Abstract
Death-associated protein kinase (DAPK3) is a serine/threonine kinase involved in various signaling pathways important to tissue homeostasis and mammalian biology. Considered to be a putative tumor suppressor, the molecular mechanism by which DAPK3 exerts its suppressive function is not fully understood and the field lacks an appropriate mouse model. To address these gaps, an in vitro 3D tumorigenesis model was used and a constitutive DAPK3 knockout mouse was generated. In the 3D morphogenesis model, loss of DAPK3 through lentiviral-mediated knockdown enlarged acinar size by accelerated acini proliferation and apoptosis while maintaining acini polarity. Depletion of DAPK3 enhanced growth factor-dependent mTOR activation and, furthermore, enlarged DAPK3 acini structures were uniquely sensitive to low doses of rapamycin. Simultaneous knockdown of RAPTOR, a key mTORC1 component, reversed the augmented acinar size in DAPK3-depleted structures indicating an epistatic interaction. Utilizing a validated gene trap strategy to generate a constitutive DAPK3 knockout mouse, it was demonstrated that DAPK3 is vital for early mouse development. The Dapk3 promoter exhibits spatio-temporal activity in developing mice and is actively expressed in normal breast epithelia of adult mice. Importantly, reduction of DAPK3 expression correlates with the development of DCIS and more aggressive breast cancer as observed in the Oncomine database of clinical breast cancer specimens.
INTRODUCTION
Death-associated protein kinase 3 (DAPK3 or ZIPK) is a member of the DAPK serine/ threonine protein kinase family and is known to regulate smooth muscle contraction, cell-cell adhesion, cytoskeleton dynamics, inflammation as well as cardiovascular functions and is thought to serve as a tumor suppressor through regulation of caspase-dependent and -independent apoptosis, proliferation and autophagy (1). The DAPK family contains 4 additional members, including DAPK1 (herein referred to as DAPK), DAPK2 (also DAPK-related protein 1), DRAK-1 and DRAK-2 (DAPK-related apoptosis-inducing protein kinase-1 and -2), which all share homology within their kinase domain. DAPK3 contains an N-terminal kinase domain that shares 80% amino acid homology with the prototypical DAPK, and differs from other family members by the presence of a C-terminal leucine zipper motif and absence of calmodulin-regulated (CaM)and death domains.
Similar to other family members, DAPK3 is considered to be a tumor suppressor. Overexpression of DAPK3 in mammalian cells results in cell death and cell cycle inhibition, while kinase inactivating mutations along with recurrent deleterious somatic mutations are observed in lung and breast cancers, respectively(2–4). Knockdown of DAPK3 increases proliferation of various cell lines (2). Clinically, reduced DAPK3 mRNA correlates with increased tumor invasion, metastasis and overall survival in gastric carcinoma patients (5). Abrogation of DAPK3 mRNA expression was shown to significantly decrease cisplatin sensitivity in various lung cancer cell lines and may impact overall survival of non-small cell lung cancer patients treated with platinum-based therapy(6). DAPK3 is also considered a potentially novel breast cancer gene as recurrent DAPK3 alterations were observed in both BRCA1 mutant and non-mutant breast cancers (3). Additionally, human DAPK3 regulates a variety of signaling pathways commonly deregulated in cancer. For example, DAPK3 negatively regulates the canonical Wnt/ β-catenin pathway by disrupting the interaction between Nemo-like kinase and T-cell Factor 4 in colon cancer cell lines(7). It also regulates androgen receptor-mediated transcription via ubiquitination and degradation of androgen receptor in various cancer cell lines (8). DAPK3 also interacts with and/ or phosphorylates various cancer-associated proteins in vitro, including ATF4, AATF, Daxx, Par-4 and STAT-3 (4, 9–12).
Despite these in vitro and clinical observations, the full physiological significance of DAPK3 is not well understood. Additionally, compared to the prototypical DAPK family member DAPK, relatively little is known about the functional tumor suppressive mechanisms regulated by DAPK3. These limitations are potentially exacerbated by the lack of a knockout mouse model as well as inadequate cell culture models that cannot recapitulate the physiological context of tissue development or carcinogenesis. Three-dimensional (3D) in vitro tumor systems provide the ability to functionally investigate the contribution of tumor suppressors and oncogenes to the complex development and architecture of tumor spheroids(13). Given the utility of 3D tumor systems and the clinically observed mutations of DAPK3 in breast cancer(3), we chose to further explore the functional significance of DAPK3 in a MCF10A 3D morphogenesis model. When grown on an extracellular enriched matrix (Matrigel), the immortalized MCF10A epithelial cell line forms hollow spheroids that undergo a regulated and coordinated series of biochemical and phenotypic events (14). This model has been used to investigate the contribution of loss-of-function (LOF) alterations to acini development and early events in tumor formation.
Herein, we describe the functional significance of DAPK3 in MCF10A acini morphogenesis and characterize the early lethality observed in a DAPK3 knockout mouse. We identified that DAPK3 negatively regulates MCF10A morphogenesis through a mTORC1/S6 pathway that is independent of enhanced AKT and ERK signaling. Our studies also shed light on an unexpected role for DAPK3 in mouse development as well as a spatially distinct expression pattern with potential relevancy to human development and breast cancer. We also identified that DAPK3 is downregulated in aggressive breast cancer relative to less aggressive and normal patient samples.
MATERIALS & METHODS
Cell culture and reagents
MCF10A and 293T cells were obtained directly from the American Type Culture Collection in 2011 and not further tested. MCF10A cells were cultured as previously described (14) and 293T were cultured in DMEM supplemented with 10% FBS and L-glutamine. 3D morphogenesis assays were conducted as previously described (14). Inhibitors: LY294002 (Cell Signaling, 9901), rapamycin (Sigma-Aldrich, R8781).
Plasmids viral production, and lentiviral transduction
For DAPK3 over expression, the DAPK3 ORF was PCR amplified (from Addgene plasmid 23436) and subcloned into pLVX-IRES-Hyg (Clonetech). PCR primers, forward: 5′- GAGAGACTCGAGGCCACCATGTCCACGTTCAGGCAGGAG, and reverse: 5′-GAGAGAGGATCCTTACTAGCGCAGCCCGCACTCCACGCCCTGC, were used to create the restriction enzyme sites XhoI and BamHI (in bold) that allowed for ligation into the corresponding sites in pLVX-IRES-Hyg. For RasV12 overexpression, HRasV12 was amplified and subcloned into pLVX-IRES-Hyg. PCR primers, forward: 5′-GAGAGACTCGAGGCCACCATGACGGAATATAAGCTGGTGGTGGTGG, and reverse: 5′-GAGAGAGGATCCTTATCAGGAGAGCACACACTTGCAGCTCATG, were used to create the restriction enzyme sites XhoI and BamHI (in bold) that allowed for ligation into the corresponding sites in pLVX-IRES-Hyg.
pLKO.1-puro constructs obtained from the Genome Institute at Washington University were used for RNA interference (RNAi) against DAPK3. Sequences for the short hairpin RNAs are 5′-CGTTCACTACCTGCACTCTAA (herein referred to as sh1), 5′-CCCAAGCGGAGAATGACCATT (herein referred to as sh2) and shNeg (15). For lentiviral production, 8 x 105 293T cells were co-transfected with pCMV-VSV-G, pCMVΔR8.2, and pLKO.1-puro constructs using Fugene 6 (Promega). Forty-eight hours post-transfection, viral supernatants were collected. MCF10A were subsequently incubated for 8 hours with filtered viral supernatants after which fresh media was added. The following day the media was replaced with a second round of viral supernatant after which fresh media was added for 24 hours. Stable cells were then selected using 1 μg/ mL puromycin for two passages. Stable cells were then allowed to recover from selection for one passage prior to use in 2D and 3D assays. For 2D assays, stable cells were plated at an equal density such that they would be ~75% confluent at initiation of the experiment. PLKO.1-hygro shRNA constructs against RICTOR and RAPTOR were generated by subcloning hygromycin in place of puromycin within the pLKO.1-constructs and similarly selected using 500 μg/ mL hygromycin. For double knockdown, MCF10A were simultaneously infected with both appropriate shRNA-puromycin (shRNAP) and shRNA-hygromycin (shRNAH) virus and subsequently selected with both 1 μg/ mL puromycin and 500 μg/ mL hygromycin for one passage followed by one passage in normal media.
Immunofluorescence and confocal microscopy
MCF10A acini were grown in 8 well chamber slides and at the indicated time point were fixed with 4% paraformaldehyde (PFA) in PBS for 30 minutes at room temperature. Slides were washed three times (15 minutes each wash) at room temperature in 100mM glycine in PBS and subsequently permeabilized with 0.05% Tween 20 in PBS, pH7.4 for 20 minutes. Fixed acini were blocked in IF buffer (0.2% Triton X-100, 0.1% BSA-radioimmunoassay grade from Sigma Aldrich, 0.05% Tween 20 in PBS, pH 7.4) plus 10% normal goat serum for 1.5 hours at room temperature and then blocked in secondary block containing IF buffer plus 10% goat serum and 20μ/mL goat anti-mouse IgG F(ab′)2 fragment-specific (Jackson ImmunoResearch, Cat. 115-006-006) in a humidified chamber for 30 minutes. Fixed acini were then stained with 1:100 primary antibody in IF buffer plus 10% goat serum and 20μ/mL goat anti-mouse IgG F(ab′)2. Primary antibodies were as follows: rabbit anti-Ki67 (Cell Signaling, 9129), rabbit-anti cleaved caspase 3 (Cell Signaling, 9579), rat anti-integrin alpha 6 (Millipore, MAB1378) and rabbit anti-giantin (Covance, PRB-114C). The following day, slides were washed three times in IF buffer for 20 minutes each and incubated with a secondary antibody (conjugated to Alexa Fluor 488 or 594) diluted in IF buffer plus 10% goat serum for 40 minutes at room temperature in a humidified chamber. Slides were washed three times in IF buffer for 20 minutes each and then incubated with 1μM TOPRO3 iodide (Molecular Probes) in PBS for 10 minutes at room temperature in a humidified chamber. Slides were then washed with PBS once for 10 minutes and mounted with Prolong Antifade mounting medium (Molecular Probes). Images were obtained using an Olympus FV-500 confocal microscope with a 20X water objective. Images were processed using the Olympus FLUOVIEW Ver.2.1a Viewer and ImageJ software. For Ki67 and cleaved caspase 3 evaluation, a total of 10 fields with at least 4 acini per field were acquired and then analyzed as discussed.
Acini diameter quantification, acini cell number and statistics
Size analysis was performed using a hemocytometer and ImageJ software for each Brightfield image. At least 50 acini from a single field of view were analyzed for acini diameter. Cell number was quantified by counting the number of nuclei per acini from confocal images with frames of equal thickness. The Student’s t-test was used for statistical analysis(16).
Immunoblotting and analysis
Harvested cells were re-suspended and sonicated in radioimmunoprecipitation assay (RIPA) buffer (50mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 0.5% deoxycholic acid) containing protease and phosphatase inhibitors (1mM phenylmethylsulfonyl fluoride (PMSF), 0.4 U/ml aprotinin, 10 μg/ml leupeptin, 10 μg/ml pepstatin, 1 mM β–glycerophosphate, 0.1 mM NaF, 0.1 mM NaVO4). Proteins (30 to 80 μg) were fractionated on 10% Tris-HCl, Criterion Precast Gels (Bio-Rad). Separated proteins were transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore), and probed with the following antibodies: rabbit anti-DAPK3/ ZIPK (Abcam, ab51602, or for K42A, Cell Signaling, 2928), rabbit anti-β-catenin (Santa Cruz, H-102), mouse anti-phospho-p70/p85 (Cell Signaling, 9206), rabbit anti-p70/p85 (Cell Signaling, 2708), rabbit anti-phospho-pS6 (Cell Signaling, 2215), mouse anti-S6(Cell Signaling, 2317), rabbit anti-GAPDH (Sigma-Aldrich, G9545), rabbit anti-phospho-T308 Akt (Cell Signaling, 9275), mouse anti-Akt (Cell Signaling, 2920), rabbit anti-phospho-ERK (Cell Signaling, 4370), rabbit anti-actin (Sigma-Aldrich, A2066), mouse anti-RAPTOR (Santa Cruz, sc-81537) and rabbit anti-RICTOR (Cell Signaling, 2140). Secondary horseradish peroxidase-conjugated anti-rabbit or anti-mouse antibodies (Sigma-Aldrich) were added and ECL Western Blotting Substrate or SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Scientific) were used to visualize protein bands. For protein phosphorylation densitometry analysis, phospho-protein bands were first normalized to their respective total protein controls and then normalized to GAPDH.
Generation of DAPK3 KO Mice
The pre-confirmed Bay Genomics ES line YTA407 was acquired from the International Gene Trap Consortium, further confirmed in our lab, and then injected into albino C57BL/6 mice using traditional techniques (Mouse Transgenic Core, Washington University). One initial founder chimera was chosen due to high degree chimerism as assessed by coat color and subsequently backcrossed onto a pure albino C57BL/6 background to N6 as determined by speed congenics.
Soft agar assay
MCF10A cells were first infected with RasV12, shNeg, sh1, or sh2 and then selected in puromycin (1 μg/ml) until mock-infected cells were dead. Following drug selection, 1 x 104 stable cells were seeded into 6-well plates, and cells fed with fresh media twice a week. After 3 weeks, plates were stained with crystal violet overnight, washed and colonies manually counted.
RESULTS
DAPK3 depletion augments acini morphogenesis
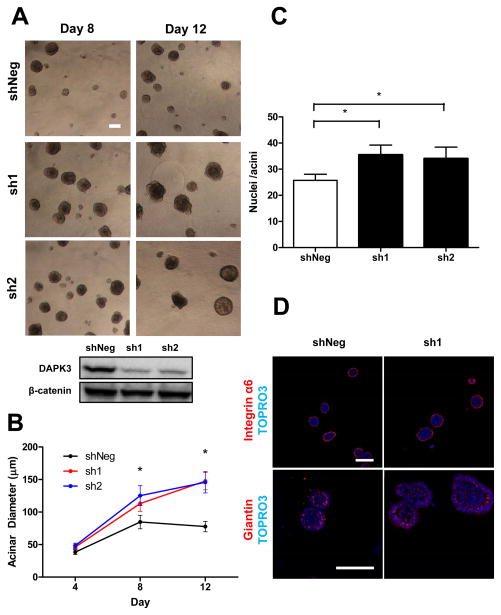
To investigate mechanisms by which DAPK3 exerts tumor suppressive functions, we performed stable shRNA knockdown of DAPK3 in MCF10A cells grown on Matrigel using two independent hairpins. As discussed previously, this model permits interrogation of acini architecture development, which undergoes a series of highly conserved temporally-concerted biochemical and phenotypic events. Loss of DAPK3 significantly enhanced acini diameter by approximately 37±10 % as compared to negative control shRNA at day 8 (Figure 1A, B). Diameter enhancement was observed as early as day 4 and continued to increase over time while the negative control plateaued at later time points. Consistent with a hyperplastic phenotype, DAPK3 depleted acini have significantly more cells per acini as evaluated using confocal microscopy and DNA staining (Figure 1C). We did not observe any alterations in acini apical or basolateral polarity as indicated by proper localization of giantin and integrin α6, respectively (Figure 1D). Interestingly, DAPK3 depletion alone was not capable of inducing anchorage-independent growth of MCF10A cells in soft agar as compared to control-positive cells expressing the HRasV12 transforming oncogene (Suppl. Figure 1). However, stable knockdown of DAPK3 in late passage HRasV12-expressing MCF10A cells displayed a trend toward augmented anchorage-independent growth (data not shown), and while not statistically significant, may merit further evaluation.
Figure 1.
Loss of DAPK3 augments MCF10A acini morphogenesis. (A) MCF10A cells stably expressing shNeg or two independent Dapk3 hairpins (sh1, sh2) were cultured on Matrigel for 12 days. Brightfield images show representative structures at specific time points. Scale bar, 100 μm.(B) Mean diameter of MCF10A acini stably expressing respective hairpins at various time points. Depicted here are representative data of three independent experiments. Error bars indicate 95% confidence intervals. *, P < 0.005 relative to control. (C) Loss of DAPK3 significantly increases the number of cells present in Day 6 acini grown on Matrigel. Error bars indicate 95% confidence intervals of three combined independent experiments.*, P <0.003 relative to control. (D) DAPK3-depleted acini (sh1) continued to show proper localization of apical-basolateral markers. Day 6 acini were analyzed for proper localization of basolateral marker integrin α6 (red, top panels) and apical Golgi marker giantin (red, bottom panels) along with DNA stain (TOPRO3, blue). Scale bars, 100 μm.
Loss of DAPK3 results in enhanced acini proliferation and apoptosis
It has been well established that size and morphogenesis of MCF10A acini are dependent on coordinated proliferation and apoptotic programs. To understand which of these processes was perturbed in DAPK3 knockdown structures, we performed confocal immunofluorescence microscopy on acini structures. DAPK3-depleted structures contained, on average, significantly more cells positive for expression of the proliferation marker Ki67, compared to negative control (Figure 2A). Interestingly, loss of DAPK3 also significantly increased the number of acini containing cells positive for apoptosis as visualized by cleaved caspase 3, relative to negative control (Figure 2B). This was additionally confirmed by ethidium bromide uptake in live MCF10A acini, which also indicated an increase in cell death in DAPK3-depleted structures (Suppl. Figure 2). Thus, while there was a net increase in acinar size, we hypothesize that as the shDAPK3 structures underwent hyperproliferation, more cells lost contact with the ECM and as a result underwent apoptosis, a phenomenon which has been observed elsewhere (17).
Figure 2.
DAPK3 depleted acini exhibit increased proliferation and apoptosis. (A) Fluorescent confocal microscopy analysis of acini stained for Ki67 (green) and DNA(TOPRO3, blue). Depicted here is a representative image of two independent experiments. Error bars indicate 95% confidence intervals. *, P < 0.001. Scale bar, 100 μm. (B) Fluorescent confocal microscopic analysis of acini stained for cleaved caspase 3 (CC3, green) and DNA (TOPRO3, blue). Depicted here is a representative image of three independent experiments. Error bars indicate 95% confidence intervals. *, P < 0.0001 relative to control. Scale bar, 100 μm.
DAPK3 overexpression disrupts normal acini formation
To determine if DAPK3 overexpression impacts acinar morphogenesis, we stably overexpressed DAPK3 fused to GFP in MCF10A cells that were subsequently grown in 3D culture. Despite a large degree of toxicity observed in the packaging cells used to generate viral particles, we were able to achieve stable populations of MCF10A cells that survived selection. However, we were not able to achieve high overexpression of DAPK3 most likely due to overexpression toxicities in both target and packaging cells, which has been commonly observed across several cell types (2, 18). Additionally, this expression was lost overtime indicating negative selection for sustained high level expression of DAPK3. Despite these technical challenges, MCF10A structures that maintained overexpression (as indicated by GFP fluorescence) displayed a dramatic and significant decrease in structure size compared to empty-GFP vector alone (Figure 3A, B). We also attempted to determine if this decrease in size was dependent on DAPK3 kinase activity but we were unable to achieve stable overexpression of a previously characterized kinase-deficient point mutant (K42A) fused to GFP (data not shown). However, confirmed stable overexpression of an unfused K42A mutant exhibited microscopic colony growth in soft agar highly similar to negative control, while a significant decrease was observed in stable cells overexpressing wildtype DAPK3 (Suppl. Figure 3A), consistent with growth inhibition dependent on kinase activity. We confirmed the signaling competency of overexpressed FLAG tagged DAPK3 in 293T cells. Specifically, transient overexpression of DAPK3 increased MLC-2 phosphorylation compared to K42A and empty vector (Suppl. Figure 3B).
Figure 3.
Stable overexpression of DAPK3 inhibits acini morphogenesis. (A) Fluorescent microscopy of live acini stably overexpressing GFP or GFP-DAPK3 at day 8. Scale bar, 100 μm.(B) Diameter analysis of day 8 GFP+ acini from GFP or GFP-DAPK3 stable acini. Error bars indicate 95% confidence intervals of three combined independent experiments.*, P < 0.0001 relative to control.
DAPK3 negatively regulates mTOR-S6K-S6 signaling with no effect on ERK or AKT activation
To identify the pathways regulated by DAPK3 in this context, we initially compared our acini phenotypes to those of previous reports. Several groups have shown that enhanced mTOR activity leads to acini with abnormalities similar to those observed upon loss of DAPK3(19-21). Indeed, knockdown of DAPK3 in MCF10A cells grown in 2D culture resulted in increased phosphorylation of S6 (P-S6) and both S6K isoforms (herein referred to as P-S6K) when stimulated with EGF or insulin (Figure 4A). We additionally observed an increase in P-S6 from MCF10A cells depleted of DAPK3 and grown in 3D culture for 6 days (Figure 4B). Of note, in 2D and 3D experiments, we did not observe a concomitant increase in the activation of ERK or AKT signaling(Suppl. Figure 4), which are upstream regulators of mTOR signaling (22).
Figure 4.
Increased activation of mTOR-specific S6K-S6 pathway upon loss of DAPK3 in stable MCF10A cells and acini. (A) Western blot analysis of overnight serum-starved MCF10A stable cells grown in 2D culture and treated with media, EGF (10ng/ mL) or insulin (10 ug/mL) for 24 hours. Shown here is a representative blot of three independent experiments. Normalized densitometry analysis is displayed under each panel. (B) Western blot analysis of MCF10A acini grown on Matrigel for 6 days. Shown here is a representative blot of two independent experiments.
Loss of DAPK3 influences proper acini morphogenesis through mTORC1
To further confirm that the observed increase in S6 signaling was functionally relevant, we treated established negative control and DAPK3-depleted acini with 100nM rapamycin once at day 4. Then, 4 days later, we observed that rapamycin had no significant effect on the diameter of the negative control. However, acini stably transduced with DAPK3 hairpins displayed a significant sensitivity to rapamycin treatment, showing a statistically significant decrease in acini diameter relative to no treatment (Figure 5A). Additionally, established shDAPK3 acini treated with increasing concentrations (100nM, 10 μM and 50 μM) of the PI3K and mTOR inhibitor LY294002 displayed similar increased sensitivity to the drug compared to negative control (Figure 5B).
Figure 5.
DAPK3 inhibits acini morphogenesis through mTORC1/ RAPTOR. (A) Analysis of MCF10A acini grown for 4 days and then treated with 100nM rapamycin for 4 days. Shown here are representative data of two independent experiments. Error bars indicate 95% confidence intervals. **, P<0.0001. *, P<0.005. (B) Analysis of MCF10A acini grown for 4 days and then treated with varying concentrations of LY294002 (vehicle, 100nM, 10 μM, or 50 μM) for an additional 4 days. Error bars indicate 95% confidence intervals of three combined independent experiments. *, P<0.026. (C) Analysis of Day 4 acini from MCF10A cells stably expressing combinations of various shRNA hairpins. MCF10A were simultaneously transduced with indicated shRNA-puromycin (shNegP, sh1P) and shRNA-hygromycin (shRAPTORH, shRICTORH) lentivirus. Shown here are representative data of two independent experiments. Error bars indicate 95% confidence intervals. *, P<0.01. (D) A proposed model for negative regulation of mTOR-S6K-S6 signaling by DAPK3.
To further delineate how DAPK3 regulates mTOR for proper acini morphogenesis, we stably knocked down DAPK3 and RICTOR or RAPTOR concurrently using previously reported hairpins (23). The mTOR catalytic subunit is shared between two distinct mTOR complexes, mTORC1 and mTORC2, which elicit the variety of functions of the mTOR pathway (22). Regulatory-associated protein of mammalian target of rapamycin (RAPTOR) and rapamycin-insensitive companion of mTOR (RICTOR) are necessary components of the mTOR complexes comprising mTORC1 and mTORC2, respectively. Interestingly, we observed that concurrent loss of DAPK3 and RAPTOR blunted the increased acini size observed with DAPK3 depletion alone, whereas concurrent loss of RICTOR did not rescue the phenotype (Figure 5C). Additionally, loss of RAPTOR by itself had a profound impact on acini formation whereas knockdown of RICTOR did not.
These results identify an epistatic interaction between DAPK3 and the mTORC1 pathway with functional consequences through regulation of S6K-S6 signaling and potentially other downstream mTORC1 effectors (Figure 5D).
Generation and characterization of embryonic lethality in DAPK3 null mice
In an attempt to understand the in vivo contribution of DAPK3 to mouse mammary development, its overall role in mouse development, and DAPK3-specific oncologic recapitulations of ontogeny, we created a constitutive DAPK3 knockout mouse using a pre-confirmed gene trap ES cell line from the International Gene Trap Consortium. As shown in Figure 6A, the gene trap (Gt) is composed of a 5′ splice acceptor site (green) followed by a β galactosidase-neomycin fusion (βgeo) (blue) and a poly A tail(red). We further confirmed that the locus and the entire gene trap were intact and incorporated into genomic DNA as a single insertion through Southern blot analysis using both external probes and an internal probe (Figure 6B). Importantly, we determined that the gene trap integrated between the second and third exon. These ES cells were then used to generate a mouse using conventional blastocyst injection and animal husbandry techniques. Depletion of Dapk3 mRNA was later confirmed using quantitative PCR on genotyped ES cell lines derived from blastocysts produced by heterozygous crosses (Suppl. Figure 5).
Figure 6.
Generation and characterization of DAPK3 knockout mice. (A) Schematic representation of the Dapk3Gt(YTA407)Byg locus that depicts the location of Southern blot probes (A, B) as well as the splice acceptor site (SA, green), β-galactosidase-neomycin fusion (βgeo, blue) and polyA tail (pA, red). (B) Southern blot evaluation of the Dapk3Gt(YTA407)Byg locus from the YTA407 ES line and mice along with representative PCR animal genotyping. (C) Brightfield images of genotype-confirmed blastocysts isolated from heterozygous crosses. (D) The appearance of increased resorbed E10.5 fetuses (red arrows) in uteri extracted from heterozygous crosses (top) versus wildtype X heterozygous crosses (bottom).(E) Distinct and localized βgal activity in the developing heart and notochord (red arrows) from genotype confirmed heterozygous embryos at E8.5 (top) and E10.5 (bottom).
Despite grossly healthy heterozygous adults, we were unable to identify any homozygous Gt/Gt(Dapk3−/−) animals on a mixed 129 Ola or backcrossed C57BL/6 (N5) background (Suppl. Table 1). We were also unable to locate homozygous Gt/Gt embryos at E12.5, E10.5, and E8.5, despite near Mendelian ratios for wild type and heterozygous embryos, indicating early homozygous lethality (Suppl. Table 1). This was further confirmed by the presence of several sites of fetal resorption on uteri extracted from heterozygous crosses versus heterozygous X wild type crosses (Figure 6D). However, note that we were able to isolate and genotype Gt/Gt homozygous blastocysts at E3.5 (Figure 6C), indicating that lethality was likely occurring post-implantation. Furthermore, there was no difference in distribution of homozygous embryos across blastocyst, morula and pre-morula stages (data not shown).
Early mouse embryonic and adult expression patterns of Dapk3
Overall, heterozygous animals appeared to develop and grow normally with no overt phenotypes under standard laboratory conditions. Taking advantage of the functional βgal expressed under the endogenous Dapk3 promoter, we characterized the expression patterns of Dapk3 during the development of heterozygous mice. We observed distinct and strong βgal activity in the developing heart of E8.5 and E10.5 embryos (Figure 6E, top panel). Additionally, E10.5 heterozygous embryos displayed localized activity within the developing notochord (Figure 6E, bottom panel). As expected, no βgal activity was observed in developing wildtype littermates (data not shown).
Mouse breast epithelium expression and clinically observed down regulation of DAPK3
To determine the potential utility of DAPK3 knockout mice for breast cancer studies we wanted to confirm Dapk3 expression in mouse mammary epithelium. Due to our inability to locate antibodies that could detect endogenous mouse DAPK3, we visualized Dapk3 expression using antibodies that recognized β-galactosidase/ LacZ as expressed from the gene trap downstream of the endogenous Dapk3 promoter. We observed strong and distinct staining in the mammary epithelium of heterozygous mouse mammary fat pads with no observable staining in wildtype littermate controls (Figure 7A). Finally, to confirm that our cell culture and murine observations mimic those seen in actual patients, we analyzed the Oncomine database (www.oncomine.org). Indeed, DAPK3 mRNA is significantly downregulated in human ductal breast carcinoma in situ (DCIS) versus normal breast and invasive breast cancer versus DCIS, respectively (Figure 7B, C).
Figure 7.
Dapk3 expression in the mouse mammary epithelium and DAPK3 mRNA downregulation in aggressive human breast cancer samples relative to normal control (Oncomine database). (A) Whole field immunofluorescence of mammary fat pads extracted from murine virgin adult littermates and stained for DNA (blue), LacZ (green) and E-cadherin (red); left, wild type; right, heterozygous Dapk3. (B) DAPK3 mRNA is significantly decreased in human ductal carcinoma in situ (DCIS) samples relative to normal patient samples. *, P<0.002. (C) DAPK3 mRNA is significantly decreased in aggressive breast cancer patient samples compared to DCIS patient samples. *, P<0.04.
DISCUSSION
Herein we have shown that loss of DAPK3 leads to increased acinar size, and enhanced cellular proliferation and apoptosis without disrupting apical-basolateral polarity of MCF10A acini grown in 3D culture. Conversely, stable overexpression of DAPK3 inhibits acini morphogenesis and is relatively toxic to cells. Loss of DAPK3 augments acini morphogenesis through mTOR-S6 signaling. This regulation appears to be specific to the mTOR pathway as loss of DAPK3 enhanced S6K-S6 phosphorylation, but not ERK or AKT. Curiously, we found an increased sensitivity of MCF10A cells to rapamycin and the mTOR/ PI3K inhibitor LY294002 upon knockdown of DAPK3. Furthermore, this regulation is specific to mTORC1 and not mTORC2 as only loss of RAPTOR (and not RICTOR) partially rescued the augmented acinar morphogenesis observed upon loss of DAPK3. Acini lacking RAPTOR alone also exhibit suppressed acini diameter similar to that of DAPK3 and RAPTOR double knockdown acini. However, loss of DAPK3 combined with loss of RAPTOR was able to significantly overcome the inhibition observed in RAPTOR knockdown alone. While this effect was statistically significant, it remains formally possible that an additional mechanism by which mTORC1/RAPTOR suppresses acini morphogenesis is independent of DAPK3 and further studies are necessary. Strikingly, DAPK3 is required for early mouse development post blastocyst implantation. Our studies also suggest that DAPK3 is expressed in mouse mammary epithelium and that down regulation correlates with neoplastic progression in breast cancer patients. Collectively, our data reveal a novel tumor suppressive mechanism for DAPK3 and highlight its fundamental requirement for mouse development.
It is well known that the mTOR pathway plays an important role in cancer cell growth, survival and proliferation(22). mTOR is the fundamental catalytic component of two distinct complexes, rapamycin-sensitive mTORC1 and rapamycin in sentitive-mTORC2, each of which is composed of distinct protein complexes that alter the protein-protein interactions, subcellular localization, activity and substrate specificity of the active complex. Genetic ablation of key scaffolding proteins RAPTOR or RICTOR effectively prevents signaling through mTORC1 and mTORC2, respectively. Activation of the mTOR pathway is controlled by several upstream tumor suppressors, including LKB1 and NF1 (24, 25). Interestingly, several other member of the DAPK tumor suppressor family are known to regulate translation and regulate or be regulated by the mTOR pathway. Conflicting reports reveal that DAPK disrupts TSC1-TSC2 association, thereby enhancing mTOR activation, and negatively regulates protein translation through an inhibitory phosphorylation at S235/236 of S6 (26, 27). However, Roux et al. revealed RSK-dependent phosphorylation at S235/ 236 actually promotes translation through assembly of the translational pre-initiation complex (28). DRAK2 phosphorylates S6K kinase in vitro and in vivo (29). DAPK3 itself is phosphorylated by DAPK in vitro and this DAPK-DAPK3 kinase cascade has been shown to inhibit transcript-specific translation through phosphorylation of the ribosomal protein L13a and activation of an translational inhibitor complex known as the IFN-gamma-activated inhibitor of translation (GAIT) complex (18, 30). Further work is required to specifically determine how DAPK3 negatively regulates mTOR-S6K-S6 signaling. We hypothesize that DAPK3 can directly or indirectly inhibit mTORC1 signaling upstream of S6 or it may directly influence S6K or S6 similar to other DAPK family members. Overall, our data provide further evidence to indicate that DAPK family members are important regulators of translation and mTOR signaling.
Moreover, through the generation and characterization of a DAPK3 knockout mouse, our data reveal that DAPK3 is crucial to early mouse development. Our analysis indicates that DAPK3-deficient blastocysts are able to implant, but subsequent development is problematic. At this point in mouse embryonic development (E4.5–E6.5), various embryonic germ layers undergo differentiation, migration and establishment of early morphologic axes(31). DAPK3 is known to regulate cellular migration and contraction and thus, DAPK3 may be crucial for these early developmental events(32, 33). Remarkably, embryonic lethality was not anticipated given that Dapk−/− mice develop normally as indicated by their overall basal health(34). This dichotomy raises a number of questions regarding signaling redundancies between DAPK and DAPK3 and highlights important developmental functions for DAPK3. Our data support the need for future development of conditional DAPK3 knockout mice, which may serve as relevant models for interrogating tumor suppressive mechanisms for DAPK3 in mouse models of breast cancer. However, it is worth noting potential functional differences between human and murine DAPK3. Specifically, murine DAPK3 exhibits C-terminal amino acid substitutions that may impart differential signaling patterns or tumor suppressive signaling mechanisms not seen in human systems (35). Nonetheless, our work identified that mouse DAPK3 exhibits localized expression in the developing heart and nervous system, and these expression patterns and developmental observations are of potential relevance to humans. Specifically, DAPK3 resides on chromosome 19p13.3, a region shared by 7 other genes in humans(ZFR2, ATCAY, NMRK2, EEF2, PIAS4, ZBTB7A, MAP2K2). Deletion of this locus correlates with facial and cardiac structural abnormalities, along with intellectual disabilities and developmental delays in affected patients (36).
Overall, the research presented herein indicates that suppression of mTOR-S6K signaling by DAPK3 maintains proper acini morphogenesis and that DAPK3 is necessary for mouse development. Furthermore, these observations have potential implications for breast cancer and human development.
Supplementary Material
Implications.
Novel cellular and mouse modeling studies of DAPK3 shed light on its tumor suppressive mechanisms and provide direct evidence that DAPK3 has relevance in early development.
Acknowledgments
The authors would like to thank colleagues of the Molecular Imaging Center for their discussions and input. This work was supported in part by the Washington University-M.D. Anderson Cancer Center Inter-Institutional Molecular Imaging Center grant (P50 CA094056) and a NIH training fellowship for stipend support to Brandon Kocher (T32 CA113275).
Footnotes
COI statement: The authors declare no conflict of interest.
References
- 1.Usui T, Okada M, Yamawaki H. Zipper interacting protein kinase (ZIPK): function and signaling. Apoptosis. 2014;19:387–91. doi: 10.1007/s10495-013-0934-3. [DOI] [PubMed] [Google Scholar]
- 2.Brognard J, Zhang YW, Puto LA, Hunter T. Cancer-associated loss-of-function mutations implicate DAPK3 as a tumor-suppressing kinase. Cancer Res. 2011;71:3152–61. doi: 10.1158/0008-5472.CAN-10-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Natrajan R, Mackay A, Lambros MB, Weigelt B, Wilkerson PM, Manie E, et al. A whole-genome massively parallel sequencing analysis of BRCA1 mutant oestrogen receptor-negative and -positive breast cancers. J Pathol. 2012;227:29–41. doi: 10.1002/path.4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kawai T, Matsumoto M, Takeda K, Sanjo H, Akira S. ZIP kinase, a novel serine/threonine kinase which mediates apoptosis. Mol Cell Biol. 1998;18:1642–51. doi: 10.1128/mcb.18.3.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bi J, Lau SH, Hu L, Rao HL, Liu HB, Zhan WH, et al. Downregulation of ZIP kinase is associated with tumor invasion, metastasis and poor prognosis in gastric cancer. Int J Cancer. 2009;124:1587–93. doi: 10.1002/ijc.24164. [DOI] [PubMed] [Google Scholar]
- 6.Tan XL, Moyer AM, Fridley BL, Schaid DJ, Niu N, Batzler AJ, et al. Genetic variation predicting cisplatin cytotoxicity associated with overall survival in lung cancer patients receiving platinum-based chemotherapy. Clin Cancer Res. 2011;17:5801–11. doi: 10.1158/1078-0432.CCR-11-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Togi S, Ikeda O, Kamitani S, Nakasuji M, Sekine Y, Muromoto R, et al. Zipper-interacting protein kinase (ZIPK) modulates canonical Wnt/beta-catenin signaling through interaction with Nemo-like kinase and T-cell factor 4 (NLK/TCF4) J Biol Chem. 2011;286:19170–7. doi: 10.1074/jbc.M110.189829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Felten A, Brinckmann D, Landsberg G, Scheidtmann KH. Zipper-interacting protein kinase is involved in regulation of ubiquitination of the androgen receptor, thereby contributing to dynamic transcription complex assembly. Oncogene. 2013;32:4981–8. doi: 10.1038/onc.2012.503. [DOI] [PubMed] [Google Scholar]
- 9.Page G, Lodige I, Kogel D, Scheidtmann KH. AATF, a novel transcription factor that interacts with Dlk/ZIP kinase and interferes with apoptosis. FEBS Lett. 1999;462:187–91. doi: 10.1016/s0014-5793(99)01529-x. [DOI] [PubMed] [Google Scholar]
- 10.Kawai T, Akira S, Reed JC. ZIP kinase triggers apoptosis from nuclear PML oncogenic domains. Mol Cell Biol. 2003;23:6174–86. doi: 10.1128/MCB.23.17.6174-6186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boosen M, Vetterkind S, Kubicek J, Scheidtmann KH, Illenberger S, Preuss U. Par-4 is an essential downstream target of DAP-like kinase (Dlk) in Dlk/Par-4-mediated apoptosis. Mol Biol Cell. 2009;20:4010–20. doi: 10.1091/mbc.E09-02-0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sato N, Kawai T, Sugiyama K, Muromoto R, Imoto S, Sekine Y, et al. Physical and functional interactions between STAT3 and ZIP kinase. Int Immunol. 2005;17:1543–52. doi: 10.1093/intimm/dxh331. [DOI] [PubMed] [Google Scholar]
- 13.Weigelt B, Bissell MJ. The need for complex 3D culture models to unravel novel pathways and identify accurate biomarkers in breast cancer. Adv Drug Deliv Rev. 2014 doi: 10.1016/j.addr.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–68. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 15.Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–98. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 16.Glantz SA. Primer of Biostatistics. 2. Mcgraw-Hill, Inc; New York: 1987. p. 379. [Google Scholar]
- 17.Partanen JI, Nieminen AI, Makela TP, Klefstrom J. Suppression of oncogenic properties of c-Myc by LKB1-controlled epithelial organization. Proc Natl Acad Sci U S A. 2007;104:14694–9. doi: 10.1073/pnas.0704677104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shani G, Marash L, Gozuacik D, Bialik S, Teitelbaum L, Shohat G, et al. Death-associated protein kinase phosphorylates ZIP kinase, forming a unique kinase hierarchy to activate its cell death functions. Mol Cell Biol. 2004;24:8611–26. doi: 10.1128/MCB.24.19.8611-8626.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anczukow O, Rosenberg AZ, Akerman M, Das S, Zhan L, Karni R, et al. The splicing factor SRSF1 regulates apoptosis and proliferation to promote mammary epithelial cell transformation. Nat Struct Mol Biol. 2012;19:220–8. doi: 10.1038/nsmb.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Debnath J, Walker SJ, Brugge JS. Akt activation disrupts mammary acinar architecture and enhances proliferation in an mTOR-dependent manner. J Cell Biol. 2003;163:315–26. doi: 10.1083/jcb.200304159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghosh S, Varela L, Sood A, Park BH, Lotan TL. mTOR signaling feedback modulates mammary epithelial differentiation and restrains invasion downstream of PTEN loss. Cancer Res. 2013;73:5218–31. doi: 10.1158/0008-5472.CAN-13-0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–93. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 24.Johannessen CM, Reczek EE, James MF, Brems H, Legius E, Cichowski K. The NF1 tumor suppressor critically regulates TSC2 and mTOR. Proc Natl Acad Sci U S A. 2005;102:8573–8. doi: 10.1073/pnas.0503224102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shaw RJ, Bardeesy N, Manning BD, Lopez L, Kosmatka M, DePinho RA, et al. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell. 2004;6:91–9. doi: 10.1016/j.ccr.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 26.Stevens C, Lin Y, Harrison B, Burch L, Ridgway RA, Sansom O, et al. Peptide combinatorial libraries identify TSC2 as a death-associated protein kinase (DAPK) death domain-binding protein and reveal a stimulatory role for DAPK in mTORC1 signaling. J Biol Chem. 2009;284:334–44. doi: 10.1074/jbc.M805165200. [DOI] [PubMed] [Google Scholar]
- 27.Schumacher AM, Velentza AV, Watterson DM, Dresios J. Death-associated protein kinase phosphorylates mammalian ribosomal protein S6 and reduces protein synthesis. Biochemistry. 2006;45:13614–21. doi: 10.1021/bi060413y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roux PP, Shahbazian D, Vu H, Holz MK, Cohen MS, Taunton J, et al. RAS/ERK signaling promotes site-specific ribosomal protein S6 phosphorylation via RSK and stimulates cap-dependent translation. J Biol Chem. 2007;282:14056–64. doi: 10.1074/jbc.M700906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mao J, Luo H, Han B, Bertrand R, Wu J. Drak2 is upstream of p70S6 kinase: its implication in cytokine-induced islet apoptosis, diabetes, and islet transplantation. J Immunol. 2009;182:4762–70. doi: 10.4049/jimmunol.0802255. [DOI] [PubMed] [Google Scholar]
- 30.Mukhopadhyay R, Ray PS, Arif A, Brady AK, Kinter M, Fox PL. DAPK-ZIPK-L13a axis constitutes a negative-feedback module regulating inflammatory gene expression. Mol Cell. 2008;32:371–82. doi: 10.1016/j.molcel.2008.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beddington RS, Robertson EJ. Axis development and early asymmetry in mammals. Cell. 1999;96:195–209. doi: 10.1016/s0092-8674(00)80560-7. [DOI] [PubMed] [Google Scholar]
- 32.Komatsu S, Ikebe M. ZIPK is critical for the motility and contractility of VSMCs through the regulation of nonmuscle myosin II isoforms. Am J Physiol Heart Circ Physiol. 2014;306:H1275–86. doi: 10.1152/ajpheart.00289.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haystead T. ZIP kinase, a key regulator of myosin protein phosphatase 1. Cellular Signaling. 2005;17:1313–22. doi: 10.1016/j.cellsig.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 34.Gozuacik D, Bialik S, Raveh T, Mitou G, Shohat G, Sabanay H, et al. DAP-kinase is a mediator of endoplasmic reticulum stress-induced caspase activation and autophagic cell death. Cell Death Differ. 2008;15:1875–86. doi: 10.1038/cdd.2008.121. [DOI] [PubMed] [Google Scholar]
- 35.Shoval Y, Pietrokovski S, Kimchi A. ZIPK: a unique case of murine-specific divergence of a conserved vertebrate gene. PLoS Genet. 2007;3:1884–93. doi: 10.1371/journal.pgen.0030180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Risheg H, Pasion R, Sacharow S, Proud V, Immken L, Schwartz S, et al. Clinical comparison of overlapping deletions of 19p13.3. Am J Med Genet A. 2013;161A:1110–6. doi: 10.1002/ajmg.a.35923. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.