenic classic H5N1 influenza virus protected mice from lethal challenge against pathogenic avian influenza virus, and the serum antibody lasted more than 6 months [83]. Other studies have reported that a plant-produced subunit influenza vaccine, HAC1, based on the recombinant HA of 2009 pandemic A/California/04/2009 (H1N1) strain, demonstrated immunogenicity and safety in Phase I clinical trials of healthy adults [82].
Compared with the full-length HA protein, the HA1 region is more immunogenic in inducing potent neutralizing antibodies and protection against influenza virus infection. As such, it is an important vaccine target. The larger N-terminal fragment, HA1 (~320 aa), forms a membrane-distal globular domain that contains the receptor-binding site and most determinants recognized by virus-neutralizing antibodies [8]. Glycan-masked HA antigens at residues 127 and 138 double mutants have been reported to induce potent neutralizing antibodies and cross-protection against heterologous H5N1 clades [84], providing useful information for the development of a broadly protective H5N1 influenza vaccine. We have also shown that a recombinant HA1 protein of A/Anhui/1/2005(H5N1) fused to the Fc of human IgG and foldon (Fd) trimeric motif (HA1-Fdc) induced strong immune-reactive responses to HA1 protein that resulted in cross-neutralization of H5N1 divergent strains [85]. In particular, we demonstrated that a critical neutralizing domain (CND) containing residues 13-263 of HA1 of A/Anhui/1/2005(H5N1) fused to Fc and Fd, HA-13-263-Fdc, in an optimal conformation elicited the strongest neutralizing antibody response and cross-protection against challenge of heterologous H5N1 strains [86], suggesting that the CND containing the RBD of H5N1 HA1 could be further developed as an effective and safe subunit vaccine against divergent strains of influenza virus from the same group. Because of sequence variation, it should be noted that the HA1 subunit typically induces strain-specific neutralizing antibodies. Thus, its ability to induce broad neutralizing antibodies against variant strains from different groups might be lower.
The influenza virus HA2 subunit is more conserved than the HA1 subunit. Consequently, immune responses induced by the HA2 subunit, particularly the stem region, are expected to elicit broad cross-protective antibodies against divergent strains from different influenza virus groups, suggesting its development as a universal vaccine [87]. Studies have shown that vaccination with a synthetic HA2 peptide-based subunit influenza vaccine protected mice from H1N1, H3N2 and H5N1 influenza virus challenge [88]. E.coli-expressed HA2 proteins were demonstrated to induce immune responses, protecting mice against lethal challenge by A/HK/68 mouse-adapted virus [89]. However, compared with HA1 subunit, HA2 subunit might be less immunogenic, and it cannot elicit strong neutralizing antibodies.
5.4. Subunit vaccines based on other viral proteins
Other proteins, such as M1 and NA, also have potential for application as targets to develop subunit influenza vaccines. While 100% protection was reached by an E.coli-expressed M1 protein formulated with chitosan adjuvant against homologous H9N2 challenge, 70% and 30% of the mice were still protected from challenge of heterologous H1N1 and H5N1 viruses, respectively [90], suggesting that M1 protein may induce broad-spectrum immunity against variant virus strains. Although immunity induced by NA is effective against homologous viruses, it is generally believed that NA contains less cross-reactivity and cross-protective ability against divergent influenza virus strains [91, 92]. However, recent studies have found that a universal monoclonal antibody targeting the conserved epitope in NA (residues 222-230) was able to inhibit all subtypes of influenza A viruses and provide heterosubtypic protection of mice against challenge from group I and II viruses [93]. In addition, NA-inhibiting antibody is correlated with cross-protection of ferrets against lethal H5N1 challenge, and purified heterologous NA protein adjuvanted with Iscomatrix has shown protection [94]. These studies highlight the importance of including NA protein as vaccine components.
5.5. Potential strategies for improving the efficacy of subunit influenza vaccines
Compared with inactivated and live-attenuated vaccine types, the immunogenicity induced by subunit vaccines might be less potent. However, a variety of approaches, such as selection of appropriate adjuvants, fusion of several viral proteins, or combination with different vaccination strategies, have been attempted to improve the immunogenicity and efficacy of subunit influenza vaccines.
In the presence of some specific adjuvants, immune responses and/or neutralization induced by influenza subunit vaccines can be enhanced significantly. For example, a saponin-derived adjuvant, GPI-0100, may improve the immunogenicity and protective efficacy of a mucosal influenza HA subunit vaccine in immunized mice [95]. Moreover, a recombinant cholera toxin B subunit (rCTB) significantly increased serum IgG and mucosal IgA immune responses and serum neutralizing antibody titers of mice intranasally immunized with a recombinant baculovirus surface-displayed HA (BacHA) compared with those from the mice administered with unadjuvanted BacHA [96]. Particularly, a MF59-adjuvanted trivalent subunit vaccine induced persistent and enhanced antibody responses against homologous and heterologous strains of influenza viruses in infants and young children, with significantly higher HI titers than the non-adjuvant vaccines [97].
In addition to adjuvants, fusing several influenza viral proteins or linking influenza viral proteins with other protein carriers may be considered as additional options to improve the immunogenicity of subunit influenza vaccines. These approaches are often revealed in the design of universal influenza vaccines, in which influenza virus M2e molecules are fused with other components to facilitate the formation and maintenance of M2e tetrameric structure and, thus, improve the immunogenicity of M2e. For example, a recombinant protein containing the TLR-5 ligand flagellin fused to four tandem copies of conserved M2e developed potent M2e-specific antibody responses, protecting immunized mice against a lethal influenza virus challenge [98]. In addition, immunization of mice with a fusion protein containing influenza HA1 globular head domain and the core fragment of respiratory syncytial virus G protein (Gcf) induced increased serum IgG and mucosal IgA responses specific to HA1 and Gcf, respectively [99].
An appropriate administration route may help to enhance host immune responses. For example, sublingual administration of a subunit influenza vaccine induces systemic antibody titers, mucosal neutralizing IgA responses, and influenza-specific Th17 cells comparable to those elicited by intramuscular immunization [100]. Other approaches, such as combinations with different vaccination strategies, can be applied to improve the efficacy of subunit vaccines. Optimal systemic antibody and T cell responses were achieved by immunization of mice with a subunit influenza vaccine containing a saponin-derived GPI-0100 adjuvant through intranasal prime-intramuscular boost strategies [101]. DNA prime and protein boost of NP is a practical way of improving cross-protective immunity against influenza A virus infection [102].
6. Clinically trialed and licensed influenza vaccines
Currently, a variety of vaccines are in clinical trials to prevent influenza A virus infection, some of which are licensed for human use [108]. The summary of these vaccines and its current stages are listed in Table 2. A variety of live attenuated and inactivated influenza vaccines are tested in healthy adults, pregnant women and children, and showed immunogenicity to induce specific antibody responses, in which the duration of antibody presences is different among different groups [37,109,110]. It is demonstrated that an inactivated AS03B-adjuvanted H5N1 vaccine was immunogenic in children from 6 months through 17 years old with antibody responses maintaining for up to one year [110]. In contrast, although an inactivated H7N9(A/Shanghai/2/13) influenza vaccine mixed with MF59 adjuvant induced seroconversion in 59% of the clinically trialed adults aged 19-64 years old, its potential application might be limited by the absence of long-term antibody responses or clinical outcomes [111]. Apart from live attenuated and inactivated vaccines, viral vector-based influenza vaccines are tested in clinical trials and show immunogenicity in healthy adults [112, 113]. For example, a modified vaccinia virus Ankara (MVA)-HA-based H5N1 vaccine, MVA-H5-sfMR, induced antibody responses in young adults aged 18-28 year old, with a single dose of higher virus titer resulting in stronger responses than did two immunizations with a lower virus dose. Also, a MVA vector vaccine encoding NP and M1 protein (MVA-NP+M1) was immunogenic in eliciting T cell responses in healthy adults [112, 113]. In addition to the above vaccine types, an increased number of protein-based subunit vaccines under clinical trials has shown good safety, tolerance and efficacy. For example, a plant-expressed recombinant protein, HAC1, which contains HA of A/California/04/2009(H1N1) strain, was shown to be effective in inducing neutralizing antibody responses in immunized healthy adults against virus infection [82]. In another case, HAI-05, a recombinant HA protein from A/Indonesia/05/2005(H5N1) was proven to be safe in healthy adults of 18-49 years old and induce specific anti-H5N1 immune responses [114].
Table 2.
Selected influenza vaccines in clinical trials and their stages
| Clinically trialed vaccines | Vaccine types | Antigen components or encoding antigens |
Stages | References |
|---|---|---|---|---|
| A/Shanghai/2/13(H7N9) | Inactivated virus-based | H7N9 HA | Phase II | [111] |
| A/Indonesia/5/05(H5N1) | Inactivated virus-based | H5N1 HA | Phase 2/3 | [110] |
| A/Vietnam/1203/04(H5N1) A/Indonesia/05/05(H5N1) |
Inactivated virus-based | H5N1 HA | Phase II | [119] |
| A/turkey/Turkey/1/05(H5N1) A/Indonesia/05/05(H5N1) |
Inactivated virus-based | H5N1 HA | Phase I (?) | [120] |
| MVA-H5-sfMR | Viral vector-based | H5N1 HA | Phase 1/2a | [113] |
| MVA-NP+M1 (A/Panama/2007/99(H3N2)) |
Viral vector-based | H3N2 NP and M protein 1 | Phase I | [112] |
| gH1-Qbeta (A/California/07/09(H1N1)) |
VLP-based | 2009H1N1 HA | Phase I | [121] |
| HAC1 (A/California/04/09(H1N1)) |
Subunit vaccine | 2009H1N1 HA | Phase I | [82] |
| HAI-05 (A/Indonesia/05/05(H5N1)) |
Subunit vaccine | H5N1 HA | Phase I | [114] |
A number of clinically trialed influenza vaccines has been licensed to pharmaceutical companies for further development, most of which are inactivated and live attenuated vaccines [115, 116]. For instance, Aflunov, an MF59-adjuvanted, egg-derived H5N1 influenza vaccine, which was proven to be safe and well-tolerated in infants, children, adolescents, adults and the elderly, and to elicit strong immunogenicity against homologous (A/Vietnam/1194/2004) and heterologous (A/Indonesia/05/2005 or A/Turkey/15/2006) viral strains [115, 117], has been licensed to Novartis Vaccines and Diagnostics. Among all licensed influenza vaccines, Flublok is the first recombinant vaccine approved by the US FDA and licensed to Protein Sciences Corporation (PSC). This insect cell-expressed, HA protein-based vaccine can be scaled up for manufacturing production [118], providing strong basis for rapid development of subunit vaccines against pandemic influenza.
7. Conclusions and perspectives
The continuing threat of pandemic arising from highly pathogenic avian influenza virus H5N1, newly emerged avian influenza virus H7N9, and other virulent influenza A viruses highlights the urgent need to develop safe and effective subunit vaccines. Compared with other vaccine types, influenza viral protein-based subunit vaccines maintain the highest safety profile. It is expected that broad cross-protective universal subunit influenza vaccines will be developed based on the highly conserved sequences of M2e, NP and stem region of HA2, and that more and more subunit vaccines will be in clinical trials and licensed for human use. Novel strategies in designing and improving efficacy of subunit vaccines are anticipated to reduce the threat of future influenza pandemics.
Acknowledgements
This study was supported by grants from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (R21AI111152 to LD) and from the Chinese Ministry of Science & Technology, Hong Kong, Macau, Taiwan Collaborative Program (201200007673 to SJ).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The authors declared no conflict of interest.
References
- [1].Cox NJ, Neumann G, Donis RO, Kawaoka Y. Orthomyxoviruses: Influenza. Topley and Wilson's Microbiology and Microbial Infections. 2010 DOI: 10.1002/9780470688618.taw0238. [Google Scholar]
- [2].Le TH, Nguyen NT. Evolutionary dynamics of highly pathogenic avian influenza A/H5N1 HA clades and vaccine implementation in Vietnam. Clin Exp Vaccine Res. 2014;3:117–27. doi: 10.7774/cevr.2014.3.2.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Webby RJ, Webster RG. Are we ready for pandemic influenza? Science. 2003;302:1519–22. doi: 10.1126/science.1090350. [DOI] [PubMed] [Google Scholar]
- [4].Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med. 2013;368:1888–97. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- [5].Steinhauer DA. Role of hemagglutinin cleavage for the pathogenicity of influenza virus. Virology. 1999;258:1–20. doi: 10.1006/viro.1999.9716. [DOI] [PubMed] [Google Scholar]
- [6].Samji T. Influenza A: understanding the viral life cycle. Yale J Bio Med. 2009;82:153–9. [PMC free article] [PubMed] [Google Scholar]
- [7].Nayak DP, Hui E, Barman S. Assembly and budding of influenza virus. Virus Res. 2004;106:147–65. doi: 10.1016/j.virusres.2004.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Gerhard W, Mozdzanowska K, Zharikova D. Prospects for universal influenza virus vaccine. Emerg Infect Dis. 2006;12:569–74. doi: 10.3201/eid1204.051020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].O'Neill RE, Talon J, Palese P. The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J. 1998;17:288–96. doi: 10.1093/emboj/17.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Noah DL, Krug RM. Influenza virus virulence and its molecular determinants. Adv Virus Res. 2005;65:121–45. doi: 10.1016/S0065-3527(05)65004-X. [DOI] [PubMed] [Google Scholar]
- [11].Lee MTM, Klumpp K, Digard P, Tiley L. Activation of influenza virus RNA polymerase by the 5′ and 3′ terminal duplex of genomic RNA. Nucleic Acids Res. 2003;31:1624–32. doi: 10.1093/nar/gkg253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Nguyen-Van-Tam JS, Hampson AW. The epidemiology and clinical impact of pandemic influenza. Vaccine. 2003;21:1762–8. doi: 10.1016/s0264-410x(03)00069-0. [DOI] [PubMed] [Google Scholar]
- [13].Taubenberger JK, Morens DM. 1918 Influenza: the mother of all pandemics. Emerg Infect Dis. 2006;12:15–22. doi: 10.3201/eid1201.050979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Reid AH, Fanning TG, Hultin JV, Taubenberger JK. Origin and evolution of the 1918 “Spanish” influenza virus hemagglutinin gene. Proc Natl Acad Sci USA. 1999;96:1651–6. doi: 10.1073/pnas.96.4.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Reid AH, Fanning TG, Janczewski TA, Taubenberger JK. Characterization of the 1918 “Spanish” influenza virus neuraminidase gene. Proc Natl Acad Sci USA. 2000;97:6785–90. doi: 10.1073/pnas.100140097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Reid A, Fanning T, Slemons R, Janczewski T, Dean J, Taubenberger J. Relationship of pre-1918 avian influenza HA and NP sequences to subsequent avian influenza strains. Avian Dis. 2003;47:921–5. doi: 10.1637/0005-2086-47.s3.921. [DOI] [PubMed] [Google Scholar]
- [17].Scholtissek C, Rohde W, Von Hoyningen V, Rott R. On the origin of the human influenza virus subtypes H2N2 and H3N2. Virology. 1978;87:13–20. doi: 10.1016/0042-6822(78)90153-8. [DOI] [PubMed] [Google Scholar]
- [18].Kawaoka Y, Krauss S, Webster RG. Avian-to-human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics. J Virol. 1989;63:4603–8. doi: 10.1128/jvi.63.11.4603-4608.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Taubenberger JK, Morens DM. Influenza: the once and future pandemic. Public Health Rep. 2010;125:16–26. [PMC free article] [PubMed] [Google Scholar]
- [20].Kilbourne ED. Perspectives on pandemics: a research agenda. J Infec Dis. 1997;176:S29–31. doi: 10.1086/514171. [DOI] [PubMed] [Google Scholar]
- [21].Neumann G, Noda T, Kawaoka Y. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature. 2009;459:931–9. doi: 10.1038/nature08157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, Balish A, et al. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science. 2009;325:197–201. doi: 10.1126/science.1176225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Dunham EJ, Dugan VG, Kaser EK, Perkins SE, Brown IH, Holmes EC, et al. Different evolutionary trajectories of European avian-like and classical swine H1N1 influenza A viruses. J Virol. 2009;83:5485–94. doi: 10.1128/JVI.02565-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Morens DM, Taubenberger JK, Fauci AS. The persistent legacy of the 1918 influenza virus. N Engl J Med. 2009;361:225–9. doi: 10.1056/NEJMp0904819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zhou NN, Senne DA, Landgraf JS, Swenson SL, Erickson G, Rossow K, et al. Genetic reassortment of avian, swine, and human influenza A viruses in American pigs. J Virol. 1999;73:8851–6. doi: 10.1128/jvi.73.10.8851-8856.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Olsen CW. The emergence of novel swine influenza viruses in North America. Virus Res. 2002;85:199–210. doi: 10.1016/s0168-1702(02)00027-8. [DOI] [PubMed] [Google Scholar]
- [27].Pensaert M, Ottis K, Vandeputte J, Kaplan MM, Bachmann PA. Evidence for the natural transmission of influenza A virus from wild ducts to swine and its potential importance for man. Bull World Health Organ. 1981;59:75–8. [PMC free article] [PubMed] [Google Scholar]
- [28].Swayne D, Suarez D. Highly pathogenic avian influenza. Revue scientifique et technique (International Office of Epizootics) 2000;19:463–82. doi: 10.20506/rst.19.2.1230. [DOI] [PubMed] [Google Scholar]
- [29].Xu X, Subbarao K, Cox NJ, Guo Y. Genetic characterization of the pathogenic influenza A/Goose/Guangdong/1/96 (H5N1) virus: similarity of its hemagglutinin gene to those of H5N1 viruses from the 1997 outbreaks in Hong Kong. Virology. 1999;261:15–9. doi: 10.1006/viro.1999.9820. [DOI] [PubMed] [Google Scholar]
- [30].Swayne DE, Beck JR, Perdue ML, Beard CW. Efficacy of vaccines in chickens against highly pathogenic Hong Kong H5N1 avian influenza. Avian Dis. 2001:355–65. [PubMed] [Google Scholar]
- [31].Meijer A, Jonges M, Abbink F, Ang W, van Beek J, Beersma M, et al. Oseltamivir-resistant pandemic A(H1N1) 2009 influenza viruses detected through enhanced surveillance in the Netherlands, 2009-2010. Antiviral Res. 2011;92:81–9. doi: 10.1016/j.antiviral.2011.07.004. [DOI] [PubMed] [Google Scholar]
- [32].Shin SY, Kang C, Gwack J, Kim JH, Kim HS, Kang YA, et al. Drug-resistant pandemic (H1N1) 2009, South Korea. Emerg Infect Dis. 2011;17:702–4. doi: 10.3201/eid1704.101467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hurt AC, Holien JK, Parker MW, Barr IG. Oseltamivir resistance and the H274Y neuraminidase mutation in seasonal, pandemic and highly pathogenic influenza viruses. Drugs. 2009;69:2523–31. doi: 10.2165/11531450-000000000-00000. [DOI] [PubMed] [Google Scholar]
- [34].Lavrov SV, Podchernyayeva RY, Blinova VK, Sokolov MI. Isolation and properties of amantadine-resistant variants of influenza A virus. Acta Virol. 1972;16:507. [PubMed] [Google Scholar]
- [35].Pabbaraju K, Wong S, Kits DK, Fox JD. Adamantane resistance in seasonal human influenza A viruses from Calgary, Alberta (January 2007 to August 2008) Can J Infect Dis Med Microbiol. 2010;21:e87–91. doi: 10.1155/2010/710149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Horimoto T, Kawaoka Y. Designing vaccines for pandemic influenza. Curr Top Microbiol Immunol. 2009;333:165–76. doi: 10.1007/978-3-540-92165-3_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Ilyushina NA, Haynes BC, Hoen AG, Khalenkov AM, Housman ML, Brown EP, et al. Live attenuated and inactivated influenza vaccines in children. J Infect Dis. 2014:jiu458. doi: 10.1093/infdis/jiu458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ambrose CS, Wu X, Caspard H, Belshe RB. Efficacy of live attenuated influenza vaccine against influenza illness in children as a function of illness severity. Vaccine. 2014;32:5546–8. doi: 10.1016/j.vaccine.2014.07.097. [DOI] [PubMed] [Google Scholar]
- [39].Peeters B, Tonnis WF, Murugappan S, Rottier P, Koch G, Frijlink HW, et al. Pulmonary immunization of chickens using non-adjuvanted spray-freeze dried whole inactivated virus vaccine completely protects against highly pathogenic H5N1 avian influenza virus. Vaccine. 2014 doi: 10.1016/j.vaccine.2014.09.048. pii: S0264-410X(14)01316-4. [DOI] [PubMed] [Google Scholar]
- [40].Richt JA, García-Sastre A. Attenuated influenza virus vaccines with modified NS1 proteins. Curr Top Microbiol Immunol. 2009;333:177–95. doi: 10.1007/978-3-540-92165-3_9. [DOI] [PubMed] [Google Scholar]
- [41].Overton ET, Goepfert PA, Cunningham P, Carter WA, Horvath J, Young D, Strayer DR. Intranasal seasonal influenza vaccine and a TLR-3 agonist, rintatolimod, induced cross-reactive IgA antibody formation against avian H5N1 and H7N9 influenza HA in humans. Vaccine. 2014;32:5490–5. doi: 10.1016/j.vaccine.2014.07.078. [DOI] [PubMed] [Google Scholar]
- [42].Pena L, Sutton T, Chockalingam A, Kumar S, Angel M, Shao H, et al. Influenza viruses with rearranged genomes as live-attenuated vaccines. J Virol. 2013;87:5118–27. doi: 10.1128/JVI.02490-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Petricciani JC. Regulatory philosophy and acceptability of cells for the production of biologicals. Dev Biol Stand. 1991;75:9–15. [PubMed] [Google Scholar]
- [44].Kistner O, Barrett P, Mundt W, Reiter M, Schober-Bendixen S, Dorner F. Development of a mammalian cell (Vero) derived candidate influenza virus vaccine. Vaccine. 1998;16:960–8. doi: 10.1016/s0264-410x(97)00301-0. [DOI] [PubMed] [Google Scholar]
- [45].Chen J, Liu Q, Chen Q, Xiong C, Yao Y, Wang H, et al. Comparative analysis of antibody induction and protection against influenza virus infection by DNA immunization with HA, HAe, and HA1 in mice. Arch Virol. 2014;159:689–700. doi: 10.1007/s00705-013-1878-1. [DOI] [PubMed] [Google Scholar]
- [46].Xu K, Ling ZY, Sun L, Xu Y, Bian C, He Y, et al. Broad humoral and cellular immunity elicited by a bivalent DNA vaccine encoding HA and NP genes from an H5N1 virus. Viral Immunol. 2011;24:45–56. doi: 10.1089/vim.2010.0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Kodihalli S, Haynes JR, Robinson HL, Webster RG. Cross-protection among lethal H5N2 influenza viruses induced by DNA vaccine to the hemagglutinin. J Virol. 1997;71:3391–6. doi: 10.1128/jvi.71.5.3391-3396.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Kodihalli S, Goto H, Kobasa DL, Krauss S, Kawaoka Y, Webster RG. DNA vaccine encoding hemagglutinin provides protective immunity against H5N1 influenza virus infection in mice. J Virol. 1999;73:2094–8. doi: 10.1128/jvi.73.3.2094-2098.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Park KS, Seo YB, Lee JY, Im SJ, Seo SH, Song MS, Choi YK, Sung YC. Complete protection against a H5N2 avian influenza virus by a DNA vaccine expressing a fusion protein of H1N1 HA and M2e. Vaccine. 2011;29:5481–7. doi: 10.1016/j.vaccine.2011.05.062. [DOI] [PubMed] [Google Scholar]
- [50].Tompkins SM, Zhao ZS, Lo CY, Misplon JA, Liu T, Ye Z, et al. Matrix protein 2 vaccination and protection against influenza viruses, including subtype H5N1. Emerg Infect Dis. 2007;13:426–35. doi: 10.3201/eid1303.061125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Bragstad K, Vinner L, Hansen MS, Nielsen J, Fomsgaard A. A polyvalent influenza A DNA vaccine induces heterologous immunity and protects pigs against pandemic A (H1N1) pdm09 virus infection. Vaccine. 2013;31:2281–8. doi: 10.1016/j.vaccine.2013.02.061. [DOI] [PubMed] [Google Scholar]
- [52].Shamlou PA. Scaleable processes for the manufacture of therapeutic quantities of plasmid DNA. Biotechnol Appl Biochem. 2003;37:207–18. doi: 10.1042/BA20030011. [DOI] [PubMed] [Google Scholar]
- [53].Prather KJ, Sagar S, Murphy J, Chartrain M. Industrial scale production of plasmid DNA for vaccine and gene therapy: plasmid design, production, and purification. Enzyme Micro Technol. 2003;33:865–83. [Google Scholar]
- [54].Hoare M, Levy MS, Bracewell DG, Doig SD, Kong S, Titchener-Hooker N, et al. Bioprocess engineering issues that would be faced in producing a DNA vaccine at up to 100 m3 fermentation scale for an influenza pandemic. Biotechnol Prog. 2005;21:1577–92. doi: 10.1021/bp050190n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Moss RB. Prospects for control of emerging infectious diseases with plasmid DNA vaccines. J Immune Based Ther Vaccines. 2009;7:3. doi: 10.1186/1476-8518-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Shen Y, Muramatsu SI, Ikeguchi K, Fujimoto KI, Fan DS, Ogawa M, et al. Triple transduction with adeno-associated virus vectors expressing tyrosine hydroxylase, aromatic-L-amino-acid decarboxylase, and GTP cyclohydrolase I for gene therapy of Parkinson's disease. Hum Gene Ther. 2000;11:1509–19. doi: 10.1089/10430340050083243. [DOI] [PubMed] [Google Scholar]
- [57].Klinman DM, Conover J, Bloom ET, Weiss W. Immunogenicity and efficacy of a DNA vaccine in aged mice. J Gerontol A Biol Sciences Med Sci. 1998;53:B281–6. doi: 10.1093/gerona/53a.4.b281. [DOI] [PubMed] [Google Scholar]
- [58].Kim SH, Kim JY, Choi Y, Nguyen HH, Song MK, Chang J. Mucosal vaccination with recombinant adenovirus encoding nucleoprotein provides potent protection against influenza virus infection. PloS One. 2013;8:e75460. doi: 10.1371/journal.pone.0075460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Li Z, Gabbard JD, Mooney A, Gao X, Chen Z, Place RJ, et al. Single-dose vaccination of a recombinant parainfluenza virus 5 expressing NP from H5N1 virus provides broad immunity against influenza A viruses. J Virol. 2013;87:5985–93. doi: 10.1128/JVI.00120-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Vemula SV, Ahi YS, Swaim AM, Katz JM, Donis R, Sambhara S, et al. Broadly protective adenovirus-based multivalent vaccines against highly pathogenic avian influenza viruses for pandemic preparedness. PloS One. 2013;8:e62496. doi: 10.1371/journal.pone.0062496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Fan X, Hashem AM, Chen Z, Li C, Doyle T, Zhang Y, et al. Targeting the HA2 subunit of influenza A virus hemagglutinin via CD40L provides universal protection against diverse subtypes. Mucosal Immunol. 2015;8:211–20. doi: 10.1038/mi.2014.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Chroboczek J, Szurgot I, Szolajska E. Virus-like particles as vaccine. Acta Biochim Pol. 2013;61:531–9. [PubMed] [Google Scholar]
- [63].Ali A, Avalos RT, Ponimaskin E, Nayak DP. Influenza virus assembly: effect of influenza virus glycoproteins on the membrane association of M1 protein. J Virol. 2000;74:8709–19. doi: 10.1128/jvi.74.18.8709-8719.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Zheng J, Ma J, Yang XF, Liu HL, Cheng HW, Si LS, et al. Highly efficient and economical baculovirus expression system for preparing human papillomavirus type16 virus-like particle. Acta Biochim Biophy Sin (Shanghai) 2004;36:548–52. doi: 10.1093/abbs/36.8.548. [DOI] [PubMed] [Google Scholar]
- [65].Green K, Lew J, Jiang X, Kapikian A, Estes M. Comparison of the reactivities of baculovirus-expressed recombinant Norwalk virus capsid antigen with those of the native Norwalk virus antigen in serologic assays and some epidemiologic observations. J Clin Microbiol. 1993;31:2185–91. doi: 10.1128/jcm.31.8.2185-2191.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Prabakaran M, Madhan S, Prabhu N, Geng GY, New R, Kwang J. Reverse micelle-encapsulated recombinant baculovirus as an oral vaccine against H5N1 infection in mice. Antiviral Res. 2010;86:180–7. doi: 10.1016/j.antiviral.2010.02.315. [DOI] [PubMed] [Google Scholar]
- [67].Mallajosyula JK, Hiatt E, Hume S, Johnson A, Jeevan T, Chikwamba R, et al. Single-dose monomeric HA subunit vaccine generates full protection from influenza challenge. Hum Vaccin Immunother. 2014;10:586–95. doi: 10.4161/hv.27567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Zebedee SL, A LR. Influenza A virus M2 protein: monoclonal antibody restriction of virus growth and detection of M2 in virions. J Virol. 1988;62:2762–72. doi: 10.1128/jvi.62.8.2762-2772.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Fiers W, De Filette M, Birkett A, Neirynck S, Min Jou W. A “universal” human influenza A vaccine. Virus Res. 2004;103:173–6. doi: 10.1016/j.virusres.2004.02.030. [DOI] [PubMed] [Google Scholar]
- [70].Zhang X, Liu M, Liu C, Du J, Shi W, Sun E, et al. Vaccination with different M2e epitope densities confers partial protection against H5N1 influenza A virus challenge in chickens. Intervirology. 2011;54:290–9. doi: 10.1159/000319440. [DOI] [PubMed] [Google Scholar]
- [71].Ma JH, Yang FR, Yu H, Zhou YJ, Li GX, Huang M, et al. An M2e-based synthetic peptide vaccine for influenza A virus confers heterosubtypic protection from lethal virus challenge. Virol J. 2013;10:227. doi: 10.1186/1743-422X-10-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Zhao G, Sun S, Du L, Xiao W, Ru Z, Kou Z, et al. An H5N1 M2e-based multiple antigenic peptide vaccine confers heterosubtypic protection from lethal infection with pandemic 2009 H1N1 virus. Virol J. 2010;7:151. doi: 10.1186/1743-422X-7-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Zhao G, Lin Y, Du L, Guan J, Sun S, Sui H, et al. An M2e-based multiple antigenic peptide vaccine protects mice from lethal challenge with divergent H5N1 influenza viruses. Virol J. 2010;7:9. doi: 10.1186/1743-422X-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Ebrahimi SM, Dabaghian M, Tebianian M, Zabeh Jazi MH. In contrast to conventional inactivated influenza vaccines, 4xM2e. HSP70c fusion protein fully protected mice against lethal dose of H1, H3 and H9 influenza A isolates circulating in Iran. Virology. 2012;430:63–72. doi: 10.1016/j.virol.2012.04.015. [DOI] [PubMed] [Google Scholar]
- [75].Alvarez P, Zylberman V, Ghersi G, Boado L, Palacios C, Goldbaum F, et al. Tandem repeats of the extracellular domain of Matrix 2 influenza protein exposed in Brucella lumazine synthase decameric carrier molecule induce protection in mice. Vaccine. 2013;31:806–12. doi: 10.1016/j.vaccine.2012.11.072. [DOI] [PubMed] [Google Scholar]
- [76].Zhao G, Du L, Xiao W, Sun S, Lin Y, Chen M, et al. Induction of protection against divergent H5N1 influenza viruses using a recombinant fusion protein linking influenza M2e to Onchocerca volvulus activation associated protein-1 (ASP-1) adjuvant. Vaccine. 2010;28:7233–40. doi: 10.1016/j.vaccine.2010.08.049. [DOI] [PubMed] [Google Scholar]
- [77].Schotsaert M, De Filette M, Fiers W, Saelens X. Universal M2 ectodomain-based influenza A vaccines: preclinical and clinical developments. Expert Rev Vaccines. 2009;8:499–508. doi: 10.1586/erv.09.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Roti M, Yang J, Berger D, Huston L, James EA, Kwok WW. Healthy human subjects have CD4+ T cells directed against H5N1 influenza virus. J Immunol. 2008;180:1758–68. doi: 10.4049/jimmunol.180.3.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].MacLeod MK, David A, Jin N, Noges L, Wang J, Kappler JW, et al. Influenza nucleoprotein delivered with aluminium salts protects mice from an influenza A virus that expresses an altered nucleoprotein sequence. PloS One. 2013;8:e61775. doi: 10.1371/journal.pone.0061775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Berkhoff E, De Wit E, Geelhoed-Mieras M, Boon A, Symons J, Fouchier R, et al. Functional constraints of influenza A virus epitopes limit escape from cytotoxic T lymphocytes. J Virol. 2005;79:11239–46. doi: 10.1128/JVI.79.17.11239-11246.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Cargnelutti DE, Sanchez M, Alvarez P, Boado L, Mattion N, Scodeller EA. Enhancement of Th1 immune responses to recombinant influenza nucleoprotein by Ribi adjuvant. New Microbiol. 2013;36:145–51. [PubMed] [Google Scholar]
- [82].Cummings JF, Guerrero ML, Moon JE, Waterman P, Nielsen RK, Jefferson S, et al. Safety and immunogenicity of a plant-produced recombinant monomer hemagglutinin-based influenza vaccine derived from influenza A (H1N1) pdm09 virus: A Phase 1 dose-escalation study in healthy adults. Vaccine. 2014;32:2251–9. doi: 10.1016/j.vaccine.2013.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Liu G, Zhang F, Shi J, Tian G, Chen H, Yu K, et al. A subunit vaccine candidate derived from a classic H5N1 avian influenza virus in China protects fowls and BALB/c mice from lethal challenge. Vaccine. 2013;31:5398–404. doi: 10.1016/j.vaccine.2013.09.009. [DOI] [PubMed] [Google Scholar]
- [84].Lin SC, Liu WC, Jan JT, Wu SC. Glycan masking of hemagglutinin for adenovirus vector and recombinant protein immunizations elicits broadly neutralizing antibodies against H5N1 avian influenza viruses. PloS One. 2014;9:e92822. doi: 10.1371/journal.pone.0092822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Du L, Zhang X, Zhou J, Chen M, He W, Zhang HY, et al. A recombinant vaccine of H5N1 HA1 fused with foldon and human IgG Fc induced complete cross-clade protection against divergent H5N1 viruses. PloS One. 2011;6:e16555. doi: 10.1371/journal.pone.0016555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Du L, Zhao G, Sun S, Zhang X, Zhou X, Guo Y, et al. A critical HA1 neutralizing domain of H5N1 influenza in an optimal conformation induces strong cross-protection. PloS One. 2013;8:e53568. doi: 10.1371/journal.pone.0053568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Khanna M, Sharma S, Kumar B, Rajput R. Protective immunity based on the conserved hemagglutinin stalk domain and its prospects for universal influenza vaccine development. Biomed Res Int. 2014;2014:546274. doi: 10.1155/2014/546274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Wang TT, Tan GS, Hai R, Pica N, Ngai L, Ekiert DC, et al. Vaccination with a synthetic peptide from the influenza virus hemagglutinin provides protection against distinct viral subtypes. Proc Natl Acad Sci USA. 2010;107:18979–84. doi: 10.1073/pnas.1013387107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Bommakanti G, Citron MP, Hepler RW, Callahan C, Heidecker GJ, Najar TA, et al. Design of an HA2-based Escherichia coli expressed influenza immunogen that protects mice from pathogenic challenge. Proc Natl Acad Sci USA. 2010;107:13701–6. doi: 10.1073/pnas.1007465107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Sui Z, Chen Q, Fang F, Zheng M, Chen Z. Cross-protection against influenza virus infection by intranasal administration of M1-based vaccine with chitosan as an adjuvant. Vaccine. 2010;28:7690–8. doi: 10.1016/j.vaccine.2010.09.019. [DOI] [PubMed] [Google Scholar]
- [91].Sylte MJ, Suarez DL. Influenza neuraminidase as a vaccine antigen. Curr Top Microbiol Immunol. 2009;333:227–41. doi: 10.1007/978-3-540-92165-3_12. [DOI] [PubMed] [Google Scholar]
- [92].Lu X, Liu F, Zeng H, Sheu T, Achenbach JE, Veguilla V, et al. Evaluation of the antigenic relatedness and cross-protective immunity of the neuraminidase between human influenza A (H1N1) virus and highly pathogenic avian influenza A (H5N1) virus. Virology. 2014;454:169–75. doi: 10.1016/j.virol.2014.02.011. [DOI] [PubMed] [Google Scholar]
- [93].Doyle TM, Hashem AM, Li C, Van Domselaar G, Larocque L, Wang J, et al. Universal anti-neuraminidase antibody inhibiting all influenza A subtypes. Antiviral Res. 2013;100:567–74. doi: 10.1016/j.antiviral.2013.09.018. [DOI] [PubMed] [Google Scholar]
- [94].Rockman S, Brown LE, Barr IG, Gilbertson B, Lowther S, Kachurin A, et al. Neuraminidase-inhibiting antibody is a correlate of cross-protection against lethal H5N1 influenza virus in ferrets immunized with seasonal influenza vaccine. J Virol. 2013;87:3053–61. doi: 10.1128/JVI.02434-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Liu H, Patil HP, de Vries-Idema J, Wilschut J, Huckriede A. Enhancement of the immunogenicity and protective efficacy of a mucosal influenza subunit vaccine by the saponin adjuvant GPI-0100. PloS One. 2012;7:e52135. doi: 10.1371/journal.pone.0052135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Prabakaran M, Velumani S, He F, Karuppannan AK, Geng GY, Yin LK, et al. Protective immunity against influenza H5N1 virus challenge in mice by intranasal co-administration of baculovirus surface-displayed HA and recombinant CTB as an adjuvant. Virology. 2008;380:412–20. doi: 10.1016/j.virol.2008.08.002. [DOI] [PubMed] [Google Scholar]
- [97].Nolan T, Bravo L, Ceballos A, Mitha E, Gray G, Quiambao B, et al. Enhanced and persistent antibody response against homologous and heterologous strains elicited by a MF59®-adjuvanted influenza vaccine in infants and young children. Vaccine. 2014;32:6146–56. doi: 10.1016/j.vaccine.2014.08.068. [DOI] [PubMed] [Google Scholar]
- [98].Huleatt JW, Nakaar V, Desai P, Huang Y, Hewitt D, Jacobs A, et al. Potent immunogenicity and efficacy of a universal influenza vaccine candidate comprising a recombinant fusion protein linking influenza M2e to the TLR5 ligand flagellin. Vaccine. 2008;26:201–14. doi: 10.1016/j.vaccine.2007.10.062. [DOI] [PubMed] [Google Scholar]
- [99].Park MH, Chang J. Immunogenicity and protective efficacy of a dual subunit vaccine against respiratory syncytial virus and influenza virus. Immune Netw. 2012;12:261–8. doi: 10.4110/in.2012.12.6.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Gallorini S, Taccone M, Bonci A, Nardelli F, Casini D, Bonificio A, et al. Sublingual immunization with a subunit influenza vaccine elicits comparable systemic immune response as intramuscular immunization, but also induces local IgA and TH17 responses. Vaccine. 2014;32:2382–8. doi: 10.1016/j.vaccine.2013.12.043. [DOI] [PubMed] [Google Scholar]
- [101].Liu H, Patil HP, de Vries-Idema J, Wilschut J, Huckriede A. Evaluation of mucosal and systemic immune responses elicited by GPI-0100-adjuvanted influenza vaccine delivered by different immunization strategies. PloS One. 2013;8:e69649. doi: 10.1371/journal.pone.0069649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Luo J, Zheng D, Zhang W, Fang F, Wang H, Sun Y, et al. Induction of cross-protection against influenza A virus by DNA prime-intranasal protein boost strategy based on nucleoprotein. Virol J. 2012;9:286. doi: 10.1186/1743-422X-9-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Kim MC, Lee YN, Ko EJ, Lee JS, Kwon YM, Hwang HS, et al. Supplementation of influenza split vaccines with conserved M2 ectodomains overcomes strain specificity and provides long-term cross protection. Mol Ther. 2014;22:1364–74. doi: 10.1038/mt.2014.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Kim EH, Lee JH, Pascua P, Song MS, Baek YH, Kwon H, et al. Prokaryote-expressed M2e protein improves H9N2 influenza vaccine efficacy and protection against lethal influenza a virus in mice. Virol J. 2013;10:104. doi: 10.1186/1743-422X-10-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Fiers W, De Filette M, Bakkouri KE, Schepens B, Roose K, Schotsaert M, et al. M2e-based universal influenza A vaccine. Vaccine. 2009;27:6280–3. doi: 10.1016/j.vaccine.2009.07.007. [DOI] [PubMed] [Google Scholar]
- [106].Nayak B, Rout SN, Kumar S, Khalil MS, Fouda MM, Ahmed LE, et al. Immunization of chickens with Newcastle disease virus expressing H5 hemagglutinin protects against highly pathogenic H5N1 avian influenza viruses. PLoS One. 2009;4:e6509. doi: 10.1371/journal.pone.0006509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Wu Y, Qiao C, Yang H, Chen Y, Xin X, Chen H. Immunogenicity and efficacy of a recombinant adenovirus expressing hemagglutinin from the H5N1 subtype of swine influenza virus in mice. Can J Vet Res. 2014;78:117–26. [PMC free article] [PubMed] [Google Scholar]
- [108].Belshe RB, Frey SE, Graham IL, Anderson EL, Jackson LA, Spearman P, et al. Immunogenicity of avian influenza A/Anhui/01/2005(H5N1) vaccine with MF59 adjuvant: a randomized clinical trial. JAMA. 2014;312:1420–8. doi: 10.1001/jama.2014.12609. [DOI] [PubMed] [Google Scholar]
- [109].Madhi SA, Cutland CL, Kuwanda L, Weinberg A, Hugo A, Jones S, et al. Influenza vaccination of pregnant women and protection of their infants. N Engl J Med. 2014;371:918–31. doi: 10.1056/NEJMoa1401480. [DOI] [PubMed] [Google Scholar]
- [110].Kosalaraksa P, Jeanfreau R, Frenette L, Drame M, Madariaga M, Innis BL, et al. AS03B-Adjuvanted H5N1 Influenza Vaccine in Children 6 Months Through 17 Years of Age: A Phase 2/3 Randomized, Placebo-Controlled, Observer-Blinded Trial. J Infect Dis. 2014;pii:jiu548. doi: 10.1093/infdis/jiu548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Mulligan MJ, Bernstein DI, Winokur P, Rupp R, Anderson E, Rouphael N, et al. Serological responses to an avian influenza A/H7N9 vaccine mixed at the point-of-use with MF59 adjuvant: a randomized clinical trial. JAMA. 2014;312:1409–19. doi: 10.1001/jama.2014.12854. [DOI] [PubMed] [Google Scholar]
- [112].Berthoud TK1, Hamill M, Lillie PJ, Hwenda L, Collins KA, Ewer KJ, et al. Potent CD8+ T-cell immunogenicity in humans of a novel heterosubtypic influenza A vaccine, MVA-NP+M1. Clin Infect Dis. 2011;52:1–7. doi: 10.1093/cid/ciq015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Kreijtz JH, Goeijenbier M, Moesker FM, van den Dries L, Goeijenbier S, De Gruyter HL, et al. Safety and immunogenicity of a modified-vaccinia-virus-Ankara-based influenza A H5N1 vaccine: a randomised, double-blind phase 1/2a clinical trial. Lancet Infect Dis. 2014;14:1196–207. doi: 10.1016/S1473-3099(14)70963-6. [DOI] [PubMed] [Google Scholar]
- [114].Chichester JA1, Jones RM, Green BJ, Stow M, Miao F, Moonsammy G, et al. Safety and immunogenicity of a plant-produced recombinant hemagglutinin-based influenza vaccine (HAI-05) derived from A/Indonesia/05/2005 (H5N1) influenza virus: a phase 1 randomized, double-blind, placebo-controlled, dose-escalation study in healthy adults. Viruses. 2012;4:3227–44. doi: 10.3390/v4113227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Del Giudice G1, Fragapane E, Della Cioppa G, Rappuoli R. Aflunov®: a vaccine tailored for pre-pandemic and pandemic approaches against influenza. Expert Opin Biol Ther. 2013;13:121–35. doi: 10.1517/14712598.2013.748030. [DOI] [PubMed] [Google Scholar]
- [116].Isai A, Durand J, Le Meur S, Hidalgo-Simon A, Kurz X. Autoimmune disorders after immunisation with Influenza A/H1N1 vaccines with and without adjuvant: EudraVigilance data and literature review. Vaccine. 2012;30:7123–9. doi: 10.1016/j.vaccine.2012.09.032. [DOI] [PubMed] [Google Scholar]
- [117].Gasparini R, Amicizia D, Lai PL, Panatto D. Aflunov(®): a prepandemic influenza vaccine. Expert Rev Vaccines. 2012;11:145–57. doi: 10.1586/erv.11.170. [DOI] [PubMed] [Google Scholar]
- [118].Buckland B, Boulanger R, Fino M, Srivastava I, Holtz K, Khramtsov N, et al. Technology transfer and scale-up of the Flublok recombinant hemagglutinin (HA) influenza vaccine manufacturing process. Vaccine. 2014;32:5496–502. doi: 10.1016/j.vaccine.2014.07.074. [DOI] [PubMed] [Google Scholar]
- [119].Belshe RB, Frey SE, Graham I, Mulligan MJ, Edupuganti S, Jackson LA, et al. Safety and immunogenicity of influenza A H5 subunit vaccines: effect of vaccine schedule and antigenic variant. J Infect Dis. 2011;203:666–73. doi: 10.1093/infdis/jiq093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Langley JM, Frenette L, Jeanfreau R, Halperin SA, Kyle M, Chu L, et al. Immunogenicity of heterologous H5N1 influenza booster vaccination 6 or 18 months after primary vaccination in adults: A randomized controlled clinical trial. Vaccine. 2014 doi: 10.1016/j.vaccine.2014.11.018. doi: 10.1016/j.vaccine.2014.11.018. [DOI] [PubMed] [Google Scholar]
- [121].Low JG, Lee LS, Ooi EE, Ethirajulu K, Yeo P, Matter A, et al. Safety and immunogenicity of a virus-like particle pandemic influenza A (H1N1) 2009 vaccine: results from a double-blinded, randomized Phase I clinical trial in healthy Asian volunteers. Vaccine. 2014;32:5041–8. doi: 10.1016/j.vaccine.2014.07.011. [DOI] [PubMed] [Google Scholar]