Abstract
We describe an approach for targeted genome resequencing, called oligonucleotide-selective sequencing (OS-Seq), in which we modify the immobilized lawn of oligonucleotide primers of a next-generation DNA sequencer to function as both a capture and sequencing substrate. We apply OS-Seq to resequence the exons of either 10 or 344 cancer genes from human DNA samples. In our assessment of capture performance, >87% of the captured sequence originated from the intended target region with sequencing coverage falling within a tenfold range for a majority of all targets. Single nucleotide variants (SNVs) called from OS-Seq data agreed with >95% of variants obtained from whole-genome sequencing of the same individual. We also demonstrate mutation discovery from a colorectal cancer tumor sample matched with normal tissue. Overall, we show the robust performance and utility of OS-Seq for the resequencing analysis of human germline and cancer genomes.
Next-generation sequencing has improved our ability to identify human genetic variation1–3. For many research studies and clinical applications, targeted resequencing is a practical and cost-effective approach for identifying candidate somatic or germline mutations, rare variants and polymorphisms of interest. Targeted resequencing also has the potential to become an important diagnostic tool. Academic and commercial groups have developed a variety of capture methods for enriching or selectively amplifying subsets of the genome for targeted resequencing4–10. These approaches are generally based on either PCR amplification, target-specific DNA circularization or hybridization-based selection of DNA targets. Despite the success of current targeted-resequencing approaches, substantial technical issues are associated with each approach11. Simplex PCR, even when run in massively parallel reactions, requires specialized equipment, which can increase costs for large-scale targeted resequencing analysis11. Targeted circularization methods have not been expanded to capture exomes11. Hybridization-based methods involve complex workflows that require additional PCR and sample preparation steps.
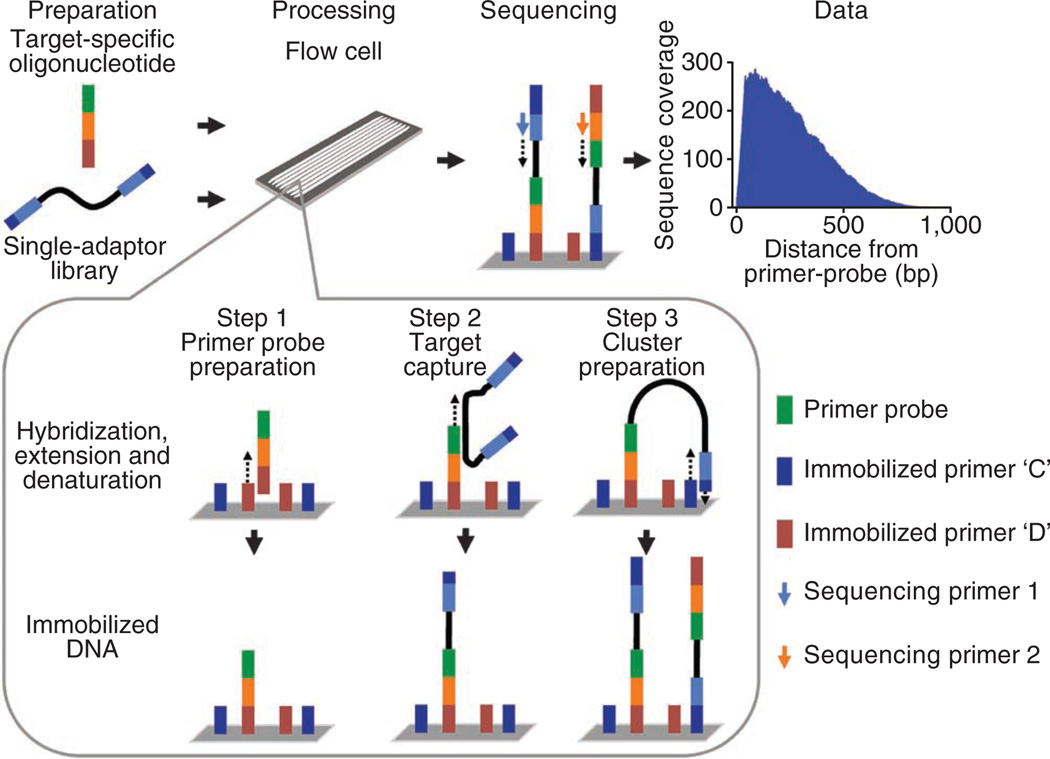
Here we report OS-Seq, a targeted resequencing approach that involves capturing and sequencing genomic targets on a sequencer’s solid-phase support, such as the Illumina flow cell (Fig. 1), thereby overcoming many of the limitations encountered in targeted resequencing of human normal and cancer genomes. Briefly, target-specific oligonucleotides were first synthesized and immobilized on the flow cell; these ‘primer probes’ served as both capture probes and sequencing primers (Supplementary Fig. 1). Second, a single-adaptor library prepared from genomic DNA was added to the flow cell, where the desired targets were captured by the immobilized primer probes. Third, the captured library fragments were prepared for bridge amplification, clustered and sequenced.
Figure 1.
Overview of OS-Seq. Capture of targets, processing and sequencing are performed on an Illumina next-generation sequencer. Reads originating from each primer probe are target and strand specific. Shown here is the median coverage profile for OS-Seq-366. For step 1, target-specific oligonucleotides are used to modify flow cell primers to create ‘primer probes’. Hybridized oligonucleotides are used as a template for DNA polymerase and D primers are extended. After denaturing, target-specific primer probes are randomly immobilized on the flow cell. For step 2, genomic targets in a single-adaptor library are captured using primer probes. These adapters incorporate sites for sequencing primers and immobilized flow cell primers. Targets in the single-adaptor library are captured during a high-heat hybridization step to their complementary primer probes. Captured single-adaptor library fragments are used as a template for DNA polymerase, and primer probes are extended. Denaturation releases template DNA from immobilized targets. For step 3, immobilized captured targets are rendered to be compatible for DNA sequencing. During a low-heat hybridization step the single-adaptor tails of the immobilized targets hybridize to type C primers on the flow cell surface, which stabilizes a bridge structure. The 3′ ends of immobilized targets and C primers are extended using DNA polymerase, which creates two molecules capable of bridge PCR. After denaturation, bridge amplification and cluster processing, paired-end sequencing can be conducted.
To prepare the capture substrate, we reengineered the Illumina flow cell by modifying a subset of the existing primer lawn to become target-specific primer probes. To create these primer probes, we hybridized the 3′ universal sequence of a complex pool of oligonucleotides to its complement on the flow cell and extended the immobilized primer using a DNA polymerase extension reaction. The result was a set of randomly placed, target-specific primer probes that were fixed onto the flow cell surface. During a high-heat incubation, the primer probes hybridized to complementary target sequences within the single-adaptor genomic DNA library; after hybridization, the primer probes then functioned as primers for another DNA polymerase extension reaction. The extension step effectively captured the target sequence. After extension, we performed a denaturation step followed by low-heat hybridization to stabilize the sequencing library adaptor to its complement on the flow cell, which creates a bridge structure. A third DNA polymerase extension reaction incorporates additional sequence to the 3′ ends, creating two molecules capable of solid phase amplification. Captured molecules were bridge amplified, processed and sequenced using the standard sequencing protocol. A detailed description of the molecular biology steps in OS-Seq is given in Supplementary Methods and related programs for OS-Seq are provided in Supplementary Data Files.
As a proof-of-principle demonstration, we developed two capture assays. First, we designed 366 OS-Seq primer probes to flank the exons of ten cancer genes (OS-Seq-366) (Supplementary Fig. 2). This assay was intended to test the OS-Seq method and not to definitively cover each exon. We synthesized OS-Seq-366 oligonucleotides using column-based methods. Second, to demonstrate scalability, we designed and synthesized 11,742 primer probes to capture the exons of 344 cancer genes (OS-Seq-11k). These primer probes avoided repeats and were tiled across large exons for improved exon coverage. For high-throughput production of OS-Seq-11k, we synthesized the oligonucleotides on a programmable microarray. These array-synthesized oligonucleotides required amplification for processing and for obtaining sufficient material for OS-Seq (Supplementary Fig. 3). After processing, OS-Seq oligonucleotides contained a target-specific 40-mer complementary to the 5′ end of the targeted region (Supplementary Fig. 4). These oligonucleotides also contained sequence required for annealing the paired-end sequencing primer and for hybridization to the immobilized primer lawn on the flow cell.
To assess capture performance of the OS-Seq-366 (10 genes) and OS-Seq-11k (344 genes) assays, we prepared and sequenced DNA from a Yoruban individual (NA18507). Paired-end sequencing was conducted on all targeting assays. The first read (read 1) is derived from targeted genomic DNA, and the second read (read 2) comes from the synthetic target-specific primer probes (Fig. 1). For each OS-Seq-366 experiment, we used a single lane of an Illumina GAIIx. We developed an indexing scheme for multiplex sequencing using adapters with a unique barcode sequence (Supplementary Fig. 4c) positioned between the read 1 sequencing primer and the genomic DNA fragment. Using this system, the barcode is sequenced as the first seven bases of read 1. With this indexing scheme, each sample of OS-Seq-11k was run on the equivalent of 1.3 lanes. Overall, 87.6% of OS-Seq-366 reads and 91.3% of OS-Seq-11k reads were mapped to the human genome reference after indexing (Table 1). In comparison, 58% of reads derived using a previously reported hybrid selection method could be mapped to the human genome reference9.
Table 1.
A performance analysis of OS-Seq
| Sample | NA18507 | NA18507 | Normal | Tumor |
|---|---|---|---|---|
| Number of primer probes | 366 | 11,742 | 11,742 | 11,742 |
| Total reads | 1,969,091 | 1,602,825 | 2,038,270 | 1,551,279 |
| Mapped reads (percentage of total reads) | 1,725,215 (87.6%) | 1,463,782 (91.3%) | 1,897,967 (93.1%) | 1,415,388 (91.2%) |
| Captured on-target readsa (percentage of mapped reads) | 1,499,052 (86.9%) | 1,365,305 (93.3%) | 1,747,192 (92.1%) | 1,316,563 (93.0%) |
| Captured on-target exon readsb (percentage of mapped reads) | 518,318 (30.0%) | 624,937 (42.7%) | 725,072 (38.2%) | 608,458 (43.0%) |
| Captured off-target reads (percentage of mapped reads) | 226,163 (13.1%) | 98,477 (6.7%) | 150,775 (7.9%) | 98,825 (7.0%) |
| On-target regiona | 233 kb | 7,296 kb | 7,296 kb | 7,296 kb |
| Captured on-target region used for SNV callinga,c (percentage of on-target region) | 191 kb (82.0%) | 1,541 kb (21.1%) | 1,754 kb (24.0%) | 1,476 kb (20.2%) |
| OS-Seq SNVs called from captured on-target region | 105 | 985 | 871 | 727 |
| OS-Seq SNPs that are reported | 97%d | 95.7%d | - | - |
| OS-Seq SNPs concordant with array genotype | - | - | 99.8%e | 99.5%e |
| Exon regionsb | 31 kb | 959 kb | 959 kb | 959 kb |
| Captured exon regionsb,f (percentage of exon regions) | 26 kb (83.9%) | 917 kb (95.6%) | 901 kb (94.0%) | 909 kb (94.8%) |
| Average fold-coverage on captured exonsb,f | 729 | 31 | 38 | 31 |
Within 1 kb from primer probes.
Within exons.
Filtered insert size ≥40+ read 1 length. Fold-coverage ≥10. Phred-like quality score >50.
Merged variant bases from ref. 3 and dbSNP131.
Positions genotyped using Affymetrix SNP 6.0 arrays.
Fold-coverage ≥1.
To assess overall coverage of each primer probe, we determined the number of reads originating from the read 1 data that fell within 1 kb from the 3′ end of the primer probe. OS-Seq primer probes are strand specific and only capture the 5′ ends of the DNA targets (Supplementary Fig. 5). As an example, the median coverage profile of all primer probes in OS-Seq-366 (Fig. 1) illustrates how sequence is captured up to 1 kb downstream from the primer probe. Generally, we detected a bias toward smaller insert sizes; for OS-Seq-366, 50% of targeted reads mapped within 283 bases from the primer probes. In both assays, we identified additional reads beyond the 1 kb interval and as far distant as 1.7 kb. The sequence reads beyond 1 kb represent the tail end of the capture distribution from any given primer probe and were <0.15% of the overall sequence data for both OS-Seq-366 and OS-Seq-11k. We have observed that the coverage distribution characteristics are correlated with the fragment size introduced during library creation and from size constraints inherent to bridge formation and solid-phase PCR (Supplementary Fig. 5). Also, coverage along the target can be increased by introducing a higher molar concentration of the single-adaptor library, sequencing additional lanes or using longer reads.
On-target reads were defined as read 1 sequences that mapped within 1 kb of a primer probe. Using these on-target coverage criteria, 86.9% of 40-base reads in OS-Seq-366 (10 genes) and 93.3% of 53-base reads in OS-Seq-11k (344 genes) were on target (Table 1). The improved specificity of OS-Seq-11k is likely a result of our effort to refine the in silico design of the primer probes by avoiding repeat sequences . In comparison, 89% of 76-base reads and 50% of 36-base reads mapped in proximity of a probe in a published hybrid selection method9, suggesting that on-target specificity is similar between methods and that moving toward longer reads may improve the on-target specificity of OS-Seq. On-exon specificity of OS-Seq was also similar to the published hybrid selection method. Using OS-Seq-11k, we observed that 42.7% of reads mapped within exons (Table 1), whereas a hybrid selection capture technology reported 42% of reads mapped to exons9.
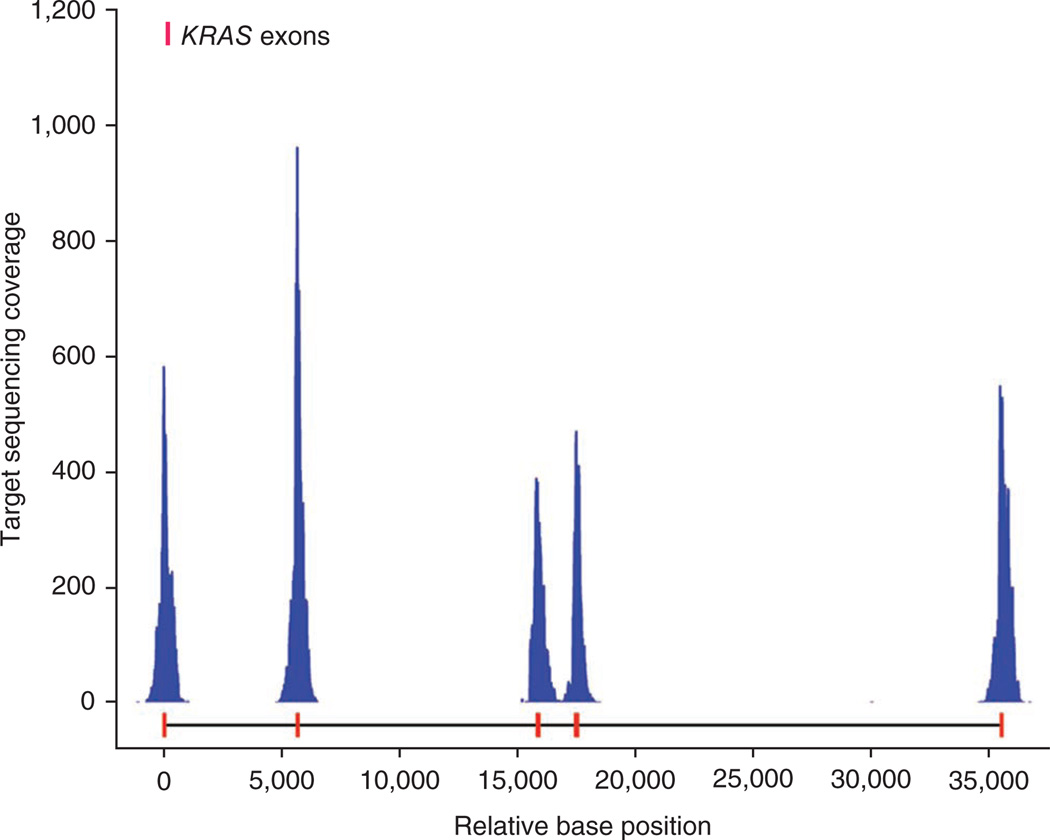
As an example of a profile of typical capture gene coverage, we show the target sequence data for the KRAS gene (Fig. 2). The exon targets were sequenced at high fold-coverage relative to the off-target adjacent regions. We report the average fold coverage for exons in Table 1 and detailed breakdowns of coverage classes (e.g., 10× and 20×) in Supplementary Table 1. Overall, 83.9% of exon bases in the OS-Seq-366 were covered with at least one read, with a portion of the remainder not having been intentionally targeted in this pilot assay. Similarly, among the three samples analyzed with OS-Seq-11k, 94–95.6% of exon bases were covered with at least one read. Compared with OS-Seq-366, the tiling-based OS-Seq-11k assay showed increased sequence coverage on exons.
Figure 2.
Targeted sequencing coverage profile along the KRAS gene from the OS-Seq-366 assay. Base positions relative to the start of exon 1 are presented on the x axis and KRAS exons are marked in red on the x axis. Sequencing fold coverage is listed on the y axis.
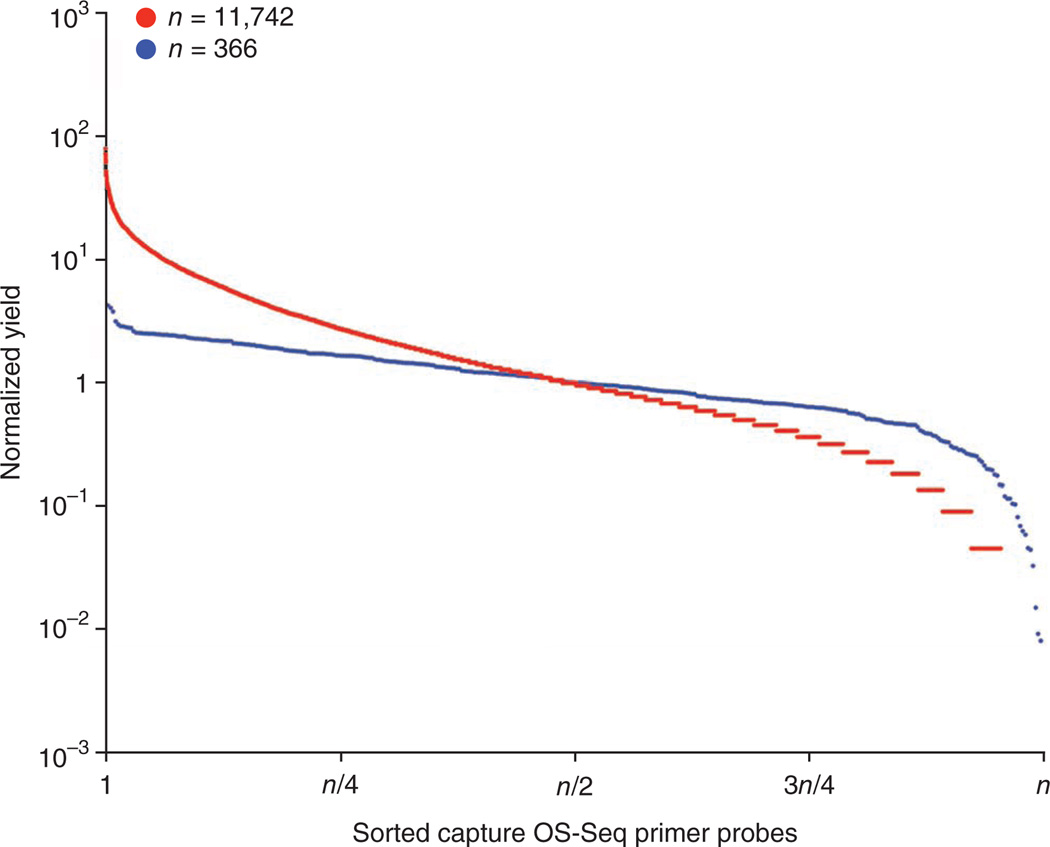
We evaluated the uniformity of targets selected by OS-Seq by binning read 1 data by its associated primer probe and counting reads aligning to its target. After sorting the OS-Seq-366 primer probes based on the observed capture yields, we observed that 100% of the primer probes had a yield minimum of one sequence read and the yield of 89.6% of the primer probes were within a tenfold range. Similarly, for OS-Seq-11k, 95.7% of primer probes had a capture yield minimum of one sequence read and 54% of the primer probes had a yield within a tenfold range (Fig. 3). OS-Seq-366 oligonucleotides were column-synthesized and in equimolar concentration in the primer probe pooling. Higher variance in primer probe yields for OS-Seq-11k is most likely attributable to amplification bias introduced during PCR amplification of the microarray-synthesized oligonucleotides used for primer probe pools.
Figure 3.
Coverage assessment of OS-Seq. Uniformity assessment of primer-probe yields within column- and array-synthesized oligonucleotides. We compared the uniformity of capture between column-synthesized (blue, n = 366) and array-synthesized (red, n = 11,742) oligonucleotides. On the x axis, oligonucleotides are sorted by sequence capture yields, on the y axis is the normalized primer probe yield. To calculate normalized yield, we divided the yield of each oligonucleotide by the median yield from all oligonucleotides.
We evaluated the technical reproducibility of OS-Seq by comparing the sequence yields of individual primer probes from the OS-Seq-11k assay (Supplementary Fig. 6). We pooled multiplexed libraries (NA18507, normal and tumor) and performed the capture and sequencing on two independent Illumina GAIIx lanes. We compared the sequence yields of each individual primer probe between the technical replicates and calculated the correlation coefficient, R2 = 0.986. For evaluation of biological reproducibility, two different multiplexed sequencing libraries were run in the same lane. The correlation coefficient of biological replicates was R2 = 0.90.
To assess the variant calling performance of OS-Seq-366 and OS-Seq-11k assays, we conducted a targeted sequencing analysis on NA18507, whose genome has undergone whole genome sequencing analysis3. For SNV calling with either OS-Seq assay, we analyzed only on-target positions with genotype quality scores >50 and a minimum of 10× coverage (Table 1). For OS-Seq-366 and OS-Seq-11k data, a total of 191 kb and 1,541 kb fulfilled these criteria, respectively. We called 105 SNVs from OS-Seq-336 and 985 SNVs from OS-Seq-11k (Table 1). We extracted the published NA18507 SNVs and other reported single-nucleotide polymorphisms (SNPs) that occurred in these same high-quality regions. In comparison, 97% of the OS-Seq-366 and 95.7% of the OS-Seq-11k had previously been reported (Table 1). For OS-Seq-366 and OS-Seq-11k the sensitivity of variant detection was 0.97 and 0.95, respectively, based on the reported SNPs (Supplementary Table 2).
To demonstrate the utility of OS-Seq in identifying somatic mutations, we applied the OS-Seq-11k assay to genomic DNA derived from a matched normal–colorectal carcinoma tumor pair. The samples were indexed, pooled and sequenced in the same lane of the flow cell to minimize technical bias. Using the same quality and coverage criteria for the analysis of NA18507, we identified 871 SNVs from the normal sample and 727 from the tumor (Supplementary Table 3). For comparison, we genotyped the two samples with the Affymetrix SNP 6.0 array. Genotyping accuracy using Affymetrix SNP 6.0 arrays and the Birdseed algorithm is high, as the average successful call rate for SNPs is 99.47% and called SNPs have a 99.74% concordance with HapMap genotypes from other platforms12. In comparing the OS-Seq SNVs to Affymetrix SNPs, we observed a high concordance of 99.8% for the normal and 99.5% for the tumor. By filtering normal tissue variants and considering novel cancer-specific variants where coverage was >40, we identified and validated a clear pathogenic nonsense mutation of SMAD4 (S144*), which is frequently mutated in colorectal cancer and a colon cancer driver gene.
We investigated the capture efficiency of individual primer probes within the OS-Seq-366 and OS-Seq-11k assays and assessed the performance of each primer probe. A unique feature of OS-Seq is that captured genomic sequences can be matched to their corresponding primer probes when sequenced with paired ends. Read 1 originates from the 3′ end of the captured target and read 2 begins at the synthetic OS-Seq primer probe sequence. Thus, read 1 represents the captured genomic DNA sequence, and read 2 functionally serves as a molecular barcode for a distinct primer probe. This enables the identification of the exact OS-Seq primer probe that mediated the targeting, and facilitates the assessment of the performance of individual primer probes. For example, we observed a strong relationship between the GC content of primer probes and target sequence yield. Extremely low GC (<20%) or high GC content (>70%) was associated with failure of a primer probe to capture its target sequence (Supplementary Fig. 7).
We developed OS-Seq technology for streamlined and highly scalable targeted resequencing of human normal and cancer genomes. Our proof-of-principle study shows that the OS-Seq assay effectively and reproducibly captured target genomic regions with good uniformity and high specificity. Variant analysis of the NA18507 reference genome demonstrated high specificity and low false-discovery rate for SNV determination. Targeted resequencing of matched colorectal normal and tumor samples demonstrated the applicability of OS-Seq to high-throughput genetic analysis of cancer genomes.
The OS-Seq technology enables users to create custom targeted-resequencing assays. The design and production of the primer probe oligonucleotides is relatively straightforward, and target regions can be selected simply by using balanced GC and nonrepetitive sequence. Although our largest targeting assay covered the exons and adjacent sequence of 344 genes, we believe that OS-Seq will scale to target many more genes. We estimated that there was >2,000-fold excess of primer probes compared to target fragments in the hybridization mix inside the flow cell (Supplementary Methods). We extrapolated that during a 20-h hybridization 4.9% of all potential targets within the library were captured for sequencing.
OS-Seq sample preparation does not require a gel excision or other narrow size purifications and therefore can be completed in one day and is readily automated (Supplementary Fig. 8). Only a single adaptor needs to be added to the 5′ ends of a genomic DNA fragment. This feature allows straightforward sample multiplexing of sequencing assays and has many potential applications, such as matched normal and cancer genome sequencing. Overall, OS-Seq is particularly useful for translational studies and clinical diagnostics by enabling high-throughput analysis of candidate genes and identification of clinically actionable target regions.
ONLINE METHODS
Genomic DNA samples
Genomic DNA for NA18507 was obtained from the Coriell Institute. Fresh frozen tissue samples were obtained from a colorectal cancer patient. Patient material was obtained with informed consent from the Stanford Cancer Center and the study was approved by the institutional review board at Stanford University School of Medicine. Frozen tissue sections were prepared and stained with hematoxylin-eosin, and the tumor composition of each sample was determined by pathological examination. Samples representing tumor and normal tissues were dissected from areas where cellular composition was 90% tumor or purely normal, respectively. Genomic DNA was extracted using E.Z.N.A. SQ DNA/RNA Protein Kit (Omega Bio-Tek). Standard protocols for DNA preparation, array hybridization and scanning were used to analyze samples using SNP 6.0 arrays (Affymetrix). Data analysis was performed using the Genotyping Console software and Birdseed V2 algorithm (Affymetrix). Thirteen additional microarray data sets were analyzed in concert with the studied samples to assess the quality of the SNP calls. SNP 6.0 array data were filtered using P-value threshold of 0.01.
Target selection and in silico OS-Seq oligonucleotide design
We used CCDS build release 20090902 (ftp://ftp.ncbi.nlm.nih.gov/pub/CCDS/current_human/), human genome build NCBI 37 – hg19 (ftp://ftp.ncbi.nlm.nih.gov/genomes/H_sapiens/) and dbSNP Build ID 131 (http://www.ncbi.nlm.nih.gov/SNP/) as the polymorphism reference data set. For gene selection, the GeneRanker annotation database was used to choose 344 cancer genes prioritized by importance13. To find target-specific sequences of oligonucleotides, we took the exon definitions for the candidate genes from CCDS14. For most targeted exons (<500 bp), the 40-mer target-specific sequences were ten bases outside of the 5′ end of the exon boundary (Supplementary Fig. 2a). Both strands of the exons were targeted using individual primer probes. OS-Seq-366 only covered the flanks of exons. In the OS-Seq-11k assay, exons >500 bp were treated by tiling target-specific sequences until the entire exonic region was covered (Supplementary Fig. 2b). To improve the on-target specificity of OS-Seq-11k, we used Repbase to identify and eliminate oligonucleotide sequences that targeted highly repetitive sequences15.
Oligonucleotide synthesis
Two strategies were applied for oligonucleotide synthesis. For OS-Seq-366, we designed 366 101-mer oligonucleotides (Supplementary Fig. 4a), which were then column-synthesized (Stanford Genome Technology Center) (Supplementary Fig. 3a). Oligonucleotides were quantified and pooled in equimolar concentration. For OS-Seq-11k, an in situ microarray synthesis (LC Sciences) approach was used to synthesize the 11,742 precursor oligonucleotides (Supplementary Fig. 4b). The sequences of target-specific oligonucleotides are in Supplementary Table 4 (OS-Seq-366) and Supplementary Table 5 (OS-Seq-11k).
Amplification of microarray-synthesized oligonucleotides
We used three 25 µl subpools of precursor 80-mer oligonucleotides (587, 638 and 415 nM) (Supplementary Fig. 4b). We used a PCR approach to amplify the precursor, low-concentration oligonucleotides (Supplementary Fig. 3b). The array-synthesized oligonucleotide subpools were diluted to 10 fM/oligo and used as a template for PCR amplification. PCR was done using Taq DNA polymerase (New England Biolabs), and dNTPs (1 mM dATP, 1 mM dCTP, 1 mM cGTP and 1 mM dTTP) in standard reaction conditions. After denaturation in 95 °C for 30 s, 20 amplification cycles (95 °C, 30 s; 55 °C, 30 s; 68 °C, 30 s) were carried out. Amplification primer 1 contained uracil at the 3′ end, whereas amplification primer 2 incorporated additional functional sequences (Supplementary Fig. 4b). Amplified oligonucleotides were purified to remove excess primer (Fermentas), then processed using 0.1 U/µl Uracil DNA-excision Mix (Epicentre) in 37 °C for 45 min to detach the universal amplification primer site and cleave the mature 101-mer coding strands of the oligonucleotides. The oligonucleotides require the 5′ ends to be functional and free to have accurate extension of the target-specific site during primer probe immobilization. After heat inactivation of the enzymes (65 °C, 10 min), the oligonucleotide preparations were purified (Fermentas). Finally, we quantified the three oligonucleotide subpools and created a single pool with equimolar concentration of each subpool.
Preparation of OS-Seq primer probes by modification of the flow cell primer lawn
In the Illumina Genome Analyzer IIx (Illumina) system, the solid phase support (that is, the flow cell) has two primers (C and D), which are randomly immobilized on a polyacrylamide layer at extremely high density3,16. For OS-Seq experiments, a subset of the D primers was specifically modified using the Illumina Cluster station (Supplementary Methods). Prior to the NGS primer modification, 133 nM oligonucleotide pools were heat denatured at 95 °C for 5 min. We used heat denaturing (95 °C for 5 min) to free the coding strand of the OS-Seq oligucleotides. Additional strand purification was not required as the second strand is inactive on the flow cell and is washed away after hybridization. Denatured oligonucleotides were diluted with 4× Hybridization buffer (20× SSC, 0.2% Tween-20). The resulting 100 nM oligonucleotides were used in the flow cell modification experiments. The oligonucleotide (30 µl) was dispensed into each lane of the flow cell. During a temperature ramp (from 96 °C to 40 °C in 18 min) oligonucleotides annealed specifically to the immobilized primer D. Then, DNA polymerase was used to extend the ‘D’ primer with the annealed oligonucleotide as a template. After extension, the original oligonucleotide template was denatured from the extended D primer and washed from the solid phase support. Standard Illumina v4 reagents were used for extension, wash and denaturation steps. The modification of primer D caused immobilization of the primer probes.
Sequencing library preparation
We outline the general scheme of genomic DNA fragmentation, end repair, A-tailing, adaptor ligation and PCR used in the preparation of the OS-Seq sequencing library in Supplementary Fig. 1. We used 1 µg of genomic DNA from NA18507 and flash frozen colorectal cancer and normal colon samples as starting material. Genomic DNA was fragmented using Covaris E210R (Covaris) to obtain a mean fragment size of 500 bp (duty cycle 5%, intensity 3, 200 cycles per burst and 80 s). The randomly fragmented DNA was end-repaired using 0.25 U of Klenow large fragment, 7.5 U of T4 DNA polymerase, 400 µM of each dNTP, 25 U of T4 Polynucleotide kinase and T4 DNA ligase buffer with ATP (all from New England Biolabs) in a 50 µl reaction volume at 25 °C for 45 min. After end repair, adenines were added to the 3′ ends of the template DNA using 3.2 U of Taq DNA polymerase (New England Biolabs), 100 µM dATP (Invitrogen) and Taq buffer with 1.5 mM MgCl2 in 80 µl reaction in 72 °C for 15 min. Before adaptor ligation, reactions were purified using PCR purification kit (Fermentas).
We developed an indexing system for OS-Seq. The sequencing library adapters contain an optional 6-base indexing sequence, a sequencing primer 1 site and a 12-mer sequence for primer C hybridization (Supplementary Methods and Supplementary Fig. 4c). Sequence information for the basic oligonucleotides is provided in the Supplementary Methods. Adaptor oligonucleotides were synthesized at the Stanford Genome Technology Center. Before ligation, adaptor oligonucleotides were annealed during a slow temperature ramp down from 85 °C to 20 °C, followed by a fast cool down to 4 °C to avoid short hairpin structures. For the targeted resequencing of NA18507, we used both a singleplex adaptor as well as a multiplex adaptor with ‘AACCTG’ tag. For the indexing of the matched normal tumor sample, we used a ‘TGCTAA’ barcode for the normal tissue, whereas the tumor sample was tagged with ‘AGGTCA’. Double-strand DNA adapters with T-overhang were ligated to the A-tailed templates using 2,000 U of T4 DNA ligase (New England Biolabs) and T4 DNA ligase buffer in 25 °C for 1 h. After adaptor ligation, reactions were purified using PCR purification kit (Fermentas) and libraries were amplified using PCR. We prepared 50 µl reactions of 1 U of Phusion Hot Start DNA polymerase (Finnzymes), 1 µM library amplification primer (Supplementary Methods), Phusion HF buffer and 200 µM of each dNTP (NEB). Reactions were denatured in 98 °C for 30 s. After that, 20–22 PCR cycles were carried out (98 °C for 10 s, 65 °C for 30 s and 72 °C for 30 s) followed by 72 °C for 7 min and 4 °C. Thereafter, PCR reactions were purified using PCR purification kit (Fermentas) and quantified. Multiplexed libraries were pooled in equal concentrations.
Capture of targets using primer probes
Targets were captured on the flow cell using OS-Seq primer probes (Fig. 1 and Supplementary Methods). An Illumina Cluster Station was used to carry out the primer probe hybridization and extension steps (Supplementary Methods). Prior to hybridization to primer probes, 22.5 µl of sequencing libraries (40–56.6 ng/µl) was denatured at 95 °C for 5 min. After heat denaturing, the genomic DNA libraries were diluted to a total volume of 30 µl using 4× hybridization buffer. The final DNA concentrations of sequencing libraries ranged from 30 to 41.7 ng/µl. We injected 30 µl of the genomic sequencing libraries into the flow cell. Target DNA was hybridized to the primer probes by incubating the sequencing libraries in the flow cell at 65 °C for 20 h. Due to the high concentration of the sequencing libraries, the hybridization volume was kept at minimum. Therefore, a custom Cluster Station program was developed to allow reproducible low-volume hybridization. The following extension, wash and denaturation steps were performed using Illumina v4 reagents. These programs are available in the Supplementary Data Files.
Flow cell processing and sequencing
After capture and extension of the targets, the temperature of the flow cell was raised to 96 °C and cooled to 40 °C for 18 min to allow the 12 bases in the 3′ end of the captured genomic DNA library fragments to hybridize to primer C (Fig. 1 and Supplementary Methods). In the bridge formation, the library fragment and primer C were extended using DNA polymerase to finalize and replicate the captured DNA fragment. Afterwards, bridge PCR was carried out to generate the clonally amplified sequencing clusters. Samples were sequenced using 40-by-40 (OS-Seq-366) or 60-by-60 (OS-Seq-11k) paired-end cycles on an Illumina Genome Analyzer IIx using standard version 4 sequencing reagents and recipes (Illumina). Image analysis and base calling were done using the SCS 2.8 and RTA 2.8 software (Illumina).
Sequence analysis and variant detection
Sequence reads were aligned to the human genome version human genome build NCBI 37 – hg19 using Burrows-Wheeler Aligner (BWA)17. After alignment, on-target reads (read 1) were defined as being within 1 kb of the 5′ end of the primer probe. Off-target reads were defined as aligning outside 1 kb of the 5′ end of the primer probe or mapping on a different chromosome from the location of the associated primer probe. For the de-multiplexing of indexed lanes, we used a perl script to generate an index of the 7-base tags using the base-call files (Supplementary Data Files). This index file and another perl script were used to de-multiplex either the combined base-call file (so that separate fastq files can be generated for further processing) or the aligned file.
To eliminate any synthetic primer probe sequences for variant calling, we applied insert size filtering on the mate pairs. We determined the insert size by comparing alignment of paired sequence reads. For variant calling, extracted sequences were required to have an insert size greater than (40 + the length of read 1). After insert size filtering, variant calling was performed using SAMtools and BCFtools. A sequence pileup was performed against the human genome (hg19) using SAMtools mpileup with a mapping quality threshold of 50. BCFtools view was used to genotype base positions and data were filtered using vcfutils.pl, a variant filter perl script provided in the SAMtools package. The vcfutils varFilter conditions were (i) coverage of 10 or greater, (ii) removal of the strand bias filter (because OS-Seq is a strand-specific capture method), (iii) forcing the script to output both reference and nonreference positions. Reference and nonreference calls were used for comparisons with the Affymetrix SNP 6.0 array data. Genotyped positions were filtered to have a Phred-like quality score >50. We used BEDtools intersectBed to define target regions for each primer probe and combinations where probes overlap in their targets.
Variant comparison
For quality assessment of extracted variants, variant calls of the NA18507 data were compared to calls from variants identified from a complete genome sequence analysis3 and HapMap genotyping data. Comparisons of OS-Seq data and Affymetrix SNP 6.0 array data were made using MATLAB scripts. We used dbSNP131 for SNP annotation.
Supplementary Material
Acknowledgments
This work was supported by the US National Institutes of Health grants K08CA96879 (H.P.J.), DK56339 (H.P.J.), P01HG000205 (J.D.B., J.M.B. and H.P.J.), RC2HG005570 (G.N., J.M.B. and H.P.J.), R21CA140089 (G.N., J.M.B. and H.P.J.), the Sigrid Jusélius Foundation Fellowship (S.M.), the Academy of Finland Grant (S.M.), Doris Duke Clinical Foundation Clinical Scientist Development Award (H.P.J.), the Howard Hughes Medical Foundation Early Career Grant (H.P.J.), the Reddere Foundation Award (H.P.J.), the Liu Bie Ju Cha and Family Fellowship in Cancer (H.P.J.), and the Wang Family Foundation Research Grant (G.N. and H.P.J.).
Footnotes
Accession codes. Short read sequence data have been deposited at the NCBI Sequence Read Archive (SRA) under the accession number SRA036669. Programs and related scripts as cited in the Supplementary Methods are also available at http://dna-discovery.stanford.edu/.
Note: Supplementary information is available on the Nature Biotechnology website.
AUTHOR CONTRIBUTIONS
The project was conceived and experiments planned by S.M., J.D.B. and H.P.J. S.M. and J.D.B. carried out all experiments. J.D.B., S.M., J.M.B., G.N. and H.P.J. performed data analysis. J.D.B., S.M., J.M.B. and H.P.J. wrote the manuscript, and all authors reviewed it. All aspects of the study were supervised by H.P.J.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shendure J, et al. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- 3.Bentley D, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albert TJ, et al. Direct selection of human genomic loci by microarray hybridization. Nat. Methods. 2007;4:903–905. doi: 10.1038/nmeth1111. [DOI] [PubMed] [Google Scholar]
- 5.Hodges E, et al. Genome-wide in situ exon capture for selective resequencing. Nat. Genet. 2007;39:1522–1527. doi: 10.1038/ng.2007.42. [DOI] [PubMed] [Google Scholar]
- 6.Okou DT, et al. Microarray-based genomic selection for high-throughput resequencing. Nat. Methods. 2007;4:907–909. doi: 10.1038/nmeth1109. [DOI] [PubMed] [Google Scholar]
- 7.Porreca GJ, et al. Multiplex amplification of large sets of human exons. Nat. Methods. 2007;4:931–936. doi: 10.1038/nmeth1110. [DOI] [PubMed] [Google Scholar]
- 8.Turner EH, Lee C, Ng SB, Nickerson DA, Shendure J. Massively parallel exon capture and library-free resequencing across 16 genomes. Nat. Methods. 2009;6:315–316. doi: 10.1038/nmeth.f.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gnirke A, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat. Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Natsoulis G, et al. A flexible approach for highly multiplexed candidate gene targeted resequencing. PLoS ONE. 2011;6:e21088. doi: 10.1371/journal.pone.0021088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mamanova L, et al. Target-enrichment strategies for next-generation sequencing. Nat. Methods. 2010;7:111–118. doi: 10.1038/nmeth.1419. [DOI] [PubMed] [Google Scholar]
- 12.Korn JM, et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat. Genet. 2008;40:1253–1260. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonzalez G, Uribe JC, Armstrong B, McDonough W, Berens ME. GeneRanker: an online system for predicting gene-disease associations for translational research. Summit on Translat. Bioinforma. 2008;2008:26–30. [PMC free article] [PubMed] [Google Scholar]
- 14.Pruitt KD, et al. The consensus coding sequence (CCDS) project: Identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 2009;19:1316–1323. doi: 10.1101/gr.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jurka J, et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 16.Fedurco M, Romieu A, Williams S, Lawrence I, Turcatti G. BTA, a novel reagent for DNA attachment on glass and efficient generation of solid-phase amplified DNA colonies. Nucleic Acids Res. 2006;34:e22. doi: 10.1093/nar/gnj023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.