Abstract
The Arf tumor suppressor represents one of several genes encoded at the Cdkn2a and Cdkn2b loci in the mouse. Beyond its role blunting the growth of incipient cancer cells, the Arf gene also plays an essential role in development: Its gene product, p19Arf, is induced by Tgfβ2 in the developing eye to dampen proliferative signals from Pdgfrβ, which effect ultimately fosters the vascular remodeling required for normal vision in the mouse. Mechanisms underlying Arf induction by Tgfβ2 are not fully understood. Using the chr4Δ70kb/Δ70kb mouse, we now show that deletion of the coronary artery disease (CAD) risk interval lying upstream of the Cdkn2a/b locus represses developmentally-timed induction of Arf resulting in eye disease mimicking the persistent hyperplastic primary vitreous (PHPV) found in Arf-null mice and in children. Using mouse embryo fibroblasts, we demonstrate that Arf induction by Tgfβ is blocked in cis to the 70 kb deletion, but Arf induction by activated RAS and cell culture “shock” is not. Finally, we show that Arf induction by Tgfβ is derailed by preventing RNA polymerase II recruitment following Smad 2/3 binding to the promoter. These findings provide the first evidence that the CAD risk interval, located at a distance from Arf, acts as a cis enhancer of Tgfβ2-driven induction of Arf during development.
Keywords: 9p21, Arf, Tgfβ, PHPV
Introduction
The mouse Cdkn2a and Cdkn2b genetic loci contain three genes serving as important mammalian tumor suppressors (Figure 1A). Cdkn2a includes Ink4a encoding p16Ink4a from three exons, and this protein inhibits Cyclin-dependent kinases (Cdk) 4 and 6, thereby activating the Retinoblastoma tumor suppressor (Rb) and arresting cell proliferation (Serrano et al., 1993). Arf shares exons 2 and 3 with Ink4a, but its first exon (1β) resides approximately 12 kb upstream of exon 1α of Ink4a, and the protein encoded by Arf, p19Arf, is translated in an alternate reading frame (Quelle et al., 1995). Bearing no structural resemblance to p16Ink4a, p19Arf enhances the functional activity of p53 – the other major mammalian tumor suppressor pathway (Levine, 1989; Levine et al., 1991). Cdkn2b encodes the p15Ink4b Cdk4/6 inhibitor and this gene resides 12 kb further upstream of Arf exon 1β (Hannon and Beach, 1994). This unusual genomic organization in which a single locus contains three genes regulating the two major mammalian tumor suppressors is conserved in known mammalian genomes (Gil and Peters, 2006). Gross chromosomal deletions involving Cdkn2a and Cdkn2b or epigenetic silencing of the locus is relatively common in many human cancers (Baghdassarian and Ffrench, 1996; Dreyling et al., 1998; Gil and Peters, 2006; Heyman and Einhorn, 1996; Sharpless and DePinho, 1999). Mouse lines engineered to lack Arf, Ink4a, or Cdkn2b are susceptible to a wide range of cancers as they age (Kamijo et al., 1997; Krimpenfort et al., 2001; Latres et al., 2000; Serrano et al., 1996; Sharpless et al., 2001).
Figure 1.
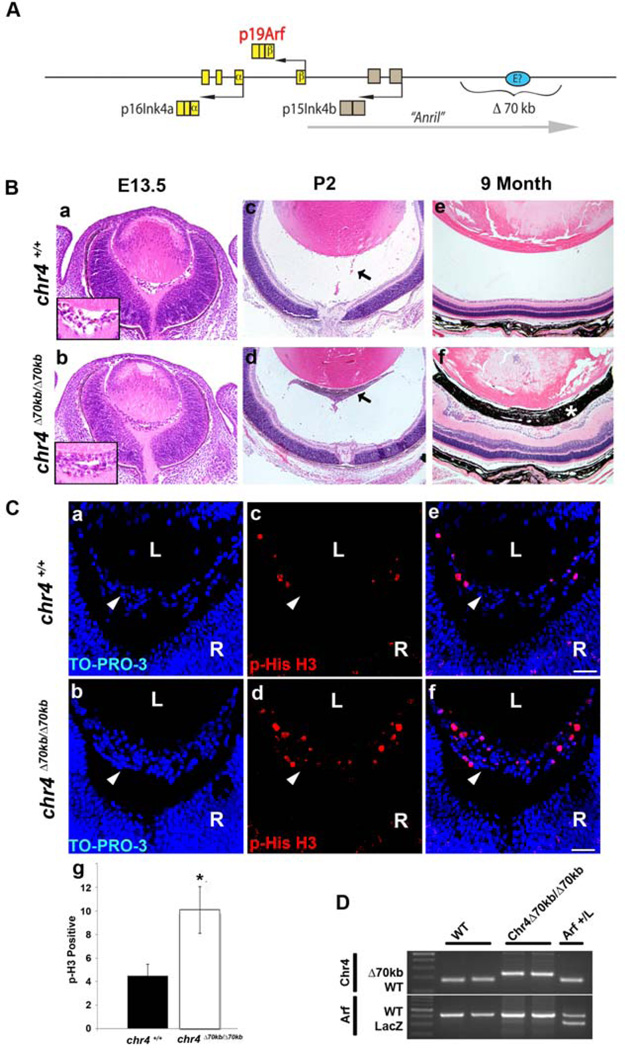
PHPV-like eye phenotype in chr4Δ70kb/Δ70kb mice. (A) Schematic diagram showing Cdkn2a and Cdkn2b genetic loci, which encodes p16Ink4a, p19Arf and p15Ink4b. A long non-coding RNA (lncRNA), AK14831 (putative mouse “Anril”), is transcribed starting proximal to and proceeding upstream of Cdkn2a. A putative enhancer element (E?) may lie within the deleted 70kb segment orthologous to human chromosome 9p21 (see Discussion). (B) Representative photomicrographs of hematoxylin- and eosin-stained slides of E13.5 embryos, P2 and 9 months showing the primary vitreous hyperplasia in chr4Δ70kb/Δ70kb mice (b, d, f) but not in its wild type littermates (a, c, e). Inset (a and b) shows higher magnification of vitreous area. Arrows (c and d) show hyperplastic retrolental mass in mutant eye. Asterisk (f) shows pigmented cells in retrolental mass. Photos were taken at 20×. (C) Representative photomicrographs of immunofluorescence-stained slides of E13.5 embryos showing TO-PRO-3 (a, b), phospho-Histone H3 (c, d) and the overlay (e, f) of primary vitreous hyperplasia in chr4Δ70kb/Δ70kb embryos (b, d, f) and wild type littermates (a, c, e). Photos were taken at 40× magnification. (g) Quantification of the phospho-histone H3 stained E13.5 embryos showed 2-fold increase of phospho-histone H3 staining from chr4Δ70kb/Δ70kb embryos versus those from wild type littermates. L, lens. R, retina. Arrowhead, vitreous. (D) Representative agarose gel showing the genotyping of wild type, chr4Δ70kb/Δ70kb, and Arf+/lacZ mice.
Some mammalian tumor suppressors also serve pivotal roles in embryo or cellular development. This is true for Arf, which is essential for vascular remodeling that accompanies later stages of eye development (McKeller et al., 2002; Silva et al., 2005). Despite the aforementioned genomic complexity, we can attribute the developmental defect to Arf because it is only evident when exon 1β or exon 2 is disrupted, but not in mice lacking exon 1α of the Ink4a gene (Martin et al., 2004). In this developmental capacity, p19Arf is expressed between mouse embryonic day (E) 12.5 and postnatal day (P) 5 to repress Pdgfrβ (Silva et al., 2005; Widau et al., 2012), a receptor tyrosine kinase required for pericyte accumulation in the developing mouse (Hoch and Soriano, 2003). Mouse genetic studies demonstrate that deregulated Pdgfrβ in the Arf −/− embryo drives excess perivascular cell accumulation around the hyaloid vessels in the developing vitreous space (Silva et al., 2005; Widau et al., 2012). The hyaloid vessels normally involute between P5 and P10 in the mouse and in late stages of human eye development (Martin et al., 2004), but they fail to do so when embraced by overgrowing perivascular cells (Silva et al., 2005). Hyperplasia in the primary vitreous and persistence of the hyaloid vessels leads to secondary pathological changes in the lens and retina, mimicking a human eye disease known as Persistent Hyperplastic Primary Vitreous (PHPV) (Haddad et al., 1978; Shastry, 2009) or Persistent Fetal Vasculature (PFV) (Goldberg, 1997), and rendering Arf −/− animals sightless (Martin et al., 2004).
Of note, PFV was suggested as a more unifying term to account for the fact that what had historically been called PHPV can have a broad range of manifestations from relatively small remnants of the hyaloid vessels in the anterior or posterior vitreous space to truly hyperplastic lesions (Goldberg, 1997). This disease spectrum is also reflected in mouse models in which the primary defect seems to be in pro-apoptotic events needed to eliminate hyaloid vessel endothelial cells, such as BALB/cOlaHsd mice lacking p53 (Reichel et al., 1998), mice lacking Ang2 (Hackett et al., 2002), or mice with defective hyalocyte-mediated signaling from Wnt7b to FZD4 and Lrp5 (Kato et al., 2002; Lang and Bishop, 1993; Lobov et al., 2005). These models truly reflect persistence of fetal vasculature (PFV). In contrast, primary vitreous hyperplasia is the major defect in animals with deregulated expression of Vegf-A (Rutland et al., 2007) or the immediate early protein IE180 of Pseudorabies Virus (Taharaguchi et al., 2005), or in the absence of Tgfβ2 (Freeman-Anderson et al., 2009) (discussed more below). The Arf−/− phenotype described above also principally represents primary vitreous hyperplasia, hence our reference to the disease as PHPV.
With an essential role for Arf in development and the general importance of the Cdkn2a/b locus in cancer biology, understanding how its expression is controlled represents a fundamental question. Several points are already clear. First, the entire locus can be regulated as a single unit by epigenetic silencing by Polycomb repressor complexes (Gil and Peters, 2006). Certain transcriptional repressors, such as Bmi1 (Jacobs et al., 1999) and Cbx7 (Gil et al., 2004), work to silence Cdkn2a/b expression throughout much of the developing embryo, and perhaps as a mechanism to evade their tumor suppressive effects. Second, Arf is induced by a variety of oncogenic stimuli – such as expression of activated forms of RAS (Lin and Lowe, 2001; Palmero et al., 1998), Adenovirus E1a (de Stanchina et al., 1998), and Myc (Zindy et al., 1998), and supra-physiologic mitogenic signals from cell culture “shock” (Sherr and DePinho, 2000; Zindy et al., 1998). Of course, these general mechanisms cannot account for the tightly controlled expression of Arf in the developing eye. In the normal developmental context, Tgfβ2 is required for Arf expression in the hyaloid vasculature, and also in the internal umbilical vessels; Tgfβ2 deficient mice develop PHPV like Arf−/− mice (Freeman-Anderson et al., 2009). Tgfβ2 signals through TβrII to activate Smads 2 and 3 and p38 Mapk, which are essential for Arf induction (Zheng et al., 2010); in mouse embryo fibroblasts (MEFs), these effects are limited to Arf, not the entire Cdkn2a/b locus – implying a regulatory mechanism in normal development that is distinct from its control in response to pathological states such as oncogenic stress.
We have focused on understanding the molecular mechanisms that drive Tgfβ-dependent control of Arf during eye development. We are considering the general hypothesis that additional cis-acting elements are needed to explain the temporal response to Tgfβ (i.e., delayed transcriptional induction despite immediate Smad 2/3 binding to the Arf promoter) and the temporally- and spatially-restricted expression pattern of Arf despite wide spread effects of Tgfβ. We became interested in a 70 kb segment of genomic DNA lying about 100 kb upstream of the Cdkn2b/Cdkn2a locus (Figure 1A). This region demonstrates substantial sequence similarity and synteny with a 58 kb intragenic region that is strongly linked to coronary artery disease (CAD) risk in humans (Helgadottir et al., 2007; McPherson et al., 2007). Single nucleotide polymorphisms in the so-called CAD risk allele at human chromosome 9p21, and knockout of the orthologous 70 kb region in the mouse genome, can influence the expression of flanking Cdkn2a and Cdkn2b genes (Visel et al., 2010). Here we report that this genomic locus contains a cis-acting element that is required for Tgfβ2-driven Arf expression and proper eye development, and we provide mechanistic insight into how it operates in a manner that is distinct from oncogene-driven Arf induction.
Materials and Methods
Animals and cell culture
Chr4Δ70kb/Δ70kb mice (Visel et al., 2010) were obtained from NCI Mouse Models of Human Cancer Consortium repository. ArflacZ/+ mice generated in the Skapek laboratory (Freeman-Anderson et al., 2009) were maintained in a mixed C57BL/6 × 129/Sv genetic background. The University of Texas Southwestern Medical Center Animal Care and Use Committee approved all laboratory animal studies.
Primary MEFs from wild type, ArflacZ/+, chr4Δ70kb/Δ70kb, chr4Δ70kb/+, and ArflacZ/+, chr4+/Δ70kb mice were derived and cultivated by us as previously described (Zindy et al., 1997). Tgfβ1 was obtained from R&D Systems, Inc (Minneapolis, MN). Of note, Tgfβ1 was used throughout as we previously established that Tgfβ1, 2, and 3 have similar effects on Arf expression (Zheng et al., 2010). β-galactosidase activity was measured in cultured cells using a commercial kit (Applied Biosystems; Foster City, CA). MSCV-based retrovirus vector encoding human H-RASV12 and GFP, or just GFP, were produced in our laboratory using vectors from Addgene (Cambridge, MA). Antibodies used in Western blotting experiments were directed against Hsc70 (Santa Cruz Biotechnology, Inc; Santa Cruz, CA); and p19Arf (Abcam Inc; Cambridge, MA).
Histology studies
Mouse embryos were obtained at E13.5 from females euthanized by CO2 and processed for paraffin or cryostat sectioning and routine staining as described previously (Martin et al., 2004; McKeller et al., 2002). Phospho-histone H3 immunofluorescence staining was used to assess proliferation using 8 µm cryostat sections as follows: sections were incubated in 96°C citrate buffer (pH 6) for 30 minutes for antigen retrieval, blocked in 10% goat serum with 0.1% Triton X-100 in PBS, and stained using rabbit anti-phospho-histone H3 (ser 10) antibody (1:500, Millipore; Temecula, CA) followed by Cy3-conjugated goat anti-rabbit antibody (1:250, Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) and counterstaining with TO-PRO-3 (Life Technologies). The fraction of positively stained cells was determined using paired littermates from five litters and verified by two individuals who were blinded to the genotypes. Pdgfrβ was detected using goat anti-Pdgfrβ (AF1042, R&D Systems) as previously described (Silva et al., 2005). All immunofluorescence staining was visualized using a Zeiss 510 NLO multi-photon/confocal laser-scanning microscope at 20× or 40× magnification.
Laser-capture microdissection (LCM)
LCM was accomplished as previously described (Widau et al., 2012). Briefly, mouse embryos were harvested at E13.5 for LCM. Embryo heads were immediately embedded in OCT freezing medium without fixation. 14 µm sections were cut on a CryoStar NX70 cryostat, mounted on PEN Membrane Metal Slides (Applied Biosystems), and stained with hematoxylin and eosin (Molecular Machines & Industries AG; Glattbrugg, Switzerland). LCM was carried out with the Arcturus Veritas Microdissection System. At least ten, micro-dissected sections from the vitreous, lens, and retina were pooled from each embryo. Total RNA was extracted using the Arcturus PicoPure LCM RNA isolation kit (Applied Biosystems) and the expression of specific genes was analyzed with real time RT-PCR as described below.
RT-PCR
Total RNA was extracted from cell pellets or LCM tissue dissolved in Trizol (Invitrogen), according to the manufacturer’s recommendations. Total RNA (2 µg) was use to synthesize cDNA with Superscript III RT kits (Invitrogen) and quantitative RT-PCR (qRT-PCR) was performed using Fast SYBR Green Master mix and a model 7900HT Fast Cycler instrument (both from Applied Biosystems). The primers were as follows: Arf: 5’-TTCTTGGTGAAGTTCGTGCGATCC (forward) and 5’-CGTGAACGTTGCCCATCAT CATCA (reverse); Pdgfrβ: 5’-AACAGAAGACAGGGAGGTGG-3’ (forward) and 5’-TGGTATCACTCCTGGAAGCC-3’ (reverse); p21Cip1: 5’-CAGATCCACAGCGATATCCA (forward) and 5’-ACGGGACCGAAGAGACAAC (reverse); p15Ink4b: 5’-GATCCCAACGCCCTGAAC (forward) and 5’-TGGTAAGGGTGGCAGGGT (reverse); Pai-1: 5’-GCCTCCTCATCCTGCCTAA (forward) and 5’-TGCTCTTGGTCGGAAAGACT- (reverse); Gapdh: 5’-TCAACAGCAACTCCCACTCTTCCA (forward) and 5’- ACCCTGTTGCTGTAGCCGTATTCA (reverse). Results are pooled from three separate experiments.
Chromatin immunoprecipitation (ChIP)
ChIP was performed as previously described (Zheng et al., 2010). Briefly, MEFs (5×106/ChIP) were treated with Tgfβ1 (5 ng/ml) or vehicle 24 hours. Cells were cross-linked, sonicated, and subjected to immunoprecipitation using anti-Smad2/3 antibody (sc6033, Santa Cruz Biotechnology), or anti-RNA polymerase II (sc899, Santa Cruz). Rabbit IgG (sc2027, Santa Cruz) or goat IgG (AB-108-C, R&D Systems) was used as a negative control. Protein A/G sepharose beads (sc2003, Santa Cruz) were used to collect the protein-chromatin complexes. The beads were washed sequentially with low salt, high salt, LiCl and TE buffers (Upstate ChIP Kit, Millipore) and eluted in 0.1 M NaHCO3, 1% SDS. Cross-linking was reversed by incubation at 67°C overnight and the genomic DNA was extracted using Qiagen PCR Purification Kit. Quantitative analysis of the precipitated and input DNA were carried out using specific primer sets and Fast SYBR green master mix on a model 7900HT Fast Cycler instrument (both from Applied Biosystems). The primer sets for Arf promoter regions were as follows: 5’-AGATGGGCGTGGAGCAAAGAT (forward) and 5’-ACTGTGACAAGCGAGGTGAGAA (reverse).
Statistical Analysis
Quantitative data are presented as the mean ± S.D. from three or more representative experiments. Statistical significance (p value <0.05) was calculated using Student’s t test.
Results
PHPV-like eye phenotype in the chr4Δ70kb/Δ70kb mouse
We evaluated the developmental importance of the 70 kb interval lying upstream of Cdkn2b/Cdkn2a (Figure 1A), which is orthologous to the human chromosome 9p21 CAD risk interval (Visel et al., 2010), by examining eyes taken from chr4Δ70kb/Δ70kb and wild type mice at embryonic and postnatal points. Deleting the 70 kb CAD risk interval led to primary vitreous hyperplasia at E13.5 (Figure 1B, b versus a); a retrolental fibrovascular mass in the immediate postnatal period (Figure 1B, d versus c); and progressive accumulation of pigmented cells within this mass as animals aged (Figure 1B, f versus e). These pathologic features, perfectly mimicking the PHPV-like phenotype in Arf−/− mice (McKeller et al., 2002), were observed in all chr4Δ70kb/Δ70kb we examined (n > 6), and none of the heterozygous (n>3) or wild type littermates (n > 6). Of note, the progressive accumulation of pigmented cells within the retrolental mass has occasionally been observed in human eyes with severe PHPV (Haddad et al., 1978) and also in the Arf−/− eyes, which are also severely affected (Martin et al., 2004). In that model, their progressive accumulation with time, occasional absence even in severely affected eyes, and anatomic evidence suggesting their migration from the hyaloid artery stalk, all suggested to us that the cells might represent retinal pigment epithelial cells reacting as a secondary event to the severe vitreoretinopathy (Martin et al., 2004).
The primary vitreous hyperplasia in Arf−/− animals (McKeller et al., 2002), and in Tgfβ2−/− mice where Arf mRNA is diminished (Freeman-Anderson et al., 2009), is primarily driven by excess proliferation evident as early as E13.5. Immunostaining for phospho-histone H3 demonstrated an approximately 2-fold increase in the relative number of cells proliferating in the vitreous of chr4Δ70kb/Δ70kb mice eyes as compared to wild type controls (Figure 1C). Because the morphological changes and this functional assay mirrored the Arf−/− phenotype, we used PCR to confirm that the Arf exon 1β was not inadvertently deleted in the chr4Δ70kb/Δ70kb animals (Figure 1D). Taken together, these results indicate that loss of the CAD risk allele in the chr4Δ70kb/Δ70kb mouse blocks normal eye development leading to a PHPV-like eye disease.
Failed Arf induction and Pdgfrβ repression in chr4Δ70kb/Δ70kb mouse
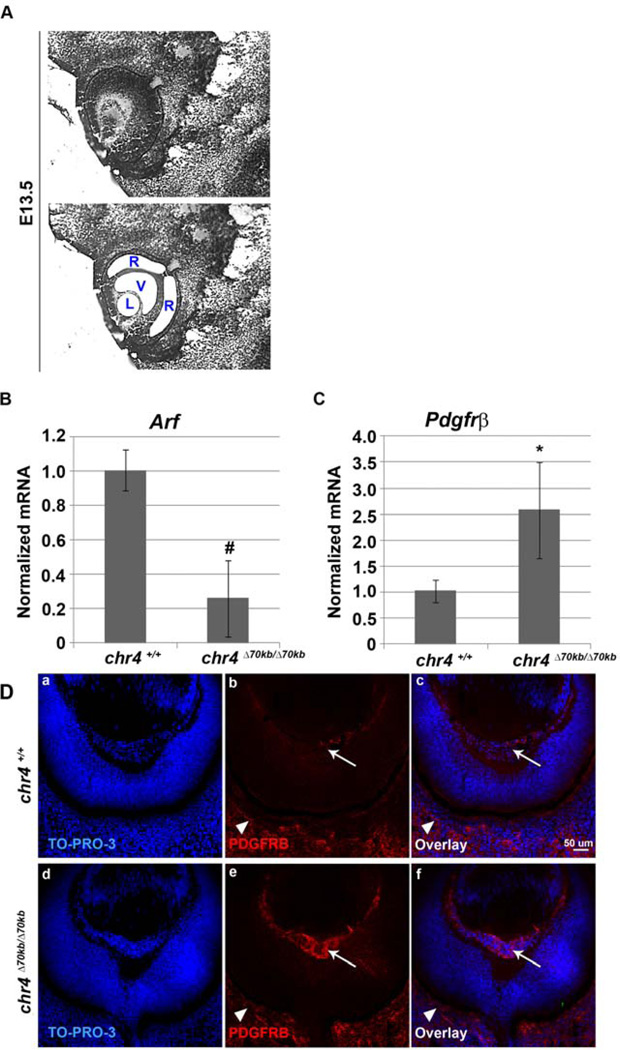
Given the fact that the Arf deficient developmental defect occurs in chr4Δ70kb/Δ70kb mouse, we examined whether Arf expression was influenced as the primary vitreous develops. To accomplish this, we utilized laser-capture microdissection (LCM) to specifically assess Arf expression in the lens, the retina, and the vitreous space at E13.5 (Figure 2A). This approach confirmed that Arf expression was limited to the vitreous, as we previously established (Silva, Thornton et al. 2005) (YZ and SXS, additional data not shown), and that its expression was dramatically diminished in chr4Δ70kb/Δ70kb embryo eyes (Figure 2B). p19Arf controls Pdgfrβ mRNA and protein expression in both p53-dependent and independent mechanisms, respectively (Silva et al., 2005; Widau et al., 2012), and deregulated Pdgfrβ is required for the primary vitreous hyperplasia in Arf−/− mice (Silva et al., 2005). Consistent with diminished Arf representing the key defect in chr4Δ70kb/Δ70kb embryos, Pdgfrβ mRNA and protein were both increased in the vitreous of the mutant mice as compared to wild type littermates (Figure 2C and 2D, e versus b). Finding similar Pdgfrβ immunostaining in the choroid/sclera (Figure 2D, arrowhead) demonstrated that the increased expression of this receptor was not a global change but was limited to the anatomic sites where p19Arf normally resides.
Figure 2.
Loss of the 70kb CAD risk interval decreases Arf mRNA but increases Pdgfrβ expression in vitreous of developing eye. (A). LCM of the vitreous (V), lens (L) and retina (R) from E13.5 WT mouse embryos. qRT-PCR analysis of (B) Arf and (C) Pdgfrβ were carried out using total RNA isolated from the vitreous from E13.5 chr4Δ70kb/Δ70kb embryos and wild type littermate embryos. Expression was normalized to that of Gapdh. The results were average of 3 independent experiments. # (decrease), * (increase), p<0.05. (D) Representative photomicrographs of immunofluorescence-stained sections from E13.5 embryos showing TO-PRO-3 (a, d), Pdgfrβ (b, e) and the overlay (c, f) of primary vitreous hyperplasia in chr4Δ70kb/Δ70kb embryos (d, e, f) and wild type littermates (a, b, c). Arrow (b, e) depicts Pdgfrβ staining in vitreous; arrowhead (b, e) depicts such staining in choroid/sclera.
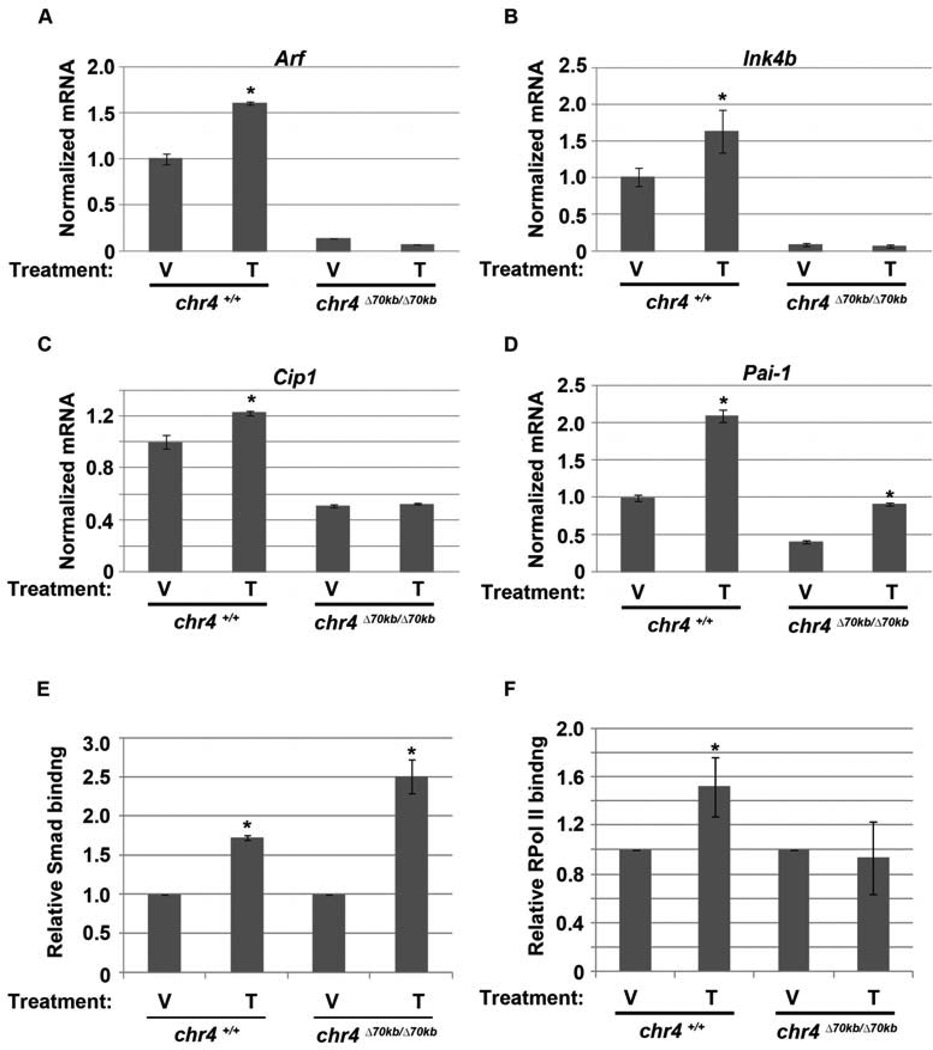
We previously established that Tgfβ2 is an essential regulator of Arf in the developing eye and Tgfβ1, 2, and 3 can each induce p19Arf in cultured MEFs and in HeLa cells (Freeman-Anderson et al., 2009; Zheng et al., 2010) **(additional data not shown). As expected, mice lacking Tgfβ2 develop primary vitreous hyperplasia that resembles the Arf−/− eye disease (Freeman-Anderson et al., 2009). The fact that Arf mRNA is low in chr4Δ70kb/Δ70kb mice suggested that the normal Tgfβ2-driven induction of this transcript might be blocked by deletion of the CAD risk interval. We investigated this concept further using cultured MEFs. First, quantitative RT-PCR found basal Arf expression in chr4Δ70kb/Δ70kb MEFs to be much lower than in similarly passed wild type MEFs (Figure 3A), consistent with a previous report (Visel et al., 2010) and our in vivo studies (Figure 2B). Tgfβ1 increased Arf mRNA and p19Arf protein expression in wild type MEFs but failed to do so in chr4Δ70kb/Δ70kb MEFs (Figure 3A; and additional data not shown). Similar findings were observed for Cdkn2b and Cdkn1a mRNA, encoding p15Ink4b and p21Cip1, respectively (Figures 3B and C). Of note, we recognize that failed Cdkn1a induction might be secondary to failed p19Arf-dependent activation of p53. Moreover, Tgfβ still augmented the expression of plasminogen activator inhibitor-1 (PAI-1) in these MEFs, even though the basal levels for this transcript were lower than in the wild type cells (Figure 3D).
Figure 3.
Tgfβ fails to activate Arf, Ink4b and Cdkn1a (Cip1). Quantitative analysis of Arf (A), Ink4b (B), Cip1 (C) and Pai-1 (D) by RT-PCR using total RNA isolated from chr4Δ70kb/Δ70kb and wild type MEFs exposed to vehicle (V) or Tgfβ (Τ) for 48 hours. Differences in transcript level between Tgfβ- and vehicle-treated wild type MEFs are significant [p <0.05 (*)]. (E, F) Quantitative analysis of representative chromatin immunoprecipitation (ChIP) assays of using chr4Δ70kb/Δ70kb and wild type MEFs exposed to vehicle (V) or Tgfβ (T) for 24 hours. ChIP assay was carried out using antibodies specific to (E) Smad2/3, (F) RPol II; expressed relative to IgG and normalized against vehicle control. Immunoprecipitated and input DNA were amplified with primers specific for the proximal Arf promoter. *, p <0.05 for Tgfβ versus corresponding vehicle.
In response to Tgfβ signaling, Smads 2/3 bind to the Arf promoter and they are required for Tgfβ induction of p19Arf (Zheng et al., 2010). Indeed, Smads 2/3 were recruited to the Arf promoter following Tgfβ stimulation in chr4Δ70kb/Δ70kb MEFs at least as well as in wild type cells (Figure 3E). Following Smad 2/3 binding, delayed recruitment of RNA polymerase II (RPol II) to the Arf promoter coincides with increased Arf mRNA (Zheng et al., 2010). Even though Smad 2/3 binding to the promoter was enhanced, RPol II was not recruited in chr4Δ70kb/Δ70kb MEFs (Figure 3F). Of note, the lower basal Arf expression in the chr4Δ70kb/Δ70kb could not be attributed to differences in Smad 2/3 and RPolII binding, both of which were low in the vehicle treated MEFs regardless of genotype (YZ and SXS, negative data not shown).
The 9p21 CAD risk interval contains a cis-acting enhancer that cooperates with Tgfβ, but not oncogenic RAS, to induce Arf transcription
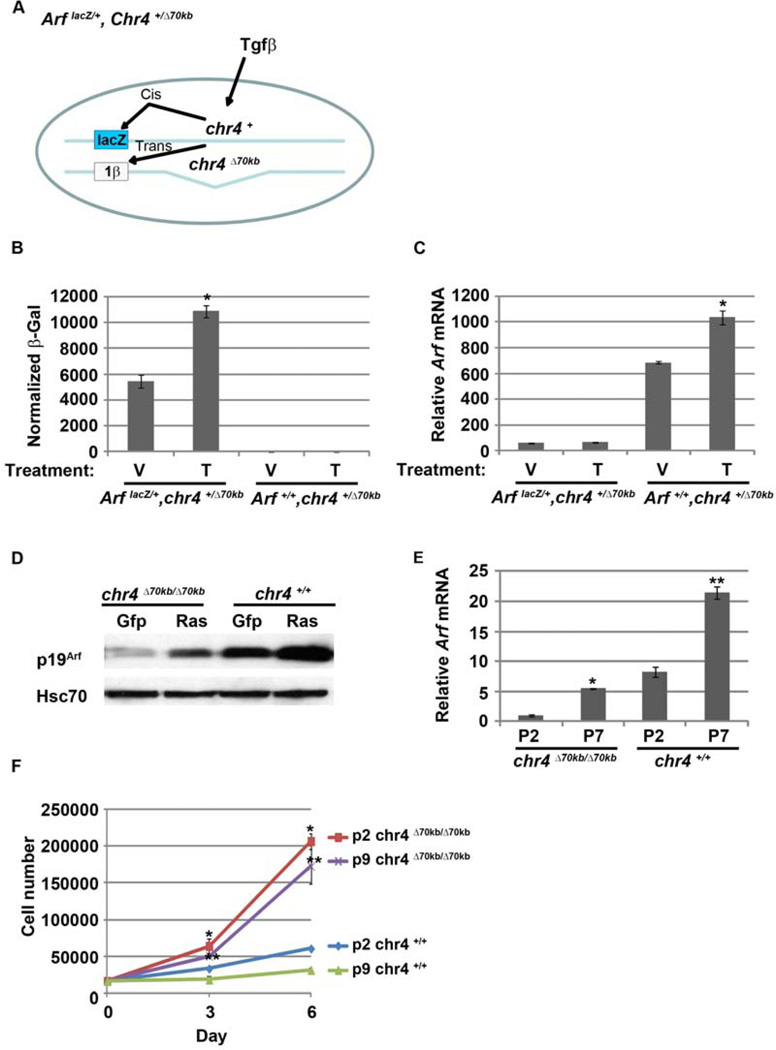
Deletion of the CAD risk allele in the mouse represses Cdkn2b in cis in the heart and several other organs (Visel et al., 2010). To test whether Arf induction by Tgfβ was similarly affected, we derived MEFs from ArflacZ/+, chr4+/Δ70kb mouse embryos. In these cells, native Arf lies in cis to the 70 kb deletion whereas the lacZ cDNA driven by the Arf promoter resides on a chromosome retaining the putative upstream regulatory elements (Figure 4A). Exposure of the MEFs to Tgfβ1 induced β-galactosidase activity but not the endogenous Arf mRNA in ArflacZ/+, chr4+/Δ70kb MEFs, whereas the native Arf mRNA was induced in the Arf+/+, chr4+/Δ70kb MEFs (Figure 4B and C). Of note, some evidence from human cells supports the concept that ANRIL, a long non-coding RNA extending into the CAD risk allele, serves to recruit Polycomb repressor complexes in cis to Cdkn2a and Cdkn2b promoters (Kotake et al., 2011; Yap et al., 2010). Tgfβ exposure did not measurably change the expression of AK148321, the mouse ANRIL ortholog (YZ and SXS, negative data not shown).
Figure 4.
Cis elements in the 70kb CAD interval are required for Arf induction by Tgfβ but not other stimuli. (A) Schematic diagram showing potential cis and trans activation of β-galactosidase and Arf mRNA, respectively, in ArflacZ/+, Chr4+/Δ70kb MEFs treated with Tgfβ. (B and C). β-galactosidase activity (B) and qRT-PCR analysis of Arf mRNA (C) in ArflacZ/+, chr4+/Δ70kb and Arf+/+, chr4+/Δ70kb MEFs. mRNA expression, normalized to Gapdh, expressed relative to that in vehicle treated ArflacZ/+, Chr4+/Δ70kb MEFs. (D) Oncogenic signal from human H-RASV12 induce p19Arf in chr4Δ70kb/Δ70kb MEFs. Representative western blot for the indicated proteins using lysates from chr4Δ70kb/Δ70kb and wild type MEFs after transduction with Gfp- or H-RASV12-expressing retrovirus. (E) qRT-PCR analysis of Arf mRNA using chr4Δ70kb/Δ70kb and wild type MEFs from different passage. * and **, p <0.05 for P7 versus P2 in chr4Δ70kb/Δ70kb and wild type, respectively. (F) Cell counting assay of chr4Δ70kb/Δ70kb and wild type MEFs from different passage after cultured for 6 days. *, and ** p <0.05 for chr4Δ70kb/Δ70kb versus wild type for P2 and P9, respectively.
Ectopic expression of human H-RASV12 and serial passage in culture also induces Arf transcription in MEFs and murine keratinocytes (Lin and Lowe, 2001; Palmero et al., 1998; Zindy et al., 1998). Because it is not yet clear whether mechanisms underlying Arf induction by Tgfβ are also utilized in these scenarios, we investigated the role that this intergenic region plays in Arf induction by other stimuli. As expected, mutant RAS induced p19Arf in wild type MEFs (Figure 4D). It also induced p19Arf expression in chr4Δ70kb/Δ70kb MEFs, although the basal expression was lower (Figure 4D). Similarly, Arf mRNA and p19Arf protein were induced by serial passage of both wild type and chr4Δ70kb/Δ70kb MEFs (Figure 4E; additional data not shown). Of note, Arf induction may be relevant to diminished cell accumulation that we observed in early versus later passage wild type or chr4Δ70kb/Δ70kb MEFs (Figure 4F); however, the much greater accumulation of chr4Δ70kb/Δ70kb MEFs, independently of cell passage number, strongly implies that differential induction of other cell cycle regulators such as p16Ink4a or p15Ink4b also contribute to differential proliferation in these cells. Viewed together, the simplest interpretation of our findings is that a cis-acting element resides within the deleted CAD risk interval and it is required for Arf promoter activation in MEFs by Tgfβ but not by oncogenic RAS or cell culture “shock”.
Discussion
Several recent genome-wide association studies (GWAS) have linked common sequence variants in a 58 kb gene-poor region of human chromosome 9p21 to coronary artery disease and type 2 diabetes (Helgadottir et al., 2007; McPherson et al., 2007; Saxena et al., 2007; Scott et al., 2007; Zeggini et al., 2007). Due to the importance of this so-called coronary artery disease (CAD) risk allele, the orthologous region was deleted to generate the chr4Δ70kb/Δ70kb mouse (Visel et al., 2010). Although the mutant mice seem to suffer slightly increased embryonic and neonatal lethality, they were thought to be overtly normal at birth. More importantly, they displayed increased weight gain and mortality, especially when fed high-fat, high-cholesterol diets (Visel et al., 2010). Increased tumor susceptibility was noted in the chr4Δ70kb/Δ70kb animals, but how this related to the increased mortality with a “western” diet and the physiological derangements underlying the increased weight gain were not clear. Indeed, the only molecular changes in the animal related to decreased Cdkn2a and Cdkn2b in the heart and several other organs. Again, how these molecular defects translated to disease was not established.
We now provide definitive evidence for a developmental defect in these animals, and we demonstrate a molecular mechanism that can account for the disease: chr4Δ70kb/Δ70kb mice suffer a developmental defect in the eye that perfectly mimics that observed in the absence of the Arf tumor suppressor in a model for severe PHPV (McKeller et al., 2002). Arf expression in the primary vitreous at E13.5 – the critical initial phase of primary vitreous development – is extremely low in chr4Δ70kb/Δ70kb mice and, consequently, Pdgfrβ is elevated to drive the proliferation of pericyte-like cells, which is fundamental to the PHPV in the Arf−/− model (Silva et al., 2005; Widau et al., 2012). We previously established that Tgfβ2 is the dominant signal that controls Arf expression in the eye (Freeman-Anderson et al., 2009; Zheng et al., 2010). Although Tgfβ can still activate Smad proteins and enhance the expression of at least one target gene, it fails to increase Arf transcription in chr4Δ70kb/Δ70kb MEFs because RPolII is not recruited to the promoter. In summary, our findings provide the first mechanistic link between molecular derangements due to deletion of the CAD risk allele and disease in the mouse.
Two important caveats should be emphasized. First, our mechanistic studies were carried out in MEFs, whereas Arf is expressed in a limited number of perivascular cells flanking the hyaloid and internal umbilical vessels and their relationship to MEFs is not clear (Freeman-Anderson et al., 2009; Martin et al., 2004). We know the Arf expressing cells in the eye are derived from a Wnt1-expressing lineage because targeted inactivation of Arf in that lineage recapitulates the Arf−/− eye phenotype (Zheng et al., 2010). In vivo, the cells express NG-2, Pdgfrβ, and smooth muscle α-actin to some extent (Martin et al., 2004; Silva et al., 2005; Thornton et al., 2007), all of which can be expressed in pericytes; indeed, this swayed us to refer to them as “pericytes” in a recent manuscript (Widau et al., 2012). But it is important emphasize that true pericytes are defined by their anatomic location within the vascular basement membrane and their intimate, “peg and socket” interactions with endothelial cells (Armulik et al., 2005; Hirschi and D'Amore, 1996), and we have not carried out the studies needed to precisely define the Arf expressing cells as pericytes.
Regardless of whether the Arf expressing cells represent true pericytes, features of our MEFs model do reflect the in vivo biology, though, such as their expression of Pdgfrβ and its repression by p19Arf (Silva et al., 2005; Widau et al., 2012), Tgfβ2 induction of Arf (Freeman-Anderson et al., 2009; Zheng et al., 2010), and diminished Arf mRNA in chr4Δ70kb/Δ70kb cells shown here. Ideally, we would study the very cells that express Arf in vivo. Our preliminary findings indicate that we can use FACS to sort the Gfp-expressing cells from the postnatal eye in ArfGfp/Gfp animals, and we are striving to derive them from ArfGfp/+, chr4+/Δ70kb animals (mimicking experiments shown in Figure 4A–C). But even this system may suffer from two potential pitfalls. Tgfβ2 has already activated the Arf promoter in the cells we purify, and this may cast doubt on mechanistic studies focused on the initial Arf induction. More importantly for this work, even if we can derive ArfGfp/Gfp, chr4Δ70kb/Δ70kb animals by careful screening for homologous recombination placing the two mutant alleles in cis, we may not be able to use FACS to isolate them because the Gfp reporter will not likely be expressed.
Second, our studies have focused on the hyperplasia in the primary vitreous. As discussed above, PHPV/PFV represents a spectrum of diseases that affect different aspects of the hyaloid vasculature, from the hyaloid artery and vasa hyaloidea propria in the vitreous to the tunica vasculosa lentis (TVL) embracing the lens and the pupillary membrane (PM) (Goldberg, 1997). We previously demonstrated that the TVL and PM regress normally in the absence of Arf (Martin et al., 2004), which finding is consistent with the fact that Arf expressing perivascular cells are not found in the more anterior elements of the hyaloid vasculature (Freeman-Anderson et al., 2009; Martin et al., 2004; McKeller et al., 2002). Based on the fact that Arf expression is dampened in the vitreous of chr4Δ70kb/Δ70kb mouse embryo, we assume that the regression of anterior elements of the hyaloid vessels will not be affected by deletion of the CAD risk interval. This is supported by our analysis of several mature chr4Δ70kb/Δ70kb animals (CCD, YZ and SXS, unpublished data), but we cannot exclude a more subtle delay that is unrelated to the regulation of Arf.
Others have previously linked alterations in the expression of genes in the Cdkn2a/b locus to polymorphisms in the CAD risk interval (Liu et al., 2009) and, as mentioned above, in the mouse model (Visel et al., 2010). Mechanisms to account for how this region influences gene expression at a distance are still being unraveled. This genomic segment includes exons in a long non-coding RNA (lncRNA), ANRIL, the transcription of which actually starts just proximal to and proceeding upstream of Cdkn2a (Figure 1A) (Pasmant et al., 2007). In cultured human cells, the ANRIL transcript interacts with CBX7 and SUZ12, key components of Polycomb repressor complexes PRC1 and 2, respectively, and recruits them to silence the entire CDKN2B/ARF/CDKN2A locus (Kotake et al., 2011; Yap et al., 2010). It is assumed that specific polymorphisms correlating with changes in Cdkn2a/b gene expression may influence the known differential splicing of ANRIL and this somehow influences promoter activation.
This proposed model is somewhat difficult to reconcile with findings in the mouse. The region deleted in the chr4Δ70kb/Δ70kb mouse includes several exons in the putative mouse “Anril” (formally: AK148321). We did not detect Tgfβ-driven changes in mouse AK148321 expression in MEFs – which might be expected if it were forming a cis-repressor complex like human ANRIL. Nor did we observe significantly different expression of AK148321 in wild type versus chr4Δ70kb/Δ70kb mice (YZ and SXS, negative data not shown). While it is formally possible that deletion of the CAD risk allele in the mouse may facilitate transcriptional silencing from a mutated form of this lncRNA, we favor an alternative hypothesis that this segment contains a previously unrecognized cis-activating element, as proposed by the Pennacchio laboratory (Visel et al., 2010). Interestingly, their search for such elements is reported to have been limited to E11.5 mouse embryos, which is two days before robust Arf promoter activity is evident in the embryo (Silva et al., 2005). Further, our finding that the CAD risk element is required for Tgfβ-driven activation of Arf but not for induction by other stimuli also speaks to specific interactions that such a cis-enhancer may make with Smad 2/3 binding elements at this promoter.
Finally, our results might help to explain the other pathological findings in the chr4Δ70kb/Δ70kb. Obviously, the enhanced tumor susceptibility might merely be due to decreased expression of Arf and other genes at this locus. However, the fact that an oncogenic form of RAS can enhance Arf expression in MEFs does cast some doubt on this potential explanation. As mentioned above, a comprehensive search for metabolic or pathological changes, including atherogenic plaques, in the original description of the mouse did not unveil a precise cause for enhanced coronary artery disease risk either. Our developmental studies of the mouse eye implicate p19Arf as an essential regulator of perivascular cell proliferation and Pdgfrβ expression in the primary vitreous (Silva et al., 2005; Widau et al., 2012). One might consider whether such failed induction of Arf in perivascular cells in other vessels might contribute to the disease. It is important to emphasize that we have only found Arf expressing perivascular cells within the hyaloid and internal umbilical vessels in the embryo and early postnatal period; however, we have also not studied this extensively in older animals, nor in animals stressed by a high-fat and high-cholesterol diet.
Conclusions
Our studies demonstrated that the CAD risk interval acts as a cis enhancer of Tgfβ2-driven induction of Arf during development. Loss of the CAD risk interval represses the developmentally timed induction of Arf, resulting in eye disease resembling PHPV found in Arf-deficient mice.
Highlights.
Deleting intergenic DNA upstream of mouse Cdkn2a causes eye disease mimicking PHPV.
Arf induction by Tgfβ, but not activated RAS, is blunted in chr4Δ70kb/Δ70kb cells.
Tgfβ fails to recruit RNA polymerase II to the Arf promoter in chr4Δ70kb/Δ70kb MEFs.
Intergenic DNA upstream of mouse Cdkn2a acts as a cis enhancer of Arf.
Acknowledgement
We gratefully acknowledge A. Visel and L. A. Pennacchio (Lawrence Berkeley National Laboratory) for sharing mouse tissue specimens from chr4Δ70kb/Δ70kb animals for an initial analysis; C. J. Sherr (St. Jude Children’s Research Hospital) for helpful initial discussions; K. Bachmeyer and L. Roach (both at the University of Chicago) for technical assistance; and Syann Lee and Joel Elmquist (both at UT Southwestern Medical Center) for assistance with laser capture microdissection. This research is supported by grants to SXS from the National Eye Institute (EY 014368 and EY 019942).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Armulik A, Abramsson A, Betsholtz C. Endothelial/pericyte interactions. Circ Res. 2005;97:512–523. doi: 10.1161/01.RES.0000182903.16652.d7. [DOI] [PubMed] [Google Scholar]
- Baghdassarian N, Ffrench M. Cyclin-dependent kinase inhibitors (CKIs) and hematological malignancies. Hematol Cell Ther. 1996;38:313–323. doi: 10.1007/s00282-996-0313-4. [DOI] [PubMed] [Google Scholar]
- de Stanchina E, McCurrach ME, Zindy F, Shieh SY, Ferbeyre G, Samuelson AV, Prives C, Roussel MF, Sherr CJ, Lowe SW. E1A signaling to p53 involves the p19(ARF) tumor suppressor. Genes Dev. 1998;12:2434–2442. doi: 10.1101/gad.12.15.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyling MH, Roulston D, Bohlander SK, Vardiman J, Olopade OI. Codeletion of CDKN2 and MTAP genes in a subset of non-Hodgkin's lymphoma may be associated with histologic transformation from low-grade to diffuse large-cell lymphoma. Genes Chromosomes Cancer. 1998;22:72–78. [PubMed] [Google Scholar]
- Freeman-Anderson NE, Zheng Y, McCalla-Martin AC, Treanor LM, Zhao YD, Garfin PM, He TC, Mary MN, Thornton JD, Anderson C, Gibbons M, Saab R, Baumer SH, Cunningham JM, Skapek SX. Expression of the Arf tumor suppressor gene is controlled by Tgfbeta2 during development. Development. 2009;136:2081–2089. doi: 10.1242/dev.033548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil J, Bernard D, Martinez D, Beach D. Polycomb CBX7 has a unifying role in cellular lifespan. Nat Cell Biol. 2004;6:67–72. doi: 10.1038/ncb1077. [DOI] [PubMed] [Google Scholar]
- Gil J, Peters G. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nat Rev Mol Cell Biol. 2006;7:667–677. doi: 10.1038/nrm1987. [DOI] [PubMed] [Google Scholar]
- Goldberg MF. Persistent fetal vasculature (PFV): an integrated interpretation of signs and symptoms associated with persistent hyperplastic primary vitreous (PHPV). LIV Edward Jackson Memorial Lecture. Am J Ophthalmol. 1997;124:587–626. doi: 10.1016/s0002-9394(14)70899-2. [DOI] [PubMed] [Google Scholar]
- Hackett SF, Wiegand S, Yancopoulos G, Campochiaro PA. Angiopoietin-2 plays an important role in retinal angiogenesis. J Cell Physiol. 2002;192:182–187. doi: 10.1002/jcp.10128. [DOI] [PubMed] [Google Scholar]
- Haddad R, Font RL, Reeser F. Persistent hyperplastic primary vitreous. A clinicopathologic study of 62 cases and review of the literature. Surv Ophthalmol. 1978;23:123–134. doi: 10.1016/0039-6257(78)90091-7. [DOI] [PubMed] [Google Scholar]
- Hannon GJ, Beach D. p15INK4B is a potential effector of TGF-beta-induced cell cycle arrest. Nature. 1994;371:257–261. doi: 10.1038/371257a0. [DOI] [PubMed] [Google Scholar]
- Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, Sigurdsson A, Baker A, Palsson A, Masson G, Gudbjartsson DF, Magnusson KP, Andersen K, Levey AI, Backman VM, Matthiasdottir S, Jonsdottir T, Palsson S, Einarsdottir H, Gunnarsdottir S, Gylfason A, Vaccarino V, Hooper WC, Reilly MP, Granger CB, Austin H, Rader DJ, Shah SH, Quyyumi AA, Gulcher JR, Thorgeirsson G, Thorsteinsdottir U, Kong A, Stefansson K. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- Heyman M, Einhorn S. Inactivation of the p15INK4B and p16INK4 genes in hematologic malignancies. Leuk Lymphoma. 1996;23:235–245. doi: 10.3109/10428199609054826. [DOI] [PubMed] [Google Scholar]
- Hirschi KK, D'Amore PA. Pericytes in the microvasculature. Cardiovasc Res. 1996;32:687–698. [PubMed] [Google Scholar]
- Hoch RV, Soriano P. Roles of PDGF in animal development. Development. 2003;130:4769–4784. doi: 10.1242/dev.00721. [DOI] [PubMed] [Google Scholar]
- Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- Kamijo T, Zindy F, Roussel MF, Quelle DE, Downing JR, Ashmun RA, Grosveld G, Sherr CJ. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- Kato M, Patel MS, Levasseur R, Lobov I, Chang BH, Glass DA, 2nd, Hartmann C, Li L, Hwang TH, Brayton CF, Lang RA, Karsenty G, Chan L. Cbfa1-independent decrease in osteoblast proliferation, osteopenia, and persistent embryonic eye vascularization in mice deficient in Lrp5, a Wnt coreceptor. J Cell Biol. 2002;157:303–314. doi: 10.1083/jcb.200201089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, Xiong Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30:1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimpenfort P, Quon KC, Mooi WJ, Loonstra A, Berns A. Loss of p16Ink4a confers susceptibility to metastatic melanoma in mice. Nature. 2001;413:83–86. doi: 10.1038/35092584. [DOI] [PubMed] [Google Scholar]
- Lang RA, Bishop JM. Macrophages are required for cell death and tissue remodeling in the developing mouse eye. Cell. 1993;74:453–462. doi: 10.1016/0092-8674(93)80047-i. [DOI] [PubMed] [Google Scholar]
- Latres E, Malumbres M, Sotillo R, Martin J, Ortega S, Martin-Caballero J, Flores JM, Cordon-Cardo C, Barbacid M. Limited overlapping roles of P15(INK4b) and P18(INK4c) cell cycle inhibitors in proliferation and tumorigenesis. EMBO J. 2000;19:3496–3506. doi: 10.1093/emboj/19.13.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine AJ. The p53 tumor suppressor gene and gene product. Princess Takamatsu Symp. 1989;20:221–230. [PubMed] [Google Scholar]
- Levine AJ, Momand J, Finlay CA. The p53 tumour suppressor gene. Nature. 1991;351:453–456. doi: 10.1038/351453a0. [DOI] [PubMed] [Google Scholar]
- Lin AW, Lowe SW. Oncogenic ras activates the ARF-p53 pathway to suppress epithelial cell transformation. Proc Natl Acad Sci U S A. 2001;98:5025–5030. doi: 10.1073/pnas.091100298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Sanoff HK, Cho H, Burd CE, Torrice C, Mohlke KL, Ibrahim JG, Thomas NE, Sharpless NE. INK4/ARF transcript expression is associated with chromosome 9p21 variants linked to atherosclerosis. PLoS One. 2009;4:e5027. doi: 10.1371/journal.pone.0005027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobov IB, Rao S, Carroll TJ, Vallance JE, Ito M, Ondr JK, Kurup S, Glass DA, Patel MS, Shu W, Morrisey EE, McMahon AP, Karsenty G, Lang RA. WNT7b mediates macrophage-induced programmed cell death in patterning of the vasculature. Nature. 2005;437:417–421. doi: 10.1038/nature03928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AC, Thornton JD, Liu J, Wang X, Zuo J, Jablonski MM, Chaum E, Zindy F, Skapek SX. Pathogenesis of persistent hyperplastic primary vitreous in mice lacking the arf tumor suppressor gene. Invest Ophthalmol Vis Sci. 2004;45:3387–3396. doi: 10.1167/iovs.04-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeller RN, Fowler JL, Cunningham JJ, Warner N, Smeyne RJ, Zindy F, Skapek SX. The Arf tumor suppressor gene promotes hyaloid vascular regression during mouse eye development. Proc Natl Acad Sci U S A. 2002;99:3848–3853. doi: 10.1073/pnas.052484199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, Hinds DA, Pennacchio LA, Tybjaerg-Hansen A, Folsom AR, Boerwinkle E, Hobbs HH, Cohen JC. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmero I, Pantoja C, Serrano M. p19ARF links the tumour suppressor p53 to Ras. Nature. 1998;395:125–126. doi: 10.1038/25870. [DOI] [PubMed] [Google Scholar]
- Pasmant E, Laurendeau I, Heron D, Vidaud M, Vidaud D, Bieche I. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007;67:3963–3969. doi: 10.1158/0008-5472.CAN-06-2004. [DOI] [PubMed] [Google Scholar]
- Quelle DE, Zindy F, Ashmun RA, Sherr CJ. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell. 1995;83:993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- Reichel MB, Ali RR, D'Esposito F, Clarke AR, Luthert PJ, Bhattacharya SS, Hunt DM. High frequency of persistent hyperplastic primary vitreous and cataracts in p53-deficient mice. Cell Death Differ. 1998;5:156–162. doi: 10.1038/sj.cdd.4400326. [DOI] [PubMed] [Google Scholar]
- Rutland CS, Mitchell CA, Nasir M, Konerding MA, Drexler HC. Microphthalmia, persistent hyperplastic hyaloid vasculature and lens anomalies following overexpression of VEGF-A188 from the alphaA-crystallin promoter. Mol Vis. 2007;13:47–56. [PMC free article] [PubMed] [Google Scholar]
- Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ, Hughes TE, Groop L, Altshuler D, Almgren P, Florez JC, Meyer J, Ardlie K, Bengtsson Bostrom K, Isomaa B, Lettre G, Lindblad U, Lyon HN, Melander O, Newton-Cheh C, Nilsson P, Orho-Melander M, Rastam L, Speliotes EK, Taskinen MR, Tuomi T, Guiducci C, Berglund A, Carlson J, Gianniny L, Hackett R, Hall L, Holmkvist J, Laurila E, Sjogren M, Sterner M, Surti A, Svensson M, Tewhey R, Blumenstiel B, Parkin M, Defelice M, Barry R, Brodeur W, Camarata J, Chia N, Fava M, Gibbons J, Handsaker B, Healy C, Nguyen K, Gates C, Sougnez C, Gage D, Nizzari M, Gabriel SB, Chirn GW, Ma Q, Parikh H, Richardson D, Ricke D, Purcell S. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU, Prokunina-Olsson L, Ding CJ, Swift AJ, Narisu N, Hu T, Pruim R, Xiao R, Li XY, Conneely KN, Riebow NL, Sprau AG, Tong M, White PP, Hetrick KN, Barnhart MW, Bark CW, Goldstein JL, Watkins L, Xiang F, Saramies J, Buchanan TA, Watanabe RM, Valle TT, Kinnunen L, Abecasis GR, Pugh EW, Doheny KF, Bergman RN, Tuomilehto J, Collins FS, Boehnke M. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- Serrano M, Lee H, Chin L, Cordon-Cardo C, Beach D, DePinho RA. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- Sharpless NE, Bardeesy N, Lee KH, Carrasco D, Castrillon DH, Aguirre AJ, Wu EA, Horner JW, DePinho RA. Loss of p16Ink4a with retention of p19Arf predisposes mice to tumorigenesis. Nature. 2001;413:86–91. doi: 10.1038/35092592. [DOI] [PubMed] [Google Scholar]
- Sharpless NE, DePinho RA. The INK4A/ARF locus and its two gene products. Curr Opin Genet Dev. 1999;9:22–30. doi: 10.1016/s0959-437x(99)80004-5. [DOI] [PubMed] [Google Scholar]
- Shastry BS. Persistent hyperplastic primary vitreous: congenital malformation of the eye. Clin Experiment Ophthalmol. 2009;37:884–890. doi: 10.1111/j.1442-9071.2009.02150.x. [DOI] [PubMed] [Google Scholar]
- Sherr CJ, DePinho RA. Cellular senescence: mitotic clock or culture shock? Cell. 2000;102:407–410. doi: 10.1016/s0092-8674(00)00046-5. [DOI] [PubMed] [Google Scholar]
- Silva RL, Thornton JD, Martin AC, Rehg JE, Bertwistle D, Zindy F, Skapek SX. Arf-dependent regulation of Pdgf signaling in perivascular cells in the developing mouse eye. EMBO J. 2005;24:2803–2814. doi: 10.1038/sj.emboj.7600751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taharaguchi S, Yoshida K, Tomioka Y, Yoshino S, Uede T, Ono E. Persistent hyperplastic primary vitreous in transgenic mice expressing IE180 of the pseudorabies virus. Invest Ophthalmol Vis Sci. 2005;46:1551–1556. doi: 10.1167/iovs.04-0743. [DOI] [PubMed] [Google Scholar]
- Thornton JD, Swanson DJ, Mary MN, Pei D, Martin AC, Pounds S, Goldowitz D, Skapek SX. Persistent hyperplastic primary vitreous due to somatic mosaic deletion of the arf tumor suppressor. Invest Ophthalmol Vis Sci. 2007;48:491–499. doi: 10.1167/iovs.06-0765. [DOI] [PubMed] [Google Scholar]
- Visel A, Zhu Y, May D, Afzal V, Gong E, Attanasio C, Blow MJ, Cohen JC, Rubin EM, Pennacchio LA. Targeted deletion of the 9p21 non-coding coronary artery disease risk interval in mice. Nature. 2010;464:409–412. doi: 10.1038/nature08801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widau RC, Zheng Y, Sung CY, Zelivianskaia A, Roach LE, Bachmeyer KM, Abramova T, Desgardin A, Rosner A, Cunningham JM, Skapek SX. p19Arf represses platelet-derived growth factor receptor beta by transcriptional and posttranscriptional mechanisms. Mol Cell Biol. 2012;32:4270–4282. doi: 10.1128/MCB.06424-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou MM. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, Timpson NJ, Perry JR, Rayner NW, Freathy RM, Barrett JC, Shields B, Morris AP, Ellard S, Groves CJ, Harries LW, Marchini JL, Owen KR, Knight B, Cardon LR, Walker M, Hitman GA, Morris AD, Doney AS, McCarthy MI, Hattersley AT. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Zhao YD, Gibbons M, Abramova T, Chu PY, Ash JD, Cunningham JM, Skapek SX. Tgfbeta signaling directly induces Arf promoter remodeling by a mechanism involving Smads 2/3 and p38 MAPK. J Biol Chem. 2010;285:35654–35664. doi: 10.1074/jbc.M110.128959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zindy F, Eischen CM, Randle DH, Kamijo T, Cleveland JL, Sherr CJ, Roussel MF. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 1998;12:2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zindy F, Quelle DE, Roussel MF, Sherr CJ. Expression of the p16INK4a tumor suppressor versus other INK4 family members during mouse development and aging. Oncogene. 1997;15:203–211. doi: 10.1038/sj.onc.1201178. [DOI] [PubMed] [Google Scholar]