Abstract
Rotavirus (RV) A is a very common cause of acute diarrhoea in infants and young children worldwide. Most human strains are classified into two major Wa-like and DS-1-like genotype constellations, whilst a minor third strain, AU-1, was described in 1989 among human RV isolates from Japan. AU-1 demonstrates a high degree of homology to a feline RV, FRV-1, which suggests interspecies transmission of feline RV. However, there has been no subsequent report of RVs possessing the AU-1 genotype throughout all 11 genes of the genome. Between March 1997 and December 1999, 157 RV-positive stool samples were collected from Brazilian children, and 16 of the RVs (10.2 %) were P[9] genotype. We analysed eight strains by almost full-genome sequencing. These eight strains were divided into two groups: five AU-1-like and three Wa-like strains. Four of the five AU-1-like strains had the AU-1-like genotype constellation throughout the 11 genes. The remaining AU-1-like strain was considered to be a reassortant strain comprosed of nine, two and one genes from the AU-1-like, Wa-like and G9 strains, respectively. The three Wa-like strains were considered to be reassortants comprising seven to eight genes and three to four genes from Wa-like and non-Wa-like strains, respectively. This report of human G3P[9] RV strains possessing the AU-1 genotype constellation throughout all genes demonstrates the stability and infectivity of the AU-1-like strain with its original genotype over distance and time.
Introduction
Group A rotaviruses (RVAs) are important aetiological agents of severe diarrhoea in infants and young children worldwide (Estes & Kapikian, 2007). Rotaviruses (RVs) are non-enveloped, icosahedral viruses of the family Reoviridae possessing a genome with 11 segments of dsRNA. The two outer capsid proteins, VP7 (G types) and VP4 (P types), independently elicit neutralizing antibodies and are the basis of a dual classification system defined by G and P types, respectively (Estes & Kapikian, 2007).
Recently, a novel RV genotyping classification system based on all 11 genome segments was introduced (Matthijnssens et al., 2008). This comprehensive classification system revealed that most human RV strains can be classified into either a Wa-like (I1-R1-C1-M1-A1-N1-T1-E1-H1) genotype constellation with G1P[8], G3P[8], G4P[8] and G9P[8] specificities, or a DS-1-like (I2-R2-C2-M2-A2-N2-T2-E2-H2) genotype constellation with G2P[4] specificity.
In addition, AU-1 and AU-1-like RVA strains make up a small third group. AU-1 was identified in 1982 in Japan as the first example of interspecies transmission from cat to human (Nakagomi & Nakagomi, 1989) and had the I3-R3-C3-M3-A3-N3-T3-E3-H3 genotype constellation with G3P[9] specificity. After AU-1 was discovered, limited numbers of AU-1-like strains, which mostly had less conserved genomes compared with AU-1 and/or had at least partially reassortant genomes, have been isolated sporadically in humans and cats (Iizuka et al., 1994; Gentsch et al., 1996; Gollop et al., 1998; Santos et al., 2001; Santo & Hoshino, 2005; Rahman et al., 2007; Grant et al., 2011; Matthijnssens et al., 2011; Wang et al., 2013; Gauchan et al., 2014; Theamboonlers et al., 2014). Furthermore, there have been no reports of human RVs possessing the AU-1 genotype throughout all 11 genes (Rahman et al., 2007; Wang et al., 2013; Theamboonlers et al., 2014).
Santos et al. (2001) screened diarrhoea stool samples from Brazilian children between 1997 and 1999, and found that 16 out of 157 RVs (10.2 %) were P[9] genotype by reverse transcription (RT)-PCR genotyping. In the present study, we analysed eight of these 16 strains by whole-genome sequencing, and examined whether these were also less conserved and/or reassortant strains like those isolated so far or were unusually stable and infective genuine descendants of the AU-1 strain.
Results
Complete genotype constellation of P[9] strains
The complete genotype constellations and nucleotide identity of the eight P[9] strains and previously reported five AU-1-like strains (FRV-1, L621, E2451, CU365-KK/08 and T152) compared with the AU-1 strain are shown in Table 1 (Gauchan et al., 2014; Wang et al., 2013; Theamboonlers et al., 2014; Rahman et al., 2007). Our eight strains were divided into two groups: AU-1-like and Wa-like. The AU-1-like group consisted of five strains: R47, R55, R57, R142 and R138. Surprisingly, the genotypes of all 11 segments of four of the five strains (with the exception of R138) were very closely related to those of the AU-1 strain (91.3–99.3 % nucleotide identity). R138 had a somewhat complicated genotype and was considered to be a reassortant strain: nine genes (VP1–VP4, VP6, NSP1, and NSP3–NSP5) were AU-1-like, two genes (NSP2 and NSP3) were Wa-like and the VP7 gene was G9. Moreover, R138 consisted of a mixed population of AU-1-like and Wa-like genotypes, which was shown by NSP3 gene analysis with plaque-purified strains.
Table 1. Complete genotype constellations and nucleotide identity of the eight human P[9] strains of this study and of five previously reported AU-1-like RVs compared with the AU-1 strain.
The Wa-like, DS-1-like and AU-1-like genotypes are shown in bold, bold italic and italic, respectively. The G9 genotypes G12 and A12 are shown in underlined italic and underlined bold, respectively. The GenBank accession numbers of the other five RVs are listed in Table S1.
| Strain | Genomic constellation | ||||||||||
| VP7 | VP4 | VP6 | VP1 | VP2 | VP3 | NSP1 | NSP2 | NSP3 | NSP4 | NSP5 | |
| AU-1 | G3 (100) | P[9] (100) | I3 (100) | R3 (100) | C3 (100) | M3 (100) | A3 (100) | N3 (100) | T3 (100) | E3 (100) | H3 (100) |
| R47 | G3 (98.9) | P[9] (98.6) | I3 (98.9) | R3 (98.3) | C3 (98.9) | M3 (98.5) | A3 (93.0) | N3 (98.8) | T3 (99.3) | E3 (98.9) | H3 (99.0) |
| R55 | G3 (91.3) | P[9] (98.6) | I3 (98.9) | R3 (98.3) | C3 (98.8) | M3 (98.5) | A3 (93.0) | N3 (97.9) | T3 (99.2) | E3 (98.9) | H3 (99.0) |
| R57 | G3 (98.8) | P[9] (98.5) | I3 (98.9) | R3 (98.3) | C3 (98.8) | M3 (98.5) | A3 (93.0) | N3 (98.8) | T3 (99.3) | E3 (99.0) | H3 (99.0) |
| R142 | G3 (98.8) | P[9] (98.5) | I3 (98.9) | R3 (98.3) | C3 (98.9) | M3 (98.5) | A3 (92.9) | N3 (98.8) | T3 (99.3) | E3 (99.0) | H3 (99.0) |
| R138 | G9 | P[9] (98.6) | I3 (98.9) | R3 (98.3) | C3 (98.9) | M3 (98.4) | A3 (93.0) | N1 | T1/T3 (99.1) | E3 (99.0) | H3 (99.0) |
| R135 | G9 | P[9] (98.5) | I1 | R1 | C1 | M1 | A1 | N1 | T2 | E1 | H1 |
| R70 | G1 | P[9] (98.5) | I1 | R1 | C1 | M3 (98.4) | A1 | N1 | T2 | E1 | H1 |
| R49 | G1 | P[9] (98.4) | I1 | R1 | C1 | M2 | A1 | N2 | T2 | E1 | H1 |
| FRV-1 | G3 (93.7) | P[9] (98.8) | I3 (92.6) | R3 (99.0) | C3 (88.6) | M3 (97.6) | A3 (97.9) | N3 (97.8) | T3 (98.4) | E3 (99.2) | H3 (99.4) |
| L621 | G3 (92.3) | P[9] (97.3) | I3 (92.5) | R3 (86.0) | C3 (87.3) | M3 (85.7) | A3 (95.9) | N3 (87.1) | T3 (84.4) | E3 (92.8) | H6 |
| E2451 | G3 (92.7) | P[9] (97.6) | I3 (91.9) | R3 (85.2) | C3 (92.6) | M3 (85.6) | A3 (95.1) | N3 (86.9) | T3 (98.0) | E3 (97.2) | H6 |
| CU365-KK/08 | G3 (91.9) | P[9] (98.7) | I3 (91.9) | R3 (92.9) | C3 (87.5) | M3 (87.4) | A3 (96.3) | N3 (85.8) | T3 (84.4) | E3 (92.0) | H6 |
| T152 | G12 | P[9] (89.8) | I3 (85.0) | R3 (94.0) | C3 (93.7) | M3 (87.4) | A12 | N3 (86.0) | T3 (85.5) | E3 (90.8) | H6 |
The remaining three strains – R135, R70 and R49 – formed the other group, and were considered to be reassortants with seven to eight Wa-like and three to four non-Wa-like genes.
Highly conserved nucleotide sequence of AU-1 genotypes
Using the mVISTA module (http://genome.lbl.gov/vista/index.shtml) (Frazer et al., 2004), we aligned whole-genome nucleotide sequences of our AU-1-like strains (R47, R55, R57, R142 and R138) and previously reported AU-1-like strains (FRV-1, L621, E2451, CU365-KK/08 and T152) and compared them with the AU-1 strain (Fig. 1) (Gauchan et al., 2014; Wang et al., 2013; Theamboonlers et al., 2014; Rahman et al., 2007). All genome segments of our four AU-1-like strains (R47, R55, R57 and R142) shared a much higher nucleotide identity (97.9–99.3 %) with the AU-1 strain, except for the VP7 gene of R55 (91.3 %) and NSP1 gene of all strains (92.9–93.0 %) than that of the previously reported four human AU-1-like strains (L621, E2451, CU365-KK/08 and T152) (Table 1 and Fig. 1). Eight of 11 genome segments of the R138 strain (VP1–VP4, VP6 and NSP3–NSP5 genes) that had the AU-1 genotype were also highly conserved (98.3–99.1 % nucleotide identity). Furthermore, each of the 11 genome segments of our four AU-1-like strains (R47, R55, R57 and R142) was extremely highly conserved (99.4–100 % nucleotide identity) among them except for the VP7 gene of R55 (91.6–91.7 % nucleotide identity).
Fig. 1.
mVISTA whole-genome nucleotide alignment comparing the AU-1 strain with the five AU-1-like strains (R47, R55, R57, R142 and R138) analysed in this study and five previously reported AU-1-like strains (FRV-1, L621, E2451, CU365-KK/08 and T152). Percentage (right y-axis) indicates sequence identity, and shading indicates the level of conservation.
Sequence and phylogenetic analyses of the P[9] strains
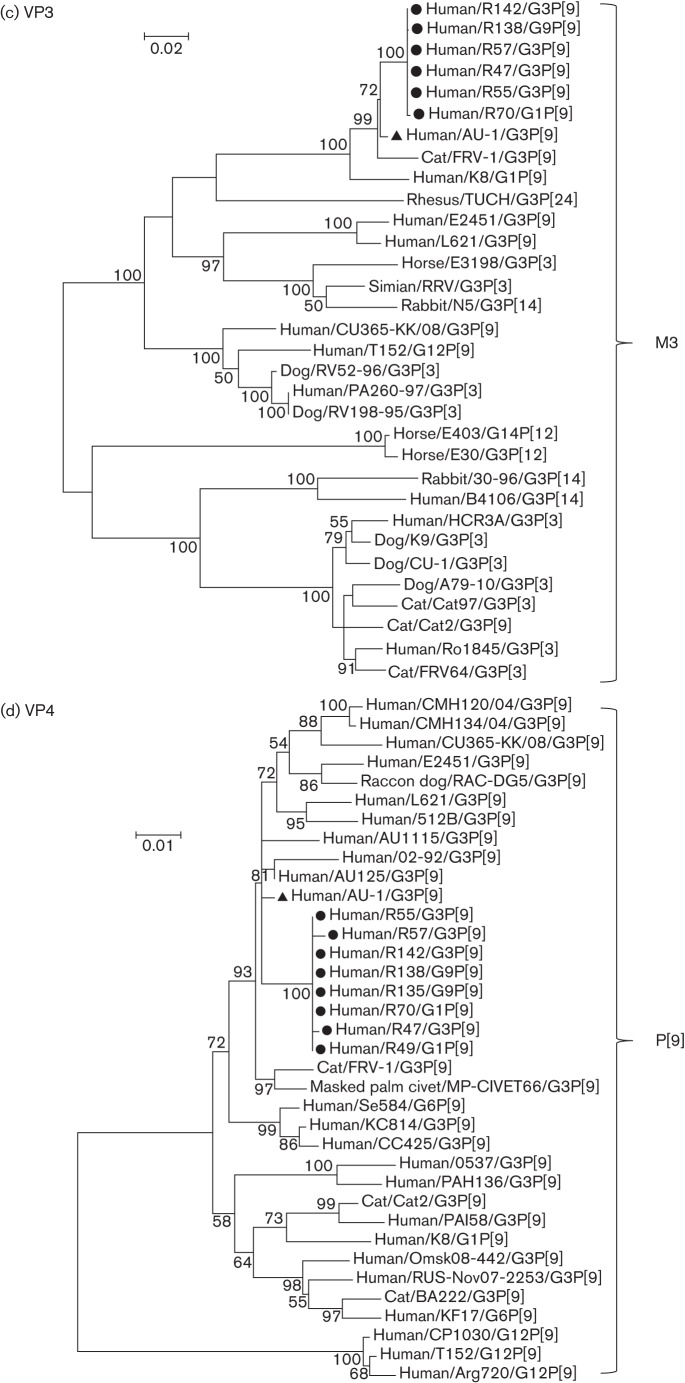
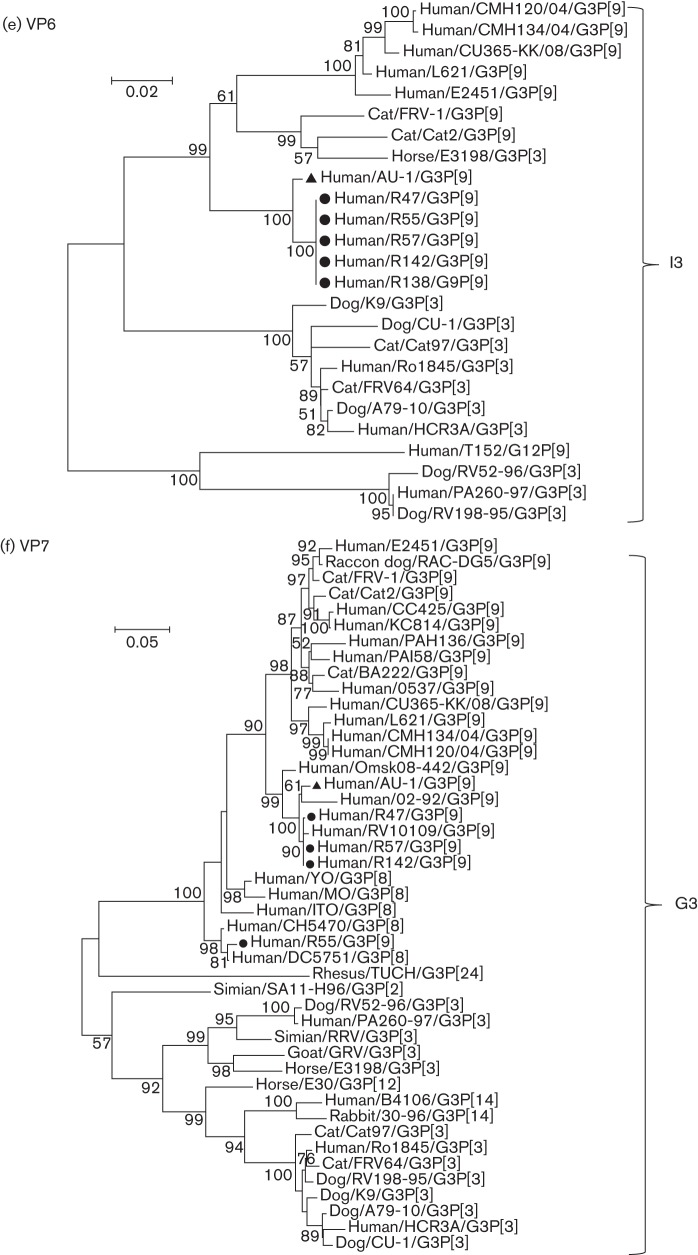
Phylogenetic trees were reconstructed with the nucleotide sequences of structural genes (VP1–VP4, VP6 and VP7) (Fig. 2a–f) and non-structural genes (NSP1–NSP5) (Fig. 3a–e) of AU-1, the AU-1-like strains of this study and other AU-1-like strains.
Fig. 2.
Phylogenetic trees reconstructed from the nucleotide sequences of the structural genes VP1 (a), VP2 (b), VP3 (c), VP4 (d), VP6 (e) and VP7 (f) of the AU-1-like genotypes. Phylogenetic analysis was performed using the maximum-likelihood method provided by mega v.5.2.2 software. Bootstrap values (500 replicates) above 50 % are shown. Filled circles and filled triangles indicate AU-1-like strains analysed in this study and the AU-1 strain, respectively. The GenBank accession numbers of sequences of the reference strains are listed in Table S1 available in the online Supplementary Material. Bars, nucleotide substitutions per site.
Fig. 3.
Phylogenetic trees reconstructed from the nucleotide sequences of the non-structural genes NSP1 (a), NSP2 (b), NSP3 (c), NSP4 (d) and NSP5 (e) of the AU-1-like genotypes. Phylogenetic analysis was performed using the maximum-likelihood method provided by mega v.5.2.2 software. Bootstrap values (500 replicates) above 50 % are shown. Filled circles and filled triangles indicate the AU-1-like strains analysed in this study and the AU-1 strain, respectively. The GenBank accession numbers of sequences of the reference strains are listed in Table S1. Bars, nucleotide substitutions per site.
Each of four genes (VP1–VP3 and VP6) of the AU-1-like strains (R47, R55, R57, R142 and R138) shared extremely high nucleotide identities (99.96–100 %) and formed a distinct phylogenetic cluster with the AU-l strain (Fig. 2a, b, e). Strain R70 also showed high nucleotide identity (99.84–99.88 %) with the five AU-1-like strains in the VP3 gene (Fig. 2c).
VP4 of our five AU-1-like strains and three Wa-like strains (R135, R70 and R49) had almost identical nucleotide sequences (99.74–99.96 % nucleotide identity) and formed a distinct phylogenetic cluster with the other human/feline G3P[9] RV strains, including human AU-1 and feline FRV-1 RV strains (Fig. 2d).
With regard to VP7, four of our five AU-1-like strains were classified into genotype G3. Three strains (R47, R57 and R142) shared almost identical nucleotide sequences (99.90–100 % nucleotide identity) and formed a distinct phylogenetic cluster with AU-1 and other human G3P[9] strains (Fig. 2f). Although the R55 strain displayed a moderately high nucleotide identity (91.60–91.70 %) with the three above-mentioned strains, it clustered in a different branch from the conventional human G3P[8] strains. Thus, the VP7 of R55 was considered to be an intra-genogroup reassortant between AU-1/AU-1-like (G3P[9]) strains and conventional human G3P[8] strains.
NSP1 of our five AU-1-like strains shared almost identical nucleotide sequences (99.93–100 %), displayed a moderately high nucleotide identity (92.88–93.01 %) with AU-1 and clustered into a different branch from AU-1 (Fig. 3a). Similarly, each of four cognate genes (NSP2–NSP5) of our four to five AU-1-like strains shared almost identical nucleotide sequences (99.32–100 %) and formed a distinct phylogenetic cluster with the AU-l strain (Fig. 3b–e).
Discussion
Whole-genome sequence analysis of the eight unusual human P[9] strains detected in Brazil in 1997/1999 revealed that four of the strains (R47, R55, R57 and R142) had the AU-1-genotype constellations throughout the 11 genes by full-genome sequencing. The AU-1 strain was isolated in 1982 in Japan. Therefore, it can be stated that these AU-1-like strains conserved all 11 genotypes of AU-1 across time (1982 vs 1997–1999) and location (Japan vs Brazil) (Tables 1 and 2).
Table 2. Clinical information on the eight human P[9] RVs analysed in this study.
| Strain | VP7 genotype | Date of isolation | Area of isolation | In-/outpatient | Age of patient |
| R47 | G3 | 10/4/1997 | Niterói | Outpatient | <5 years |
| R49 | G1 | 22/4/1997 | Rio de Janeiro | Outpatient | <5 years |
| R55 | G3 | 14/7/1997 | Rio de Janeiro | Outpatient | <5 years |
| R57 | G3 | 14/7/1997 | Rio de Janeiro | Outpatient | <5 years |
| R70 | G1 | 14/7/1997 | Rio de Janeiro | Outpatient | <5 years |
| R135 | G9 | 21/7/1998 | Rio de Janeiro | Outpatient | <5 years |
| R138 | G9 | 8/9/1998 | Rio de Janeiro | Outpatient | <5 years |
| R142 | G3 | 6/1/1999 | Rio de Janeiro | Inpatient | 2 years |
To date, only four human AU-1-like strains (L621, E2451, CU365-KK and T152) have been described, and their genotypes did not entirely resemble that of the AU-1 strain (Rahman et al., 2007; Wang et al., 2013; Theamboonlers et al., 2014). Namely, NSP5 of all four strains had changed from H3 to H6, and VP7 and NSP1 of T152 were replaced by G12 and A12 from G3 and A3, respectively (Table 1). Furthermore, these four strains were isolated in different countries (China or Thailand vs Japan) and at different times (2006, 2011, 2008 or 1998 vs 1982) compared with AU-1, and were less conserved compared with AU-1 than four Brazilian AU-1-like strains (Table 1 and Fig. 1). Although the reason is unknown as to why the nucleotide sequences of the NSP1 genes of the four Brazilian AU-1-like strains were less conserved compared with AU-1 (92.9–93.1 % nucleotide identity) than the other 10 genes (97.9–99.3 % nucleotide identity) except for the VP7 gene of R55 (91.3 % nucleotide identity), it might be possible that the NSP1 genes had biological properties associated with virulence (e.g. anti-IFN agent) and affected selective pressure from the host. In future, full-genome sequencing of AU-1-like strains must be continued to determine whether AU-1 genotype constellations are still highly conserved like the Brazilian strains or not (i.e. L621, E2451, CU365-KK and T152).
The discovery of the AU-1 strain in 1982 was the initial confirmation of suspected interspecies transmissions from cat to human (Nakagomi & Nakagomi, 1989). Previously, RNA–RNA hybridization and haemagglutination assays had demonstrated the existence of two genogroups among feline RVs: the AU-1-like genogroup (human–feline group: human AU-1 and feline FRV-1) and Cat97-like genogroup (feline–canine: feline Cat97 and FRV64, and canine CU-1 and K9) (Nakagomi et al., 1990; Nakagomi & Nakagomi, 1991; Mochizuki et al., 1992, 1997). Subsequently, we also confirmed these two genogroups by full-genome sequencing in selected human, feline and canine RVs (Tsugawa & Hoshino, 2008). Recently, a third genogroup of feline RVs (BA222-05-like) was proposed by full-genome sequencing (Matthijnssens et al., 2011). To date, only five feline strains (Cat97, Cat2, BA222-05, FRV-1 and FRV64) have had their whole genomes sequenced (Tsugawa & Hoshino, 2008; Martella et al., 2011; Gauchan et al., 2014), and there is only one report of an AU-1-like feline strain (FRV-1) being fully sequenced (Table 1 and Fig. 1) (Gauchan et al., 2014). Further full-genome sequence data on many feline strains are needed to understand the molecular epidemiology of feline strains and the relationship between human AU-1-like and feline strains with regard to zoonotic transmission. Full-genome sequencing of RVs from other animals closely interacting with humans (e.g. cows, pigs, dogs and horses) is also necessary for an understanding of interspecies transmission and/or reassortant events of RVs.
Seroepidemiological studies have revealed that (i) 91 % (20 of 22) of cat sera had neutralizing antibodies to G3 in Germany during 1991–1992, and (ii) 23 % (7 of 30) of cat sera had neutralizing antibodies to P[9] in Japan during 1989–1994 (Streckert et al., 1993; Mochizuki et al., 1997). Therefore, it was concluded that G3P[9] (AU-1-like) strains have been circulating within cats for a considerable time.
Although P[9] strains have been detected in humans at very low rates of approximately 1 % globally (Kaga et al., 1994; Gentsch et al., 1996; Santos & Hoshino, 2005), they were continuously detected (16/157 , 10.2 %) in the Rio de Janeiro area between 1997 and 1999 (Santos et al., 2001). In the present study, we analysed eight P[9] strains and revealed that they had AU-1-like genotype constellations with high nucleotide identities in up to 11 segments. Although it was unknown whether the AU-1-like strains were circulating within cat and/or human populations, we concluded that these strains were genetically rather stable and were infectious to humans.
The effectiveness of current RV vaccines against Wa-like and DS-1-like strains is well established, but effectiveness against AU-1-like RVs remains unclear (Braeckman et al., 2012; Payne et al., 2013). Although AU-1-like RV infections remain relatively infrequent in humans, the results of the current study reveal that it preserves the potential for prevalence with its original or reassortant genotype. RV surveillance by whole-genome sequencing must continue to monitor changes in dominant genotypes and the emergence of reassortant RVs after widespread use of RV vaccines. Novel G1P[8] double-reassortant strains (DS-1-like genotype) have been isolated predominantly in Japan since 2012 (Fujii et al., 2014; Kuzuya et al., 2014; Yamamoto et al., 2014). This information could be critical for the development of RV vaccine strategies.
Methods
Viruses.
Between March 1997 and December 1999, 157 of 678 (23.2 %) diarrhoeal stool samples collected from Brazilian children under 5 years of age were RV positive, and 16 of the 157 (10.2 %) were P[9] genotype (Santos et al., 2001). We analysed six of the 16 P[9] strains (R47, R55, R70, R135, R138 and R142) and two other strains (R49 and R57) passaged by primary African green monkey kidney (AGMK) and/or MA104 cell culture for almost full-genome sequencing, excluding the 5′- and 3′-end primer-binding regions (Table 2) (Ward et al., 1984; Tsugawa et al., 2014).
RNA extraction and RT-PCR.
The RT-PCR and nucleotide sequencing were performed as described previously (Tsugawa & Hoshino, 2008) with primers listed in Table S2. Briefly, dsRNA was extracted with TRIzol (Invitrogen) from infected cell-culture lysates. Two microlitres of extracted dsRNA was added to 1 µl DMSO. The mixture was heated at 94 °C for 3 min and placed on ice. Reverse transcription was performed at 45 °C for 45 min, followed by 94 °C for 3 min. PCR was carried out under the following conditions: initial denaturation at 95 °C for 15 min; 40 cycles of 94 °C for 45 s, 50 °C for 45 s and 70 °C for 2.5 min; and a final extension at 70 °C for 7 min. The PCR products were analysed by electrophoresis in a 1 % agarose gel in 1× Tris/acetate/EDTA buffer containing 0.2 µg ethidium bromide ml−1.
Plaque purification of the mixed population.
To sequence the mixed population of the NSP3 gene of the R138 strain, plaque purification was performed as described previously (Hoshino et al., 2003). After individual plaques were picked and passaged twice by primary AGMK cell roller-tube cultures, they were amplified by RT-PCR and sequenced.
Nucleotide sequencing.
The PCR amplicons were purified with a Wizard SV Gel and PCR Clean-up system (Promega) and sequenced with a BigDye terminator v.3.1 cycle sequencing reaction kit (Applied Biosystems) on an ABI 3730 DNA analyser (Applied Biosystems).
Sequence analysis.
Sequence files were analysed using Sequencher v.4.7 (Gene Codes Corp.) and MacVector v.9.5.2 (Accelrys). The genotyping of each of the 11 segments for RV strains was determined using the RotaC online classification tool (http://rotac.regatools.be/) (Maes et al., 2009). Multiple sequence alignments and/or phylogenetic analyses (maximum likelihood) were performed using mega v.5.2.2 software (http://www.megasoftware.net/) and mVISTA software (http://genome.lbl.gov/vista/index.shtml) (Frazer et al., 2004; Tamura et al., 2011).
Acknowledgements
This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health, USA. There is no conflict of interest to declare. The authors thank N. Santos for providing us with the RV strains used in this study and Y. Hoshino for continuing support. We also thank P. Olley, Emeritus Professor, University of Alberta, for English language advice.
Footnotes
Two supplementary tables are available with the online Supplementary Material.
References
- Braeckman T., Van Herck K., Meyer N., Pirçon J. Y., Soriano-Gabarró M., Heylen E., Zeller M., Azou M., Capiau H. & other authors (2012). Effectiveness of rotavirus vaccination in prevention of hospital admissions for rotavirus gastroenteritis among young children in Belgium: case–control study. BMJ 345, e4752. 10.1136/bmj.e4752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes M. K., Kapikian A. Z. (2007). Rotaviruses. In Fields Virology, 5th edn, pp. 1917–1974 Edited by Knipe D. M. Philadelphia: Lippincott, Williams & Wilkins. [Google Scholar]
- Frazer K. A., Pachter L., Poliakov A., Rubin E. M., Dubchak I. (2004). vista: computational tools for comparative genomics. Nucleic Acids Res 32 (Web Server issue), W273–W279. 10.1093/nar/gkh458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii Y., Nakagomi T., Nishimura N., Noguchi A., Miura S., Ito H., Doan Y. H., Takahashi T., Ozaki T. & other authors (2014). Spread and predominance in Japan of novel G1P[8] double-reassortant rotavirus strains possessing a DS-1-like genotype constellation typical of G2P[4] strains. Infect Genet Evol 28, 426–433. 10.1016/j.meegid.2014.08.001 [DOI] [PubMed] [Google Scholar]
- Gauchan P., Sasaki E., Nakagomi T., Do L. P., Doan Y. H., Mochizuki M., Nakagomi O. (2014). Whole genotype constellation of prototype feline rotavirus strains FRV-1 and FRV64 and their phylogenetic relationships with feline-like human rotavirus strains. J Gen Virol [Epub ahead of print]. 10.1099/vir.0.070771-0 [DOI] [PubMed] [Google Scholar]
- Gentsch J. R., Woods P. A., Ramachandran M., Das B. K., Leite J. P., Alfieri A., Kumar R., Bhan M. K., Glass R. I. (1996). Review of G and P typing results from a global collection of rotavirus strains: implications for vaccine development. J Infect Dis 174 (Suppl. 1), S30–S36. 10.1093/infdis/174.Supplement_1.S30 [DOI] [PubMed] [Google Scholar]
- Gollop R., Nakagomi O., Silberstein I., Shulman L. M., Greenberg H. B., Mendelson E., Shif I. (1998). Three forms of AU-1 like human rotaviruses differentiated by their overall genomic constellation and by the sequence of their VP8*. Arch Virol 143, 263–277. 10.1007/s007050050285 [DOI] [PubMed] [Google Scholar]
- Grant L., Esona M., Gentsch J., Watt J., Reid R., Weatherholtz R., Santosham M., Parashar U., O’Brien K. (2011). Detection of G3P[3] and G3P[9] rotavirus strains in American Indian children with evidence of gene reassortment between human and animal rotaviruses. J Med Virol 83, 1288–1299. 10.1002/jmv.22076 [DOI] [PubMed] [Google Scholar]
- Hoshino Y., Wagner M., Yan X. Y., Perez-Schael I., Kapikian A. Z. (2003). Horizontal transmission of rhesus monkey rotavirus-based quadrivalent vaccine during a phase 3 clinical trial in Caracas, Venezuela. J Infect Dis 187, 791–800. 10.1086/368387 [DOI] [PubMed] [Google Scholar]
- Iizuka M., Chiba M., Masamune O., Kaga E., Nakagomi T., Nakagomi O. (1994). A highly conserved genomic RNA constellation of Japanese isolates of human rotaviruses carrying G serotype 3 and P serotype 9. Res Virol 145, 21–24. 10.1016/S0923-2516(07)80003-3 [DOI] [PubMed] [Google Scholar]
- Kaga E., Iizuka M., Nakagomi T., Nakagomi O. (1994). The distribution of G (VP7) and P (VP4) serotypes among human rotaviruses recovered from Japanese children with diarrhea. Microbiol Immunol 38, 317–320. 10.1111/j.1348-0421.1994.tb01784.x [DOI] [PubMed] [Google Scholar]
- Kuzuya M., Fujii R., Hamano M., Kida K., Mizoguchi Y., Kanadani T., Nishimura K., Kishimoto T. (2014). Prevalence and molecular characterization of G1P[8] human rotaviruses possessing DS-1-like VP6, NSP4, and NSP5/6 in Japan. J Med Virol 86, 1056–1064. 10.1002/jmv.23746 [DOI] [PubMed] [Google Scholar]
- Maes P., Matthijnssens J., Rahman M., Van Ranst M. (2009). RotaC: a web-based tool for the complete genome classification of group A rotaviruses. BMC Microbiol 9, 238. 10.1186/1471-2180-9-238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martella V., Potgieter A. C., Lorusso E., De Grazia S., Giammanco G. M., Matthijnssens J., Bányai K., Ciarlet M., Lavazza A. & other authors (2011). A feline rotavirus G3P[9] carries traces of multiple reassortment events and resembles rare human G3P[9] rotaviruses. J Gen Virol 92, 1214–1221. 10.1099/vir.0.027425-0 [DOI] [PubMed] [Google Scholar]
- Matthijnssens J., Ciarlet M., Heiman E., Arijs I., Delbeke T., McDonald S. M., Palombo E. A., Iturriza-Gómara M., Maes P. & other authors (2008). Full genome-based classification of rotaviruses reveals a common origin between human Wa-like and porcine rotavirus strains and human DS-1-like and bovine rotavirus strains. J Virol 82, 3204–3219. 10.1128/JVI.02257-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijnssens J., De Grazia S., Piessens J., Heylen E., Zeller M., Giammanco G. M., Bányai K., Buonavoglia C., Ciarlet M. & other authors (2011). Multiple reassortment and interspecies transmission events contribute to the diversity of feline, canine and feline/canine-like human group A rotavirus strains. Infect Genet Evol 11, 1396–1406. 10.1016/j.meegid.2011.05.007 [DOI] [PubMed] [Google Scholar]
- Mochizuki M., Nakagomi O., Shibata S. (1992). Hemagglutinin activity of two distinct genogroups of feline and canine rotavirus strains. Arch Virol 122, 373–381. 10.1007/BF01317199 [DOI] [PubMed] [Google Scholar]
- Mochizuki M., Nakagomi T., Nakagomi O. (1997). Isolation from diarrheal and asymptomatic kittens of three rotavirus strains that belong to the AU-1 genogroup of human rotaviruses. J Clin Microbiol 35, 1272–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagomi T., Nakagomi O. (1989). RNA–RNA hybridization identifies a human rotavirus that is genetically related to feline rotavirus. J Virol 63, 1431–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagomi O., Nakagomi T. (1991). Genetic diversity and similarity among mammalian rotaviruses in relation to interspecies transmission of rotavirus. Arch Virol 120, 43–55. 10.1007/BF01310948 [DOI] [PubMed] [Google Scholar]
- Nakagomi O., Ohshima A., Aboudy Y., Shif I., Mochizuki M., Nakagomi T., Gotlieb-Stematsky T. (1990). Molecular identification by RNA–RNA hybridization of a human rotavirus that is closely related to rotaviruses of feline and canine origin. J Clin Microbiol 28, 1198–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne D. C., Boom J. A., Staat M. A., Edwards K. M., Szilagyi P. G., Klein E. J., Selvarangan R., Azimi P. H., Harrison C. & other authors (2013). Effectiveness of pentavalent and monovalent rotavirus vaccines in concurrent use among US children <5 years of age, 2009–2011. Clin Infect Dis 57, 13–20. 10.1093/cid/cit164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M., Matthijnssens J., Yang X., Delbeke T., Arijs I., Taniguchi K., Iturriza-Gómara M., Iftekharuddin N., Azim T., Van Ranst M. (2007). Evolutionary history and global spread of the emerging G12 human rotaviruses. J Virol 81, 2382–2390. 10.1128/JVI.01622-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos N., Hoshino Y. (2005). Global distribution of rotavirus serotypes/genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Rev Med Virol 15, 29–56. 10.1002/rmv.448 [DOI] [PubMed] [Google Scholar]
- Santos N., Volotão E. M., Soares C. C., Albuquerque M. C., da Silva F. M., de Carvalho T. R., Pereira C. F., Chizhikov V., Hoshino Y. (2001). Rotavirus strains bearing genotype G9 or P[9] recovered from Brazilian children with diarrhea from 1997 to 1999. J Clin Microbiol 39, 1157–1160. 10.1128/JCM.39.3.1157-1160.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streckert H. J., Kappes M., Olivo M., Schulze-Lammers J. (1993). [Cat sera neutralize rotaviruses of serotype G3]. Dtsch Tierarztl Wochenschr 100, 223–225 (in German). [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). mega5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28, 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theamboonlers A., Maiklang O., Thongmee T., Chieochansin T., Vuthitanachot V., Poovorawan Y. (2014). Complete genotype constellation of human rotavirus group A circulating in Thailand, 2008–2011. Infect Genet Evol 21, 295–302. 10.1016/j.meegid.2013.11.020 [DOI] [PubMed] [Google Scholar]
- Tsugawa T., Hoshino Y. (2008). Whole genome sequence and phylogenetic analyses reveal human rotavirus G3P[3] strains Ro1845 and HCR3A are examples of direct virion transmission of canine/feline rotaviruses to humans. Virology 380, 344–353. 10.1016/j.virol.2008.07.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugawa T., Tatsumi M., Tsutsumi H. (2014). Virulence-associated genome mutations of murine rotavirus identified by alternating serial passages in mice and cell cultures. J Virol 88, 5543–5558. 10.1128/JVI.00041-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. H., Pang B. B., Zhou X., Ghosh S., Tang W. F., Peng J. S., Hu Q., Zhou D. J., Kobayashi N. (2013). Complex evolutionary patterns of two rare human G3P[9] rotavirus strains possessing a feline/canine-like H6 genotype on an AU-1-like genotype constellation. Infect Genet Evol 16, 103–112. 10.1016/j.meegid.2013.01.016 [DOI] [PubMed] [Google Scholar]
- Ward R. L., Knowlton D. R., Pierce M. J. (1984). Efficiency of human rotavirus propagation in cell culture. J Clin Microbiol 19, 748–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S. P., Kaida A., Kubo H., Iritani N. (2014). Gastroenteritis outbreaks caused by a DS-1-like G1P[8] rotavirus strain, Japan, 2012–2013. Emerg Infect Dis 20, 1030–1033. 10.3201/eid2006.131326 [DOI] [PMC free article] [PubMed] [Google Scholar]