Abstract
The emergence of catalysis in early genetic polymers like RNA is considered a key transition in the origin of life1, predating the appearance of protein enzymes. DNA also demonstrates the capacity to fold into three-dimensional structures and form catalysts in vitro2. However, to what degree these natural biopolymers comprise functionally privileged chemical scaffolds3 for folding or the evolution of catalysis is not known. The ability of synthetic genetic polymers (XNAs) with alternative backbone chemistries not found in nature to fold into defined structures and bind ligands4 raises the possibility that these too might be capable of forming catalysts (XNAzymes). Here we report the discovery of such XNAzymes, elaborated in four different chemistries (ANA (arabino nucleic acids)5, FANA (2′-fluoroarabino nucleic acids)6, HNA (hexitol nucleic acids) and CeNA (cyclohexene nucleic acids)7 directly from random XNA oligomer pools, exhibiting in trans RNA endonuclease and ligase activities. We also describe an XNA-XNA ligase metalloenzyme in the FANA framework, establishing catalysis in an entirely synthetic system and enabling the synthesis of FANA oligomers and an active RNA endonuclease FANAzyme from its constituent parts. These results extend catalysis beyond biopolymers and establish technologies for the discovery of catalysts in a wide range of polymer scaffolds not found in nature8. Evolution of catalysis independent of any natural polymer has implications for the definition of chemical boundary conditions for the emergence of life on earth and elsewhere in the universe9.
Life is dependent on catalysis as many chemical transformations essential for cellular function are kinetically sluggish and/or thermodynamically disfavoured under ambient conditions. The emergence of a catalyst (or catalytic system) for RNA self-replication is considered to have been a key event in the origin of life. Thus the development of molecular heredity itself not only depends on the capacity of nucleic acids for genetic information storage and retrieval but also to form catalysts1. Although proteins have largely supplanted this role in present day biology, nucleic acid-mediated catalysis remains crucial, notably in RNA processing10 and translation11. Furthermore a range of RNA and DNA enzymes (ribozymes / DNAzymes) have been discovered by in vitro evolution12.
Catalysis by nucleic acids (and by biopolymers in general) minimally requires the presence of chemically functional groups and a framework for their precise arrangement. Synthetic genetic polymers (XNAs) with backbones based on congeners of the canonical ribofuranose share with RNA and DNA a capacity for heredity, evolution and the ability to fold into defined three-dimensional structures, forming ligands (aptamers)4. We therefore sought to establish whether XNAs could also support the evolution of catalysts.
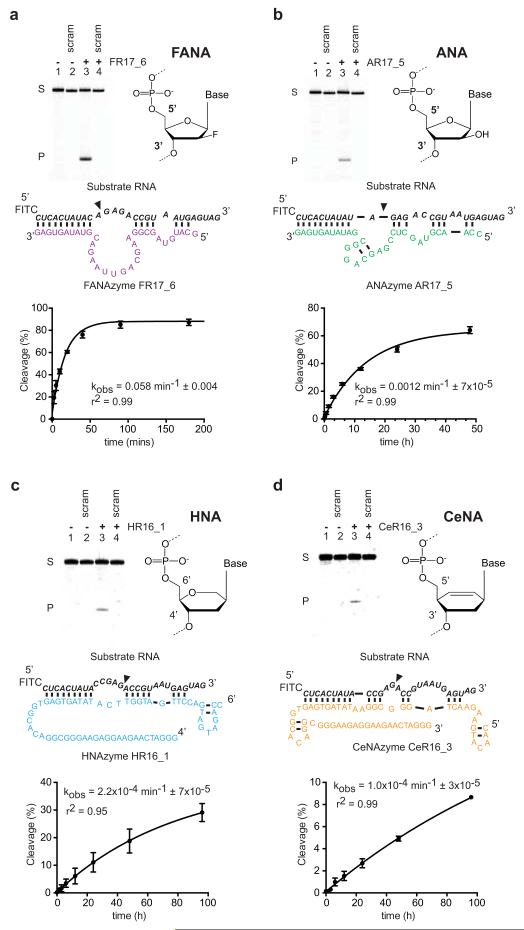
Leveraging XNA replication technology developed previously4, we devised a strategy for the discovery of RNA endonuclease XNAzymes by cleavage of an internal RNA sequence (Extended Data Fig. 1). Chimeric RNA-XNA libraries were prepared by RNA-primed XNA synthesis in four scaffolds using mutant polymerases: D4K4 for arabinonucleic acid (ANA)5 and fluoro-arabinonucleic acid (FANA)13, 6G124 for cyclohexenyl nucleic acid (CeNA)7 and a newly engineered 6G12 I521L variant (see Methods) for 1,5 anhydrohexitol nucleic acid (HNA)7. After 13 - 17 rounds of selection, polyclonal pools showed RNA endonuclease activity and were deep sequenced (Extended Data Fig. 2). Abundant sequences across all four XNAs were tested for intramolecular (in cis) endonuclease activity and a subset of active clones for bimolecular (in trans) activity. We further examined one RNA endonuclease XNAzyme for each scaffold (FR17_6 (FANA), AR17_5 (ANA), HR16_1 (HNA) and CeR16_3 (CeNA)) (Fig. 1). All showed site-specific sequence-dependent (Extended Data Fig. 3) RNA cleavage with a range of catalytic rates (kobs: 0.06 min−1 – 1×10−4 min−1 (25°C)). While the rate of the FR17_6 XNAzyme is comparable to analogous ribozymes and DNAzymes, ANA and in particular HNA and CeNA catalysts are 20 - 600-fold slower. Nevertheless, all four catalyze RNA cleavage through a classic transesterification mechanism (as seen in e.g. the ‘hammerhead’ or ‘hairpin’ ribozymes14), yielding products with 2′,3′ cyclic phosphate and 5′ hydroxyl groups (Extended Data Fig. 4).
Figure 1. RNA endonuclease XNAzymes elaborated in four synthetic genetic polymer chemistries.
Gel electrophoresis, putative secondary structures and pre-steady state reaction rates (kobs) (n=3, error bars=sd, 25°C) of a, FANA, b, ANA, c, HNA, d, CeNA enzymes. Urea-PAGE gels show bimolecular cleavage in trans of cognate RNA substrates (NucSR variants, see Extended Data Fig. 3) (lanes 1 & 3), but not scrambled RNA (NucSR SCRAM1) (lanes 2 & 4), catalyzed by XNAzymes (lanes 3 & 4) (17°C).
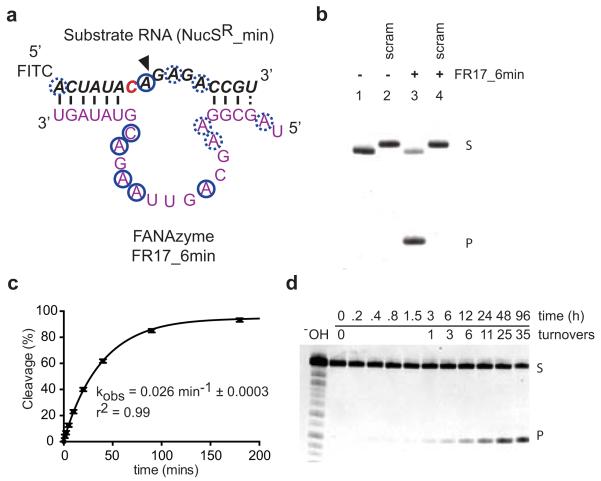
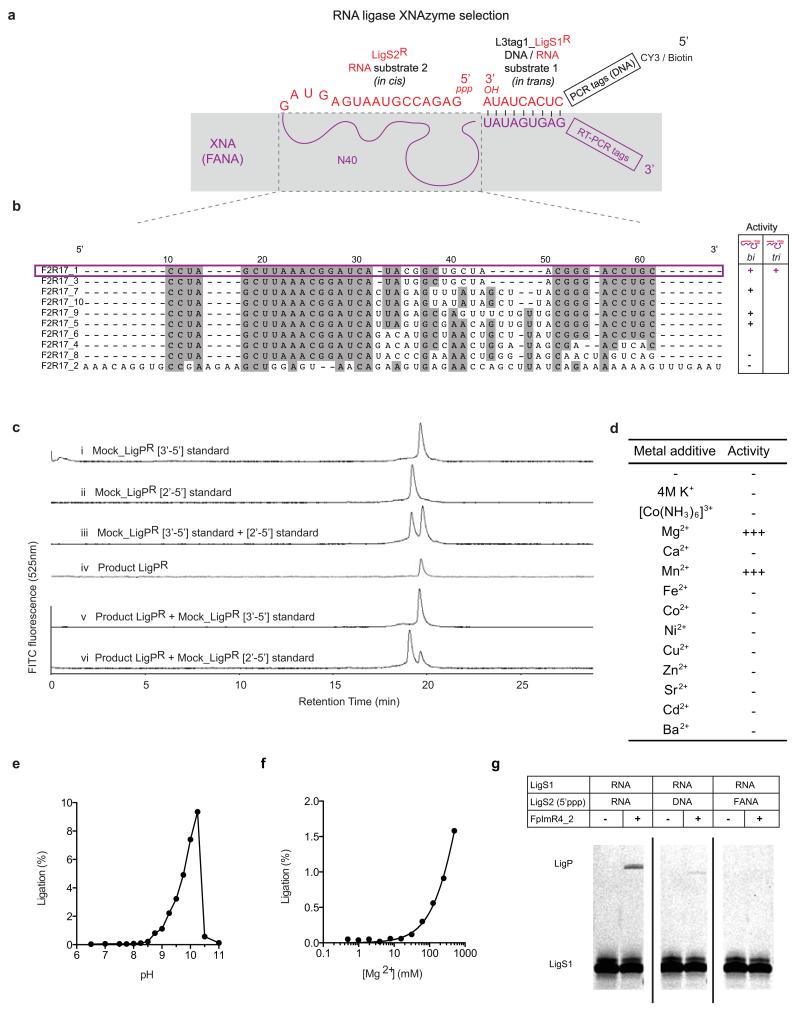
We dissected contributions of individual nucleotides in the FR17_6 XNAzyme, defining a 26nt catalytic core (FR17_6min). As all four FANA nucleotide phosphoramidites are commercially available, this minimized XNAzyme could be prepared by solid-phase synthesis (see Methods) and was found to retain full activity (Fig. 2a-c) (kobs: 0.06 min−1 (25°C)), including multiple turnover catalysis (Fig. 2d). FR17_6min shows a pH optimum (pHopt) of 9.25 (Extended Data Fig. 4h), consistent with a mechanism involving deprotonation of the cleavage site-proximal 2′ hydroxyl. A screen of Irving-Williams divalent metals reveals that FR17_6min is Mg2+-dependent with apparent Km ≈ 30 mM (Extended Data Fig. 4i), with only Mn2+ able to partially restore activity (Extended Data Fig. 4g).
Figure 2. Chemical synthesis yields an active RNA endonuclease XNAzyme.
a, Secondary structure of truncated FANAzyme FR17_6 (FR17_6min, purple), determined by RNA-SHAPE & RNA/FANA-DMS mapping (derived from a larger construct, Extended Data Fig. 5), red indicates SHAPE reactive residues (RNA), blue circles indicate DMS-reactive (preferentially A or C) residues (RNA or FANA). Dashed circles indicate marginal reactivity. b, FR17_6min synthesized using FANA phosphoramidites (Extended Data Fig. 10) cleaves a minimized cognate RNA substrate (NucSR_min) (lanes 1 & 3), but not a scrambled RNA (NucSR SCRAM2) (lanes 2 and 4), with, c, essentially unchanged catalytic rate (kobs) (n=3, error bars=sd, 25°C). d, FR17_6min (10 nM) can perform multiple turnover cleavage of RNA NucSR_min (1μM).
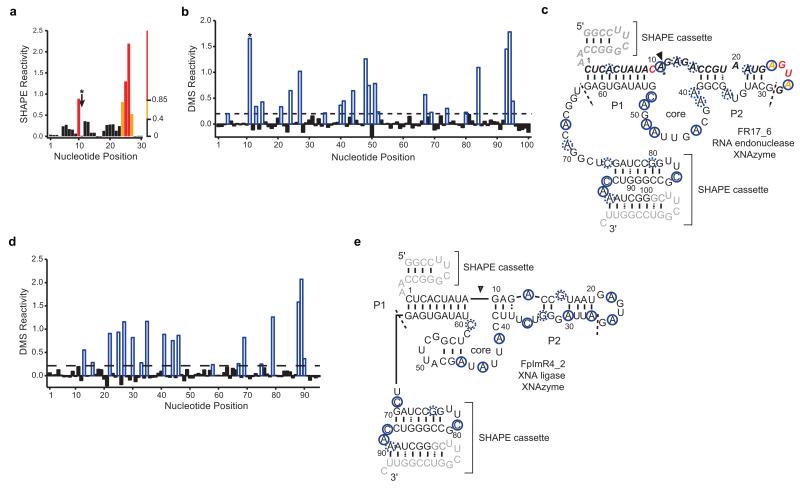
The secondary structure of FR17_6, including an inert RNA substrate modified with 2′O-Me at the cleavage site (Extended Data Fig. 5), was probed by Selective 2′hydroxyl Acylation analysed by Primer Extension (SHAPE)15 (for RNA, modifying 2′OH at flexible regions) and/or Dimethyl Sulfate (DMS)16 (for FANA, modifying primarily unpaired adenine and cytosine), revealing a structure broadly similar to other RNA-acting nucleic acid catalysts, with a central domain flanked by substrate-binding arms (P1, P2), albeit with a three nucleotide bulge in P2 (Extended Data Fig. 5c).
In general, the RNA endonuclease XNAzymes are novel sequences, although some in the ANA system (Extended Data Fig. 2c) retain partial sequences from the 8-17 and 10-23 DNAzymes17 used in library design (in addition to N40 sequences) (see Methods). The AR17_5 ANAzyme shares 12 of the 14 core residues of the 8-17 DNAzyme, as well as A′G > N′G cleavage preference (Extended Data Fig. 3b). However, conversion of the complete 8-17 sequence into ANA (or indeed FANA, HNA or CeNA) yields no activity (Extended Data Fig. 3e), indicating the acquisition or rearrangement of key residues during selection. Nevertheless, topological similarities (without sequence homology) between FR17_6 and this family of DNAzymes18 suggests the possibility that for XNAs that form DNA-like B-form duplexes, such as ANA and FANA (albeit with a non-canonical O4′-endo (east) sugar conformation)19, catalysts may reside in the structural or sequence vicinity of extant DNAzymes.
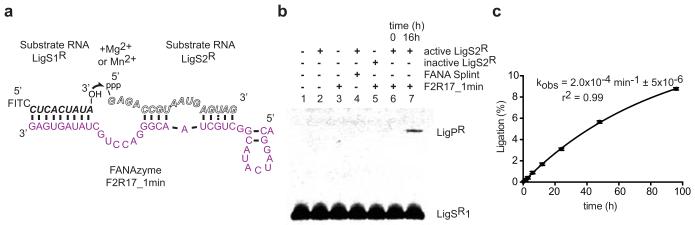
Having established the capacity for catalysis in four different XNA backbones, we wondered whether XNAzymes could be evolved to ligate RNA as well as cleave20-22. We selected for RNA-RNA ligase activity using a bi-molecular strategy: 5′-RNA-XNA libraries carrying 5′ triphosphate moieties (5′ppp) (Extended Data Fig. 6) were challenged to ligate to DNA-RNA-3′ substrates. We identified RNA ligase XNAzymes (FANA) by deep sequencing and screening (Extended Data Fig. 7), and chose one (F2R17_1) for further characterization. A minimized (39nt), chemically-synthesized version (F2R17_1min) was capable of ligating two RNA substrates (LigS1R (3′OH) + LigS2R (5′ppp)) in a tri-molecular reaction (Fig. 3) with ‘natural’ regioselectivity (3′-5′ rather than 2′-5′), as judged by comparison with ‘mock’ RNA using Strong Anion Exchange Chromatography (SAX-HPLC) (Extended Data Fig. 7c). The reaction rate is low (kobs: 2×10−4 min−1 (25°C)), but represents an enhancement (kobs / kuncat) of 104-fold compared to the uncatalysed background reaction (RNA substrates LigS1R + LigS2R hybridized to a complementary FANA splint; kuncat: 2×10−8 min−1). Like the RNA endonuclease FANAzyme FR17_6, the activity of F2R17_1min is enhanced at basic pH (pHopt = 10.25) (Extended Data Fig. 7e), as well as by Mg2+ (Extended Data Fig. 7f), for which only Mn2+ can be substituted (Extended Data Fig. 7d), consistent with a mechanism involving deprotonation and nucleophilic attack of the 3′ hydroxyl of LigS1R on the α–phosphate of 5′ppp –LigS2R, analogous to e.g. the R3C ligase ribozyme23.
Figure 3. An RNA ligase XNAzyme (FANA).
a, Putative secondary structure of truncated chemically synthesized FANAzyme (F2R17_1min, purple) that ligates RNA substrate LigS1R to LigS2R, activated with 5′ triphosphate (ppp), in a trimolecular reaction in trans. b, Urea-PAGE gel showing no significant product (LigPR) observed with: substrate LigS1R alone (lane 1), no XNAzyme (lane 2), no LigS2R (lane 3), complementary FANA splint (lane 4), or LigS2R lacking 5′ppp (lane 5); product formation is dependent on LigS1R, activated LigS2R and XNAzyme (lanes 6 and 7). No product was detectable with combinations of RNA, DNA or FANA versions of LigS1 and (5′ppp)LigS2, except DNA LigS1 and RNA LigS2, which showed ~1.5% ligation after 20 h (Extended Data Fig. 7g). c, Pre-steady state trimolecular reaction rate (kobs) (n=3, error bars=sd, 25°C).
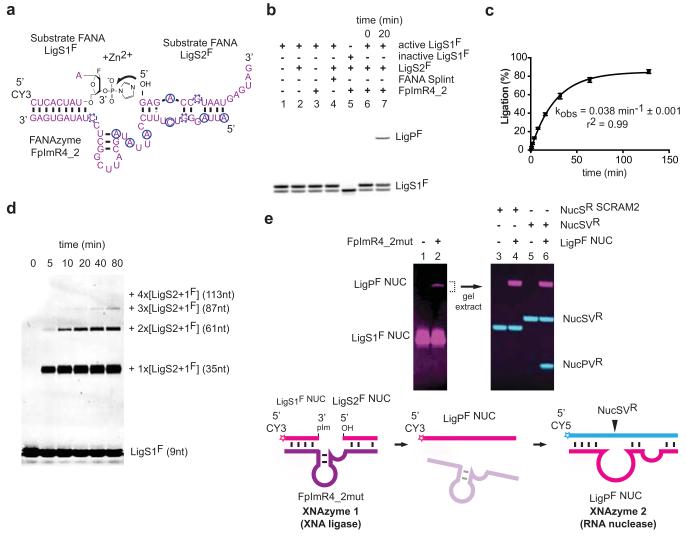
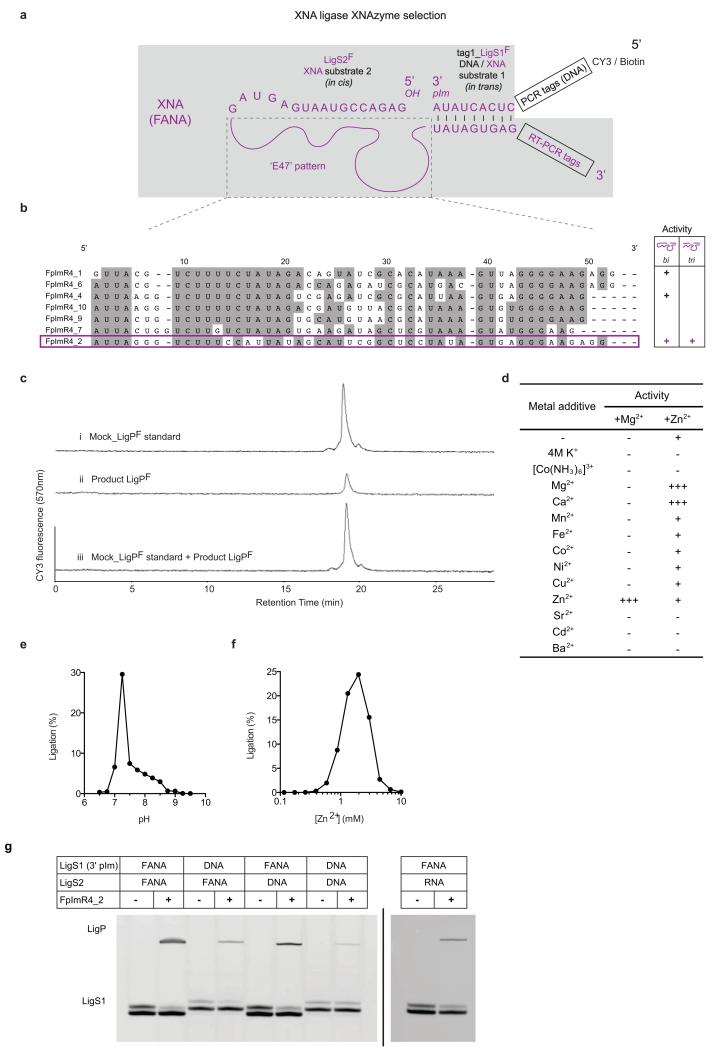
In the above examples, XNA catalysts cleave or ligate natural substrates (RNA, DNA). Next we sought to discover whether XNA catalysts could act on XNA substrates establishing a fully synthetic catalytic system. We chose to select for XNA-XNA ligase activity with a view to its potential synthetic utility for the assembly of larger XNA oligomers (Extended Data Fig 8). Again exploiting solid-phase FANA synthesis for substrate and primer strands, we synthesized an all-FANA library loosely patterned on the secondary structure of the DNA-ligase DNAzyme E4724 (see Methods) and selected for ligation of the library 5′ hydroxyl group to a substrate activated with 3′ phosphorylimidazolide (pIm). After 4 rounds, we identified multiple FANA ligase FANAzymes (Extended Data Fig. 9). One of these, FpImR4_2 (41nt), was found to be a Zn2+-dependent metalloenzyme capable of XNA-XNA (FANA-FANA) ligation in a trimolecular reaction (LigS1F + LigS2F + FpImR4_2 -> LigPF + FpImR4_2) (Fig. 4). The product (LigPF) shows an identical SAX-HPLC profile to a ‘mock’ product synthesized by D4K polymerase (Extended Data Fig. 9c), suggesting the ligation proceeds with the 3′-5′ regioselectivity. Despite the higher reactivity of the activating group, uncatalyzed FANA reactions with or without a complementary FANA splint yielded no detectable ligation of LigS1F and LigS2F (Fig. 4c), even after incubation for several days.
Figure 4. XNA-XNA ligase XNAzyme (FANA) demonstrates catalysis without natural nucleic acids.
a, Secondary structure (determined by DMS mapping, Extended Data Fig. 5) of chemically-synthesised FANAzyme FpImR4_2, which ligates FANA LigS1F, activated with 3′ phosphorylimidazolide (pIm), to LigS2F in trans. b, Urea-PAGE gel showing no product with: substrate LigS1F alone (lane 1), no XNAzyme (lane 2), no LigS2F (lane 3), splint (lane 4), or LigS1F lacking 3′pIm (lane 5); product formation is dependent on LigS2F, activated LigS1F and XNAzyme (lanes 6 and 7). c, Pre-steady state trimolecular reaction rate (kobs) (n=3, error bars=sd, 35°C). d, FpImR4_2-catalysed oligomerisation of XNA (FANA) substrates. e, XNAzyme-catalysed assembly of an active XNAzyme. A variant XNA ligase (FpImR4mut) catalyses ligation (lane 2) of FANA substrates LigS1F NUC and LigS2F NUC. The product (LigPF NUC) is a variant of XNAzyme FR17_6min (Fig. 2), which cleaves RNA substrate NucSVR (lanes 5 and 6), but not scrambled RNA (NucSR SCRAM2) (lanes 3 and 4).
Despite no apparent sequence and structural homology, as judged by DMS probing (Extended Data Fig. 5), FpImR4_2 and DNAzyme E47 may employ analogous catalytic strategies as they display a similar pH optimum (pHopt: 7.25) (Extended Data Fig. 9e), metal ion dependence (Zn2+) (Extended Data Fig. 9f) and catalytic rate (FpImR4_2 kobs: 0.04min−1 vs. E47 kobs: 0.06min−1 (35°C))24. However, unlike with E47, in the FpImR4_2 reaction, Cu2+ cannot substitute Zn2+, and Mg2+ (or Ca2+) enhances activity (Extended Data Fig. 9d). Unlike RNA ligase FANAzyme F2R17_1, FpImR4_2 displays a relaxed recognition of substrate chemistry; although most efficient at FANA × FANA ligation, it can also ligate FANA × DNA/RNA, DNA × FANA as well as DNA × DNA (Extended Data Fig. 9g).
Finally, in order to explore the synthetic potential of XNA ligation, the substrate strands and the XNA ligase FANAzyme were adapted to perform novel reactions. Modification of LigS2F substrate to include the LigS1F sequence (i.e. LigS2+1F) and a 3′-phosphorylimidazolide activation group enabled iterative substrate addition, thus synthesizing FANA oligomers up to 100nt long (Fig. 4d). Modification of the FpImR4_2 substrate-binding strands allowed ligation of a variant of the FR17-6 RNA endonuclease from constituent fragments (see Methods), enabling XNAzyme-catalysed synthesis of another XNAzyme (Fig. 4e).
Synthesis, replication (via a DNA intermediate) and evolution of synthetic genetic polymers (XNAs) not found in nature has opened up new sequence spaces for exploration, but their phenotypic richness remains to be determined. Here we show the discovery of catalysts (RNA endonucleases) in four such XNA sequence spaces (ANA, FANA, HNA, CeNA) and the elaboration of three different catalytic activities (RNA endonuclease, RNA ligase and XNA ligase) in one (FANA). These results indicate that properties such as catalysis (as well as heredity and evolution) are generalizable to a range of nucleic acid scaffolds and are likely to be emergent properties of many synthetic genetic polymers. This argues against a strong functional imperative for the chemistry of life’s genetic systems.
Limitations in current XNA technology (e.g. XNA-specific sequence biases, lower fidelity and sensitivity) contribute to library undersampling, genetic drift and reduced selection stringency, complicating comparisons of phenotypic richness of the respective XNAs with DNA and RNA sequence spaces. Nevertheless, we note that the FANA framework, with similar hybridization energetics and conformational analogy to DNA19, yielded the most active XNAzymes, while catalysts in other XNAs, which exhibit reduced (ANA)13 or enhanced (HNA and CeNA) duplex stability, as well as divergent helical conformations and dynamics7,25, showed slower rates. Substrate binding that is too weak or too strong, or conformational dynamics that are either too rapid or too slow, will reduce catalytic power by slowing conformational transitions required for catalysis and stabilizing inactive XNAzyme conformers. The evolutionary landscape of structurally more divergent XNAs may extend beyond the narrow parameters of DNA and RNA suggesting that e.g. more effective HNA- or CeNAzymes might be discovered under non-physiological conditions. More work will be needed to resolve the question of whether life’s reliance on RNA and DNA reflects a potential functional privilege of the natural polymers over unnatural XNAs in an ambient terrestrial environment9.
Future advances in methodologies for the synthesis, replication and evolution of chemically ever more divergent genetic polymers should help to resolve these questions, providing a growing database of the molecular limits of chemical encoding and replication of information, while also yielding XNA catalysts (and ligands) that fully exploit their expanded range of physicochemical properties and biostability4,26-28 with potential applications ranging from medicine to nanotechnology.
Methods
Nucleotides and oligonucleotides
Triphosphates of HNA (hNTPs), CeNA (ceNTPs) and ANA aGTP were synthesised and analysed as described previously4. Triphosphates of ANA (aATP, aCTP, aUTP) were obtained from TriLink BioTechnologies (USA), FANA (faNTPs) from Metkinen Chemistry (Finland) and DNA (Illustra dNTPs) from GE Life Sciences (USA). Oligonucleotides were synthesized by Integrated DNA technologies (Belgium) or Sigma Aldrich (USA), unless stated otherwise. Triphosphorylated RNA (LigS2R) was obtained from Trilink BioTechnologies (USA). Mock_LigPR[2′-5′] and Mock_LigPR[3′-5′] RNA standards were obtained from ChemGenes (USA). All oligonucleotides were purified by denaturing Urea-PAGE and ethanol-precipitated from filtrates of freeze-thawed gel mash as described previously4.
Synthesis of XNAs
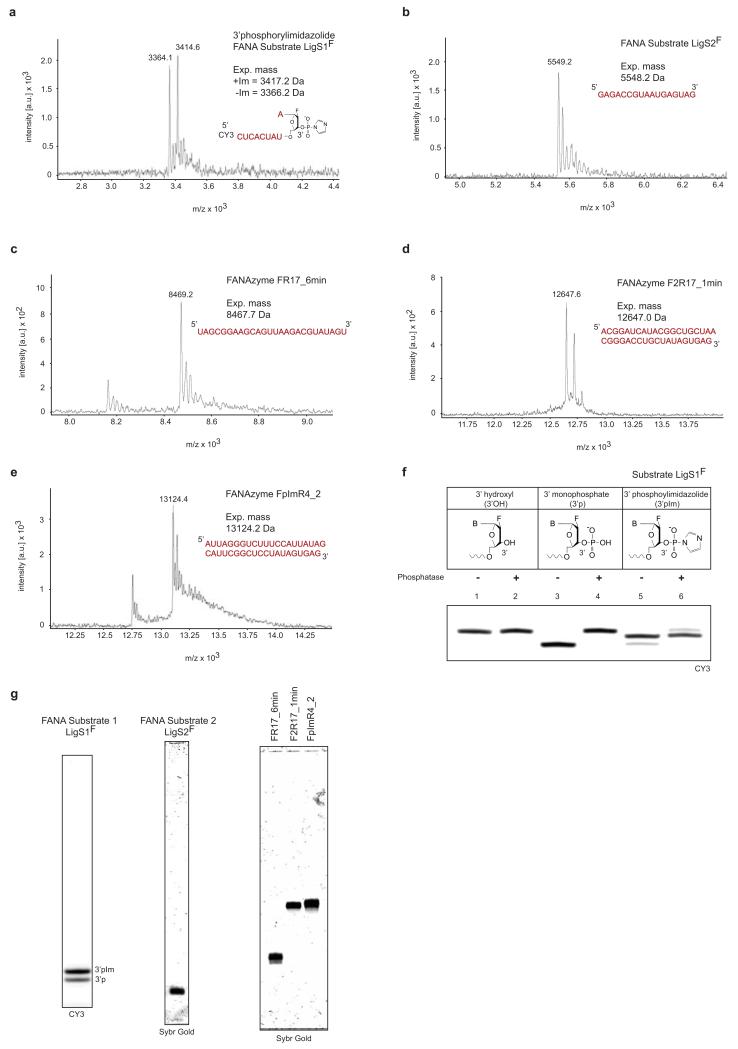
FANA and chimeric DNA-FANA oligonucleotides were prepared either enzymatically (see below) or by solid-phase chemical synthesis using a Mermade4 instrument (Bio-Automation, USA) with 1 μmole scale 3′ phosphate (Synbase 1000, Link Technologies, UK) or universal (UnySupport 1000, Glen Research, USA) CPG supports. Phosphoramidites of DNA and all synthesis reagents were obtained from Link Technologies (UK), unless stated otherwise. The solid-phase synthesis method was adapted from Deleavey et al.29. Phosphoramidites of 2′ fluoro-arabinonucleosides (FANA), cyanine 3 fluorophore (CY3) or biotin-triethyleneglycol (BiotinTEG) were obtained from Glen Research (USA) and prepared as 0.15M solutions in anhydrous acetonitrile (ACN) (Sigma Aldrich, USA), those of DNA were prepared as 0.1M solutions. Phosphoramidites were activated with 0.3M BTT (5-benzylthio-1H-tetrazole in ACN), deblocking was performed with 3% trichloroacetic acid in dichloromethane, capping of failure sequences was perfomed with pyridine acetic anhydride and 10% methylimidazole in tetrahydrofuran, and oxidation was performed with 0.02M Iodine oxidiser (Proligo series, Sigma Aldrich, USA). Coupling times were 600 s for all phosphoramidites, with the exception of CY3, BiotinTEG and the FANA guanosine phosphoramidite, which were allowed to couple for 900 s. Deprotection and cleavage from CPG support was achieved by incubation in 3:1 NH4OH:EtOH for 48 h at room temperature, then dried by speedvac. PAGE purified chemically synthesised FANA substrates and XNAzymes were analysed by mass spectrometry (Extended Data Fig. 10). For triphosphate addition to synthesized FANA (and DNA), the method described by Zlatev et al.30 was followed prior to deprotection and cleavage from the solid support.
All other XNAs were prepared enzymatically using polymerase mutants as described previously4; polymerases D4K for ANA and FANA, 6G12 for CeNA and 6G12 I521L (see below) for 1,5 anhydrohexitol nucleic acid (HNA), with the addition of 4% ET-SSB (NEB, USA). For preparation of all-XNA strands using polymerases (e.g. for in trans XNAzyme reactions), either the appropriate FANA primer was used, or an RNA primer was used to synthesis an RNA-XNA chimeric strand, which was then incubated in 0.8M NaOH at 65°C for 1 h to completely hydrolyse the RNA portion. All XNA oligonucleotides were purified by denaturing Urea-PAGE.
Preparation of single-stranded DNA, RNA & XNA using streptavidin beads
Biotinylated oligos were captured using Dynabeads® MyOne™ Streptavidin C1 beads (Invitrogen / Life Technologies, USA) in BWBS (10 mM Tris·HCl pH 7.4, 1 M NaCl, 0.1% v/v Tween20, 1 mM EDTA) for 1-2h at room temperature or overnight at 4°C. Denaturation/elution of unbiotinylated strands was achieved by three washes in BWBS followed by rapid (<1min) incubation in 0.1M NaOH at room temperature. Where eluted strand was being prepared, NaOH supernatant was immediately neutralized in 1M Tris pH 7.4. Elution of biotinylated oligos from beads was achieved by three washes in H2O, then incubation in either H2O for 2 × 2min at 80°C, or PAGE loading buffer (95% formamide, 10 mM EDTA, 0.05% bromophenol blue) for 2 × 2 min at 95°C.
Preparation of 6G12 I521L polymerase
We introduced the I521L mutation to the 6G12 backbone by iPCR using primers RT520fo and RT521ba. PCR was carried out using Expand High Fidelity polymerase (Roche Diagnostics GmbH, Germany) as an initial incubation of 2 min at 95°C followed by 25 × of (30 s 95°C, 30 s 50°C, 18 min 68°C,) followed by a final extension of 10 min 68°C. Amplified DNA was purified (QIAquick PCR purification kit, QIAGEN GmbH, Germany) according to the manufacturer’s recommendations and restricted with BsaI and DpnI (New England Biolabs Inc., Massachusetts, USA). Reactions were again purified (QIAquick PCR purification kit) and ligated with T4 DNA ligase (NEB, USA). Ligated plasmids were transformed into E. coli NEB 10-β cells (NEB, USA), and isolated transformants were checked by DNA sequencing (Source Biosciences, UK).
A transformant with the correct sequence was expressed and purified as previously described4 and used to determine the impact of the additional mutation on the fidelity and processivity of 6G12. The resulting 6G12 I521L polymerase had a different divalent cation optimum and could synthesise HNA in the absence of Mn2+ ions, in reactions carried out with 3 mM Mg2+. 6G12 I521L was more processive than 6G12 alone and could synthesise HNA at higher fidelities (aggregate DNA->HNA->DNA fidelity: 3.0×10−3 – experiment carried out as described previously4).
Preparation of FANA phosphorylimidazolide oligonucleotides
Preparation phosphorylimidazolide oligonucleotides was adapted from a method used by Orgel and others31. 3′ phosphorylated FANA or DNA was prepared by solid-phase chemical synthesis (see above) and re-suspended to 100 μM in 0.5 M imidazole (pH 6.0). 50 μl oligo/imidazole solution was added to 6.5 μmol solid 1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride (EDC) (Pierce Biotechnology / Thermo, USA) and incubated at room temperature for 2 h. Oligos were desalted using Amicon 3,000 MW cut off spin filters (Merck Millipore, USA). Purification of (and analysis of reactions involving) all phosphorylimidazolide oligos were perfomed using Tris-free Urea-PAGE gels run using 10 mM NaOH, pH adjusted to 8.5 with Boric acid32. FANA phorphorylimidazolides were analysed by mass spectrometry, phosphatase protection and Urea-PAGE mobility (Extended Data Fig. 10).
XNAzyme selections
General schemes for selections are shown in Extended Data Figs. 1, 6 and 8. Purified single-stranded libraries and substrates were annealed by incubation at 80°C for 60 s, then allowed to cool to room temperature over 5 min, except for phosphorylimidazolide oligos, which were not annealed. Substrates and enzymes were incubated separately in reaction buffer for 5 min at reaction temperature, then mixed to start reactions. For selection of RNA ligase and endonuclease XNAzymes, reactions were performed at 17°C in 30 mM HEPES (pH 8.5), 150 mM KCl, 25mM MgCl2 and 0.5 U/ul RNasein RNase inhibitor (Promega, USA). For selection of XNA (FANA) ligase XNAzymes, reactions were performed at 35°C in 30 mM HEPES (pH 7.2), 150 mM KCl, 25mM MgCl2 and 1mM ZnCl2. In general, ~1 nmol of starting library was prepared and reacted at 10 μM with equimolar substrate for 5 days. For rounds 2-17, 10-50 pmol XNA pools were prepared and reacted at 1μM over steadily decreasing reaction times, settling on 30 mins in rounds 15-17. In RNA endonuclease XNAzyme selections, the three libraries for each XNA (fully degenerate N40 library, and the ‘8-17’ and ‘10-23’ patterned libraries) were synthesised separately, but pooled after round 5.
All XNA reverse transcriptions (using polymerase RTI521L) were performed as described previously for HNA aptamer selections4, but without a polyA tailing step and using 0.2 μM RT primer Tag4test7_2Me (or a version with 5′ BiotinTEG), which contains 2′O-methyl RNA modifications to improve annealing. First-stand cDNA was amplified by a two-step nested PCR strategy (see Extended Data Figs. 1, 6 and 8). The first ‘out-nested’ RT + PCRs used 0.5 μM primers and a mixture of OneTaq Hot Start (NEB, USA) (itself a mix of Taq and Deep VentR™) and 0.15 U/ul Thermoscript (Invitrogen / Life Technologies, USA) polymerases, which is able to transcribe 2′O-methyl RNA, in 20 mM Tris-HCl (pH 8.9 at 25°C), 22mM NH4Cl, 22mM KCl, 0.06% IGEPAL CA-630, 0.05% Tween20, 4mM MgCl2 and 200 μM dNTPs. Cycling conditions were 80°C for 30 sec, 52°C for 30 sec, 72°C for 15 min, 94°C for 1 min, 20-35×[94°C for 30 sec, 54°C for 30 sec, 72°C for 30 sec], 72°C for 2 min. Following the first PCR, primers were digested using ExoSAP (Ambion / Life Technologies, USA), which was then heat inactivated, according to the manufacturer’s instructions. Second step (‘in-nest’) PCRs used using 1 μl of unpurified out-nest PCR product as template in a 50 μl reaction using OneTaq Hot Start master mix (NEB, USA) and cycling conditions 94°C for 1 min, 10-20×[94°C for 30 sec, 54°C for 30 sec, 72°C for 30 sec], 72°C for 2 min. Reactions were analysed by electrophoresis on 4% NGQT-1000 agarose (Thistle Scientific, UK) gels containing GelStar stain (Lonza, Switzerland). Bands of appropriate size were purified using a gel extraction kit (Qiagen, Netherlands) as per manufacturer’s instructions. Purified DNA was used as the polyclonal template for either sequencing library PCR (see below) or large scale preparative PCR (2ml) for generation of DNA templates for XNA synthesis. Prep PCR were performed with 1 μM primers using 0.05 U/ul SUPER Taq in 1× buffer (HT Biotechnology, UK) with 0.125 μM dNTPs. Cycling conditions were the same as the second step PCR above. Single-stranded DNA templates were isolated using streptavidin beads (see above) and ethanol-precipitated before further use.
XNAzyme reactions
Purified XNAzymes and substrates were annealed as described above and reacted under selection conditions unless stated otherwise, in DNA- or protein- (for 3′pIm reactions) LoBind tubes (Eppendorf, Germany). In pH titration experiments, buffer was substituted for 50 mM EPPS (pH 6.5-8.75), CHES (pH 9.0-10.0) or CAPS (pH 10.25-11.0). For determination of pseudo first-order reaction rate (kobs) under single-turnover presteady state (Km/kcat) conditions, a five-fold excess of enzyme (5μM) was incubated in trans with either fluorophore-labeled 1μM NucS (nuclease substrate), or fluorophore-labeled 1μM LigS1 (ligase substrate 1) and 5μM LigS2 (ligase substrate 2). RNA endonuclease and ligase reactions were performed in 30 mM EPPS (pH 8.5), 150 mM KCl, 50 mM MgCl2 at 25°C. XNA ligase reactions were performed in 30 mM HEPES (pH 7.5), 150 mM KCl, 50 mM MgCl2, 1mM ZnCl2 at 35°C. Reactions were stopped at different time points by addition of 95% formamide, 20 mM EDTA and cooling on dry ice. Reactions were separated by Urea-PAGE and fluorophores visualized using a Typhoon Trio imager (GE Life Sciences, UK). The fraction of reaction product to substrate was quantified using ImageQuant TL software (GE Life Sciences, UK) and mean data from three independent reactions (except for CeNAzyme CeR16_3, for which only two data sets were collected) were fit to equation 1 using Prism 6.0b (GraphPad, USA):
| [1] |
Where P(t) is the percentage of cleaved or ligated RNA or XNA (FANA) at time t, P∞ is the apparent reaction end point and kobs is the observed rate constant. For magnesium titration experiments, data were fit to equation 2.
| [2] |
Where P(t) is the percentage of cleaved or ligated RNA or XNA (FANA) at time t, P∞ is the apparent reaction end point and Km is the apparent Michaelis constant.
For FR17_6min multiple turnover catalysis, 1 μM NucSR_min(AG) was reacted with 10 nM FR217_6min at 25°C in 30 mM HEPES (pH 8.5), 150 mM KCl, 50 mM MgCl2. DNAzyme 8-17 synthesised as XNAs (5 μM) was reacted with substrate NucSR_AG (1 μM) for 1 h at 37°C in 30 mM HEPES (pH 8.5), 150 mM NaCl, 50 mM MgCl2. For characterisation of bivalent metal ion requirements of selected XNAzymes, bimolecular (FR17_6min) or trimolecular (F2R17_1min and FpImR4_2) reactions were performed under conditions used to determine kobs with MgCl2 and/or ZnCl2 substituted by chlorides of the metals of the Irving-Williams series (100 μM-50 mM).
Deep sequencing
Amplified polyclonal cDNA from XNA selections was prepared for deep sequencing by the Illumina Miseq method by appending the bridge-amplification sequences by PCR. Sequencing library generating PCR reactions were performed with OneTaq Hot Start master mix (NEB, USA) with 10 ng/50ul gel-purified polyclonal template DNA (see above), 0.1 μM primers (P5_P2 and P3_Test7-2) and cycling conditions 94°C for 1 min, 10×[94°C for 30 sec, 56°C for 30 sec, 72°C for 30 sec], 72°C for 2 min. Sequencing library DNA was purified using a PCR purification kit (Qiagen, Netherlands), then a 12pM sample of pooled libraries plus 20% PhiX control (Illumina, UK) was denatured and sequenced (single-end read, 75 cycles) using a MiSeq reagent kit and instrument (Illumina, UK) according to manufacturer’s instructions. Libraries were barcoded using variants of the P5_P2 primer containing 6nt sequences from the NEXTflex series (Illumina, UK). Data was analysed using the Galaxy server33-35 and sequences ordered by abundance.
Analysis of oligonucleotide phosphorylation
The presence or absence of 3′ or 5′ phosphates on RNA cleavage products, and the protection of 3′ phosphates on FANA ligase substrates by formation of phosphorylimidazolides, was assayed by Urea-PAGE gel shift following incubation in rAPid alkaline phosphatase (Roche, Switzerland) or T4 polynucleotide kinase (NEB, USA) in manufacturer’s buffers for 30 min at 37°C. Hydrolysis of cyclic phosphates was achieved by incubation in 10 mM Glycine pH 2.5 for 10mins at room temperature. Partial alkaline hydrolysis of RNA substrates (denoted by −OH) was achieved by incubation at 90°C in 50 mM sodium carbonate buffer (pH 9.2) for 10 mins. Partial RNase T1 digestion was was achieved by incubation at 55°C in 0.1 U/μl RNase T1 (Invitrogen / Life Technologies, USA) in 30 mM sodium acetate (pH 5) for 10 mins, then stopped in 7M Urea 1.5mM EDTA.
Analysis of oligonucleotide mass by MALDI-ToF mass spectrometry
Oligo samples, 0.75 μl in water were spotted onto MALDI target followed by 0.75 μl of 3-hydroxypicolinic acid. Some oligo samples were vacuum dried, resuspended in 25 μl, 0.1 M TEAA (Triethylammonium acetate) and desalted using a zip-tip C18 (Merck Millipore, USA). The zip-tip C18 was washed 3 × 10 μl 0.1 M TEAA and then 3 × 10 μl water. Next, the oligo was eluted directly onto a MALDI target with 5 μl of 3-hydroxypicolinic acid. All mass spectrometric measurements were carried out in positive ion mode on an Ultraflex III TOF-TOF instrument (Bruker Daltonik, Bremen, Germany).
Analysis of oligonucleotide linkage isomers by SAX-HPLC
RNA and FANA ligation products were analysed by strong anion exchange chromatography (SAX-HPLC) using a Varian Prostar system (Agilent, USA) with a DNAPac PA200 column (Dionex/Thermo, USA) under conditions sufficient to resolve linkage regioisomers36: 10nM sodium phosphate buffer (pH 11.5), gradient 0.4M to 1.4M NaCl over 30min, flow rate 1.5 ml/min. Fluorescence was detected using a 122 fluorometer (Gilson, USA) set to excitation 488 nm, emission 520 nm for carboxyfluorescein(6FAM)-labeled RNA, and emission 550 nm, excitation 570 nm for cyanine 3 (CY3)-labeled FANA.
Chemical probing of XNAzymes and secondary structure prediction
Selective 2′hydroxyl Acylation analysed by Primer Extension (SHAPE) structure probing experiments were performed on RNA endonuclease XNAzyme (FANA) FR17_6 using a chimeric RNA-FANA construct, FR17_6wilk, prepared using primer SHAPE_Nucprim and template FR17_6wilk_temp. The construct contains sequences of NucSR (RNA) and FR17_6 (FANA) flanked by 5′ and 3′ structure cassette sequences from Wilkinson et al.15, with a 2′-O-methyl RNA modification at adenine 11. For the XNA ligase XNAzyme (FANA) FpImR4_2, an analogous construct, FpImR4_2wilk was prepared by ligation of 1 μM modified FANA substrate LigS1wilkF to equimolar concentration of a version of FpImR4_2 with LigS2F in cis (prepared by LigS2F-primed FANA synthesis on template FpImR4wilk_temp) for 2 h at 35°C in 30 mM HEPES (pH 7.2), 150 mM KCl, 25mM MgCl2 and 1mM ZnCl2. FANA constructs (1 μM in 8 μL H2O) were denatured at 80 °C for 1 minute, incubated at room temperature for 5 minutes, treated with 1 μl 10× SHAPE folding buffer [500 mM EPPS (pH 8.2), 1.5 M KCl, 250 mM MgCl2], and allowed to fold at 17 °C for 20 minutes. After folding, FANA constructs were treated with 1-methyl-7-nitroisatoic anhydride (1 μl, 100 mM in neat DMSO) and incubated at 17°C for 15 minutes. No-reagent control reactions were performed with 1 μl neat DMSO. Denaturing control reactions were performed as described previously37. After modification, FANA constructs were purified with a G-50 spin column (GE Healthcare). cDNA was generated using SuperScript II reverse transcriptase (Life Technologies) under SHAPE-MaP conditions37.
Dimethyl sulfate (DMS) modification was adapted from the RING-MaP approach (P. Homan, K. Weeks et al., manuscript in preparation). FANA constructs (nuclease: 1 μM in 5 μL H2O, ligase: 0.5 μM in 5 μL H2O) were annealed as described above and treated with 4 μl 2.5× DMS folding buffer [750 mM cacodylate (pH 7.0), 25 mM MgCl2]. Folded FANA constructs were treated with DMS (1 μl, 1.7 M in absolute ethanol), incubated at 17 °C for 6 minutes, quenched with 10 μl neat 2-mercaptoethanol, and purified with a G-50 spin column. No-reagent control reactions were performed with 1 μl absolute ethanol. cDNA was generated using RT521K polymerase as described previously4. Briefly, 5 pmol FANA construct and 10 pmol primer were denatured for 1 min at 95 °C, chilled on ice, and incubated with ~2 μg/ml RT521K and 0.2 mM dNTPs in 1× ThermoPol buffer (New England Biolabs) at 65 °C for 4 hours. cDNA from reverse transcription reactions was purified with G-50 spin columns. SHAPE- and DMS-MaP sequencing libraries were created using the targeted gene-specific approach37, with minor changes: PCR 1 was performed for 23 cycles, 98°C for 30 sec, 23×[98°C for 10 sec, 68°C for 30 sec, 72°C for 20 sec], 72°C for 2 min, and PCR 2 was performed for 7 cycles, using 1 μl of unpurified PCR 1 product as template in a 50 μl reaction. Purified libraries were pooled and sequenced with an Illumina MiSeq, generating datasets of 2×150 paired-end reads. Sequencing reads were aligned to reference sequences and per-nucleotide mutation rates, excluding primer-binding sites, were calculated using the SHAPE-MaP analysis pipeline. SHAPE reactivities were calculated for RNA nucleotides37; FANA nucleotides were excluded from SHAPE analysis. DMS reactivities for all nucleotides were calculated by subtracting the mutation rate of the no-reagent control from the mutation rate of DMS-modified FANAzymes at each position. SHAPE and DMS reactivity profiles were normalized by the “2%-8%” method38. The FR17_6 FANAzyme secondary structure model is the only structure predicted using ShapeKnots39, incorporating pseudo-free energy constraints derived from SHAPE reactivities. All other XNAzyme secondary structure models were predicted using ViennaRNA (version 2.1.6)40 or mfold41. The FpImR4_2 structure was further manually curated using DMS reactivity data.
Extended Data
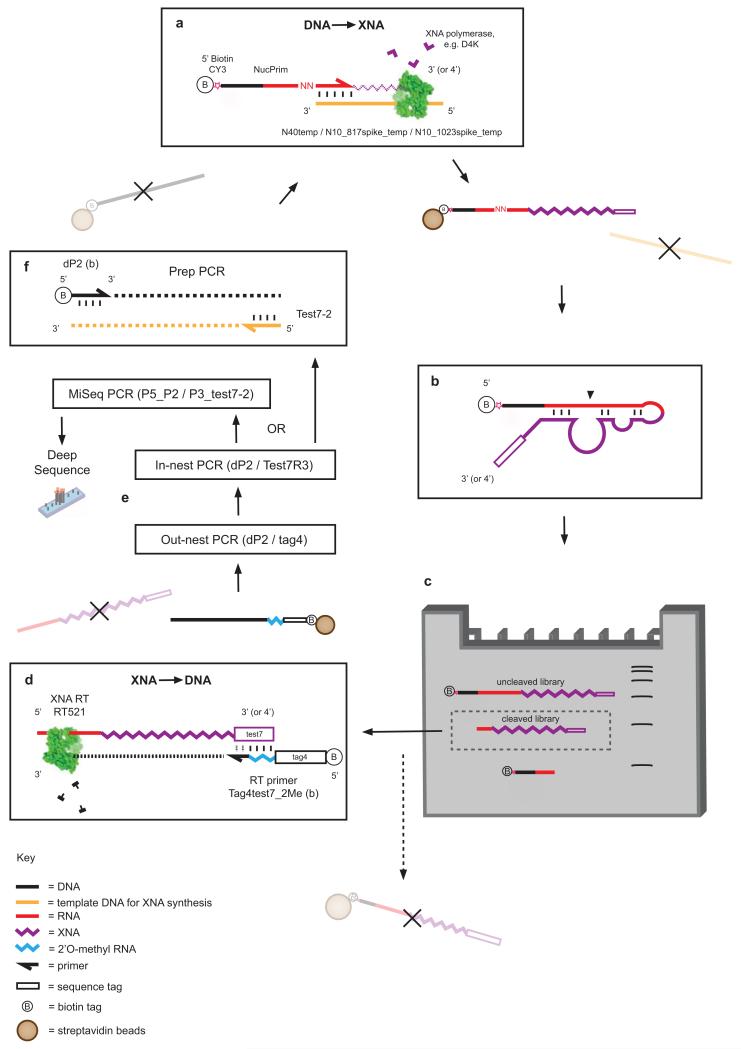
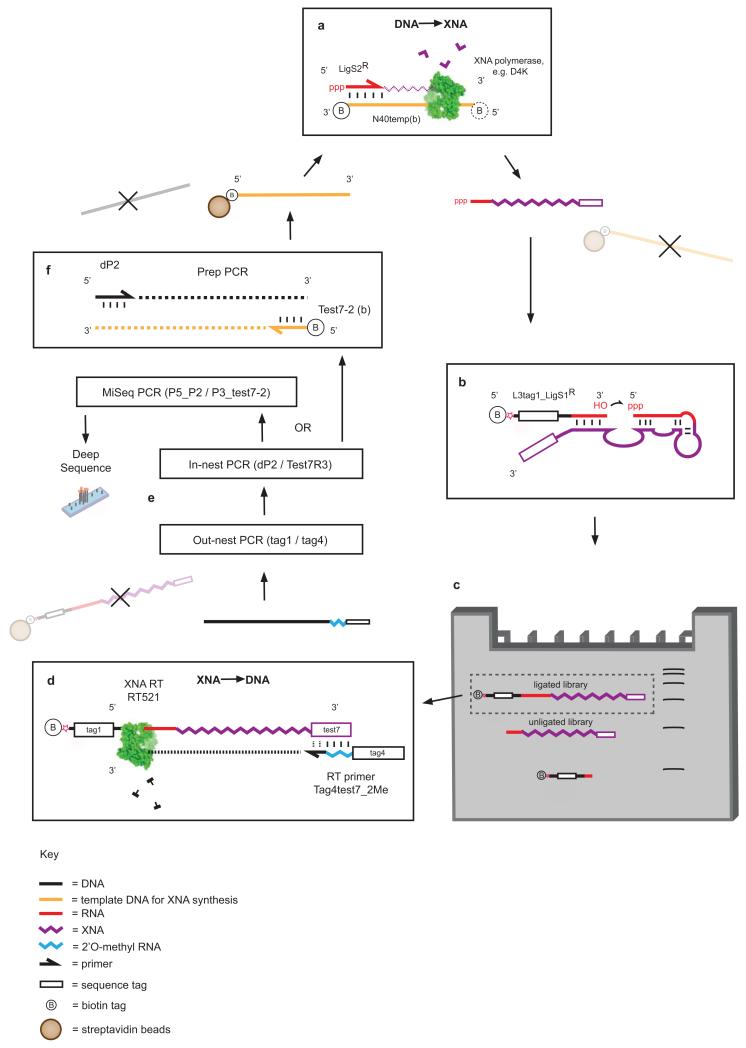
Extended Data Figure 1. Selection scheme for RNA endonuclease XNAzymes.
a, XNA library preparation using DNA-dependent XNA polymerases, primed by a biotinylated chimeric DNA-RNA primer (NucPrim), which serves as substrate for RNA cleavage in cis. Libraries are captured by streptavidin beads, allowing denaturation and removal of DNA templates. b, Single-stranded libraries are annealed and incubated in reaction buffer (see Methods), successful XNAzymes cleave the biotinylated RNA substrate in cis. c, Size separation of reacted XNA pools using denaturing polyacrylamide electrophoresis (Urea-PAGE). Cleaved XNA pools are gel-extracted from the gel and incubated with streptavidin beads to deplete any uncleaved carry-over. d, Reverse transcription of isolated, cleaved XNA pools using XNA-dependent DNA polymerase RT521L (i.e. XNA -> cDNA). e, Amplification of transcribed cDNA by successive PCR reactions. f, PCR reaction generating templates for XNA synthesis for further rounds of selection.
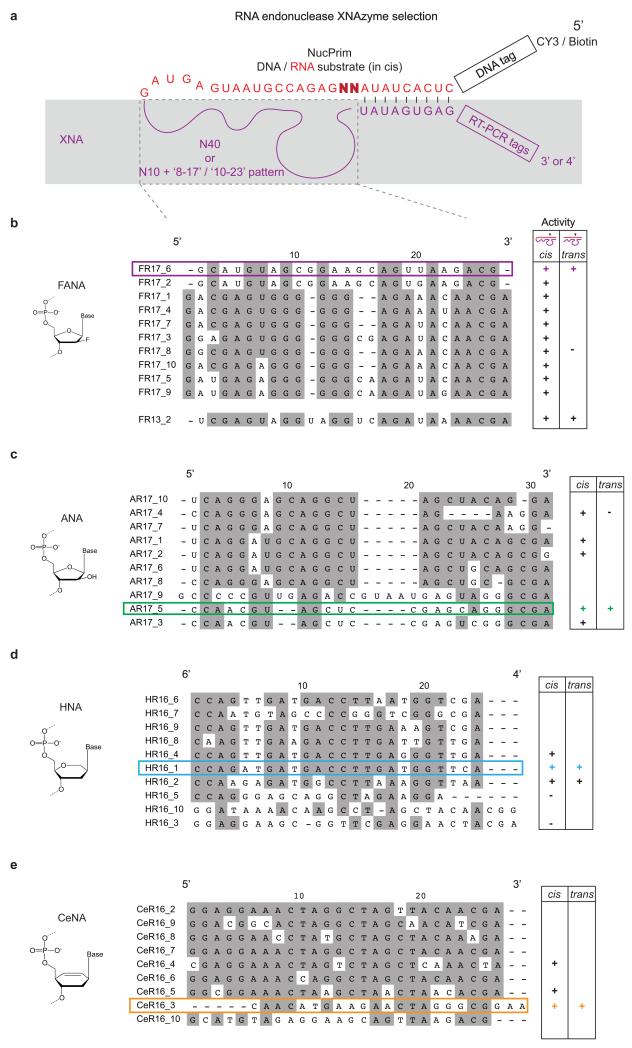
Extended Data Figure 2. Sequences of RNA endonuclease XNAzymes.
a, Schema showing DNA-RNA-(red)-XNA(purple) chimeric library setup for selection of in cis RNA-cleaving XNAzymes. The sequences of the XNA region under selection (dashed box) of the most abundant clones revealed by deep sequencing are shown for selections using b, FANA, c, ANA, d, HNA, and e, CeNA. The top 10 sequences, or representatives of sequence families, were screened by Urea-PAGE gel shift for activity in cis (unimolecular reaction, as selected) and in trans (bimolecular reaction). Sequences chosen for further characterization are highlighted.
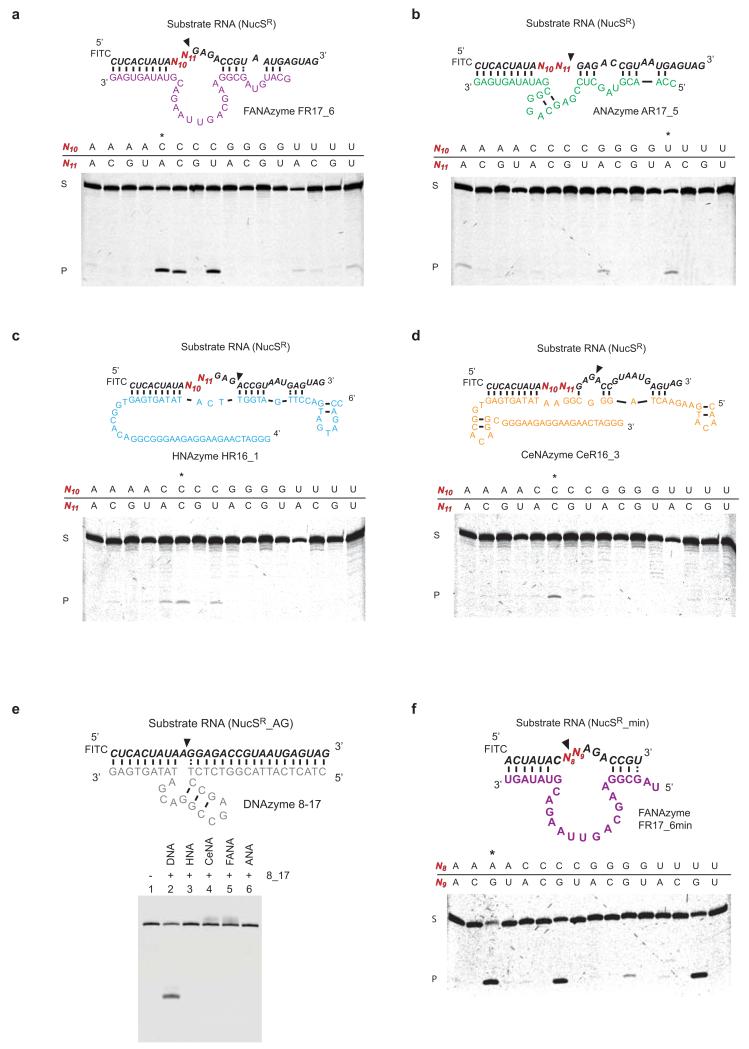
Extended Data Figure 3. Sequence dependence of RNA endonuclease XNAzyme cleavage.
XNAzymes were selected with degeneracy in the RNA substrate (see Extended Data Fig. 2a). The sequence requirements at these positions (upstream of the cleavage sites (▼)) in the RNA substrate (N10 and N11 shown in red) were determined by Urea-PAGE gel shift using all 16 variants of the substrate NucSR with each XNAzyme in trans: a, FR17_6 (FANA), b, AR17_5 (ANA), c, HR16_1 (HNA), d, CeR16_3 (CeNA). e, RNA substrate NucSR_AG (lane 1) was reacted in trans with RNA endonuclease DNAzyme 8-1717 synthesized as DNA (lane 2), HNA (lane 3), CeNA (lane 4), FANA (lane 5) or ANA (lane 6). Activity of 8-17 is lost upon conversion to the XNAs described. f, Sequence requirements for RNA cleavage by FANAzyme FR17_6min proximal to the cleavage site (▼), positions 8 and 9 (highlighted in red) of minimized RNA substrate NucSR_min. Substrate sequences used for further characterization of each XNAzyme are indicated by (*)
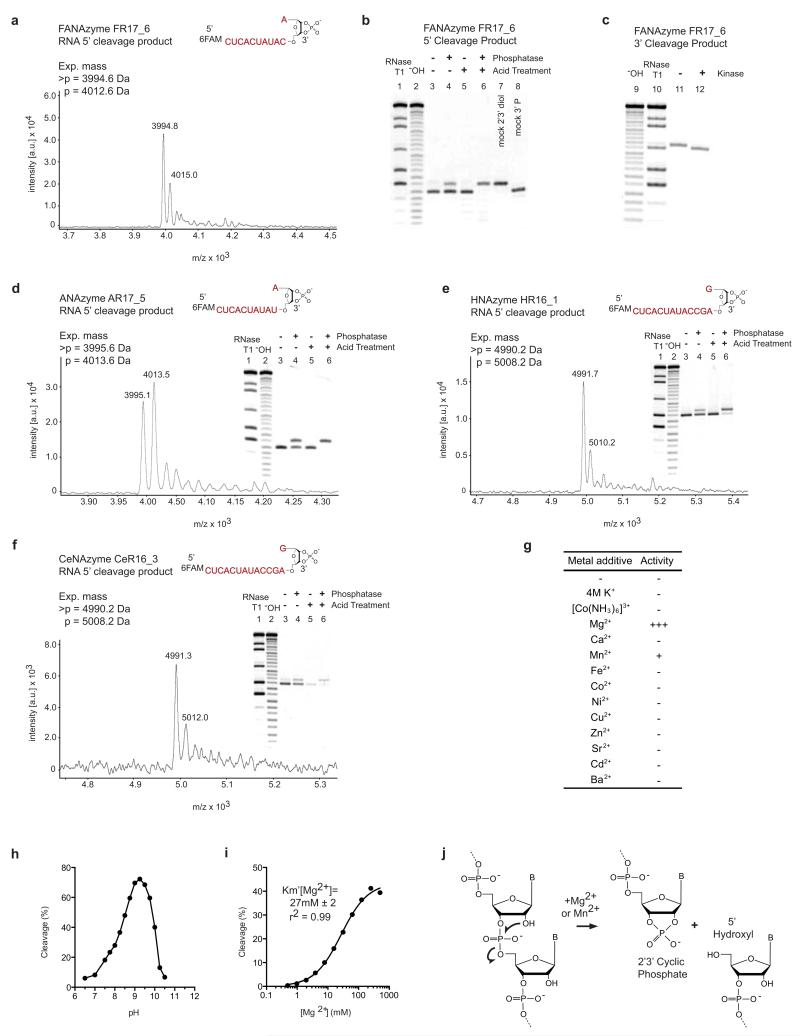
Extended Data Figure 4. Analysis of RNA endonuclease XNAzyme cleavage products.
a, 5′ cleavage product of FANAzyme FR17_6 reaction shows expected mass for a 2′,3′ cyclic phosphate (>p) using Matrix-Assisted Laser Desorption/Ionization-Time of Flight mass spectrometry (MALDI-ToF). b, Hydrolysis of 5′ FR17_6 cleavage product >p in low pH and dephosphosphorylation with calf intestinal phosphatase (removes 2′p or 3′p, but not >p). c, Phosphorylation of 3′ FR17_6 cleavage product with T4 polynucleotide kinase (adds 5′p). Mass spectra and dephosphorylation assays of 5′ cleavage products of d, ANAzyme AR17_5, e, HNAzyme HR16_1 and f, CeNAzyme CeR16_3 reveal all RNA endonuclease XNAzymes yield products with 2′,3′ cyclic phosphates. (RNase T1) and (−OH) indicate partial hydrolysis reactions of the RNA substrates used. g, Bivalent metal ion requirements and titration of, h, pH or i, MgCl2, of FANAzyme FR17_6min reaction with NucSR_min. j, Reaction catalyzed by RNA endonuclease XNAzymes.
Extended Data Figure 5. Chemical probing of XNAzyme secondary structures.
a, Chemical probing using Selective 2′hydroxyl Acylation analysed by Primer Extension (SHAPE) (RNA)15 or, b, d, Dimethyl sulfate (DMS)16 (RNA and FANA) footprinting, used to inform secondary structure predictions of c, RNA endonuclease FANAzyme FR17_6 or e, FANA ligase FANAzyme FpImR4_2, embedded in structural cassettes15, with RNA substrate (inactivated by a 2′O-methyl modification at position 11, indicated by (*) in all panels) or FANA product in cis. SHAPE analyses RNA 2′OH. Black, orange and red solid bars and bases indicate low, moderate, and high SHAPE reactivity, respectively, for positions 1-28 in FR17_6 construct, corresponding to RNA substrate NucSR. DMS reacts predominantly with A & C bases (RNA and FANA). Positions were defined as reactive (blue open bars and circles) if reactivity was greater than a cutoff (dashed line in b and d) of one half standard deviation above the median. Dashed circles indicate positions with marginal reactivity. Site of cleavage (in unmodified RNA) or ligation is indicated by (▼). Primer-binding regions (no structural data) are shown in grey.
Extended Data Figure 6. Selection scheme for RNA ligase XNAzymes.
a, XNA library preparation using DNA-dependent XNA polymerases, primed by a 5′ triphosphorylated (5′ppp) RNA primer (LigS2R), which serves as one of the substrates for RNA ligation in cis. Libraries are synthesized with 3′ biotinylated DNA template, allowing capture and removal by streptavidin beads. b, Single-stranded libraries (unbiotinylated) are annealed and incubated in reaction buffer (see Method) together with a biotinylated chimeric DNA-RNA substrate (tag1_LigS1R), which successful XNAzymes ligate to RNA substrate LigS2R in cis. c, Size separation of reacted XNA pools using Urea-PAGE. Ligated XNA pools are gel-extracted and captured by streptavidin beads. d, Reverse transcription of XNA pools using XNA-dependent DNA polymerase RT521L, which is also able to transcribe RNA across the ligation junction (i.e. [RNA-RNA-XNA] -> cDNA). e, Amplification of transcribed cDNA by successive PCR reactions; out-nest reaction depends on priming site (tag1) from ligated substrate tag1_LigS1R. f, PCR reaction generating templates for XNA synthesis (now 5′ biotinylated) for further rounds of selection.
Extended Data Figure 7. Sequences and analyses of RNA ligase XNAzymes.
a, Schema showing RNA (red)-XNA(purple) chimeric library setup for selection of FANAzymes capable of catalyzing a bimolecular RNA ligation. b, Sequences of the FANA region under selection (dashed box) of the most abundant clones revealed by deep sequencing. Representatives of sequence families were screened for activity in bimolecular (LigS2R attached to XNAzyme) or trimolecular (XNAzyme separate from both substrates). Sequence F2R17_1 (highlighted) was chosen for further characterization. c, Regiospecificity of RNA product (LigPR) of ligation catalyzed by XNAzyme F2R17_1min (see Fig. 3), analyzed by Strong Anion Exchange Chromatography (SAX-HPLC)36. Mock RNA ligation product (i-iii) containing a single 2′-5′ (Mock_LigPR[2′-5′]) or 3′-5′ linkage (Mock_LigPR[3′-5′]) at a position analogous to the ligation site were compared to the XNAzyme-catalysed RNA product LigPR (iv-vi). The XNAzyme product gives an identical elution profile to the natural (3′-5′) linkage standard. d, Bivalent metal ion requirements and titration of, e, pH or f, MgCl2, of FANAzyme F2R17_1min reaction. g, Substitution of RNA ligase substrates with DNA and XNA (FANA) versions in F2R17_1min reaction shows that 5′-RNA-RNA-3′ ligation is preferred, but some ligase activity can be seen with 5′-RNA-DNA-3′.
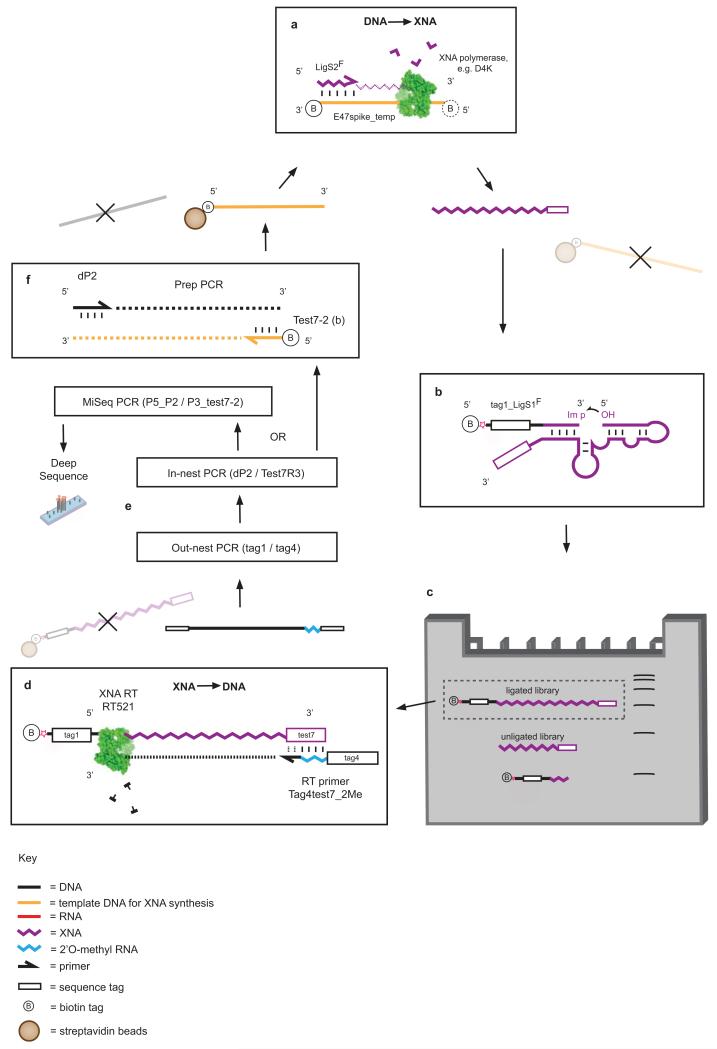
Extended Data Figure 8. Selection scheme for XNA ligase XNAzymes.
a, XNA library preparation using DNA-dependent XNA polymerases, primed by an all-XNA (FANA) primer (LigS2F), which serves as one of the substrates for FANA ligation in cis. Libraries are synthesized with 3′ biotinylated DNA template, allowing capture and removal by streptavidin beads. b, Single-stranded libraries (unbiotinylated) are annealed and incubated in reaction buffer (see Methods) together with a biotinylated chimeric DNA-XNA (FANA) substrate (tag1_LigS1F), activated with a 3′ phosphorylimidazolide (Extended Data Fig. 10), which successful XNAzymes ligate to XNA (FANA) substrate LigS2F in cis. c, Size separation of reacted XNA pools using Urea-PAGE. Ligated XNA pools are gel-extracted and captured by streptavidin beads. d, Reverse transcription of XNA pools using XNA-dependent DNA polymerase RT521L (i.e. XNA -> cDNA). e, Amplification of transcribed cDNA by successive PCR reactions; out-nest reaction depends on priming site (tag1) from ligated substrate tag1_LigS1F. f, PCR reaction generating templates for XNA synthesis (now 5′ biotinylated) for further rounds of selection.
Extended Data Figure 9. Sequences and analyses of XNA ligase XNAzymes (FANA).
a, Schema showing all-FANA library setup for selection of FANAzymes capable of catalyzing a bimolecular XNA (FANA) ligation. b, FANA sequences of the region under selection (dashed box) of the most abundant clones revealed by deep sequencing. Representatives of sequence families were screened for activity in bimolecular (LigS2F attached to XNAzyme) or trimolecular (XNAzyme separate from both substrates). Sequence FpImR4_2 (highlighted) was chosen for further characterization. c, Regiospecificity of XNA (FANA) product (LigPF) of ligation catalyzed by XNAzyme FpImR4_2 (see Fig. 4), analyzed by Strong Anion Exchange Chromatography (SAX-HPLC). Mock FANA ligation product (Mock_LigPF) (i), prepared by polymerase (D4K) gives an identical elution profile to the XNAzyme-catalysed FANA product (LigPF) (ii and iii). d, Bivalent metal ion requirements and titration of, e, pH or f, MgCl2, of FANAzyme FpImR4_2 reaction. g, Substitution of XNA ligase substrates with RNA and DNA versions in FpImR4_2 reaction. Although 5′-FANA × FANA-3′ is preferred, ligase activity can be seen with 5′-FANA × DNA-3′, and × RNA-3′, as well as 5′-DNA × FANA-3′, × DNA-3′ and × RNA-3′.
Extended Data Figure 10. Analysis of XNA (FANA) substrates and enzymes prepared by solid-phase synthesis.
MALDI-ToF mass spectra showing expected masses of a, XNA (FANA) ligase substrate LigS1F-3′phosphorylimidazolide (prepared by solid-phase synthesis of the 3′ phosphorylated (3′p) oligonucleotide, followed by reaction with carbodiimide and imidazole (see Methods)), b, XNA (FANA) ligase substrate LigS2F, and XNAzymes c, FR17_6min, d, F2R17_6min, and e, FpImR4_2. f, Dephosphorylation assay of of versions of LigS1F (3′ hydroxyl, lanes 1 and 2, 3′ phosphate, lanes 2 and 3, or 3′ phorphorylimidazolide, lanes 5 and 6) with calf intestinal phosphatase (lanes 2, 4 and 6). The majority of the LigS1F preparation shown (~70%) is protected from dephosphorylation, consistent with formation of the 3′pIm. g, Urea-PAGE analyses of purified FANA substrates and XNAzymes.
Supplementary Material
Acknowledgements
This work was supported by the Medical Research Council (MRC) programme grant U105178804 (PH, AIT, VBP, ASM, SPC, CC) and by grants from the European Science Foundation (ESF) and the Biotechnology and Biological Sciences Research Council (BBSRC) UK (09-EuroSYNBIO-OP-013) (PH, AIT), the European Union Framework (FP7/2007-2013 (PtH), the European Research Council (ERC-2012 ADG_20120216/320683 (PtH)), the US National Science Foundation (MCB-1121024 (KMW)) and by an NSF Graduate Research Fellowship (DGE-1144081 (MJS)).
Footnotes
Oligonucleotide sequences can be found in Supplementary Table 1.
Competing financial interests: The authors declare no competing financial interests.
References
- 1.Atkins JF, Gesteland RF, Cech TR, editors. RNA Worlds. Cold Spring Harbor Laboratory; 2012. [Google Scholar]
- 2.Breaker RR, Joyce GF. A DNA enzyme that cleaves RNA. Chem. Biol. 1994;1:223–229. doi: 10.1016/1074-5521(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 3.Eschenmoser A. Chemical Etiology of Nucleic Acid Structure. Science. 1999;284:2118–2124. doi: 10.1126/science.284.5423.2118. [DOI] [PubMed] [Google Scholar]
- 4.Pinheiro VB, et al. Synthetic Genetic Polymers Capable of Heredity and Evolution. Science. 2012;336:341–344. doi: 10.1126/science.1217622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noronha AM, et al. Synthesis and Biophysical Properties of Arabinonucleic Acids (ANA): Circular Dichroic Spectra, Melting Temperatures, and Ribonuclease H Susceptibility of ANA·RNA Hybrid Duplexes. Biochemistry. 2000;39:7050–7062. doi: 10.1021/bi000280v. [DOI] [PubMed] [Google Scholar]
- 6.Wilds CJ. 2′-Deoxy-2“-fluoro-beta-D-arabinonucleosides and oligonucleotides (2”F-ANA): synthesis and physicochemical studies. Nucleic Acids Research. 2000;28:3625–3635. doi: 10.1093/nar/28.18.3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herdewijn P. Nucleic acids with a six-membered ‘carbohydrate’ mimic in the backbone. Chem. Biodivers. 2010;7:1–59. doi: 10.1002/cbdv.200900185. [DOI] [PubMed] [Google Scholar]
- 8.Pinheiro VB, Holliger P. The XNA world: progress towards replication and evolution of synthetic genetic polymers. Current Opinion in Chemical Biology. 2012;16:245–252. doi: 10.1016/j.cbpa.2012.05.198. [DOI] [PubMed] [Google Scholar]
- 9.Benner SA, Ricardo A, Carrigan MA. Is there a common chemical model for life in the universe? Current Opinion in Chemical Biology. 2004;8:672–689. doi: 10.1016/j.cbpa.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 10.Valadkhan S, Manley JL. Splicing-related catalysis by protein-free snRNAs: Abstract: Nature. Nature. 2001;413:701–707. doi: 10.1038/35099500. [DOI] [PubMed] [Google Scholar]
- 11.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 12.Joyce GF. Directed evolution of nucleic acid enzymes. Annu. Rev. Biochem. 2004;73:791–836. doi: 10.1146/annurev.biochem.73.011303.073717. [DOI] [PubMed] [Google Scholar]
- 13.Martin-Pintado N, et al. The solution structure of double helical arabino nucleic acids (ANA and 2′F-ANA): effect of arabinoses in duplex-hairpin interconversion. Nucleic Acids Research. 2012;40:9329–9339. doi: 10.1093/nar/gks672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lilley DMJ. Mechanisms of RNA catalysis. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 2011;366:2910–2917. doi: 10.1098/rstb.2011.0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilkinson KA, Merino EJ, Weeks KM. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 16.Tijerina P, Mohr S, Russell R. DMS footprinting of structured RNAs and RNA-protein complexes. Nat Protoc. 2007;2:2608–2623. doi: 10.1038/nprot.2007.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santoro SW, Joyce GF. A general purpose RNA-cleaving DNA enzyme. Proc. Natl. Acad. Sci. U.S.A. 1997;94:4262–4266. doi: 10.1073/pnas.94.9.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santoro SW, Joyce GF. Mechanism and utility of an RNA-cleaving DNA enzyme. Biochemistry. 1998;37:13330–13342. doi: 10.1021/bi9812221. [DOI] [PubMed] [Google Scholar]
- 19.Minasov G, Teplova M, Nielsen P, Wengel J, Egli M. Structural Basis of Cleavage by RNase H of Hybrids of Arabinonucleic Acids and RNA. Biochemistry. 2000;39:3525–3532. doi: 10.1021/bi992792j. [DOI] [PubMed] [Google Scholar]
- 20.Bartel DP, Szostak JW. Isolation of new ribozymes from a large pool of random sequences [see comment] Science. 1993;261:1411–1418. doi: 10.1126/science.7690155. [DOI] [PubMed] [Google Scholar]
- 21.Paul N, Springsteen G, Joyce GF. Conversion of a ribozyme to a deoxyribozyme through in vitro evolution. Chem. Biol. 2006;13:329–338. doi: 10.1016/j.chembiol.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 22.Ekland EH, Szostak JW, Bartel DP. Structurally complex and highly active RNA ligases derived from random RNA sequences. Science. 1995;269:364–370. doi: 10.1126/science.7618102. [DOI] [PubMed] [Google Scholar]
- 23.Rogers J, Joyce GF. The effect of cytidine on the structure and function of an RNA ligase ribozyme. RNA. 2001;7:395–404. doi: 10.1017/s135583820100228x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cuenoud B, Szostak JW. A DNA metalloenzyme with DNA ligase activity. Nature. 1995;375:611–614. doi: 10.1038/375611a0. [DOI] [PubMed] [Google Scholar]
- 25.Lescrinier E, et al. Solution structure of an HNA-RNA hybrid. Chemistry and Biology. 2000;7:719–731. doi: 10.1016/s1074-5521(00)00017-x. [DOI] [PubMed] [Google Scholar]
- 26.Dowler T, Bergeron D, Tedeschi AL. Improvements in siRNA properties mediated by 2′-deoxy-2′-fluoro-β-d-arabinonucleic acid (FANA) Nucleic Acids Research. 2006;34:1669–75. doi: 10.1093/nar/gkl033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hendrix C, et al. 1′,5′-Anhydrohexitol Oligonucleotides: Hybridisation and Strand Displacement with Oligoribonucleotides, Interaction with RNase H and HIV Reverse Transcriptase. Chem. Eur. J. 1997;3:1513–1520. [Google Scholar]
- 28.Nauwelaerts K, Fisher M, Froeyen M. Structural Characterization and Biological Evaluation of Small Interfering RNAs Containing Cyclohexenyl Nucleosides. J. Am. Chem. Soc. 2007;129:9340–8. doi: 10.1021/ja067047q. [DOI] [PubMed] [Google Scholar]
- 29.Deleavey GF, Damha MJ. Designing chemically modified oligonucleotides for targeted gene silencing. Chem. Biol. 2012;19:937–954. doi: 10.1016/j.chembiol.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 30.Zlatev I, Manoharan M, Vasseur J-J, Morvan F. Solid-Phase Chemical Synthesis of 5′-Triphosphate DNA, RNA, and Chemically Modified Oligonucleotides. Curr. Protoc. Nucleic Acid Chem. 2012;Chapter 1(Unit 1.28) doi: 10.1002/0471142700.nc0128s50. doi:10.1002/0471142700.nc0128s50. [DOI] [PubMed] [Google Scholar]
- 31.Chu BC, Wahl GM, Orgel LE. Derivatization of unprotected polynucleotides. Nucleic Acids Res. 1983;11:6513–6529. doi: 10.1093/nar/11.18.6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brody JR, Kern SE. Sodium boric acid: a Tris-free, cooler conductive medium for DNA electrophoresis. BioTechniques. 2004;36:214–216. doi: 10.2144/04362BM02. [DOI] [PubMed] [Google Scholar]
- 33.Goecks J, Nekrutenko A, Taylor J, Galaxy Team Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blankenberg D, et al. Galaxy: a web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol. 2010;Chapter 19(Unit 19.10-21) doi: 10.1002/0471142727.mb1910s89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giardine B, et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bowler FR, et al. Prebiotically plausible oligoribonucleotide ligation facilitated by chemoselective acetylation. Nat. Chem. 2013;5:383–389. doi: 10.1038/nchem.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siegfried NA, Busan S, Rice GM, Nelson JAE, Weeks KM. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP) Nat. Methods. 2014;11 doi: 10.1038/nmeth.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mortimer SA, Weeks KM. A Fast-Acting Reagent for Accurate Analysis of RNA Secondary and Tertiary Structure by SHAPE Chemistry. J. Am. Chem. Soc. 2007;129:4144–4145. doi: 10.1021/ja0704028. [DOI] [PubMed] [Google Scholar]
- 39.Hajdin CE, et al. Accurate SHAPE-directed RNA secondary structure modeling, including pseudoknots. Proc. Natl. Acad. Sci.USA. 2013;110:5498–5503. doi: 10.1073/pnas.1219988110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lorenz R, et al. ViennaRNA Package 2.0. Algorithms Mol Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.