Abstract
Crimean–Congo hemorrhagic fever virus (CCHFV) is a member of the genus Nairovirus of the family Bunyaviridae, that can cause severe haemorrhagic fever in humans, with mortality rates above 30 %. CCHFV is the most widespread of the tick-borne human viruses and it is endemic in areas of central Asia, the Middle East, Africa and southern Europe. Its viral genome consists of three negative-sense RNA segments. The large segment (L) encodes a viral RNA-dependent RNA polymerase (L protein), the small segment (S) encodes the nucleocapsid protein (N protein) and the medium segment (M) encodes the envelope proteins. The N protein of bunyaviruses binds genomic RNA, forming the viral ribonucleoprotein (RNP) complex. The L protein interacts with these RNP structures, allowing the initiation of viral replication. The N protein also interacts with actin, although the regions and specific residues involved in these interactions have not yet been described. Here, by means of immunoprecipitation and immunofluorescence assays, we identified the regions within the CCHFV N protein implicated in homo-oligomerization and actin binding. We describe the interaction of the N protein with the CCHFV L protein, and identify the N- and C-terminal regions within the L protein that might be necessary for the formation of these N–L protein complexes. These results may guide the development of potent inhibitors of these complexes that could potentially block CCHFV replication.
Introduction
Crimean–Congo hemorrhagic fever virus (CCHFV), a member of the genus Nairovirus of the family Bunyaviridae, is transmitted to humans by ticks and is the causative agent of Crimean–Congo haemorrhagic fever (CCHF), a severe disease with a case fatality rate of up to 30 % (Bente et al., 2013; Hoogstraal, 1979; Whitehouse, 2004). The remarkably wide geographical range of CCHF reflects the broad distribution of the tick vector, which extends throughout Africa, Asia, the Middle East and southern Europe (Ergönül, 2006). The virus is transmitted to humans through tick bites or following exposure to blood and tissues of infected animals (Mardani & Keshtkar-Jahromi, 2007). Different domestic and wild animals have been identified as a reservoir for CCHFV, including cattle, sheep, goats, hedgehogs and hares (Albayrak et al., 2010). Clinical manifestations of CCHF are often characterized by sudden onset of fever and myalgia, and in the most severe cases, haemorrhages and multiple organ failure. In the absence of a vaccine and effective antivirals, only palliative treatment is available for this disease (Soares-Weiser et al., 2010a, b). Therefore, the World Health Organization has indicated that CCHFV should be handled only in Biosafety Level 4 facilities (Flick & Whitehouse, 2005).
The genome of CCHFV consists of three segments. The large segment (L) encodes the L protein, a multifunctional 448 kDa protein containing a viral RNA-dependent RNA polymerase (RdRp) domain and an ovarian tumour (OTU) domain in the N-terminal region. The OTU domain has been shown to contain deubiquitinating and deISGylating activities, which were proven to be important for the evasion of the innate immune response and therefore could potentially be targeted for development of antiviral approaches (Frias-Staheli et al., 2007). The medium segment (M) encodes the envelope proteins Gc and Gn and the non-structural proteins GP38 and NSm (Bergeron et al., 2007; Erickson et al., 2007; Sanchez et al., 2002, 2006), and the small segment (S) encodes the nucleoprotein or nucleocapsid protein (N protein) as has now been nominated by the Ninth Report of the International Committee on Taxonomy of Viruses (Plyusnin et al., 2012).
It has been shown for other bunyaviruses that the N protein binds the genomic viral RNA, forming the viral ribonucleoprotein (vRNP) complex, which is recognized by the viral RdRp, allowing the initiation of the viral replication (Osborne & Elliott, 2000). The vRNP complexes associate with the viral glycoproteins, a process that is crucial for packaging of the genome and egress of the virion from the infected cell (Overby et al., 2007).
Recently, three independent groups (Carter et al., 2012; Guo et al., 2012; Wang et al., 2012) have determined the crystal structure of the N protein of CCHFV, unravelling critical information about its structure–function relationships and elucidating intramolecular interactions. Carter et al. (2012) showed that the N protein monomer of the Baghdad-12 strain resembles the structure of the nucleoprotein of Lassa virus, a member of the family Arenaviridae, suggesting that nairoviruses and arenaviruses might share a common ancestor. Interestingly, the structure described for the N protein of this strain revealed surfaces that may be implicated in RNA binding and N–N interactions. However, comparison of the crystal structure of the N protein of the Baghdad-12 strain with the crystal structure of the N protein of the YL04057 strain revealed differences in spatial positioning of domains (Carter et al., 2012; Guo et al., 2012). In addition, Guo et al. (2012) showed that the monomeric form of the CCHFV N protein has low RNA-binding affinity but a strong metal-dependent DNA-endonuclease activity. Wang et al. (2012) reported that the N protein of the IbAr10200 strain possesses two different conformations: one resembling an oligomerized RNP and a monomeric form with an exposed caspase-3 cleavage site. Furthermore, the disruption of the caspase-3 cleavage site resulted in an enhancement of the CCHFV polymerase activity during vRNA transcription. Also, a head to tail arrangement of the N oligomers was described, and key residues that stabilize this arrangement were identified (Wang et al., 2012).
Previous studies have demonstrated the interaction of the N protein of CCHFV with actin, which is necessary for the perinuclear localization of the N protein (Andersson et al., 2004). The actin cytoskeleton plays a role in the assembly and budding of viruses such as human immunodeficiency virus particles, at the cell surface (Taylor et al., 2011). Moreover, the interaction of the viral proteins with actin filaments has been shown to enhance replication of paramyxoviruses such as Newcastle disease virus and Sendai virus (Taylor et al., 2011).
Here, we identified the functional interaction domains of the N and L proteins of CCHFV. We found that the N protein interacts with both the C- and N-terminal regions of the L protein. In addition, we demonstrated that a region harbouring a newly recognized coiled-coil motif located in the central region of the N protein is essential for N proteins self-interactions. Finally, we mapped the domain that mediates the interaction of the N protein with actin filaments to the N-terminal region of the N protein adjacent to the coiled-coil motif. Altogether, these results provide critical information about the interacting domains of the N protein. The identification of the regions involved in protein interactions will contribute to the understanding of the multiple functions of the N protein during viral replication, and could identify potential targets for the development of antivirals against CCHFV.
Results
Analysis of the N–L interaction
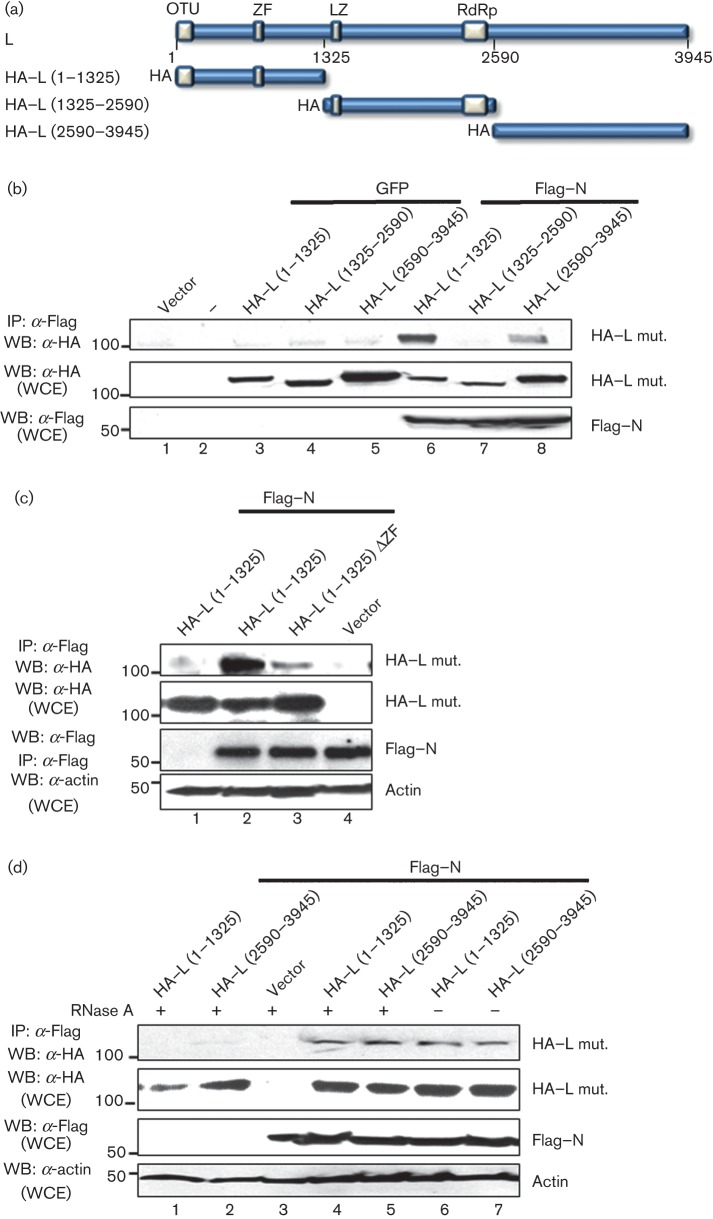
The interaction of the N protein with the L protein of CCHFV is most likely essential for viral RNA replication. Therefore we attempted to characterize this interaction and determine which regions within the L protein mediate the N–L association. For this purpose, we utilized three plasmids expressing haemagglutinin (HA)-tagged L protein truncation mutants (Frias-Staheli et al., 2007) (Fig. 1a). These L protein expression plasmids were individually co-transfected with the plasmids expressing either Flag-tagged N protein (Flag–N) or GFP into HEK 293T cells. At 24 h post-transfection, lysates were collected and subjected to immunoprecipitation with anti-Flag M2 resin, followed by Western blot analysis to detect co-immunoprecipitated HA-tagged L protein. To monitor protein expression, aliquots of the cell lysates were subjected to SDS-PAGE and Western blot using anti-HA and anti-Flag antibodies. The results indicated that the HA–L polypeptides (aa 1–1325 or aa 2590–3945) were specifically co-precipitated with Flag–N. The L protein internal region (aa 1325–2590), containing the RdRp domain, did not interact with the N protein in this assay (Fig. 1b, lane 7). HA-tagged L protein did not co-immunoprecipitate with GFP from lysates of control-transfected cells (Fig. 1b, lanes 6 and 8). These results revealed that the N- and C-terminal regions of the L protein are able to independently associate with the N protein and suggest that the central region of the L protein is not involved in this interaction. Interestingly, a putative zinc-finger (ZF) domain is located within the L protein (aa 1–1325) truncation mutant. ZF domains have been shown to mediate protein–protein interactions in other negative-strand RNA viruses (Levingston Macleod et al., 2011; Wang et al., 2010). To study the contribution of this domain to the CCHFV N–L interaction, a ZF truncation mutant lacking the ZF domain [HA–L (1–1325) ΔZF] was constructed as indicated in Methods. Co-immunoprecipitation experiments showed that this mutant was considerably impaired in interacting with Flag–N (Fig. 1c, compare lanes 2 and 3).
Fig. 1.
CCHFV N and L protein interaction. (a) Scheme of the L protein truncated mutants used in our experiments. The predicted domains are indicated with grey boxes. OTU, ovarian tumour domain; ZF, zinc finger; LZ, leucine zipper; and RdRp, RNA-dependent RNA polymerase. All the proteins were HA tagged. (b) HEK 293T cells were transfected with the indicated Flag–N, HA–L, GFP or empty vector. Twenty-four hours later, the cells were harvested and lysed, and proteins were immunoprecipitated with anti-Flag (IP: α-FLAG). Proteins were detected by Western blotting (WB) using the indicated antibodies. (c) HEK 293T cells were transfected with Flag–N, HA–L (1–1325), HA–L (1–1325) ΔZF or empty vector, as indicated. Twenty-four hours later, cells were harvested and lysed, and proteins were immunoprecipitated with anti-Flag. After SDS-PAGE, proteins were detected by WB using the indicated antibodies. (d) Aliquots of the cytoplasmic extracts were treated with RNase A at a final concentration of 0.1 mg ml−1 for 30 min at room temperature, prior to immunoprecipitation with anti-Flag. Molecular masses of markers are indicated on the left in kDa. WCE, Whole-cell extract.
Since N and L are both RNA-binding proteins, we then tested whether RNA mediates their interaction. Treatment of the co-immunoprecipitated N–L complexes with RNase A did not disrupt the interaction between the N protein and the L protein truncation mutants HA–L (1–1325) and HA–L (2590–3945) (Fig. 1d, compare lanes 4 and 5 with 6 and 7), suggesting that, under these conditions, RNA does not mediate this interaction or that the RNA is protected from the RNase treatment by this interaction.
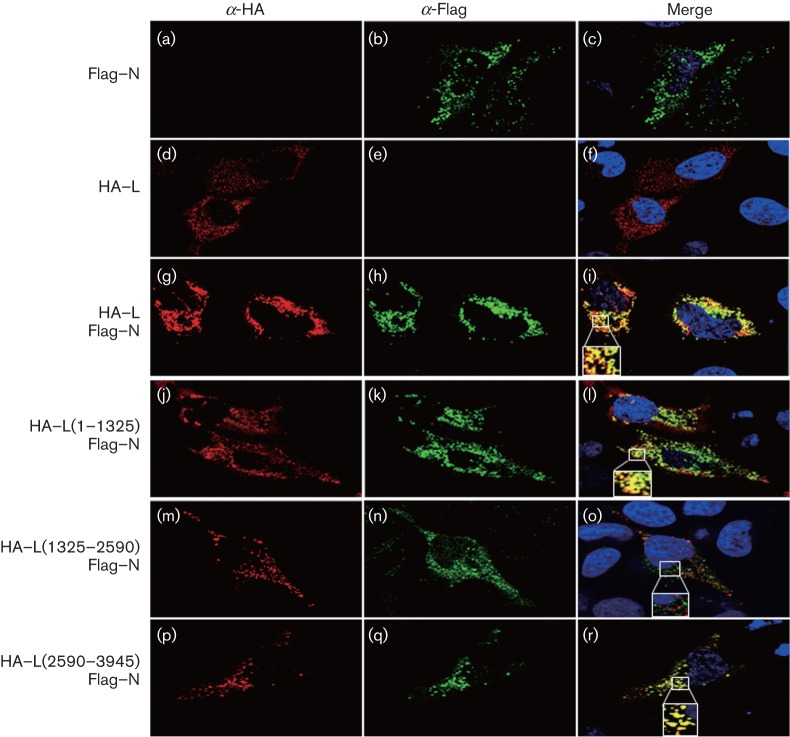
To further support our results, we examined the subcellular co-localization of the L protein mutants with Flag–N in HeLa (Fig. 2) or HEK 293T (data not shown) cells by confocal microscopy. As shown previously (Andersson et al., 2004), CCHFV N displayed a cytoplasmic and perinuclear distribution when expressed alone (Fig. 2b). Essentially, the same pattern was observed when the N protein was co-expressed with either the WT L protein or the truncated mutants. The full-length L protein localized in the cytoplasm, where the two proteins co-localized (Fig. 2f, i). Consistent with the biochemical data, the HA–L (1–1325) and HA–L (2590–3945) truncation mutants co-localized with the N protein in the cytoplasm (Fig. 2l, r); however, no co-localization was seen between the N protein and HA–L (1325–2590) (Fig. 2o).
Fig. 2.
Intracellular distribution of L protein mutants and N protein. HeLa cells expressing Flag–N and/or L-HA or each of the HA-tagged L mutants were fixed with 4 % paraformaldehyde 24 h post-transfection, permeabilized with Triton X-100 (0.1 % v/v) and probed with a mouse anti-Flag and rabbit anti-HA antibodies. The samples were then incubated with a mix of Alexa Fluor 488-conjugated anti-mouse and Alexa Fluor 568-conjugated anti-rabbit antibodies. The nuclei were stained with DAPI (blue). The merged image shows co-localization in yellow. Imaging and processing settings were kept identical for all fields to allow comparisons. The insets are further magnified to show the localization of the different proteins.
Oligomerization of the N protein in transfected cells
Homo-oligomerization of the N protein plays a central role in RNA encapsidation and in the formation of the nucleocapsid structure of bunyaviruses (Ruigrok et al., 2011). We investigated whether the N protein oligomerizes when expressed in mammalian cells. For this, HEK 293T cells were transfected with WT N–Flag; cell extracts were prepared 24 h post-transfection and then treated with RNase A. The cell lysates were divided into aliquots, which were heated at increasing temperatures to disrupt non-covalent bonds. Samples were resolved by SDS-PAGE and analysed by Western blotting using anti-Flag antibody to detect precipitated N–Flag (Fig. 3a). Regardless of the incubation temperature, the N protein was predominantly found in a monomeric form of approximately 54 kDa. Oligomeric forms of the N protein were only detected upon overexposure of the film; two high-molecular-mass bands were detected, one of approximately 110 kDa and another of 150 kDa, consistent with the migration of dimeric and trimeric forms of the N protein, respectively. Incubation of the cell extracts at temperatures higher than 80 °C resulted in the disappearance of the oligomeric forms (Fig. 3a, lane 5). These results suggested that the N protein forms oligomers, which are most likely stabilized by non-covalent bonds.
Fig. 3.

The region between residues 240 and 482 of the CCHFV N protein is important for N–N interactions. (a) Denaturation of CCHFV N protein. HEK 293T cells were transfected with Flag–N, as indicated in Methods. At 48 h post-transfection, cells were collected in lysis buffer containing protease inhibitors and cell lysates were clarified at 16 000 g for 20 min at 4 °C. Aliquots of the cytoplasmic extracts were treated with RNase A at a final concentration of 0.1 mg ml−1 for 20 min at room temperature. The different samples were incubated at the indicated temperatures in SDS-PAGE sample buffer for 5 min prior to being subjected to SDS-PAGE. Proteins were analysed by Western blotting (WB) using anti-Flag antibody. (b) N–N interaction was examined in co-immunoprecipitation experiments. HEK 293T cells co-expressing N–HA and Flag–N, or each of the Flag-tagged N mutants, were lysed and cell extracts were immunoprecipitated using anti-Flag antibody (IP: α-Flag). Precipitated proteins were analysed by Western blotting, as indicated on the left of each panel. Expression levels of WT Flag–N, each N protein mutant or N–HA in whole-cell extracts (WCE) were examined by Western blotting, as indicated. Molecular masses of markers are indicated on the left in kDa.
The central region (aa 250–300) of the N protein is required for homotypic interactions
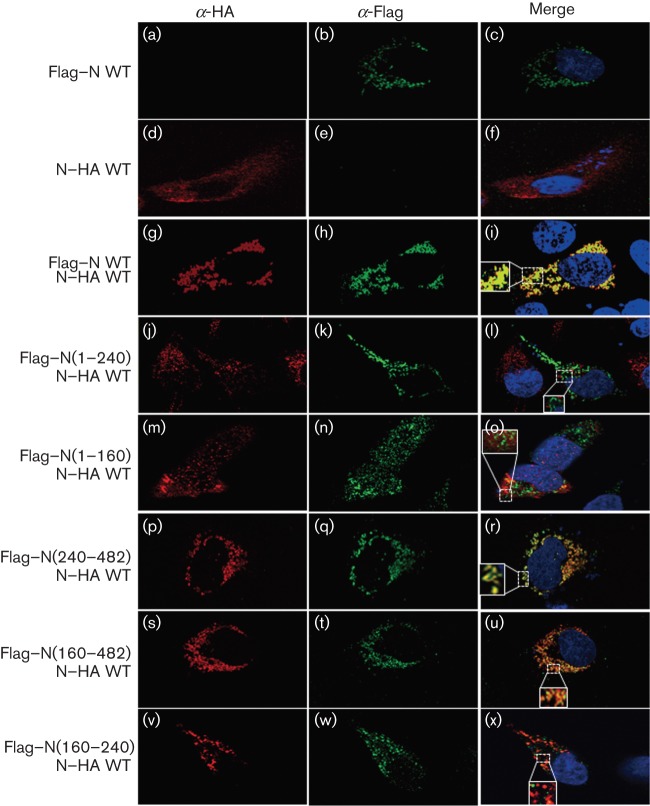
In order to identify the regions of the N protein responsible for this homotypic oligomerization, we performed co-immunoprecipitation assays between HA-tagged full-length CCHFV N protein and various Flag-tagged CCHFV N protein polypeptides (Fig. 3b). The mutants were designed to contain different structural regions according to the structure published by Wang et al. (2012). This structure revealed two domains in the N protein: the N-terminal residues 1–183 and the C-terminal residues 295–482 combine to form a globular head domain, while residues 195–294 form a stalk protruding from the globular head. We generated the following N protein truncation mutants: the N-terminal half of the head domain [mutant N (1–160)], the N-terminal part plus half of the stalk domain [mutant N (1–240)], the C-terminal fraction of the stalk plus the C-terminal part of the head domain [mutant N (240–482)] and the whole stalk with the C-terminal part of the head domain [mutant N (160–482)]. Also, a mutant was generated containing only the N-terminal part of the stalk domain [mutant N (160–240)]. Western blot analysis of cytoplasmic extracts revealed comparable levels of full-length and N mutant proteins (Fig. 3b, WCE, WB: α-Flag). Likewise, N–HA protein levels were similar in all transfection conditions (Fig. 3b, WCE, WB: α-HA). Immunoprecipitation of cell lysates with anti-Flag M2 resin, followed by Western blotting with anti-HA antibody, revealed that full-length N–HA protein co-immunoprecipitated with Flag–N (Fig. 3b, panel IP: α-Flag, lane 2), whereas N–HA was not co-immunoprecipitated by anti-Flag antibody from control cell lysates containing N–HA alone or GFP (Fig. 3b, lanes 8 and 1, respectively), indicating that N–HA and Flag–N specifically interact. These data confirmed our results that CCHFV N protein is able to self-associate (Fig. 3a). In addition, the deletion of the last 242 residues of the N protein diminished the N–N interaction to undetectable levels, as mutants Flag–N (1–160), Flag–N (1–240) and Flag–N (160–240) did not co-immunoprecipitate with N–HA (Fig. 3b, lanes 3, 4 and 7). In contrast, Flag–N (160–482) and Flag–N (240–482) co-immunoprecipitated with N–HA (Fig. 3b, lanes 5 and 6). These results indicate that residues 240 to 482 mediate N–N homo-oligomerization.
In order to validate these results by a second approach, we investigated the co-localization of the full-length N–HA protein in the presence of either the full-length Flag–N or the indicated Flag-tagged N protein truncation mutants by confocal microscopy (Fig. 4). Consistent with the previous findings on the localization of CCHFV N protein (Andersson et al., 2004), both N–HA and Flag–N proteins co-localized within the cytoplasm of transfected HeLa cells (Fig. 4b–d, f, i). Co-localization was also observed between the full-length N protein and the mutants Flag–N (240–482) and Flag–N (160–482) (Fig. 4r, u) but not with the deletion mutants Flag–N (1–160), Flag–N (1–240) or Flag–N (160–240) (Fig. 4l, o, x). In summary, the confocal microscopy data confirmed the biochemical results that residues 240–482 of the N protein mediate the oligomerization of the CCHFV N protein.
Fig. 4.
Intracellular distribution of CCHFV N protein mutants co-expressed with N–HA. After 24 h transfection, HeLa cells co-expressing either N–HA, Flag–N WT or N–Flag mutants were fixed with paraformaldehyde (4 % v/v), permeabilized with Triton X-100 (0.1 % v/v) and then probed with a mouse anti-Flag and a rabbit anti-HA antibody for 1 h at room temperature. Then, the samples were washed as described in Methods and incubated with a mix of Alexa Fluor 488-conjugated anti-rabbit and Alexa Fluor 568-conjugated anti-mouse antibodies at room temperature. After 1 h, the cells were washed as indicated in Methods. The nuclei were stained with DAPI (blue). The merged image shows co-localization in yellow. Magnified insets are included.
A putative N protein coiled-coil motif is essential for the N–N self-interaction
In order to identify functional motifs within residues 240–482 that mediate CCHFV N protein homo-oligomerization, we performed a comparative analysis of N protein sequences from a representative set of nairoviruses. We searched for conserved motifs that could be associated with homo-oligomerization within the region spanning residues 240–482. We identified a well-conserved 50 aa region in the C-terminal half of the stalk central region of the CCHFV N protein, where a coiled-coil motif was predicted (Fig. 5a, b). The coiled-coil motif contains the characteristic repeating heptad pattern of amino acids (abcdefg), with the occurrence of hydrophobic residues preferentially in the first (a) and, in most cases, the fourth (d) positions of the heptads (Lupas et al., 1991). A coiled-coil domain has already been shown to mediate hantavirus nucleoprotein oligomerization (Alminaite et al., 2008). Within the CCHFV N protein, this motif extends over positions 254–300, comprising four consecutive heptads, in which the first positions are occupied by the hydrophobic residues V254, L261 and V268 (Fig. 5b). Remarkably, these residues are conserved as non-polar among the nairovirus N protein sequences accessible through the NCBI server (Fig. 5b). The coiled-coil motif corresponds to the last two α-helices of the N stalk domain according to the crystal structure (Fig. 5c), which form an anti-parallel arm protruding from the globular body (Wang et al., 2012).
Fig. 5.
CCHFV N–N interaction requires a predicted coiled-coil motif. (a) Output from analysis of CCHFV N protein by the Lupas program. The y-axis represents the probability (1 = 100 %) that an amino acid is within a coiled-coil region and the x-axis represents the 482 aa sequence of CCHFV N. (b) Multiple alignment of N protein sequences of representatives nairoviruses. The region shown, corresponding to residues 250–300 of CCHFV N protein, was predicted to form an α-helical coiled-coil structure by using the algorithm of Lupas et al. (1991), as indicated in Methods. The GenBank accession numbers of the N protein sequences used are as follows: CCHFV, NC_005302.1; Hazara virus (HAZV), P27318.1; Nairobi sheep disease virus (NSDV), HQ286608.1; Dugbe virus (DUGV), NC_004157.1; kupe virus (KUPV), gb|ABY82500.1|. Strictly conserved amino acids are marked with asterisks, and the ones with similar properties are highlighted with double dots. Blue arrows indicate hydrophobic amino acids that were mutated in subsequent experiments; black arrows indicate other hydrophobic residues. (c) Structure of the CCHFV N protein PDB: 4AQF (Wang et al., 2012). The predicted coiled-coil, from residues 250 to 300, is in pink, and the non-polar residues that are conserved in the multiple sequence alignment are visible in the structure as sticks. (d) Schematic representation of WT (Flag–N) and coiled-coil deletion mutant (Flag–NΔCC) form of CCHFV N protein. (e, f) HEK 293T cells were transfected with the indicated plasmids; 24 h later, cells were harvested and lysed, and the proteins were immunoprecipitated with anti-Flag (IP: α-Flag). After SDS-PAGE, Western blot (WB) analysis was performed with the indicated antibodies. Molecular masses of markers are indicated on the left in kDa. WCE, Whole-cell extracts.
To analyse whether the predicted coiled-coil motif was important for CCHFV N protein self-interaction, the sequence composing the whole motif in the Flag–N plasmid was replaced by a linker of eight glycines (Fig. 5d). The resulting plasmid, referred to as Flag–NΔCC, was co-transfected along with the plasmid expressing full-length HA-tagged CCHFV N protein (N–HA) into HEK 293T cells, and the homotypic interactions were investigated by co-immunoprecipitation. Flag–NΔCC protein did not co-immunoprecipitate with N–HA (Fig. 5e), implying that the deletion of residues 250–300 completely abolished the ability of the N protein to self-interact. These results indicate that the 50-residue region harbouring the predicted coiled-coil motif is essential for maintaining the ability of the N protein to self-interact. Furthermore, these results are consistent with the main chain interactions seen in the crystal structure of the N protein oligomers, in which residues 266 and 269 on the coiled-coil domain and at the loop between the two anti-parallel α-helices of this domain interact with residues 352–354, located at the C-terminal part of the globular head (Wang et al., 2012).
We next investigated which residues of the coiled-coil were important for the N protein homo-oligomerization. We generated a battery of point mutants in which the non-polar residues V254, L261, V268 and I277 were exchanged for the polar amino acid glutamine (Q) (Fig. 5b, blue arrows). Double point mutants around the non-polar residues were also included to disrupt the coiled-coil structure. Western blot analysis of lysates prepared from cells expressing each of the HA-tagged N protein mutants and N–HA full-length proteins confirmed the steady state levels of the proteins. Immunoprecipitation assays of the cellular lysates with the anti-Flag M2 resin revealed that none of the expressed mutants was able to interact with N–HA (Fig. 5f).
Based on the 4AQF trimeric structure of the N protein (Wang et al., 2012), residues I210 and D219 were shown to mediate interactions of one N protein molecule with the C-terminal head domain of a second N protein molecule. However, these residues were not required for the interaction of the N protein with the N (240–482) deletion mutant (Figs 3 and 4). Nevertheless, we generated the mutants Flag–N I210Q and Flag–N D219Q and tested their interaction with full-length N protein by co-immunoprecipitation assays. We found that these mutants where defective in interacting with each other (Fig. 5f). Thus, while it appears that the core N–N interacting domain is located within the last 240–482 residues, upstream residues are also important for full-length N–N interactions.
Analysis of the interaction of CCHFV N protein with actin
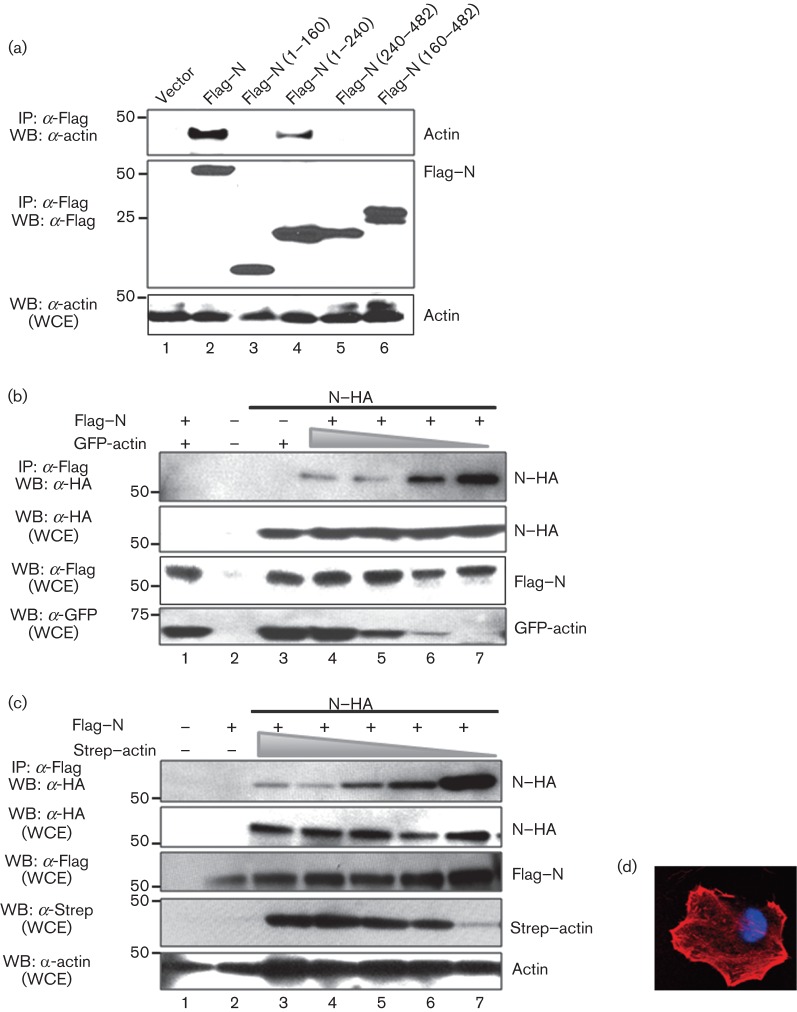
It has been shown that the CCHFV N protein directly interacts with actin filaments, and this interaction seems to be important for its localization to the perinuclear region of mammalian cells (Andersson et al., 2004). Since it is unknown which domains within the CCHFV N protein mediate the interaction with actin, we decided to take advantage of our truncated mutants and map this interaction by co-immunoprecipitation and Western blot analyses. We detected co-immunoprecipitation of actin with the full-length N protein and, to a lesser extent, with the deletion mutant Flag–N (1–240) (Fig. 6a). Although the first 240 aa of the N protein were required for N protein–actin interaction, the polypeptide comprising residues 1–160 did not bind to actin (Fig. 6a), suggesting that CCHFV N protein amino acid residues 160–240 provide a critical contribution to this interaction. Taken together, these results indicate that the N-terminal region of the CCHFV N protein is implicated in the interaction of the N protein with actin.
Fig. 6.
Mapping of the CCHFV N–actin interaction. (a) HEK 293T cells were transfected with the CCHFV N protein or N protein Flag-tagged truncated mutants, as indicated. Cells were harvested 24 h post-transfection and lysed, and the overexpressed proteins were immunoprecipitated with anti-Flag (IP: α-Flag). Expression levels of WT Flag–N, each N protein mutant and actin were examined by Western blotting (WB), using the antibodies indicated. (b) HEK 293T cells were transfected with the indicated plasmids expressing HA-N, Flag–N or various concentrations of GFP–actin (5, 1 and 0.1 µg). After 24 h, cells were harvested and lysed, and the overexpressed proteins were immunoprecipitated with anti-Flag. Expression levels of WT Flag–N, each N protein mutant and actin were examined by Western blotting, using the antibodies indicated. (c) HEK 293T cells were transfected with the indicated plasmids expressing HA-N, Flag–N or various concentrations of Strep–actin (10, 5, 1 and 0.1 µg). After 24 h, the cells were harvested and lysed, and the overexpressed proteins were immunoprecipitated with anti-Flag. Expression levels of WT Flag–N, each N protein mutant and actin were examined by Western blotting, using the antibodies indicated. Molecular masses of markers are indicated on the left in kDa. WCE, Whole-cell extracts. (d) HeLa cells expressing Strep–actin were visualized with phalloidin 568 by confocal microscopy as described in Methods.
We further investigated if the interaction of the N protein with actin would prevent its self-interaction. To test this hypothesis, HEK 293T cells were transfected with increasing amounts of a plasmid expressing GFP–actin and a constant amount of plasmids expressing N–HA and/or Flag–N. The lysates were analysed by co-immunoprecipitation assays using anti-Flag M2 resin followed by Western blot using anti-HA antibody. We detected a reduction in the amount of N–HA co-immunoprecipitated with Flag–N when increasing amounts of GFP–actin were present in the lysates (Fig. 6b). This result indicates that the increase in the concentration of the GFP–actin negatively affects the oligomerization of the CCHFV N protein. To rule out an effect of GFP in impairing N–N interactions, we replaced GFP–actin with Strep–actin under similar experimental conditions and observed that increasing amounts of Strep–actin interfered with the ability of the N protein to self-interact, confirming our previous finding (Fig. 6c). As expected, the Strep–actin protein conserved the same phenotypic localization as untagged actin (Fig. 6d).
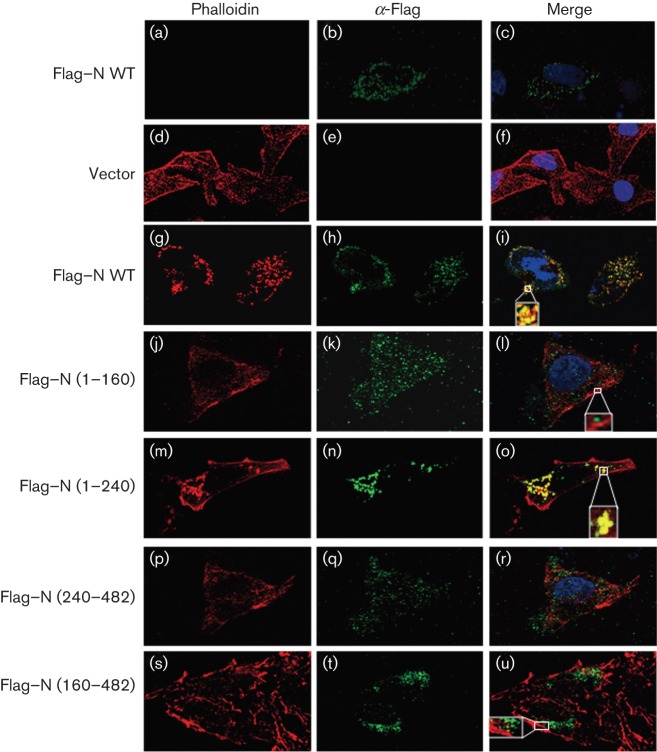
The interaction between CCHFV N and actin was also analysed by confocal microscopy (Fig. 7). In accordance with the biochemical data, the mutants that failed to co-immunoprecipitate with actin, Flag–N (1–160), Flag–N (240–482) and Flag–N (160–482) did not show extensive co-localization with actin filaments (Fig. 7l, r, u, respectively). By contrast, full-length N protein and truncated Flag–N (1–240) extensively co-localized with actin (Fig. 7i, o).
Fig. 7.
Intracellular distribution of CCHFV N protein mutants and actin. HeLa cells expressing N–Flag mutants were fixed 24 h post-transfection with 4 % paraformaldehyde, permeabilized with Triton X-100 (0.1 % v/v) and probed with mouse anti-Flag, followed by phalloidin 568 and/or Alexa 488 anti-mouse antibody as indicated in Methods. The nuclei were stained with DAPI (blue). The merged image shows co-localization in yellow. Magnified insets are included.
Using the server patchdock (Schneidman-Duhovny et al., 2005), we generated a predicted interaction model between the N protein and actin (Fig. 8a). Interestingly, the predicted actin-interacting domain was localized within the central region of the N protein, in the stalk structure, although other regions were also implicated in the stabilization of the complex. According to this molecular docking model, the only residue implicated both in the N protein self-interaction and in the N–actin interaction was D219. In order to test whether D219 contributes significantly to these interactions of the N protein, we co-expressed the N–HA protein with either the WT Flag–N protein or the Flag–ND219Q mutant, in which the negative charge of the residue was abolished. N proteins mutated in residues within or outside the predicted N–actin-binding region that were also important for N protein self-oligomerization, were also used. All N protein mutants were expressed at similar levels (Fig. 8b). Co-immunoprecipitation assays revealed that, of the N protein mutants examined, only Flag–N D219Q was unable to co-immunoprecipitate with actin, indicating that D219 is a key residue responsible for the interaction between the N protein of CCHFV and actin, and is also required for N–N interaction.
Fig. 8.
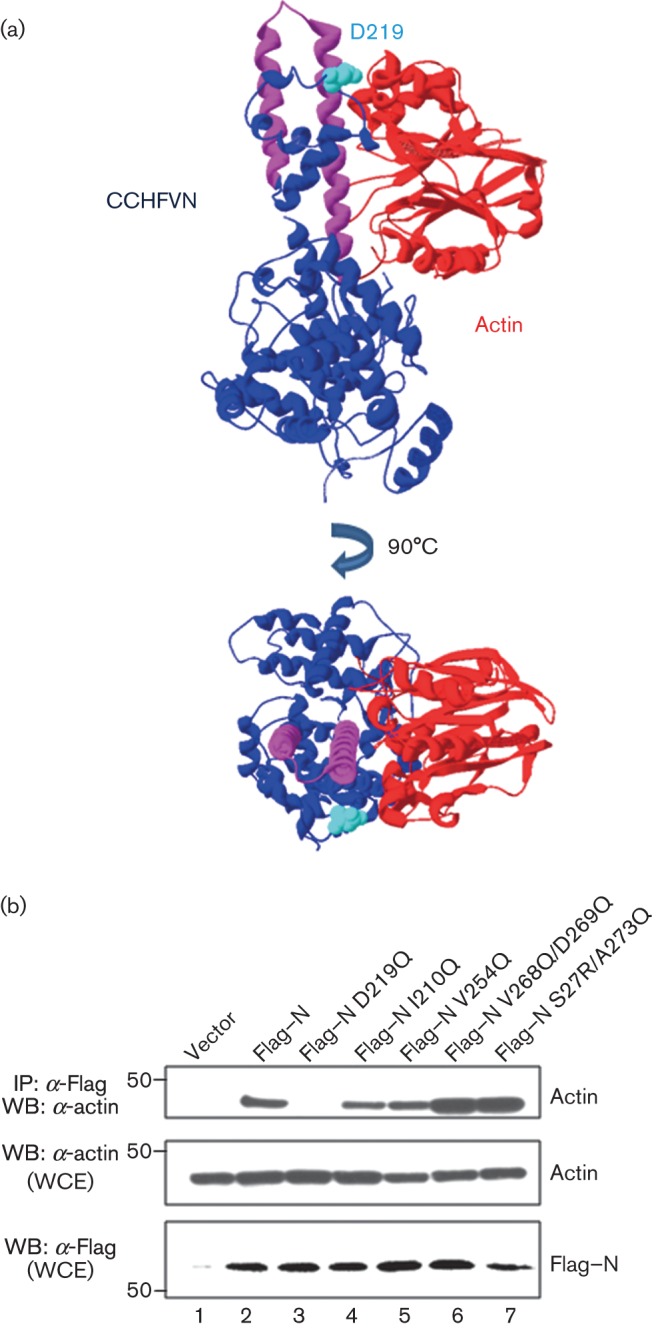
Molecular modelling of the CCHFV N–actin interaction. (a) The model was constructed with the structures of CCHFV N protein PDB: 4AQF (Wang et al., 2012) (in blue) and human actin PDB: 1ATN (in red), using the program PatchDock as described in Methods. The coiled-coil motif is in pink. The aspartate (D) in position 219 is highlighted in cyan in the CCHFV N protein structure. (b) HEK 293T cells were transfected with CCHFV N protein or N protein Flag-tagged point mutants, as indicated. After 24 h, the cells were harvested and lysed, and the overexpressed proteins were immunoprecipitated with anti-Flag (IP: α-Flag). After SDS-PAGE, expression levels of WT Flag–N, each N protein mutant and actin were examined by Western blotting (WB) using the antibodies indicated. Molecular masses of markers are indicated on the left in kDa. WCE, Whole-cell extracts.
Discussion
The nucleoproteins of negative-strand RNA viruses play important and diverse functions in the life cycle of the virus and therefore constitute an attractive target to study their biological functions, which might ultimately lead to the development of antiviral approaches. These functions are mediated by multiple interactions with viral and cellular factors. Recently, three independent groups reported the crystal structure of the CCHFV N protein (Carter et al., 2012; Guo et al., 2012; Wang et al., 2012). These crystal structures provided hints on the spatial positions of structural features of the N protein, and enabled studies on structure–function relationships, including its oligomerization and RNA-binding domains. Based on the crystal structure determined by Wang and colleagues for the IbAr10200 strain (same strain examined herein), a model was proposed in which the structure of the N protein is drastically modified by binding of RNA (Wang et al., 2012). Binding to RNA triggers a conformational change in the CCHFV N protein superhelical polymer in which the stalk domain of the N protein monomers rotates away from its position in the polymer, releasing the monomeric N protein. Monomeric CCHFV N protein is speculated to participate in viral RNA replication by interacting with the L protein and/or by binding to viral RNAs to prevent degradation of genomic RNA.
By assessing the interaction of truncation mutants of the N protein with the full-length protein in co-immunoprecipitation assays, we identified the region enclosing a coiled-coil motif (residues 250–300) in the central stalk region responsible for these N–N interactions. This coiled-coil motif contains the characteristic repeating heptad pattern of amino acids. The conserved non-polar residues of the heptads face towards the internal interaction surface and likely stabilize the structure of the N protein. Deletion of this region prevents N protein self-interactions. This is consistent with the structure presented by Wang and colleagues, in which N–N interactions are shown to involve extensive hydrogen bonding and Van der Waals contacts of residues 320–354 with residues 210–219 and 260–272 (Wang et al., 2012). Similarly, the N–N interaction of Tacaribe virus, an arenavirus, also required the N-terminal coiled-coil motif (Levingston Macleod et al., 2011; Wang et al., 2010). Moreover, in hantaviruses, the coiled-coil is also located within the N-terminal region of the nucleocapsid protein (Alminaite et al., 2008).
The interaction of the N protein with the L protein is likely to be essential for viral replication. By assessing the interaction of truncation mutants of the L protein with the full-length N protein in co-immunoprecipitation assays, we determined that the putative ZF domain of the L protein mediates its interaction with the N protein. Moreover, we could not find evidence that this N–L interaction is mediated by RNA.
We have also determined that the N-terminal region of the N protein, more precisely residues 160–240 (Figs 6 and 7), mediates the interaction with actin. Based on the results obtained with GFP–actin and Strep–actin, the homo-oligomerization of the N protein appears to be regulated by its interaction with actin (Fig. 6b). Interestingly, the predicted actin-binding domain mapped within the central stalk region of the N protein; however, additional residues outside of the actin-binding domain appear to contribute to the N–N interaction (Fig. 8), possibly by stabilizing the oligomerization of the N protein. As actin competed with N oligomerization, it is possible that actin–N protein interactions only occur in the context of N protein monomers, and not with N protein oligomers. Future reverse genetics studies will be needed to understand the significance of the N–N, N–L and N–actin interactions described in our study.
In summary, we have generated a series of N- and C-terminal deletions and point mutants of the CCHFV N protein to identify and map domains involved in the interactions of the N protein with the L protein polymerase, itself and actin. The CCHFV N protein is known to participate in several steps of the bunyavirus life cycle, which would require a number of interactions with viral and cellular proteins as well as interactions with viral RNA. For example, N–N interactions are known to be important for nucleocapsid formation. We used co-immunoprecipitation assays and confocal microscopy to map interaction domains/residues of the CCHFV N protein. The mapping of functional domains that mediate the interactions of the N protein with viral and cellular proteins contributes to the understanding of the multiple functions of the N protein that are likely to be essential for the bunyavirus life cycle, and identifies potentially critical interactions that could be targeted for the development of antiviral strategies against CCHFV.
Methods
Plasmids.
The full-length sequence encoding the N protein was cloned into pCAGGS for mammalian expression. The plasmids expressing the full-length L protein or the truncation mutants HA–L (1–1325), HA–L (1325–2590) and HA–L (2590–3945) have been described previously (Frias-Staheli et al., 2007). The mutant HA–L (1–1325) ΔZF was generated by site direct mutagenesis (Stratagene), according to the manufacturer’s instructions, changing cysteine in positions 609 and 613 for alanine. Plasmid pN–HA expresses CCHFV N protein with a C-terminal HA epitope (YPYDVPDYA). Plasmid pN–Flag expresses the N protein with a C-terminal Flag epitope (DYKDDDDK). Constructs encoding the truncated versions of CCHFV N protein were generated by PCR using plasmid pCCHFV N–Flag as the template and subcloned into the pCAGGS vector. The deletion in the predicted coiled-coil motif was generated by PCR amplification of two segments of the CCHFV N protein sequence. The first PCR included the forward primer described in the construct N–Flag and the reverse primer with a BamHI restriction site followed by the complementary sequence of four Gly codons and nt 721–749. The second PCR included the forward primer containing a BamHI restriction site followed by four Gly codons and nt 900–810, and the reverse primer described in the N–Flag construct. The point mutants were generated by site direct mutagenesis (Stratagene), according to the manufacturer's instructions. The plasmid pCAG-mGFP-Actin, which encodes the GFP–actin protein, was constructed by Murakoshi et al. (2008) (Addgene). Sequences encoding the ORF of human actin were amplified by PCR from plasmid pCAG-mGFP-Actin. The actin sequence was subcloned into pcDNA4 expressing an N-terminal 2×Strep-tag sequence (kindly provided by Nevan Krogan, University of California, San Francisco). All constructs were sequenced by Sanger sequencing (Genewiz). All primer sequences and vector maps are available upon request.
DNA transfections.
HEK 293T or HeLa cell monolayers were transfected with the indicated plasmid combinations using Lipofectamine 2000 reagent (Invitrogen), according to the manufacturer’s instructions. In all transfections, the total amount of transfected DNA was kept constant by the addition of vector pCAGGS.
Analysis of protein interactions by co-immunoprecipitation.
Subconfluent monolayers of HEK 293T cells grown in 12-well plates were transfected with 1 µg of the indicated expression plasmids. At 24 h post-transfection, cell monolayers were washed twice with PBS and then lysed by lysis buffer [0.5 % v/v NP-40, 10 % v/v glycerol containing protease inhibitors (2 µg aprotinin ml−1, 20 µg phenylmethylsulfonyl fluoride ml−1, 50 µg N-α-tosyl-l-lysine chloromethyl ketone ml−1; Sigma-Aldrich)]. Cell lysates were cleared of nuclei and cellular debris by centrifugation at 16 000 g for 20 min at 4 °C. Aliquots of cytoplasmic extracts corresponding to about 0.5×105–2×105 cells were immunoprecipitated with either rabbit anti-HA polyclonal antibody (Santa Cruz Biotechnology) or a 50 % slurry of mouse anti-Flag M2 affinity gel (Sigma-Aldrich), as indicated, following the protocol previously described (Levingston Macleod et al., 2011). Immunoprecipitated proteins were resolved by SDS-PAGE and visualized by Western blotting, as described below.
Western blotting.
Immunoblotting was performed as previously described (Levingston Macleod et al., 2011). Briefly, proteins were resolved by SDS-PAGE (4–20 % polyacrylamide) and then transferred to a nitrocellulose membrane. After blocking overnight at 4 °C with PBS containing 5 % skimmed milk and 0.1 % v/v Tween 20 (Sigma-Aldrich), blots were incubated with the primary antibody overnight at 37 °C. Following incubation with the appropriate HRP-labelled secondary antibody, detection was performed with SuperSignal West Pico chemiluminescent substrate (Thermo Scientific), according to the manufacturer’s specifications. Finally, blots were exposed to X-ray films, and protein bands were quantified by densitometry, as previously described (Levingston Macleod et al., 2011). The following primary antibodies were used: rabbit anti-HA polyclonal antibody (Santa Cruz Biotechnology), HRP-conjugated mouse anti-actin (Invitrogen), rabbit anti-Strep-tag II (Abcam), mouse anti-eGFP (ClonTech) or mouse anti-Flag M2 mAb (Sigma-Aldrich). HRP-conjugated anti-mouse or anti-rabbit secondary antibodies (Jackson ImmunoResearch) were used according to the supplier’s specifications.
Immunofluorescence and confocal microscopy.
HeLa cells grown in four-well Labtek chamber slides were transfected with 1 µg of pCAGGS Flag–N, pCAGGS N–HA or pCAGGS encoding the truncated Flag–N mutants expressing plasmid, using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. After 24 h, the cells were fixed with paraformaldehyde (4 %), permeabilized with 0.1 % v/v Triton X-100 for 10 min and blocked with 5 % BSA–PBS for 1 h at 37 °C. Cells were incubated with the indicated antibodies (anti-mouse anti-Flag M2 antibody, rabbit anti-HA or rabbit anti-actin) for 1 h at room temperature, washed and labelled with secondary antibodies, a mix of anti-mouse Alexa Fluor 568 (Invitrogen) and anti-rabbit Alexa Fluor 488. Where indicated, phalloidin 568 was included in the second incubation instead of anti-rabbit Alexa Fluor 488. Confocal images were collected using a Zeiss LSM510 Meta inverted confocal microscope, equipped with spectral detection capacity to select different emission wavelengths to be detected.
Sequence analysis and molecular modelling.
Multiple-sequence alignments were done using the clustal_x program (Thompson et al., 1997). Alternatively, the non-redundant protein sequence (nr) database was searched by using the blastp (protein–protein blast) algorithm at the NCBI server (http://blast.ncbi.nlm.nih.gov/). CCHFV N protein coiled-coil predictions were obtained using Lupas et al. (1991). The predicted interaction surface was generated with the molecular docking algorithm based on shape complementarity principles, according with the server patchdocl (Schneidman-Duhovny et al., 2005). The CCHFV N protein PDB: 4AQF (Wang et al., 2012) and the actin protein PDB: 1ATN structures were acquired in the database of the Protein Data Bank (http://www.pdb.org). The structures were visualized with Swiss-Pdbviewer 4 (http://www.expasy.org/spdbv). The analysis of the protein interactions was performed in the server pisa (http://www.ebi.ac.uk/msd-srv/prot_int/cgi-bin/piserver).
Acknowledgements
We thank Richard Cadagan and Osman Lizardo for technical assistance and Randy Albrecht for the helpful editing and discussion of the manuscript. In addition, we thank the Microscopy Shared Facility at Icahn School of Medicine in Mount Sinai for assistance. This work was partially supported by NIAID grant U54 AI057158 (to A. G.-S.).
References
- Albayrak H., Ozan E., Kurt M. (2010). An antigenic investigation of Crimean-Congo hemorrhagic fever virus (CCHFV) in hard ticks from provinces in northern Turkey. Trop Anim Health Prod 42, 1323–1325. 10.1007/s11250-010-9579-1 [DOI] [PubMed] [Google Scholar]
- Alminaite A., Backström V., Vaheri A., Plyusnin A. (2008). Oligomerization of hantaviral nucleocapsid protein: charged residues in the N-terminal coiled-coil domain contribute to intermolecular interactions. J Gen Virol 89, 2167–2174. 10.1099/vir.0.2008/004044-0 [DOI] [PubMed] [Google Scholar]
- Andersson I., Simon M., Lundkvist A., Nilsson M., Holmström A., Elgh F., Mirazimi A. (2004). Role of actin filaments in targeting of Crimean Congo hemorrhagic fever virus nucleocapsid protein to perinuclear regions of mammalian cells. J Med Virol 72, 83–93. 10.1002/jmv.10560 [DOI] [PubMed] [Google Scholar]
- Bente D. A., Forrester N. L., Watts D. M., McAuley A. J., Whitehouse C. A., Bray M. (2013). Crimean-Congo hemorrhagic fever: history, epidemiology, pathogenesis, clinical syndrome and genetic diversity. Antiviral Res 100, 159–189. 10.1016/j.antiviral.2013.07.006 [DOI] [PubMed] [Google Scholar]
- Bergeron E., Vincent M. J., Nichol S. T. (2007). Crimean-Congo hemorrhagic fever virus glycoprotein processing by the endoprotease SKI-1/S1P is critical for virus infectivity. J Virol 81, 13271–13276. 10.1128/JVI.01647-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter S. D., Surtees R., Walter C. T., Ariza A., Bergeron E., Nichol S. T., Hiscox J. A., Edwards T. A., Barr J. N. (2012). Structure, function, and evolution of the Crimean-Congo hemorrhagic fever virus nucleocapsid protein. J Virol 86, 10914–10923. 10.1128/JVI.01555-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ergönül O. (2006). Crimean-Congo haemorrhagic fever. Lancet Infect Dis 6, 203–214. 10.1016/S1473-3099(06)70435-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson B. R., Deyde V., Sanchez A. J., Vincent M. J., Nichol S. T. (2007). N-linked glycosylation of Gn (but not Gc) is important for Crimean Congo hemorrhagic fever virus glycoprotein localization and transport. Virology 361, 348–355. 10.1016/j.virol.2006.11.023 [DOI] [PubMed] [Google Scholar]
- Flick R., Whitehouse C. A. (2005). Crimean-Congo hemorrhagic fever virus. Curr Mol Med 5, 753–760. 10.2174/156652405774962335 [DOI] [PubMed] [Google Scholar]
- Frias-Staheli N., Giannakopoulos N. V., Kikkert M., Taylor S. L., Bridgen A., Paragas J. J., Richt J. A., Rowland R. R., Schmaljohn C. S. & other authors (2007). Ovarian tumor domain-containing viral proteases evade ubiquitin- and ISG15-dependent innate immune responses. Cell Host Microbe 2, 404–416. 10.1016/j.chom.2007.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Wang W., Ji W., Deng M., Sun Y., Zhou H., Yang C., Deng F., Wang H. & other authors (2012). Crimean-Congo hemorrhagic fever virus nucleoprotein reveals endonuclease activity in bunyaviruses. Proc Natl Acad Sci U S A 109, 5046–5051. 10.1073/pnas.1200808109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogstraal H. (1979). The epidemiology of tick-borne Crimean-Congo hemorrhagic fever in Asia, Europe, and Africa. J Med Entomol 15, 307–417. [DOI] [PubMed] [Google Scholar]
- Levingston Macleod J. M., D’Antuono A., Loureiro M. E., Casabona J. C., Gomez G. A., Lopez N. (2011). Identification of two functional domains within the arenavirus nucleoprotein. J Virol 85, 2012–2023. 10.1128/JVI.01875-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A., Van Dyke M., Stock J. (1991). Predicting coiled coils from protein sequences. Science 252, 1162–1164. 10.1126/science.252.5009.1162 [DOI] [PubMed] [Google Scholar]
- Mardani M., Keshtkar-Jahromi M. (2007). Crimean-Congo hemorrhagic fever. Arch Iran Med 10, 204–214. [PubMed] [Google Scholar]
- Murakoshi H., Lee S. J., Yasuda R. (2008). Highly sensitive and quantitative FRET-FLIM imaging in single dendritic spines using improved non-radiative YFP. Brain Cell Biol 36, 31–42. 10.1007/s11068-008-9024-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne J. C., Elliott R. M. (2000). RNA binding properties of bunyamwera virus nucleocapsid protein and selective binding to an element in the 5′ terminus of the negative-sense S segment. J Virol 74, 9946–9952. 10.1128/JVI.74.21.9946-9952.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overby A. K., Pettersson R. F., Neve E. P. (2007). The glycoprotein cytoplasmic tail of Uukuniemi virus (Bunyaviridae) interacts with ribonucleoproteins and is critical for genome packaging. J Virol 81, 3198–3205. 10.1128/JVI.02655-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plyusnin A., Beaty B., Elliott R. M., Goldbach R., Kormelink R., Lundkvist A., Schmaljohn C. S., Tesh R. B. (2012). Family Bunyaviridae. In Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses, pp. 725–741 Edited by King A. M. Q., Adams M. J., Carstens E. B. , Lefkowitz E. J. San Diego, CA: Elsevier Academic Press,. [Google Scholar]
- Ruigrok R. W., Crépin T., Kolakofsky D. (2011). Nucleoproteins and nucleocapsids of negative-strand RNA viruses. Curr Opin Microbiol 14, 504–510. 10.1016/j.mib.2011.07.011 [DOI] [PubMed] [Google Scholar]
- Sanchez A. J., Vincent M. J., Nichol S. T. (2002). Characterization of the glycoproteins of Crimean-Congo hemorrhagic fever virus. J Virol 76, 7263–7275. 10.1128/JVI.76.14.7263-7275.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez A. J., Vincent M. J., Erickson B. R., Nichol S. T. (2006). Crimean-Congo hemorrhagic fever virus glycoprotein precursor is cleaved by furin-like and SKI-1 proteases to generate a novel 38-kilodalton glycoprotein. J Virol 80, 514–525. 10.1128/JVI.80.1.514-525.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidman-Duhovny D., Inbar Y., Nussinov R., Wolfson H. J. (2005). patchdock and symmdock: servers for rigid and symmetric docking. Nucleic Acids Res 33 (Web Server issue), W363–W367. 10.1093/nar/gki481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares-Weiser K., Maclehose H., Ben-Aharon I., Goldberg E., Pitan F., Cunliffe N. (2010a). Vaccines for preventing rotavirus diarrhoea: vaccines in use. Cochrane Database Syst Rev 2010, CD008521. [DOI] [PubMed] [Google Scholar]
- Soares-Weiser K., Thomas S., Thomson G. G., Garner P. (2010b). Ribavirin for Crimean-Congo hemorrhagic fever: systematic review and meta-analysis. BMC Infect Dis 10, 207. 10.1186/1471-2334-10-207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M. P., Koyuncu O. O., Enquist L. W. (2011). Subversion of the actin cytoskeleton during viral infection. Nat Rev Microbiol 9, 427–439. 10.1038/nrmicro2574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. (1997). The clustal_x Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Alminaite A., Vaheri A., Plyusnin A. (2010). Interaction between hantaviral nucleocapsid protein and the cytoplasmic tail of surface glycoprotein Gn. Virus Res 151, 205–212. 10.1016/j.virusres.2010.05.008 [DOI] [PubMed] [Google Scholar]
- Wang Y., Dutta S., Karlberg H., Devignot S., Weber F., Hao Q., Tan Y. J., Mirazimi A., Kotaka M. (2012). Structure of Crimean-Congo hemorrhagic fever virus nucleoprotein: superhelical homo-oligomers and the role of caspase-3 cleavage. J Virol 86, 12294–12303. 10.1128/JVI.01627-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse C. A. (2004). Crimean-Congo hemorrhagic fever. Antiviral Res 64, 145–160. 10.1016/j.antiviral.2004.08.001 [DOI] [PubMed] [Google Scholar]