Abstract
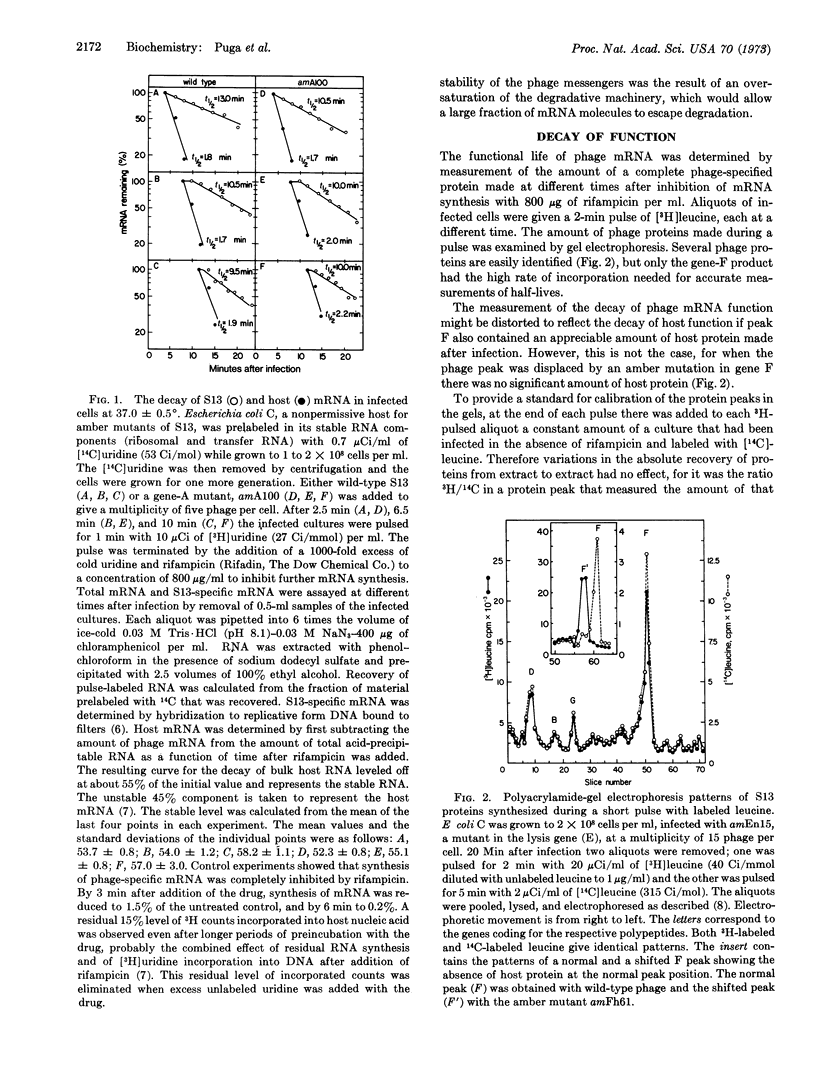
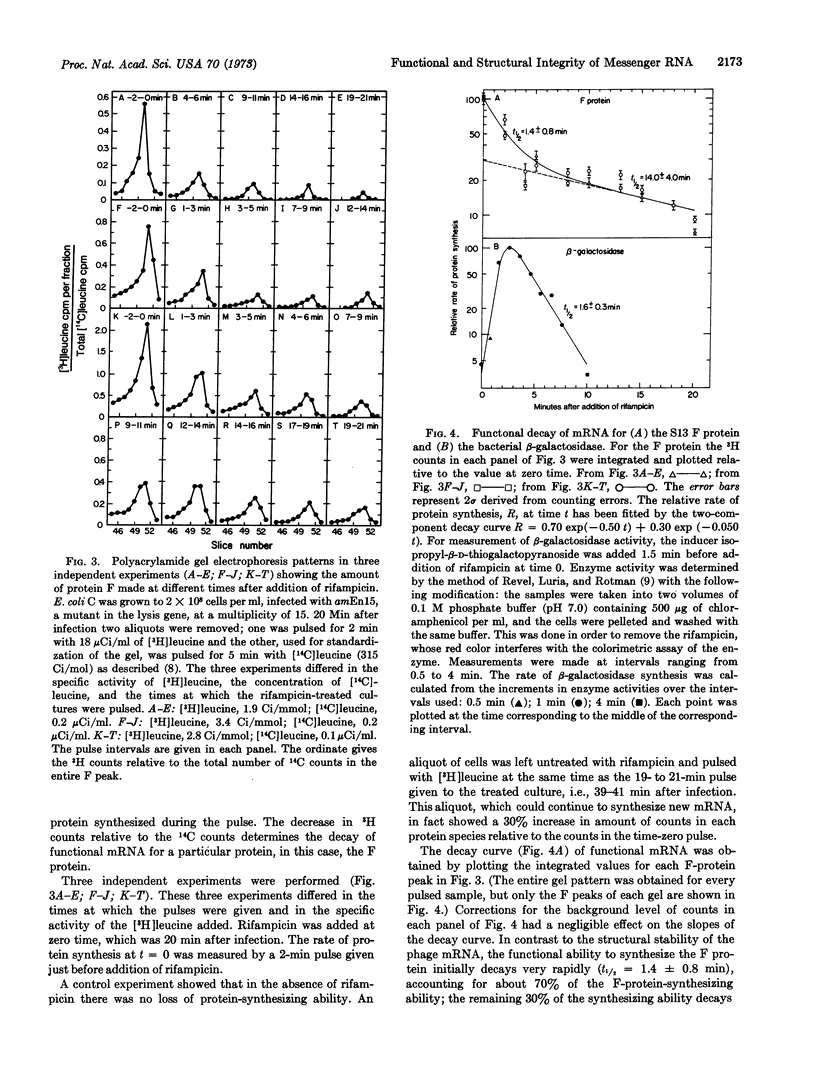
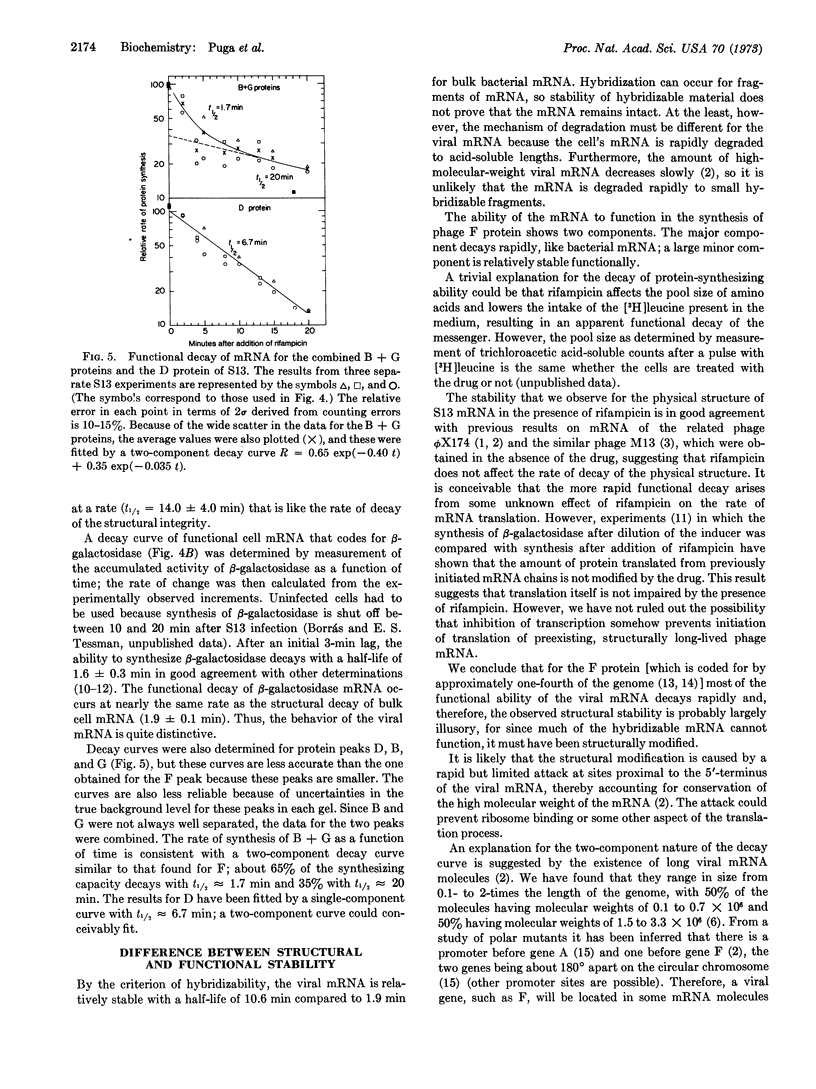
Messenger RNA molecules that are structurally stable, as measured by their ability to hybridize to DNA, may nevertheless be considerably less stable in retaining their ability to function in protein synthesis. The structure of the majority of the mRNA of phage S13 decays with a half-life of 10.6 ± 0.5 min. In contrast, much of the function of the mRNA that is involved in synthesis of a capsid protein (product of the F gene) decays rapidly with a half-life of 1.4 ± 0.8 min; a residual amount of function decays with a half-life of 14.0 ± 4.0 min. The measurements were made in the presence of rifampicin, which was used to prevent the formation of new mRNA. A proposed model for the functional decay is based on the polycistronic nature of the mRNA. Degradation of the mRNA would proceed in two steps: the first step would be a fast attack at a region near the 5′-terminus of each molecule that would eliminate the function of the proximal message; the second step would be a slow attack on the remaining messenger molecule precipitating a subsequent rapid degradation of the physical structure.
Keywords: S13 phage, Escherichia coli, long-lived mRNA, rifampicin, gel electrophoresis
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Burgess A. B., Denhardt D. T. Studies on phiX174 proteins. I. Phage-specific proteins synthesized after infection of Escherichia coli. J Mol Biol. 1969 Sep 28;44(3):377–386. doi: 10.1016/0022-2836(69)90367-2. [DOI] [PubMed] [Google Scholar]
- Carter T., Newton A. New polarity suppressors in Escherichia coli: suppression and messenger RNA stability. Proc Natl Acad Sci U S A. 1971 Dec;68(12):2962–2966. doi: 10.1073/pnas.68.12.2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelfand D. H., Hayashi M. Electrophoretic characterization of phiX174-specific proteins. J Mol Biol. 1969 Sep 28;44(3):501–516. doi: 10.1016/0022-2836(69)90376-3. [DOI] [PubMed] [Google Scholar]
- Hayashi M. N., Hayashi M. The stability of native DNA-RNA complexes during in vivo phiX-174 transcription. Proc Natl Acad Sci U S A. 1968 Nov;61(3):1107–1114. doi: 10.1073/pnas.61.3.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquet M., Kepes A. Initiation, elongation and inactivation of lac messenger RNA in Escherichia coli studied studied by measurement of its beta-galactosidase synthesizing capacity in vivo. J Mol Biol. 1971 Sep 28;60(3):453–472. doi: 10.1016/0022-2836(71)90181-1. [DOI] [PubMed] [Google Scholar]
- Jaenisch R., Jacob E., Hofschneider P. H. Replication of the small coliphage M13: evidence for long-living M13 specific messenger RNA. Nature. 1970 Jul 4;227(5253):59–60. doi: 10.1038/227059a0. [DOI] [PubMed] [Google Scholar]
- Kennell D., Bicknell I. Decay of messenger ribonucleic acid from the lactose operon of Escherichia coli as a function of growth temperature. J Mol Biol. 1973 Feb 15;74(1):21–31. doi: 10.1016/0022-2836(73)90351-3. [DOI] [PubMed] [Google Scholar]
- Kuwano M., Schlessinger D., Morse D. E. Loss of dispensable endonuclease activity in relief of polarity by suA. Nat New Biol. 1971 Jun 16;231(24):214–217. doi: 10.1038/newbio231214a0. [DOI] [PubMed] [Google Scholar]
- Marrs B. L., Yanofsky C. Host and bacteriophage specific messenger RNA degradation in T7-infected Escherichia coli. Nat New Biol. 1971 Dec 8;234(49):168–170. doi: 10.1038/newbio234168a0. [DOI] [PubMed] [Google Scholar]
- Morse D. E., Primakoff P. Relief of polarity in E. coli by "suA". Nature. 1970 Apr 4;226(5240):28–31. doi: 10.1038/226028a0. [DOI] [PubMed] [Google Scholar]
- Puga A., Tessman I. Mechanism of transcription of bacteriophage S13. II. Inhibition of phage-specific transcription by nalidixic acid. J Mol Biol. 1973 Mar 25;75(1):99–108. doi: 10.1016/0022-2836(73)90531-7. [DOI] [PubMed] [Google Scholar]
- REVEL H. R., LURIA S. E., ROTMAN B. Biosynthesis of B-D-galactosidase controlled by phage-carried genes. I. Induced beta-D-galactosidase biosynthesis after transduction of gene z-plus by phage. Proc Natl Acad Sci U S A. 1961 Dec 15;47:1956–1967. doi: 10.1073/pnas.47.12.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz T., Craig E., Kennell D. Inactivation and degradation of messenger ribnucleic acid from the lactose operon of Escherichia coli. J Mol Biol. 1970 Dec 14;54(2):299–311. doi: 10.1016/0022-2836(70)90431-6. [DOI] [PubMed] [Google Scholar]
- Summers W. C. The process of infection with coliphage T7. IV. Stability of RNA in bacteriophage-infected cells. J Mol Biol. 1970 Aug;51(3):671–678. doi: 10.1016/0022-2836(70)90015-x. [DOI] [PubMed] [Google Scholar]
- Summers W. C. Untranslated T7 phage mRNA is stabilized in suA host. Nat New Biol. 1971 Apr 14;230(15):208–208. doi: 10.1038/newbio230208a0. [DOI] [PubMed] [Google Scholar]
- Vanderbilt A. S., Borrás M. T., Germeraad S., Tessman I., Tessman E. S. A promoter site and polarity gradients in phage S13. Virology. 1972 Oct;50(1):171–179. doi: 10.1016/0042-6822(72)90357-1. [DOI] [PubMed] [Google Scholar]
- Vanderbilt A. S., Borrás M. T., Tessman E. S. Direction of translation in phage S13 as determined from the sizes of polypeptide fragments of nonsense mutants. Virology. 1971 Feb;43(2):352–355. doi: 10.1016/0042-6822(71)90307-2. [DOI] [PubMed] [Google Scholar]
- Varmus H. E., Perlman R. L., Pastan I. Regulation of lac transcription in antibiotic-treated E. coli. Nat New Biol. 1971 Mar 10;230(10):41–44. doi: 10.1038/newbio230041a0. [DOI] [PubMed] [Google Scholar]



