Abstract
Published studies have identified genetic variants, somatic mutations, and changes in gene expression profiles that are associated with hepatocellular carcinoma (HCC), particularly involving genes that encode drug metabolizing enzymes (DMEs). CYP2C9, one of the most abundant and important DMEs, is involved in the metabolism of many carcinogens and drugs and is down-regulated in HCC. To investigate the molecular mechanisms that control CYP2C9 expression, we applied integrative approaches including in silico, in vitro, and in vivo analyses to elucidate the role of microRNA hsa-miR-128-3p in the regulation of CYP2C9 expression and translation. RNA electrophoresis mobility shift assays demonstrated a direct interaction between hsa-miR-128-3p and its cognate target, the CYP2C9 transcript. Furthermore, the expression of a luciferase reporter gene containing the 3′-UTR of CYP2C9 and the endogenous expression of CYP2C9 were suppressed by transfection of hsa-miR-128-3p. Importantly, chemically-induced up- or down-regulation of hsa-miR-128-3p correlated inversely with the expression of CYP2C9. Finally, an association analysis revealed that the expression of hsa-miR-128-3p is inversely correlated with the expression of CYP2C9 in HCC tumor tissues. Altogether, the study helped to elucidate the mechanism of CYP2C9 regulation by hsa-miR-128-3p, and the inverse association in HCC.
Primary liver cancer is the fifth most common cancer and third leading cause of cancer-related death worldwide; hepatocellular carcinoma (HCC) is the predominant subtype, accounting for approximately 85%–90% of total primary liver cancer cases1. Epidemiological studies have revealed several risk factors associated with the etiology of HCC, including geographic region, socio-economic status, gender, ethnicity and environmental exposures1. It is well documented that hepatitis B virus (HBV) or hepatitis C virus (HCV) infection, dietary aflatoxin B1 contamination, chronic alcohol abuse and tobacco consumption, lack of dietary antioxidants, environmental arsenic exposure, obesity, and non-alcoholic fatty liver disease are major risk factors for HCC2. These findings indicate that chronic xenobiotic stress plays a pivotal role in the development of HCC, although the precise molecular mechanisms are poorly understood.
During the past decades, advancing technologies, such as high-throughput microarray and next-generation sequencing, have led to the exploration of the relationship between environmental stress and cancer susceptibility by identifying genetic variants, somatic mutations, gene expression profiles and specific xenobiotic exposures associated with cancer etiologies3. For example, it is now thought that the major function of drug metabolizing enzymes (DMEs) is the regulation of cell growth, apoptosis, differentiation and homeostasis, while approximately 1% of their function is devoted to the metabolism of xenobiotics4. Genetic polymorphisms in DME genes are convincingly found to be associated with risk of toxicity and/or cancer5,6, including the association with the risk of HCC7. Studies also have shown that dysregulation of DMEs, including several cytochrome P450 (CYP) family genes such as CYP2A6, CYP2C9, CYP2E1, CYP3A58,9,10, might play important roles in the development of HCC.
The human CYP enzyme superfamily is comprised of at least 57 distinct genes in 18 families that are classified as drug/xenobiotic metabolizing enzymes due to their pivotal role in protecting the organism from xenobiotics and environmental toxins by metabolizing them into inactive compounds. However, CYP450 enzyme activity also catalyzes the formation of reactive metabolites that cause DNA, RNA or protein damage11. Alteration in the expression CYP genes can affect the efficiency of xenobiotic detoxification and also the production of messenger molecules that regulate downstream signal-transduction pathways, thus having a paradoxical impact in carcinogenesis11. Therefore, it is reasonable to speculate that the dysregulation of CYP family genes might be involved in hepatocarcinogenesis. CYP2C9, one of the most abundant CYP2C proteins, accounts for ~20% of hepatic CYP content and contributes to the metabolism of many carcinogens and drugs12,13. Suppression of CYP2C9 expression has been reported as a biomarker of HCC14,15,16,17; however, the mechanism of CYP2C9 dysregulation through microRNA (miRNAs) modulation has not been investigated in HCC.
The miRNAs are small RNA molecules that modulate gene expression through translational repression of cognate mRNA targets and thus are important mediators in gene regulatory networks. Consequently, miRNA-dependent modulation of the expression of DMEs among individuals could lead to substantial changes in phenotypes. Such changes may have a significant influence on quantitative traits, including the development of cancer and other diseases18. In the last decade, many human DMEs and nuclear receptors have been reported to be regulated by miRNAs, such as CYP1B1, CYP2E1, CYP3A4, CYP 24A1, SUL1A1, PXR and VDR19. The miRNA-dependent regulation of DMEs and transporters should have potential effects in modulation of carcinogen metallization-either activation or detoxification, thus may be associated with cancer development. For example, evidences demonstrated that CYP1B1 is regulated by miR-27b, and a decreased miR-27b expression is inversely associated with an increased protein level of CYP1B1 in breast cancer patients20. Pathogenically, although the high level of CYP1B1 may decrease the estrogen activity by CYP1B1-mediated 4-hydroxylation, the increased metabolite 4-hydroxyestadiol level may contribute more significantly to breast carcinogenesis19.
There are several hundred miRNAs expressed in human tissues, and characterization of miRNA expression patterns in human tumor tissues reveal miRNA signatures that are associated with tumor initiation, progression metastasis, diagnosis, prognosis, response to treatment and survival21; however, there is a lack of conclusive information on the role for miRNA modulation of drug metabolizing genes in the pathogenesis of HCC.
In the current study, we hypothesized that hsa-miR-128-3p22,23,24 and hsa-miR-143-3p25,26, which play important roles in cancer, could influence CYP2C9 expression by targeting its mRNA transcripts based on in silico analyses of putative mRNA/miRNA complexes. To test this hypothesis, we employed a series of biochemical assays to investigate the interaction of these two miRNAs with the CYP2C9 transcript and identified hsa-miR-128-3p as an efficient suppressor of CYP2C9 expression.
Results
Bioinformatic identification of candidate miRNAs targeting CYP2C9
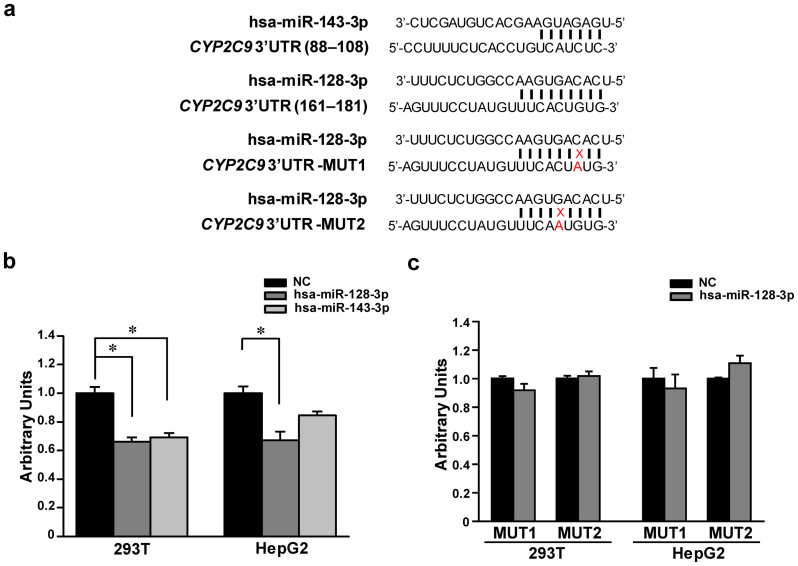
Three public databases, microRNA.org, PITA and TargetScan, applying different evaluation criteria, were used to screen for miRNAs with the potential to bind the 3′-UTR of CYP2C9. Several miRNAs, including hsa-miR-128-3p and hsa-miR-143-3p, were identified in all three databases. Due to the reported roles of hsa-miR-128-3p22,23,24 and hsa-miR-143-3p25,26 in cancer, these two miRNAs were selected as candidate miRNAs for this study (Fig. 1a).
Figure 1. The hsa-miR-128-3p and hsa-miR-143-3p suppressed reporter gene expression.
(a) Prediction of hsa-miR-143-3p targeting to the 3′-UTR of CYP2C9, and hsa-miR-128-3p targeting to the 3′-UTR of CYP2C9, as well as the “targeting” of hsa-miR128-3p to two mutated 3′UTR sequences of the CYP2C9 gene. The solid vertical line indicates base pairing and the numbering used for the hsa-miR-128-3p and hsa-miR-143-3p target sequences (88–108 or 161–181, respectively) is consistent with that given for the CYP2C9 3′-UTR in NM_000771. (b) Luciferase reporter assays to investigate the effects of the hsa-miR-128-3p and hsa-miR-143-3p on CYP2C9 3′-UTR. (c) Luciferase reporter assays to investigate the effect of hsa-miR-128-3p on mutated CYP2C9 3′-UTRs. 293T and HepG2 cells were transiently transfected with the CYP2C9- CU, CYP2C9-MUT1-CU (containing a mutated sequence in the 3′UTR of CYP2C9), or CYP2C9-MUT2-CU (containing another mutated sequence in the 3′-UTR of CYP2C9) plasmid, together with 50 nmol/L hsa-miR-128-3p mimic, hsa-miR-143-3p mimic, or miRNA negative control, respectively, and harvested 48 hours after transfection. Renilla luciferase activity was measured, and then normalized to firefly luciferase. Three independent transfection experiments were carried out, and each was performed in triplicate. Data are shown as relative activity to luciferase activity expressed by the CYP2C9-CU plasmid transfection together with miRNA negative control. * P < 0.05; NC, miRNA negative control.
hsa-miR-128-3p and hsa-miR-143-3p suppress CYP2C9 3′-UTR luciferase reporter
To test the potential effects of hsa-miR-128-3p and hsa-miR-143-3p on CYP2C9 expression, a reporter construct retaining the core region of the CYP2C9 3′-UTR that harbors the putative binding sites for hsa-miR-128-3p and hsa-miR-143-3p was constructed and then co-transfected into HepG2 and 293T cells together with the hsa-miR-128-3p mimic, hsa-miR-143-3p mimic, or miRNA negative control. Fig. 1b showed that high concentration of exogenous hsa-miR-128-3p (the amount of exogenous hsa-miR128-3p was evaluated by RT-PCR, for which please see Supplementary Fig. S1) efficiently suppressed luciferase activity in HepG2 and 293T cells (34% and 33%, respectively; all P < 0.001) as compared with the transfection of miRNA negative control. In contrast, hsa-miR-143-3p exhibited a similar significant suppression effect in 293T cells only (31%, P < 0.001), while the effect in HepG2 cells was marginally significant (14%, P = 0.062).
To investigate the specificity of hsa-miR-128-3p suppressing CYP2C9 3′UTR, CYP2C9-MUT1-CU and CYP2C9-MUT2-CU constructs (with mutated hsa-miR-128-3p targeting sequences in the CYP2C9 3′UTR), were created and applied in the reporter gene assays. As shown in Fig. 1c, transfection with the hsa-miR-128-3p mimic did not suppress luciferase activity from CYP2C9-MUT1-CU or CYP2C9-MUT2-CU construct, indicating that hsa-miR-128-3p modulates CYP2C9 expression in a sequence specific manner by targeting its non-mutated 3′UTR.
Interaction between CYP2C9 3′-UTR and hsa-miR-128-3p or hsa-miR-143-3p
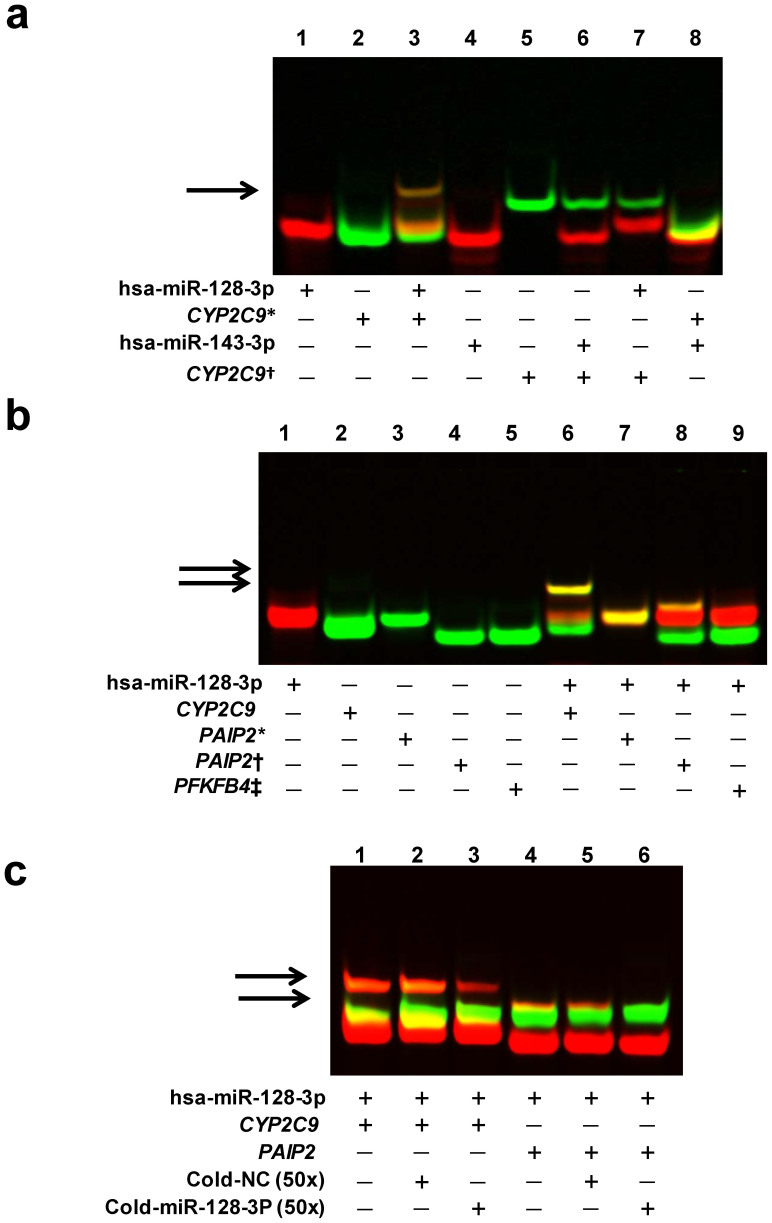
In silico analysis predicted that hsa-miR128-3p and hsa-miR143-3p might form complexes with target sequences present in the 3′-UTR of CYP2C9 to yield unique structures with different calculated free energies of binding: −23.9 kcal/mol for hsa-miR128-3p (Supplementary Fig. S2a) and −16.1 kcal/mol for hsa-143-3p (Supplementary Fig. S2b). To determine whether or not hsa-miR-128-3p or hsa-miR-143-3p is able to bind its cognate CYP2C9 3′-UTR mRNA sequence in vitro, RNA EMSA was performed. Fig. 2a shows that hsa-miR-128-3p was able to bind the corresponding target sequence of CYP2C9 3′-UTR (lane 3) and competition assays showed the binding is sequence-specific (lanes 7, 8). There was no evidence of an interaction between CYP2C9 3′-UTR and hsa-miR-143-3p (Fig. 2a).
Figure 2. Free energy directly affected the interaction between hsa-miR-128-3p and its counterparts.
(a) RNA EMSA with the cy5.5TM-labeled hsa-miR-128-3p or hsa-miR-143-3p oligonucleotides, and 2′ O-Methyl modified and IRDye®800-labeled CYP2C9 mRNA oligonucleotides. Lanes 1, 2, 4, and 5 show mobility of the labeled oligonucleotides; Lanes 3 and 6 show mobility of the labeled miRNA oligonucleotides with corresponding CYP2C9 mRNA oligonucleotides; Lanes 7 and 8 show mobility of the labeled miRNA oligonucleotides with unmatched CYP2C9 mRNA oligonucleotides (negative controls). Arrow indicates an oligonucleotide complex (yellow) in Lane 3. * or † indicates the CYP2C9 mRNA oligonucleotides retaining the hsa-miR-128-3p or hsa-miR-143-3p recognition sites, repectively. (b) RNA EMSA with the cy5.5TM-labeled hsa-miR-128-3p probe, and 2′ O-Methyl modified and IRDye®800-labeled CYP2C9, PAIP2, or PFKFB4 mRNA oligonucleotides. Lanes 1, 2, 3, 4, and 5 show mobility of the labeled oligonucleotides; Lanes 6, 7, 8 and 9 show mobility of the labeled hsa-miR-128-3p oligonucleotide with corresponding CYP2C9, PAIP2, or PFKFB4 mRNA probes with different free energy. Arrows indicate the oligonucleotide complex (yellow) in Lane 6 and 8. *, †, and ‡ indicate two PAIP2 mRNA oligonucleotides, and one PFKFB4 mRNA oligonucleotide with the binding free energy of −18.0 kcal/mol, −20.5 kcal/mol, and −15.8 kcal/mol, respectively. (c) RNA EMSA with the cy5.5TM-labeled hsa-miR-128-3p probe, and 2′ O-Methyl modified and IRDye®800-labeled CYP2C9 or PAIP2 mRNA oligonucleotides. Lanes 1 and 4 show mobility of the labeled hsa-miR-128-3p oligonucleotide with corresponding CYP2C9, and PAIP2 mRNA probes with the free energy of −23.9 kcal/mol and −20.5 kcal/mol, respectively. Lanes 2, 3, 5 or 6 show mobility of the labeled hsa-miR-128-3p oligonucleotide with corresponding CYP2C9 and PAIP2 mRNA probes, in the presence of unlabeled excess nonspecific competitor (Cold-NC) and excess specific competitor (Cold hsa-miR-128-3p), respectively. NC, nonspecific competitor. Arrows indicate the oligonucleotide complex (yellow) in Lane 1, 2, 3, 4 and 5.
Compared to the related hsa-miR-143-3p complex, the enhanced thermodynamic stability predicted for the hsa-miR-128-3p complex with its CYP2C9 3′-UTR target sequence correlated with results from RNA EMSA experiments, confirming the in vitro stability of the latter complex only. To explore the applicability of this predictive strategy further, we calculated the free energy of binding between hsa-miR-128-3p and 3 other putative target sequences (2 probes from the PAIP2 3′-UTR and 1 probe from the PFKFB4 3′-UTR). The predicted free energies of binding were −20.5 kcal/mol, −18.0 kcal/mol, and −15.8 kcal/mol for miR-128-PAIP2-target2, miR-128-PAIP2-target1 and miR-128-PFKFB4-target, respectively. EMSA assays were then conducted with these miRNAs and their targets. The hsa-miR-128-3p mimic was able to bind the probe with the free energy of −20.5 kcal/mol (Fig. 2b, lane 8), but not the ones with free energy of −18.0 kcal/mol, or −15.8 kcal/mol (Fig. 2b, lane 7 or 9), showing a correlation between the predicted free energy of binding directly and the observed interaction between miRNAs and their counterparts, which we propose should also correlate with the efficiencies of gene regulation by these miRNAs.
In addition, we found that the miRNA-mRNA complex, formed by hsa-miR-128-3p with CYP2C9 3′UTR, or with PAIP2 3′UTR, could be eliminated by adding 50× unlabeled hsa-miR-128-3p probe(Fig. 2c, lane 3 or 6), but not by adding a nonspecific probe (Cold-NC) (Fig. 2c, lane 2 or 5), suggesting the binding between hsa-miR-128-3p and its cognate 3′UTR of CYP2C9 or PAIP2 is in a sequence-specific manner.
Suppression of endogenous CYP2C9 expression/translation by exogenous hsa-miR-128-3p
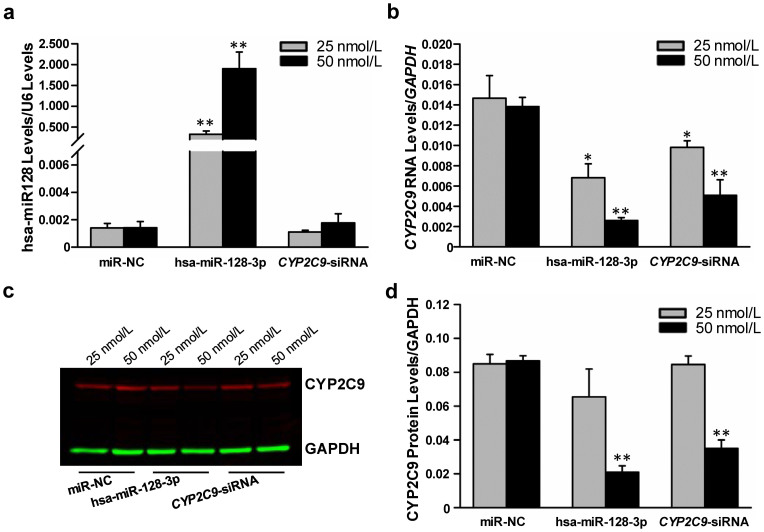
It was reported that HepaRG cells express CYP2C9 at a level similar to that found in primary hepatocytes, while the HepG2 cells barely express CYP2C9 owing to the lack of nuclear factors and other transcription factors for the expression of DMEs27. We did observed that the inherent CYP2C9 mRNA level in HepaRG cells was ~10-fold and ~20-fold greater than those in 293T cells and HepG2 cells, respectively, while the hsa-miR-128-3p was expressed in a similar level among these cell lines (Supplementary Fig. 3a). To investigate the effects of hsa-miR-128-3p on endogenous CYP2C9 transcripts and protein levels, hsa-miR-128-3p mimics, miRNA negative control or a CYP2C9-specific siRNA that served as the positive control, were transiently transfected into HepaRG cells. Fig. 3a shows that the level of hsa-miR-128-3p was dramatically increased by more than 200-fold after transfection with hsa-miR-128-3p mimics at concentrations of 25 nmol/L or 50 nmol/L. Consequently, CYP2C9 mRNA expression was significantly decreased compared to that in cells transfected with the miRNA negative control (53.7% at 25 nmol/L and 81.1% at 50 nmol/L; all P < 0.05) (Fig. 3b). In addition, the CYP2C9 protein level decreased significantly (76.7%) when the cells were transfected with 50 nmol/L hsa-miR-128-3p mimics in comparison with the transfection of the miRNA negative control (Fig. 3c and 3d). Notably, transfection of hsa-miR-128-3p was more efficient at suppressing CYP2C9 protein expression than transfection of the CYP2C9-specific siRNA positive control (Fig. 3b, 3c and 3d).
Figure 3. The hsa-miR-128-3p suppressed endogenous CYP2C9 expression in HepaRG cells.
Differentiated HepaRG cells were transiently transfected with 25 nmol/L or 50 nmol/L hsa-miR-128-3p mimic, CYP2C9-specific siRNA, or miRNA negative control, respectively, and harvested 48 hours after transfection. Each assay was done on at least 3 independent transfection experiments. NC, miRNA negative control; * P < 0.05; ** P < 0.001. (a) Unregulated expression of hsa-miR-128-3p. Data were shown as relative hsa-miR-128-3p levels versus U6. (b) Down-regulated expression of CYP2C9 mRNA by hsa-miR-128-3p or CYP2C9-specific siRNA. Data are shown as relative CYP2C9 mRNA levels versus GAPDH reference. (c) and (d) Down-regulated CYP2C9 protein levels by hsa-miR-128-3p or CYP2C9-specific siRNA.
PTEN was proved to be negatively associated with hsa-miR-128-3p level in pituitary cells, probably due to hsa-miR-128-3p targeting BMI1, one transcriptional suppressor of PTEN28. As a positive control, we observed that the PTEN mRNA level was decreased significantly after hsa-miR-128-3p transfection, compared with that in cells transfected with the miRNA negative control (40.1% at 25 nmol/L and 46.6% at 50 nmol/L; all P < 0.05) (Supplementary Fig. S4a), suggesting that our transfection experimental system is validated.
Modulation of CYP2C9 expression/translation through chemically-induced alteration of hsa-miR-128-3p levels
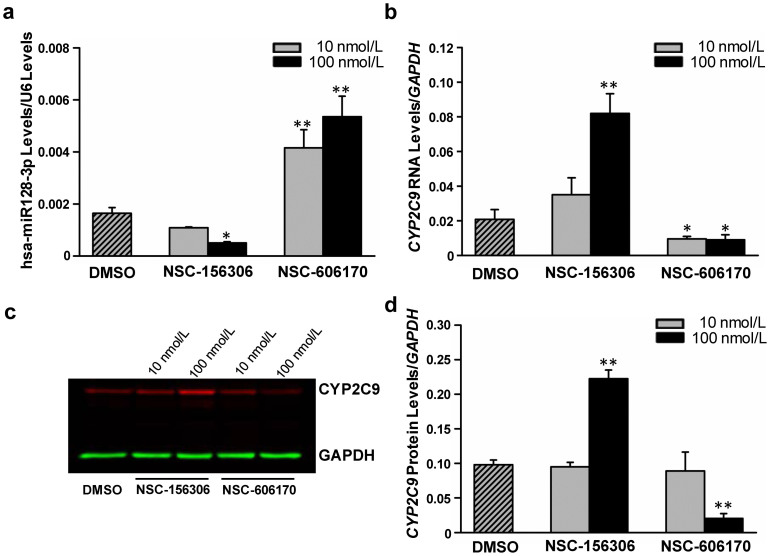
According to the CellMiner™ database (version 1.5, http://discover.nci.nih.gov/cellminer), chemical compounds NSC-156306 and NSC-606170 appear to inhibit or induce, respectively, the expression of hsa-miR-128 in human liver tissue. Therefore, we treated HepaRG cells with these compounds to determine their impact on CYP2C9 transcription and translation. Fig. 4 shows that treatment of the HepaRG cells with 100 nmol/L NSC-156306 resulted in a significant decrease in the expression of hsa-miR-128-3p and that its level was markedly increased after treating cells with NSC-606170 (Fig. 4a). The treatment-induced hsa-miR-128-3p expression changes were accompanied by inverse alterations in the expression of CYP2C9 gene expression (Fig. 4b) and protein production (Fig. 4c and 4d). Specifically, the levels CYP2C9 mRNA and protein in the NSC-156306-treated HepaRG cells were 3-fold and ~2-fold greater (Fig. 4b, 4c and 4d). In contrast, the levels of CYP2C9 mRNA and protein were dramatically decreased, by 81.1% and 79.6%, in cells treated with NSC-606170 (Fig. 4b, 4c and 4d). In addition, we also observed that the PTEN mRNA level was decreased significantly after NSC-606170 treatment in HepaRG cells (67.6% at 10 nmol/L and 75.3% at 100 nmol/L; all P < 0.05) (Supplementary Fig. S4b), suggesting that the modulation of hsa-miR-128-3p by NSC-606170 was obtained.
Figure 4. Chemical compounds NSC-156306 and NSC-606170 affected endogenous CYP2C9 expression by altering the expression of hsa-miR-128-3p in HepaRG cells.
Differentiated HepaRG cells were treated with 0, 10 nmol/L, or 100 nmol/L NSC-156306 or NSC-606170, and harvested 48 hours after treatments. Each assay was performed from at least 3 independent experiments. * P < 0.05; ** P < 0.001. (a) Dysregulated expression of hsa-miR-128-3p. Data are shown as relative hsa-miR-128-3p levels versus U6. (b) Dysregulated expression of CYP2C9 mRNA by NSC-156306 or NSC-606170 treatment. Data are shown as relative CYP2C9 mRNA levels versus GAPDH reference. (c) and (d) Dysregulated CYP2C9 protein levels by NSC-156306 or NSC-606170 treatment.
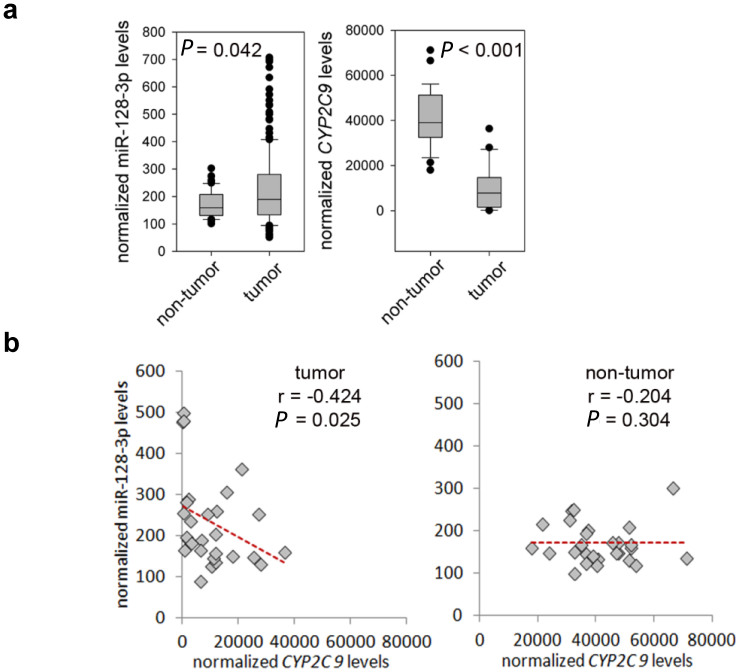
Inverse correlation between hsa-miR-128-3p and CYP2C9 expression in HCC
The expression levels of hsa-miR-128-3p and CYP2C9 mRNA in human HCC tumor tissue samples and adjacent normal liver samples were extracted from The Cancer Genome Atlas (TCGA) database. Levels of hsa-miR-128-3p measured in tumor tissues were significantly higher than those measured in matched non-tumor tissues; whereas, levels of CYP2C9 were significantly reduced in tumor tissues compared to surrounding non-tumor tissues (Fig. 5a). The relationship between hsa-miR-128-3p and CYP2C9 expression was evaluated by the Spearman Rank Order Correlation analysis in patient matched tumor and non-tumor tissues. In tumor tissues, there is a negative correlation between hsa-miR-128-3p and CYP2C9 (r = −0.424, P = 0.025, Fig. 5b), but in non-tumor tissues there is no significant correlation (r = −0.204, P = 0.304, Fig. 5b). Interestingly, no statistically significant correlation between PTEN mRNA (as a known miR-128 target) levels and hsa-miR-128-3p levels was observed in those tissues (Supplementary Fig. S5), probably due to the complexity and high heterogeneity of gene expression in tumor tissue. When hsa-miR-143-3p was examined, no statistically significant results were obtained (Data not shown).
Figure 5. Relationship between CYP2C9 mRNA expression and hsa-miR-128-3p level in HCC and paired non-tumor tissues.
(a) Expression data from HCC tumor and non-tumor samples stored in The Cancer Genome Atlas (TCGA) was downloaded and compared using the Rank Sum Test. There were 28 and 27 tumor and non-tumor samples respectively that had expression data for hsa-miR-128-3p and CYP2C9. Data has been normalized to reads per million. (b) hsa-miR-128-3p levels correlate with CYP2C9 levels in tumor tissues (r = −0.424, P = 0.025, 28 matched tumor samples), but there is no significant correlation in the non-tumor tissues (r = −0.204, P = 0.304, 27 matched non-tumor samples).
Discussion
Inter-individual variability in the expression of DMEs in humans is an important phenotypic trait that may contribute significantly to disparities in disease susceptibility and drug efficacy29,30,31. The mechanism underlying the phenotype is due, to a certain extent, to genetic polymorphisms and epigenetic variation among human populations. Genetic polymorphism(s) could alter gene transcription or change an enzyme's catalytic activity, while miRNAs regulate gene expression by targeting the mRNAs, repressing protein translation or accelerating mRNA degradation. In this study, we demonstrated that hsa-miR-128-3p plays an important role in the suppression of CYP2C9 expression and translation in human liver cells by a series of in silico analyses and in vitro and in vivo experiments. This study helped to elucidate the functional mechanism by which miRNA regulates CYP2C9 expression and translation. In addition, a modified RNA EMSA method provided direct evidence for the interaction between miRNAs and their cognate mRNA sequences that was dependent on the predicted free energy of binding.
CYP2C9 is one of the most abundant and important xenobiotic metabolizing enzymes, with substrates including commonly prescribed drugs such as warfarin, NSAIDs (non-steroidal anti-inflammatory drugs), tolbutamide, phenytoin, and torasemide13. CYP2C9 is also reported to participate in the bioactivation of carcinogens. For example, metabolism of benzo[a]pyrene by CYP2C9 results in the formation of 9-hydroxybenzo[a]pyrene-4,5-oxide and benzo[a]pyrene-7,8-diol-9,10-epoxide, which are reactive species involved in DNA adduct formation32,33,34. In addition, a high activity CYP2C9 genotype (CYP2C9*1) is associated with increased risk of colorectal cancer35 while a low activity CYP2C9 genotype (CYP2C9*2) is associated with increased risk of colorectal adenoma36 and lung cancer37. Furthermore, decreased CYP2C9 expression was reported in HCC tissue by several studies14,15,16,17, suggesting the role of CYP2C9 in detoxification may be involved in the etiology of HCC; however, the mechanisms controlling CYP2C9 expression are not fully understood.
In this study, we demonstrated that hsa-miR128-3p plays a pivotal role in the regulation of CYP2C9 expression in human liver using series of in silico, in vitro, and in vivo analyses. It is well known that the interaction between miRNAs and their cognate mRNA targets is complicated, and there are more than ten algorithms for predicting miRNA targets38. Therefore, for a given miRNA or transcript, many “putative” or “potential” interactions are predicted by in silico approaches. To reduce false positive predictions and validate true interactions, we first screened the miRNAs that could potentially target the 3′-UTR of CYP2C9 mRNA using the microRNA.org database, and then evaluated the resultant candidate miRNAs in the PITA and TargetScan databases. The candidate miRNAs that were predicted by all three database algorithms to target CYP2C9 mRNA were then experimentally examined by both in vivo and in vitro approaches for functional interaction with CYP2C9 mRNA. The selection of hsa-miR-128-3p and hsa-miR-143-3p as candidate miRNAs for further biochemical characterization was based on their reported biological significance in tumorigenesis and metastasis22,23,24,25,26. For example, hsa-miR-128 is aberrantly expressed in many types of tumors, including acute lymphoblastic leukemia, glioblastoma, and breast cancer. By targeting EGFR, Bim-1, ABCC5 and other genes, hsa-miR-128 is involved in tumor differentiation, proliferation, invasion, apoptosis and resistance to drugs39. The miRNA hsa-miR-143 was reported as a tumor suppressor in cervical cancer40 and prostate cancer41, by suppressing KRAS, ERK5, and other genes.
We first transfected the hsa-miR-128-3p or hsa-miR-143-3p mimics into liver HepG2 and kidney 293T cells, together with a reporter gene (luciferase) plasmid containing the core region of CYP2C9 3′-UTR, and found that hsa-miR-128-3p suppressed luciferase activity in both liver cells and kidney cells, while hsa-miR-143-3p exhibited a relatively smaller suppression effect only in kidney cells. RNA EMSA assays revealed that hsa-miR-128-3p bound CYP2C9 mRNA, while an interaction between hsa-miR-143-3p and the CYP2C9 3′-UTR was not detected. Further, we used RNA EMSA to test the binding efficiencies between hsa-miR-128-3p and three other target sequences with different free energies of binding predicted by the RNAhybrid software. It was observed that only the probes with free energy of less than −20 kcal/mol could bind hsa-miR-128-3p under our experimental conditions. Therefore, we postulate that the binding efficiency between miRNAs and their cognate mRNA targets is mainly dependent on the free energy state of the binding. Althogh further studies on the precise mechanisms of miRNA targeting mRNA sequences are under way, our current results provide evidence that the free energy of binding is important for accurate predictions of miRNA targeting sites.
Because of the low expression of DMEs and transporters by HepaG2 and other hepatocellular carcinoma cell lines30,42, we used HepaRG cells, which express DMEs and transporters at levels similar to primary hepatocytes, to investigate the suppression effects of hsa-miR-128-3p on endogenous CYP2C9 expression and translation. Our results showed that enforced up-regulattion of hsa-miR-128-3p reduced CYP2C9 production at both the protein and mRNA levels, indicating that hsa-miR-128-3p is at least involved in CYP2C9 mRNA degradation. Two chemicals, with strong negative or positive effect on hsa-miR-128 expression, were used to produce CYP2C9 alterations through the modulation of hsa-miR-128 expression. The results confirmed that the dramatic changes in hsa-miR-128-3p expression caused by these two compounds (Fig. 4a) indeed altered the CYP2C9 expression/production inversely (Fig. 4b, 4c and 4d), which is consistent with the transfection assays. Altogether, our results indicated that hsa-miR-128-3p is able to suppress CYP2C9 expression/production in human hepatic cells by specifically targeting the 3′-UTR of CYP2C9 mRNA molecules.
Finally, the correlation between the expression of CYP2C9 and hsa-miR-128-3p in HCC tissues was evaluated using the GSE22058 and TCGA datasets. We observed that CYP2C9 mRNA was significantly down-regulated in HCCs, consistent with findings reported by others14,15,16,17. Most importantly, this study revealed an up-regulation of hsa-miR-128-3p expression and a significant inverse correlation between CYP2C9 expression and hsa-miR-128-3p expression in HCC tissues, indicating that hsa-miR-128-3p combined with CYP2C9 are potential biomarkers for HCC diagnosis.
In summary, our study identified and experimentally confirmed that hsa-miR-128-3p is a suppressor for CYP2C9 expression in HCC, and revealed a direct interaction between a miRNA and its target mRNA sequence in vitro, which demonstrated a molecular mechanism of miRNA mediated CYP2C9 suppression.
Methods
Cell lines
HepG2 and 293T cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA) and HepaRG cells were obtained from Biopredic International (Overland Park, HS). The cells were maintained according to ATCC and Biopredic International's recommendations.
In silico analyses
The public databases microRNA.org (http://www.microrna.org/), PITA (http://genie.weizmann.ac.il/pubs/mir07/mir07_prediction.html) and TargetScan (Release 6.2, http://www.targetscan.org) were screened to identify potential miRNA response elements (MREs) located in the 3′-UTR of the CYP2C9 gene. The RNAhybrid program43 was used to calculate the free energies of binding for predicted miRNA:RNA duplexes formed between hsa-miR-128-3p and CYP2C9, PAIP2 or PFKFB4 mRNA sequences. The CellMiner™ database (version 1.5, http://discover.nci.nih.gov/cellminer), which integrates the molecular and pharmacological data sets for the NCI-60 cell lines, was used to select the chemical compounds that have demonstrated positive or negative correlations with hsa-miR-128-3p expression.
Luciferase reporter gene assay
The pGL3-Control vector (Promega, Madison, WI) was modified by adding the Universal USER Cassette (New England Biolabs, Ipswich, MA), similar to the modification of the pGL3-Promoter44. Briefly, the Xba I site was digested and blunted by using the Quick Blunting Kit (New England Biolabs). The USER Cassette sequence was then inserted, resulting in the pGL3-CU vector. Cloning primers CYP2C9-F and CYP2C9-R (All primer or oligo sequences used in this study were listed in Supplementary Table S1), with extension oligonucleotides 5′-GGA GAC AU-3′ or 5′-GGG AAA GU-3′ in their 5′ end, were designed to amplify the core region of CYP2C9 3′-UTR that harbors the putative binding sites for hsa-miR-128-3p and hsa-miR-143-3p. PCR products were digested with USER enzyme (New England Biolabs) and cloned into the linearized nicked pGL3-CU vector that was prepared following the Universal USER Cassette protocol. The DNA sequence of the resultant plasmid, designated CYP2C9-CU, was determined to confirm its identity. Besides, CYP2C9-MUT1-CU or CYP2C9-MUT2-CU construct, which with mutated hsa-miR-128-3p target sequences in the CYP2C9 3′UTR, was created by site-directed mutagenesis using CYP2C9-MUT1-F and CYP2C9-MUT1-R primers, or CYP2C9-MUT2-F and CYP2C9-MUT2-R primers, respectively.
HepG2, a human hepatoma cell line, and 293T, a human embryonic kidney line, were used for luciferase assays. HepG2 or 293T cells were cultured in Rosewell Park Memorial Institute 1640 or Dulbecco's Modified Eagle medium with 10% fetal bovine serum. Cells were seeded in 96-multiwell plates. When the cells reached 70%–80% confluence, they were transfected with the CYP2C9-CU plasmid (100 ng/well) that contains the 3′-UTR of CYP2C9 together with 50 nmol/L (final concentration) hsa-miR-128-3p mimic, hsa-miR-143-3p mimic, or miRNA negative control (Thermo Scientific, Tewksbury, MA) using the Lipofectamine reagent 2000 (Life Technologies, Carlsbad, CA). The pRL-SV40 (1 ng/well; Promega) plasmid, which expresses Renilla reniformis luciferase, was co-transfected to standardize transfection efficiency. The CYP2C9-CU plasmid co-transfected with pRL-SV40 plasmid and miRNA negative control served as a reference. Three independent transfection experiments were carried out, and each transfection was performed in triplicate.
RNA electrophoretic mobility shift assay (EMSA)
The miRNA oligonucleotides hsa-miR-128-3p and hsa-miR-143-3p were labeled with cy5.5™ dye on their 5′ ends. The 2′ O-methyl-modified mRNA oligonucleotides miR-128-CYP2C9-target, miR-128-PAIP2-target1, miR-128-PAIP2-target2, miR-128-PFKFB4-target, and miR-143-CYP2C9-target were labeled with IRDye®800 (LI-COR Biosciences, Lincoln, NE) dye on their 5′ ends. All primers and oligonucleotides used in this study were synthesized by Integrated DNA Technologies (Coraville, IA).
RNA EMSAs were performed according to the LightShift Chemiluminescent RNA EMSA Kit (Thermo Scientific) protocol. Briefly, in each 20 μL binding reaction containing 200 nmol synthetic miRNA or/and cognate mRNA binding oligonucleotides were mixed, heated for 5 minutes at 80°C, placed on ice to relax RNA secondary structures, and then incubated at 25°C for 20 min. The reaction mixtures were separated on a 12% PAGE by electrophoresis at 4°C, and the resultant mobility shifts were detected with an Odyssey CLx Infrared Imaging System (LI-COR Biosciences, Lincoln, NE).
Transfection of HepaRG cells with hsa-miR-128-3p and treatments with chemical compounds
HepaRG cells (Biopredic International, Overland Park, KS) were first incubated in Williams' E medium supplemented with growth supplement (Biopredic International) for 2 weeks and then cells were differentiated by adding the differentiation supplement (Biopredic International) for 10 additional days. Differentiated cells were then seeded into 6-well plates at a density of 70,000 cells/cm2 with 3 ml medium and incubated for another 2 days for further experiments.
The miRNA transfection was performed as previously reported45. Briefly, 25 nmol/L or 50 nmol/L (final concentration) hsa-miR-128-3p mimic, miRNA negative control, or CYP2C9-specific siRNA (positive control), was transfected into the differentiated HepaRG cells using Lipofectamine transfection reagent (Life Technology), and cells were harvested 48 hours after transfection. Chemical compounds with the NSC numbers NSC-156306 (o-amsa monomethanesulfonate) and NSC-606170 (zalcitabine) were obtained from the Developmental Therapeutics Program (DTP) of the National Cancer Institute (NCI) and diluted to 20 μmol/L using dimethyl sulfoxide (DMSO). Differentiated HepaRG cells were treated with 0, 10, or 100 nmol/L (final concentration) of NSC-156306 or NSC-606170 and cells were harvested 48 hours after treatment. Each assay was conducted using at least three independent transfection experiments or chemical treatment.
RNA isolation and quantitative reverse-transcription PCR (qRT-PCR)
Total RNA was extracted from HepaRG cells after transfection or chemical treatment using the miRNeasy Mini Kit (Qiagen, Valencia, CA) and cDNA was synthesized using QuantiTect Reverse Transcription Kit (Qiagen) or NCode™ miRNA First-Strand cDNA Synthesis Kit (Life Technologies). CYP2C9, PTEN and GAPDH RNA levels were measured by qRT-PCR on an ABI Prism7900 Sequence Detection System (Applied Biosystems) according to the QuantiFast SYBR® Green RT-PCR Kit (Qiagen) protocol using the CYP2C9-RT-F and CYP2C9-RT-R primers, PTEN-RT-F and PTEN-RT-R primers, or GAPDH-RT-F and GAPDH-RT-R primers, respectively. The miR-128-RT-F and U6-F primers, together with the Universal Reverse Primer supplied with the NCode™ miRNA First-Strand cDNA Synthesis Kit, were used to detect the hsa-miR-128-3p and U6 levels. The RNA expression levels of CYP2C9 or hsa-miR-128-3p were calculated relative to expression of GAPDH or U6, respectively.
Western blot analysis
Proteins were isolated from HepaRG cells that were harvested after transfection or chemical treatment. Quantitative Western blotting was performed following the Odyssey™ Western Blotting Kit (LI-COR Biosciences) protocol. Antibodies against CYP2C9 or GAPDH (Abcam, Cambridge, MA) were used to detect CYP2C9 or GAPDH protein levels and an Odyssey CLx Infrared Imaging System was used to perform quantitative analyses, with infrared labeled secondary antibodies.
Retrieval of data from online databases
RNA expression levels of CYP2C9 and hsa-miR-128-3p were obtained from The Cancer Genome Atlas database (TCGA, http://cancergenome.nih.gov/).
Statistical analyses
The rank sum test was used to evaluate the difference in the expression of CYP2C9 or hsa-miR-128-3p in HCCs, with P < 0.05 as the significant criterion. Spearman Rank Order Correlation analysis was used to test the correlation between CYP2C9 levels and hsa-miR-128-3p expression. Student's t-test was also used to compare results from luciferase reporter gene assays and to compare CYP2C9 or hsa-miR-128-3p protein or RNA levels between subgroups.
Author Contributions
B.N. proposed and organized the study. D.Y., I.P. and B.N. designed the study. D.Y., B.G. and A.M. performed experiments. D.Y. and B.N. wrote the manuscript. Y.G., S.K., D.L. and J.F. provided advice and revised the manuscript. All authors reviewed the manuscript.
Supplementary Material
Supplementary Information
References
- El-Serag H. B. & Rudolph K. L. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology 132, 2557–2576 (2007). [DOI] [PubMed] [Google Scholar]
- Yu M. C. & Yuan J. M. Environmental factors and risk for hepatocellular carcinoma. Gastroenterology 127, S72–78 (2004). [DOI] [PubMed] [Google Scholar]
- Ning B. et al. Toxicogenomics and cancer susceptibility: advances with next-generation sequencing. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev 32, 121–158 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebert D. W. Polymorphisms in drug-metabolizing enzymes: what is their clinical relevance and why do they exist? Am J Hum Genet 60, 265–271 (1997). [PMC free article] [PubMed] [Google Scholar]
- Bartsch H. et al. Genetic polymorphism of CYP genes, alone or in combination, as a risk modifier of tobacco-related cancers. Cancer Epidemiol Biomarkers Prev 9, 3–28 (2000). [PubMed] [Google Scholar]
- Nebert D. W., McKinnon R. A. & Puga A. Human drug-metabolizing enzyme polymorphisms: effects on risk of toxicity and cancer. DNA Cell Biol 15, 273–280 (1996). [DOI] [PubMed] [Google Scholar]
- Silvestri L. et al. CYP enzyme polymorphisms and susceptibility to HCV-related chronic liver disease and liver cancer. Int J Cancer 104, 310–317 (2003). [DOI] [PubMed] [Google Scholar]
- Raunio H. et al. Cytochrome P4502A6 (CYP2A6) expression in human hepatocellular carcinoma. Hepatology 27, 427–432 (1998). [DOI] [PubMed] [Google Scholar]
- Tsunedomi R. et al. Patterns of expression of cytochrome P450 genes in progression of hepatitis C virus-associated hepatocellular carcinoma. Int J Oncol 27, 661–667 (2005). [PubMed] [Google Scholar]
- Xu X. R. et al. Insight into hepatocellular carcinogenesis at transcriptome level by comparing gene expression profiles of hepatocellular carcinoma with those of corresponding noncancerous liver. Proc Natl Acad Sci U S A 98, 15089–15094 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebert D. W. & Dalton T. P. The role of cytochrome P450 enzymes in endogenous signalling pathways and environmental carcinogenesis. Nat Rev Cancer 6, 947–960 (2006). [DOI] [PubMed] [Google Scholar]
- Goldstein J. A. & de Morais S. M. Biochemistry and molecular biology of the human CYP2C subfamily. Pharmacogenetics 4, 285–299 (1994). [DOI] [PubMed] [Google Scholar]
- Miners J. O. & Birkett D. J. Cytochrome P4502C9: an enzyme of major importance in human drug metabolism. Br J Clin Pharmacol 45, 525–538 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. et al. Gene expression patterns in human liver cancers. Mol Biol Cell 13, 1929–1939 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. W. et al. Cancer-associated molecular signature in the tissue samples of patients with cirrhosis. Hepatology 39, 518–527 (2004). [DOI] [PubMed] [Google Scholar]
- Okabe H. et al. Genome-wide analysis of gene expression in human hepatocellular carcinomas using cDNA microarray: identification of genes involved in viral carcinogenesis and tumor progression. Cancer Res 61, 2129–2137 (2001). [PubMed] [Google Scholar]
- Smith M. W. et al. Identification of novel tumor markers in hepatitis C virus-associated hepatocellular carcinoma. Cancer Res 63, 859–864 (2003). [PubMed] [Google Scholar]
- Mattick J. S. & Makunin I. V. Non-coding RNA. Hum Mol Genet 15 Spec No 1, R17–29 (2006). [DOI] [PubMed] [Google Scholar]
- Yokoi T. & Nakajima M. microRNAs as mediators of drug toxicity. Annu Rev Pharmacol Toxicol 53, 377–400 (2013). [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y., Nakajima M., Takagi S., Taniya T. & Yokoi T. MicroRNA regulates the expression of human cytochrome P450 1B1. Cancer Res 66, 9090–9098 (2006). [DOI] [PubMed] [Google Scholar]
- Calin G. A. & Croce C. M. MicroRNA signatures in human cancers. Nat Rev Cancer 6, 857–866 (2006). [DOI] [PubMed] [Google Scholar]
- Donzelli S. et al. MicroRNA-128-2 targets the transcriptional repressor E2F5 enhancing mutant p53 gain of function. Cell Death Differ 19, 1038–1048 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godlewski J. et al. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res 68, 9125–9130 (2008). [DOI] [PubMed] [Google Scholar]
- Khan A. P. et al. Quantitative proteomic profiling of prostate cancer reveals a role for miR-128 in prostate cancer. Mol Cell Proteomics 9, 298–312 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W. et al. Deregulated expression of miR-21, miR-143 and miR-181a in non small cell lung cancer is related to clinicopathologic characteristics or patient prognosis. Biomed Pharmacother 64, 399–408 (2010). [DOI] [PubMed] [Google Scholar]
- Xu B. et al. miR-143 decreases prostate cancer cells proliferation and migration and enhances their sensitivity to docetaxel through suppression of KRAS. Mol Cell Biochem 350, 207–213 (2011). [DOI] [PubMed] [Google Scholar]
- Aninat C. et al. Expression of cytochromes P450, conjugating enzymes and nuclear receptors in human hepatoma HepaRG cells. Drug Metab Dispos 34, 75–83 (2006). [DOI] [PubMed] [Google Scholar]
- Palumbo T. et al. Functional screen analysis reveals miR-26b and miR-128 as central regulators of pituitary somatomammotrophic tumor growth through activation of the PTEN-AKT pathway. Oncogene 32, 1651–1659 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L. et al. Sex differences in the expression of drug-metabolizing and transporter genes in human liver. J Drug Metab Toxicol 3,119; 10.4172/2157-7609.1000119 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning B. et al. Systematic and simultaneous gene profiling of 84 drug-metabolizing genes in primary human hepatocytes. J Biomol Screen 13, 194–201 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L. et al. Gene expression variability in human hepatic drug metabolizing enzymes and transporters. PLoS One 8, e60368; 10.1371/journal.pone.0060368 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer E. et al. Oxidation of benzo[a]pyrene by recombinant human cytochrome P450 enzymes. Chem Res Toxicol 8, 136–142 (1995). [DOI] [PubMed] [Google Scholar]
- Ozawa S. et al. Analyses of bronchial bulky DNA adduct levels and CYP2C9, GSTP1 and NQO1 genotypes in a Hungarian study population with pulmonary diseases. Carcinogenesis 20, 991–995 (1999). [DOI] [PubMed] [Google Scholar]
- Shou M., Korzekwa K. R., Crespi C. L., Gonzalez F. J. & Gelboin H. V. The role of 12 cDNA-expressed human, rodent, and rabbit cytochromes P450 in the metabolism of benzo[a]pyrene and benzo[a]pyrene trans-7,8-dihydrodiol. Mol Carcinog 10, 159–168 (1994). [DOI] [PubMed] [Google Scholar]
- Martinez C. et al. Association of CYP2C9 genotypes leading to high enzyme activity and colorectal cancer risk. Carcinogenesis 22, 1323–1326 (2001). [DOI] [PubMed] [Google Scholar]
- Chan A. T., Tranah G. J., Giovannucci E. L., Hunter D. J. & Fuchs C. S. A prospective study of genetic polymorphisms in the cytochrome P-450 2C9 enzyme and the risk for distal colorectal adenoma. Clin Gastroenterol Hepatol 2, 704–712 (2004). [DOI] [PubMed] [Google Scholar]
- London S. J., Sullivan-Klose T., Daly A. K. & Idle J. R. Lung cancer risk in relation to the CYP2C9 genetic polymorphism among Caucasians in Los Angeles County. Pharmacogenetics 7, 401–404 (1997). [DOI] [PubMed] [Google Scholar]
- Witkos T. M., Koscianska E. & Krzyzosiak W. J. Practical Aspects of microRNA Target Prediction. Curr Mol Med 11, 93–109 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M. et al. miR-128 and its target genes in tumorigenesis and metastasis. Exp Cell Res 319, 3059–3064 (2013). [DOI] [PubMed] [Google Scholar]
- Rao Q. et al. Aberrant microRNA expression in human cervical carcinomas. Med Oncol 29, 1242–1248 (2012). [DOI] [PubMed] [Google Scholar]
- Leite K. R. et al. MicroRNA expression profiles in the progression of prostate cancer--from high-grade prostate intraepithelial neoplasia to metastasis. Urol Oncol 31, 796–801 (2013). [DOI] [PubMed] [Google Scholar]
- Guo L. et al. Similarities and differences in the expression of drug metabolizing enzymes between human hepatic cell lines and primary human hepatocytes. Drug Metab Dispos 39, 528–538 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehmsmeier M., Steffen P., Hochsmann M. & Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA 10, 1507–1517 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L. et al. Copy number variation at 6q13 functions as a long-range regulator and is associated with pancreatic cancer risk. Carcinogenesis 33, 94–100 (2012). [DOI] [PubMed] [Google Scholar]
- Yu X. et al. Functional genetic variants in the 3′-untranslated region of sulfotransferase isoform 1A1 (SULT1A1) and their effect on enzymatic activity. Toxicol Sci 118, 391–403 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information