Abstract
MITF and pigmentation play important roles in both normal melanocyte and transformed melanoma cell biology. MITF is regulated by many pathways and it also regulates many targets, some of which are still being discovered and functionally validated. MITF is involved in a wide range of processes in melanocytes, including pigment synthesis and lineage survival. Pigmentation itself plays an important role as the interface between genetic and environmental factors that contribute to melanoma.
Keywords: MITF, melanoma, melanocyte, pigmentation
Introduction
Metastatic melanoma is a frequently fatal disease and its incidence has been on the rise worldwide throughout the past 30 years, with over 132,000 individuals worldwide diagnosed with melanoma each year1,2. While melanoma is highly curable by surgical excision when detected early, it is historically notorious for its therapeutic resistance and propensity to metastasize. Although the 10-year survival rate for stage IV metastatic melanoma has been only 10-15%3,4, there is reason for optimism due to discoveries ranging from deeper understanding of melanomagenesis to new therapeutic breakthroughs. In this review, we focus on MITF and pigmentation and their roles in both normal melanocytes and transformed melanoma cells.
Melanomagenesis
Melanoma is likely to arise from a combination of genetic and environmental risk factors. Epidemiological studies have found that having a high number of nevi—neoplastic but benign melanocytic proliferations—correlates with an increased risk of melanoma1,2,5. Beyond family history, other known risk factors for melanoma include fair skin and a history of blistering sunburns during youth.
Exposure to sunlight is a widely accepted environmental carcinogen for melanoma development. However, several features have complicated a detailed understanding of its etiologic role. For example, although signature mutations typical of ultraviolet B (UVB) wavelengths (290-320 nm) are commonly present within the genomes of melanoma cells, such mutations are less commonly implicated as “drivers” among melanoma oncogenes and tumor suppressors3,4,6,7. This contrasts strikingly with non-melanoma skin cancers, for which UVB mutations are very highly implicated8,9. In addition, whereas non-melanoma skin cancers are most commonly found on sun-exposed skin, melanomas do not follow the same restricted anatomic localization9. Additional confusing aspects of sunlight's relationship to melanoma include the measurable but surprisingly modest protective effect for sunscreens against melanoma, in contrast to strong protection against cutaneous squamous cell carcinoma (although numerous potential explanations exist, including followup duration, and other variables). Finally, the Sun Protection Factor (SPF) of dark pigment (eumelanin) is relatively weak (<10) whereas darkly pigmented people exhibit profoundly diminished cutaneous melanoma risk, suggesting an unknown relationship between sunlight's carcinogenic mechanism(s) and melanoma etiology.
Activating mutations in BRAF, a serine/threonine kinase involved in the RAS/RAF/MEK/ERK signaling pathway, have been found in 60-70% of malignant melanomas10. A substitution of valine for glutamic acid at position 600 (V600E) resulting from a T1799A transversion in exon 15 accounts for over 90% of activating oncogenic BRAF mutations11-13. However, given that up to 80% of benign nevi harbor BRAFV600E, this mutation alone is thought to be insufficient for melanomagenesis12,14. In fact, BRAFV600E alone has been shown to cause classical oncogene-induced senescence in human melanocytes in vitro and also in mouse models12,15,16. Patton et al. have shown that in zebrafish, BRAFV600E is sufficient for nevus development and furthermore, in combination with a p53-deficient background, BRAFV600E contributes to melanomagenesis17. This suggests that activated BRAF may be a primary cooperating event for melanoma development11,13,17,18. In mouse models, BRAF activation along with loss of p53, p16, or PTEN significantly promotes melanomagenesis12,16,19. Treatment of BrafV600E mutant melanomas with BRAF inhibitors such as vemurafinab (PLX4032) leads to tumor regression and improved overall survival of patients although these responses are typically not indefinitely durable20.
Epidemiological evidence and Genomic Wide Association Studies have indicated that people with the red hair color (RHC) phenotype have an increased risk of melanoma21. The RHC phenotype is characterized by fair skin, red hair, tendency to burn, inability to tan, and freckles. The RHC phenotype is most often caused by polymorphisms in the melanocortin 1 receptor (MC1R), a G-protein coupled receptor that regulates pigment production in normal melanocytes. Normally, MC1R is essential for tanning. Binding of the ligand α-melanocyte stimulating hormone (αMSH) to MC1R causes cAMP levels to increase in the cell, ultimately resulting in an increase of the melanocyte master regulator microphthalmia-associated transcription factor (MITF) and induction of a switch in pigment production from red/yellow pheomelanin towards brown/black eumelanin. RHC individuals contain nonfunctional variants of MC1R, which affect multiple cAMP-mediated signaling events, including pigment production.
Although individuals with the RHC phenotype are unable to tan, D'Orazio et al. showed that in RHC (Mc1re/e) mice, the tanning pathway (and thus pigmentation) could be rescued by topically applying the adenylate cyclase agonist forskolin, thus indicating that even in the absence of functional MC1R, the pigmentation machinery downstream is still available22. In these mice, forskolin-induced eumelanin pigmentation was protective against ultraviolet light-induced DNA damage and tumorigenesis22.
Microphthalmia-associated transcription factor
Microphthalmia-associated transcription factor (MITF) is the master regulator of melanocytes, playing a key role in their development, differentiation, function, and survival. MITF belongs to the family of basic helix-loop-helix leucinezipper microphthalmia-related (MiT) transcription factors, which also includes transcription factor E3 (TFE3), transcription factor EB (TFEB), and transcription factor EC (TFEC). These transcription factors bind to DNA recognition sequences called E-boxes, which have the consensus sequence CA[T/C]GTG.
MITF is expressed in pigmented cells including melanocytes and retinal pigment epithelium cells, as well as in certain non-pigmented cell lineages including osteoclasts and mast cells23. There are at least nine different isoforms of MITF that exhibit tissue-specific expression patterns. The M isoform of MITF (MITF-M) is selectively expressed in melanocytes24.
Heterozygous mutation of MITF has been shown to cause Waardenburg syndrome Type IIA, an autosomal dominant condition in humans25. Individuals with this condition exhibit a congenital white forelock and sensorineural deafness. Complete absence of MITF in mice results in characteristics such as white fur, deafness, and small eyes (microphthalmia). Loss of pigmentation in these mice is due to an absence of melanocytes, rather than a defect in melanin synthesis within viable melanocytes.
Besides its important roles within normal melanocytes, MITF also appears to play critical roles in melanoma, where it has been shown to be a lineage-specific survival oncogene that is amplified in 5-20% of human melanomas26. In melanoma patients, MITF amplification was associated with a decreased five-year survival26. MITF has also been shown to control proliferative, invasive, and metastatic properties of melanoma cells26-28.
MITF has been suggested to have paradoxical roles, which may confer either pro-survival or anti-survival effects. A rheostat model of MITF has been proposed, wherein lower levels of MITF are associated with increased motility and invasive capacity in melanoma cells, and higher levels correspond with increased proliferative capacity29. Melanocytes expressing low levels of MITF have high levels of POU3F2 (BRN2) transcription factor, a direct repressor of MITF transcription that has been implicated in melanoma invasiveness27,30. MITF has also been shown to control both invasiveness and proliferation by regulating the expression of DIAPH1, the gene that encodes the diaphanous-related formin Dia1, which is involved in actin polymerization and coordination of the actin cytoskeleton28. Low MITF levels lead to downregulation of Dia1, a cyclin-dependent kinase inhibitor 1B (p27Kip1)-dependent G1 arrest, reorganization of the actin cytoskeleton, and increased invasiveness. Moderate MITF levels lead to actin polymerization and suppressed p27Kip1, resulting in proliferation28. Hoek et al. have suggested that melanoma cells switch between the proliferative and invasive states during tumor progression31. Prolonged MITF suppression has been suggested to trigger senescence in melanoma cells32.
Regulation of MITF
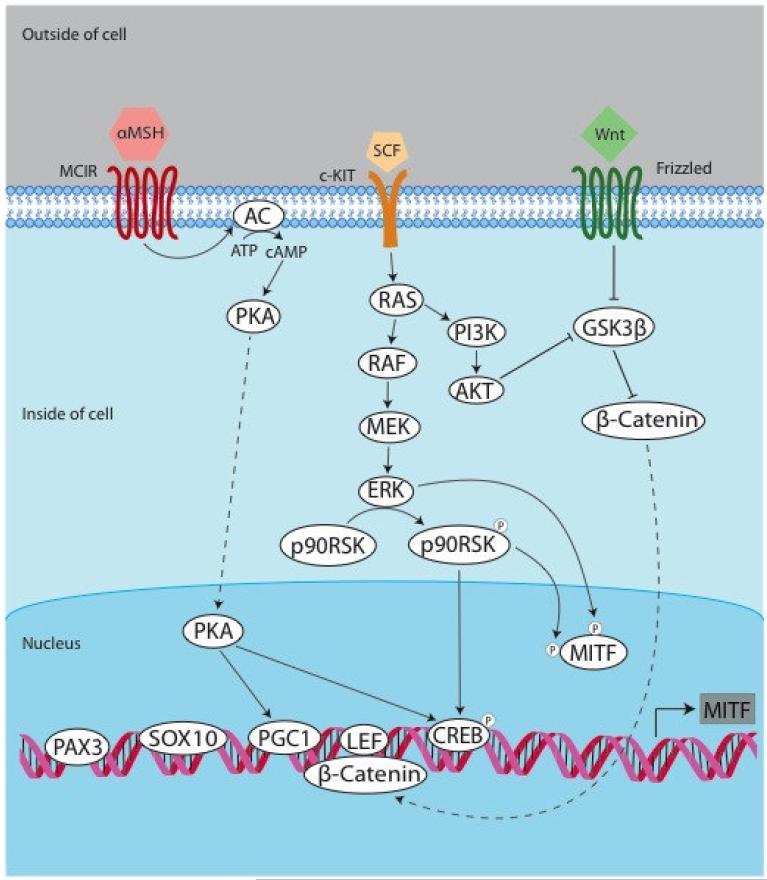
MITF is regulated both transcriptionally and post-translationally (Figure 1). At the transcriptional level, MITF-M is regulated by transcription factors including MITF itself, paired box gene 3 (PAX3), cAMP-responsive element binding protein (CREB), sex-determining region Y-box 10 (SOX10), lymphoid enhancer-binding factor 1 (LEF1/TCF), one cut domain 2 (ONECUT-2), and the mitogen-activated protein kinase (MAPK) pathway.
Figure 1. Regulators of MITF.
The regulation of MITF is complex, with multiple upstream pathways converging on MITF. The pathways thought to be relevant to both normal melanocytes and transformed melanoma cells include the tanning pathway, the MAPK pathway, and the PI3K pathway. PAX3, SOX10, PGC1, LEF, and CREB are some of the known transcription factors that bind to the MITF promoter and regulate its transcription. AC = adenylate cyclase.
Exposure of skin keratinocytes to UV light causes activation of cellular damage responses, including activation of p53, which leads to proopiomelanocortin (POMC) transcription and ultimately tanning (physiologic pigmentation). POMC can be enzymatically cleaved to produce several physiologically important derivatives including α-Melanocyte Stimulating Hormone (αMSH). During tanning, αMSH is induced and binds to MC1R on melanocytes. This induces activation of adenylyl cyclase, resulting in cAMP production and activation of Protein Kinase A (PKA). This in turn leads to phosphorylation of CREB, followed by activation of the MITF-M promoter, which contains a cAMP responsive element (CRE) site.
The transcription factors SOX10 and PAX3 are both transcriptional activators of MITF that are important for development. Tissue-specific expression of MITF-M in melanocytes is mediated by cooperation between CREB and SOX10, the latter of which is largely restricted to the neural crest lineage33. PAX3 is expressed in committed but undifferentiated melanoblasts and acts to both initiate melanogenesis and prevent terminal differentiation. PAX3 accomplishes this by activating expression of MITF (which is required for melanogenesis) while simultaneously competing with MITF for dopachrome tautomerase (DCT) enhancer occupancy34.
The Wingless-type (Wnt) pathway is critical for melanocyte development from neural crest precursors. Wnt ligand binds to cell surface Frizzled receptors, stabilizing β-catenin levels in the cell and ultimately resulting in interaction of βcatenin with T-cell transcription factor (TCF)/lymphoid enhancer binding factor (LEF) and activation of the MITF-M promoter35-37.
MITF is a target of the MAPK pathway 38. Oncogenic BRAF and ERK may both promote MITF activation and also enhance its degradation38-42. Oncogenic BRAF has been reported to upregulate MITF transcription through ERK and BRN242. Preliminary evidence has suggested that oncogenic BRAFV600E may contribute to immune escape of melanoma cells41. It is thought that blocking the activity of BRAFV600E leads to increased expression of melanocyte differentiation antigens (MDAs), the recognition of which play a key role in the immunologic response to melanomas41.
At the post-translational level, MITF is known to be phosphorylated38,39,43, sumoylated44,45, and ubiquitinated39. It is phosphorylated by MAPK, ribosomal S6 kinase (RSK), and p38. C-KIT activation in melanocytes causes MITF to be phosphorylated at Ser73 by ERK2 and at Ser409 by p90 ribosomal kinase (p90RSK). When MITF is phosphorylated at Ser73, it induces recruitment of the transcriptional coactivator p300, and also targets MITF itself for ubiquitination and proteolysis.
It is notable that most melanomas contain hyperactivated MAPK pathway activity and thus harbor phospho-MITF at Ser73, which triggers ubiquitin mediated proteosomal degradation of MITF. Conversely, a consequence of treating melanoma cells with BRAF inhibitor or MEK inhibitor is rapid stabilization and upregulation of MITF levels39,46. Correspondingly, it has been observed that certain transcriptional targets of MITF are also upregulated (via MITF) upon MAPK pathway suppression in melanoma cells. These targets include numerous pigmentation genes and PPAR-[.gamma] coactivator-1α (PGC1α), which induces a metabolic shift away from glycolysis towards oxidative phosphorylation41,46.
Interestingly, MITF has recently been shown to mediate resistance to RAFMEK-ERK inhibitor therapy47. It was seen that increased cAMP levels and PKA activation conferred resistance to MAPK pathway inhibitors in melanoma cells through induction of CREB-dependent effectors, including MITF 47.
Sumoylation of MITF, which is mediated by the protein inhibitor of activated STAT3 (PIAS3), represses its activity44,45. This suggests a mechanism whereby post-translational modification of MITF alters its target-gene specificity. PIAS3 and protein kinase C (PKC) repress MITF transcriptional activity by interacting directly with MITF and inhibiting its DNA-binding activity. The significance of the regulatory sumoylation site was demonstrated in human pedigrees prone to familial melanoma. Mutation of the sumoylation site predisposed family members to development of melanoma48,49.
Recently, it has been shown that PGC1 can activate the expression of MITF50. αMSH signaling was seen to strongly induce PGC1α and stabilize PGC1α and PGC1β proteins. It was found that the PGC1s activate the MITF promoter and that inhibiting the PGC1s results in blockage of the αMSH-mediated induction of MITF and melanogenesis.
MITF expression is reduced under hypoxic conditions through hypoxiainducible factor 1 (HIF1)-mediated induction of the transcriptional repressor basic helix-loop-helix family, member 40 (BHLHE40) 51. It was suggested by Feige et al. that this may contribute to the survival advantage of amplified MITF in metastatic melanoma51.
MITF target genes
Methods to identity MITF target genes include DNA microarray analyses on melanocytes and melanoma cells in which MITF has been overexpressed or knocked down, chromatin immunoprecipitation coupled to high throughput sequencing (ChIP-seq), and “nearest neighbor” computational analyses52-54.
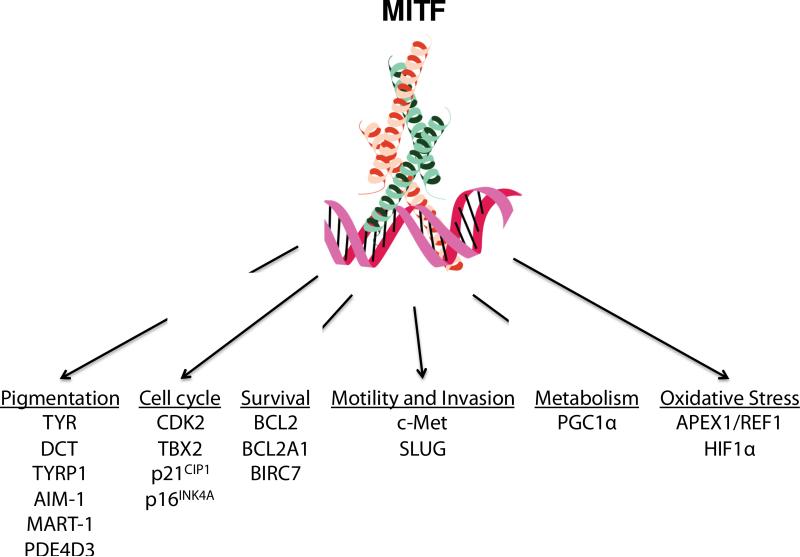
Potentially important functional transcriptional targets of MITF include regulators of the cell cycle (CDK2, p21CIP1, p16INK4A), differentiation, survival and metastasis (BCL2, BCL2A1, c-Met), cAMP levels (PDE4D3), metabolism, miRNA processing (DICER)55, DNA repair, invasiveness, and pigmentation. MITF has many targets, likely including some that remain to be identified and functionally validated. For comprehensive papers on MITF targets, please refer to recent reviews as well as Strub et al. and Hoek et al52,53. In this review, the subsets of MITF targets highlighted are those thought to be involved in pigmentation, melanomagenesis and melanoma progression (Figure 2).
Figure 2. MITF targets.
MITF is the master regulator of the melanocyte. Its many targets include those involved in a wide range of pathways that are relevant to both normal melanocytes and melanoma cells.
Pigmentation
The pigment genes tyrosinase (TYR), dopachrome tautomerase (DCT, also known as tyrosinase-related protein 2, or TYRP2), and tyrosinase-related protein 1 (TYRP1) are MITF targets that encode enzymes necessary for melanin synthesis. MITF also regulates Absent In Melanoma-1 (AIM-1), which is mutated in human oculocutaneous albinism type IV, melanoma antigen recognized by T-cells 1 (MART1) and silver/gp100, factors which are present within melanosomes, the organelles in which melanin synthesis take place.
Phosphodiesterase 4D3 (PDE4D3) has been shown to be a transcriptional target of MITF56. PDE4D3 is part of a negative feedback loop with MITF and cAMP in melanocytes, whereby chronic stimulation of the cAMP pathway leads to refractoriness. Thus, upregulation of PDE4D3 by MITF is thought to dampen activation of the cAMP pathway and stimulation of the melanin synthesis pathways in melanocytes56.
Cell cycle
Among the targets of MITF are both regulators and inhibitors of cell cycle progression. Cyclin-dependent kinase 2 (CDK2) is a MITF target gene that is involved in regulating the cell cycle. CDK2 expression appears to be essential for melanoma clonogenic growth, and in a series of melanoma cell lines and human melanoma samples, MITF and CDK2 expression levels were found to be tightly correlated57. MITF's transcriptional regulation of CDK2 is further notable because the CDK2 gene shares a promoter region with the gene encoding the melanosomal factor silver/gp100. In this manner, expression of CDK2—a ubiquitously expressed cell cycle kinase—is uniquely regulated by MITF within the melanocyte lineage.
T-Box transcription factor 2 (TBX2), another target of MITF, represses cyclin-dependent kinase inhibitor 2A (CDKN2A, also known as p16INK4A)58,59. MITF is a transcriptional activator of TBX2 and therefore plays an important role in maintaining proliferation and suppressing senescence in melanoma60.
On the flip side, MITF also has target genes that halt the cell cycle in both melanocytes and melanoma cells, including the cyclin-dependent kinase inhibitors p21CIP1 and p16INK4A, which is often mutated in melanomas61,62.
Survival
The survival genes (B-cell lymphoma 2) BCL2 and BCL2A1 have been shown to be targets of MITF54,63. Germline deletion of BCL2 results in melanocyte loss54. Disrupting MITF in melanocytes or melanoma cells triggers apoptosis that can be partially rescued by BCL2 overexpression. Analysis of expression microarrays of human primary melanomas revealed co-varied expression linkage for MITF and BCL2. MITF may regulate BCL2A1 and BCL2 expression in melanoma, thereby enhancing survival54,63. Melanoma inhibitor of apoptosis (ML-IAP, or BIRC7) is another anti-apoptotic MITF target64. BIRC7 is an apoptotic regulator that has low to undetectable levels of expression in normal adult tissues but exhibits high expression levels in melanomas and other cancers.
Motility and invasion
c-Met, a receptor tyrosine kinase, has been shown to be another transcriptional target of MITF65. c-Met and its ligand hepatocyte growth factor (HGF, also known as scatter factor) have been observed to play important roles in growth and motility in the melanocyte lineage65. c-Met is also thought to be involved in invasion and plays a key role in melanoma metastasis65. HGF has been observed to disrupt adhesion between melanocytes and keratinocytes by downregulating E-cadherin and desmoglein-1. This is thought to aid in uncontrolled proliferation and scattering in melanoma. Recent data suggest that stromal expression of HGF may contribute to resistance to BRAF inhibitor therapy in melanoma66.
SLUG (SNAI2) has also been reported to be regulated directly by MITF67. SLUG along with SNAIL (SNAI1) are key mediators of the epithelial-mesenchymal transition (EMT), a process that has a role in both normal development and also in invasion and metastasis in melanoma and other cancers.
Metabolism
MITF was recently shown to be involved in regulating metabolism and oxidative stress. It directly regulates expression of the master mitochondrial regulator peroxisome proliferator-activated receptor γ coactivator 1 α (PGC1α)46. Inhibition of oncogenic BRAF by small molecules leads to not only cell cycle arrest and apoptosis, but also induction of oxidative phosphorylation genes and mitochondrial biogenesis, thought to be caused by increased expression of PGC1α downstream of MITF46. Melanomas that have an activated BRAF/MAPK pathway exhibit lower levels of MITF and PGC1α and decreased oxidative phosphorylation. It was shown that MITF overexpression is sufficient and necessary to drive oxidative metabolism in melanocytes. When melanomas with activated BRAF are treated with BRAF inhibitors, they become addicted to oxidative phosphorylation, which in turn renders them more resistant to therapy46. This observation suggests that the combined use of BRAF/MAPK inhibition with mitochondrial antagonists may offer an attractive therapeutic opportunity.
Oxidative stress
MITF has been shown to regulate oxidative stress levels through its target apurinic/apyrimidinic endonuclease I/redox factor-1 (APEX1/REF1)68.
APEX1/REF1 is an enzyme that is involved in base excision repair of DNA damage caused by oxidating and alkylating agents. Thus, it plays a central role in the response of cells to oxidative stress. This is particularly important for melanocytes, which are exposed to environmental stresses such as UV light more than most other cells in the body.
APEX1/REF1 interacts with CREB to activate hypoxia inducible factor 1 α (HIF1α). HIF1α itself is a target of MITF and is a key regulator of transcriptional responses to oxidative stress69. Interestingly, HIF1α also regulates MITF, indicating a feedback loop between MITF and HIF1α that allows the cell to cope with hypoxia and oxidative stress.
Pigmentation and Melanoma
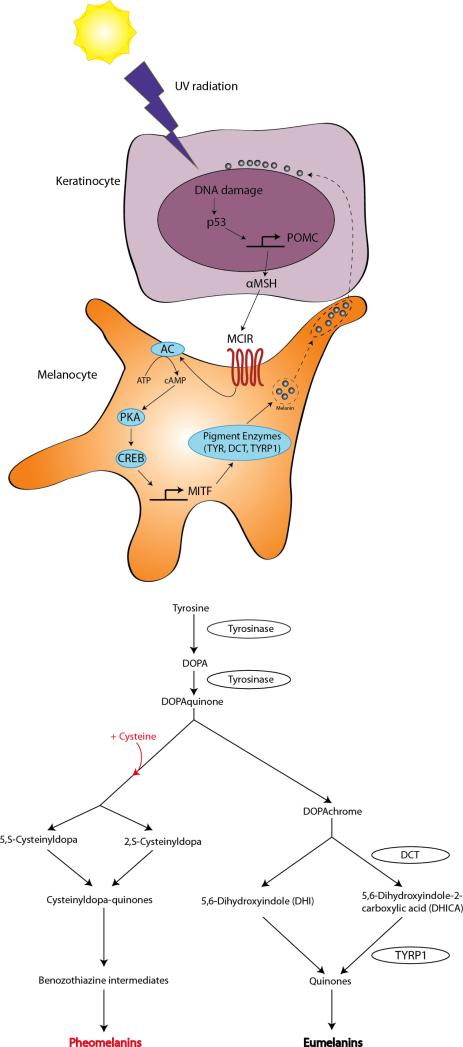
Skin and hair pigmentation are determined by the ratio of several types of melanin species: the alkali soluble yellow-red pheomelanin and insoluble brown-black eumelanin. Melanin production is an enzymatic process that converts tyrosine to melanin pigment. Synthesis of both types of melanin begins with tyrosine oxidation by tyrosinase, yielding first L-DOPA and then dopaquinone, the final common precursor. In the presence of melanosomal cysteine, pheomelanin is produced. Both eumelanin and pheomelanin synthesis require tyrosinase, but eumelanin synthesis additionally requires the enzymes TYRP1 and DCT. It is thought that initially, pheomelanin is produced, and after melanosomal cysteine is consumed, eumelanin is produced (Figure 3)70.
Figure 3. The tanning pathway (top) and melanin synthesis pathway (bottom).
The physiologic skin pigmentation response (tanning) begins with exposure to UV radiation, which causes DNA damage and thus upregulates p53, leading to POMC transcription in keratinocytes. POMC is enzymatically cleaved to produce αMSH, which binds to MC1R on melanocytes. This triggers the rest of the tanning response, ultimately resulting in transcription of MITF and its targets, which include the pigmentation enzymes tyrosinase, DCT, and TYRP1. Melanin is produced in melanosomes and transported to keratinocytes, where they form protective caps over the keratinocyte nuclei. Synthesis of both types of melanins (bottom) begins with the amino acid tyrosine. Tyrosinase catalyzes the rate-limiting step for both eumelanin and pheomelanin. Eumelanin synthesis additionally requires the enzymes DCT and TYRP1, while pheomelanin additionally requires the amino acid cysteine.
The range of pigmentation phenotypes of individuals is a result of their eumelanin to pheomelanin ratio. People with darker hair or skin have a higher eumelanin to pheomelanin ratio than people with fairer hair/skin. With the exception of albino individuals who harbor deficiencies in pigment synthesis, pheomelanin is present in all indviduals, while the levels of eumelanin increase from Fitzpatric phototype I to phototype VI (see Table 1). The Fitzpatrick phototype scale characterizes the different skin types into color, ability to tan, and tendency to burn. Melanoma risk decreases from phototype I (fair skin, red/blond) to phototype VI (dark skin).
Table 1.
Fitzpatrick phototype classification
| Phototype | Skin color | Tans | Burns |
|---|---|---|---|
| I (RHC) | Very white | Never | Always |
| II | White | Minimally | Very easily |
| III | Slightly brown | Gradually (average) | Easily |
| IV | Brown | Easily | Occasionally |
| V | Brown to black | Easily and substantially | Rarely |
| VI | Black | Maximally (readily and profusely) | Never |
Within the vast literature showing the weak UV-protection afforded by pheomelanin-containing skin, a small literature has also suggested that pheomelanin might actually be a pro-oxidant and photosensitize for DNA damage, while eumelanin may act as a photoprotective antioxidant70,71. Chemically, pheomelanin contains nitrogen and sulfur atoms, while eumelanin does not. The sulfur constituent of pheomelanin could produce and propagate reactive oxygen species (ROS)72. The pigment synthesis pathway itself may also generate byproducts including ROS. It is thought that the melanosome organelle functions to provide a measure of protection from ROS-induced damage via containment73. Leakage of toxic species out of the melanosome can be damaging to the melanocyte.
Pheomelanin synthesis has been shown to have a UV-independent, pigment-dependent effect on melanomagenesis in a BRAFV600E C57BL/6 melanoma mouse model74. In this study, four groups of mice mimicking the extremes of the Fitzpatrick phototype scale were used. Black mice with the wildtype C57BL/6 pigmentation phenotype were used to model people with dark skin who have a high eumelanin to pheomelanin ratio. Albino (white) mice with an inactivating mutation at the tyrosinase locus (Tyrc/c) were used to mimic individuals with albinism. The albino mutation does not affect melanocyte viability; rather, it affects the ability to produce pigment within viable melanocytes. In order to mimic RHC phenotype individuals, mice with a premature termination of the Mc1r transcript (Mc1re/e) were generated. These Mc1re/e mice have red-yellow fur, do not tan, and are notably predisposed to developing melanoma. All mice in this study also harbored an inducible knockin allele of BRAFV600E. In the black mice, activation of BRAFV600E normally produces primarily benign nevi. Combination of BRAFV600E activation plus a second inducible mutation (e.g. PTEN, p16INK4A, or p53 deletion) produces highly penetrant melanoma.
Unlike observations in black mice, where only a small percentage of mice developed tumors, singular activation of BRAFV600E in red Mc1re/e mice produced invasive melanomas in greater than 50% of mice. This occurred following a several month latency period, suggesting the acquisition of additional/ secondary mutational events. A particularly striking feature of these high penetrant melanomas was their occurrence in the absence of any UV exposure (or any other known carcinogen). Genetically matched albino mice demonstrated similar rates of melanoma development to black mice, indicating that lack of eumelanin is not the equivalent of increasing melanoma risk. Furthermore, the study indicated that pheomelanin synthesis was a cause for increased melanoma risk. A pigmentless mouse model that was both red-haired/fair-skinned and albino (Mc1re/e, Tyrc/c) was generated and was shown to be profoundly protected against melanoma formation. These data suggest that the pheomelanin synthesis pathway stimulates melanomagenesis via a UV-independent pathway that is mediated at least in part by synthesis of red/blond pigment.
Although the mechanism for this increased melanomagenesis in red Mc1Re/e mice is not fully understood, there are data showing that red pigment induces ROS-mediated DNA damage, which may predispose to melanoma carcinogenesis74.
Conclusion
MITF plays key roles in many key pathways within normal melanocytes and transformed melanoma cells. It is involved in the pigmentation pathway, which has both antioxidant and pro-oxidant components. The skin is subject to more environmental DNA-damaging agents than perhaps any other organ in the body. Pigmentation—particularly eumelanin—is a way to protect against these insults.
Prevention strategies against melanoma for individuals at high risk (such as those with the RHC phenotype) include minimizing exposure to UV irradiation and use of sunscreens. Although forskolin has been used to darken the skin of Mc1Re/e mice and successfully protect against UV carcinogenic challenge, in human skin forskolin exhibits poor skin penetration22,56. Thus, it would be necessary to use small molecules that better penetrate the skin. Another approach would be to use an inhibitor of PDE4D3 which should also increase cAMP levels.
Although MITF itself is difficult to “drug” because it lacks a ligand-binding pocket and a measurable catalytic activity, components of pathways upstream and downstream of MITF could potentially be drugged. Upstream of MITF, there are already some therapies targeting BRAF (e.g. vemurafinab) as well as MEK and ERK, and combination therapies against MAPK pathway components. Some combination melanoma therapies include combining MAPK inhibition along with immunotherapy, cell cycle dysregulation, histone deacetylase (HDAC) inhibitors, or use of other small molecules that target other aspects of melanoma such as metabolism (e.g. PGC1) or the stress response (e.g. HIF1).
Continued elucidation of normal melanocyte and transformed melanoma cell biology will hopefully lead to more effective ways of preventing melanomagenesis, treating metastatic melanoma, and preventing resistance and relapse.
Highlights.
The role of MITF and its targets in melanoma is reviewed
The role of pigmentation in melanoma is discussed
Regulation of MITF is reviewed
Acknowledgements
We thank Marcelo Pereira da Silva for his generous help with rendering the figures. We also thank Dr. Roydon Price and Dr. Katey Robinson for insightful comments on the manuscript. This work was supported by grants to DEF from NIH (P01 CA163222; R01 AR043369; R01CA150226) and the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest: None declared.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer Journal for Clinicians. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Kudchadkar RR, Gonzalez R, Lewis K. New targeted therapies in melanoma. Cancer Control. 2013;20:282–288. doi: 10.1177/107327481302000405. [DOI] [PubMed] [Google Scholar]
- 3.Melanoma Skin Cancer American Cancer Society. 2013 At < http://www.cancer.org/cancer/skincancer-melanoma/detailedguide/melanoma-skin-cancer-survival-rates>.
- 4.Balch CM, et al. Final Version of 2009 AJCC Melanoma Staging and Classification. Journal of Clinical Oncology. 2009;27:6199–6206. doi: 10.1200/JCO.2009.23.4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chin L. The genetics of malignant melanoma: lessons from mouse and man. Nature Reviews Cancer. 2003;3:559–570. doi: 10.1038/nrc1145. [DOI] [PubMed] [Google Scholar]
- 6.Curtin JA, et al. Distinct sets of genetic alterations in melanoma. New England Journal of Medicine. 2005;353:2135–2147. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 7.Pleasance ED, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kadekaro AL, Wakamatsu K, Ito S, Abdel-Malek ZA. Cutaneous photoprotection and melanoma susceptibility: reaching beyond melanin content to the frontiers of DNA repair. Frontiers in Bioscience. 2006;11:2157–2173. doi: 10.2741/1958. [DOI] [PubMed] [Google Scholar]
- 9.Young C. Solar ultraviolet radiation and skin cancer. Occupational Medicine. 2009;59:82–88. doi: 10.1093/occmed/kqn170. [DOI] [PubMed] [Google Scholar]
- 10.Pollock PM, Meltzer PS. A genome-based strategy uncovers frequent BRAF mutations in melanoma. Cancer Cell. 2002;2:5–7. doi: 10.1016/s1535-6108(02)00089-2. [DOI] [PubMed] [Google Scholar]
- 11.Wellbrock C, et al. V599EB-RAF is an oncogene in melanocytes. Cancer Research. 2004;64:2338–2342. doi: 10.1158/0008-5472.can-03-3433. [DOI] [PubMed] [Google Scholar]
- 12.Dhomen N, et al. Oncogenic Braf Induces Melanocyte Senescence and Melanoma in Mice. Cancer Cell. 2009;15:294–303. doi: 10.1016/j.ccr.2009.02.022. [DOI] [PubMed] [Google Scholar]
- 13.Hoeflich KP. Oncogenic BRAF Is Required for Tumor Growth and Maintenance in Melanoma Models. Cancer Research. 2006;66:999–1006. doi: 10.1158/0008-5472.CAN-05-2720. [DOI] [PubMed] [Google Scholar]
- 14.Pollock PM, et al. High frequency of BRAF mutations in nevi. Nature Genetics. 2002;33:19–20. doi: 10.1038/ng1054. [DOI] [PubMed] [Google Scholar]
- 15.Michaloglou C, et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436:720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- 16.Goel VK, et al. Melanocytic nevus-like hyperplasia and melanoma in transgenic BRAFV600E mice. Oncogene. 2009;28:2289–2298. doi: 10.1038/onc.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patton EE, et al. BRAF Mutations Are Sufficient to Promote Nevi Formation and Cooperate with p53 in the Genesis of Melanoma. Current Biology. 2005;15:249–254. doi: 10.1016/j.cub.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 18.Karasarides M, et al. B-RAF is a therapeutic target in melanoma. Oncogene. 2004;23:6292–6298. doi: 10.1038/sj.onc.1207785. [DOI] [PubMed] [Google Scholar]
- 19.Dankort D, et al. BrafV600E cooperates with Pten loss to induce metastatic melanoma. Nature Genetics. 2009;41:544–552. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flaherty KT, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. New England Journal of Medicine. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bishop DT, et al. Genome-wide association study identifies three loci associated with melanoma risk. Nature Genetics. 2009;41:920–925. doi: 10.1038/ng.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.D'Orazio JA, et al. Topical drug rescue strategy and skin protection based on the role of Mc1r in UV-induced tanning. Nature. 2006;443:340–344. doi: 10.1038/nature05098. [DOI] [PubMed] [Google Scholar]
- 23.Hershey CL, Fisher DE. Genomic analysis of the Microphthalmia locus and identification of the MITF-J/Mitf-J isoform. Gene. 2005;347:73–82. doi: 10.1016/j.gene.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 24.Fuse N, Yasumoto K, Suzuki H, Takahashi K, Shibahara S. Identification of a melanocyte-type promoter of the microphthalmia-associated transcription factor gene. Biochemical and Biophysical Research Communications. 1996;219:702–707. doi: 10.1006/bbrc.1996.0298. [DOI] [PubMed] [Google Scholar]
- 25.Tassabehji M, Newton VE, Read AP. Waardenburg syndrome type 2 caused by mutations in the human microphthalmia (MITF) gene. Nature Genetics. 1994;8:251–255. doi: 10.1038/ng1194-251. [DOI] [PubMed] [Google Scholar]
- 26.Garraway LA, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 27.Goodall J, et al. Brn-2 represses microphthalmia-associated transcription factor expression and marks a distinct subpopulation of microphthalmia-associated transcription factor-negative melanoma cells. Cancer Research. 2008;68:7788–7794. doi: 10.1158/0008-5472.CAN-08-1053. [DOI] [PubMed] [Google Scholar]
- 28.Carreira SS, et al. Mitf regulation of Dia1 controls melanoma proliferation and invasiveness. Genes & Development. 2006;20:3426–3439. doi: 10.1101/gad.406406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goding CR. A picture of Mitf in melanoma immortality. Oncogene. 2011;30:2304–2306. doi: 10.1038/onc.2010.641. [DOI] [PubMed] [Google Scholar]
- 30.Kobi D, et al. Genome-wide analysis of POU3F2/BRN2 promoter occupancy in human melanoma cells reveals Kitl as a novel regulated target gene. Pigment Cell & Melanoma Research. 2010;23:404–418. doi: 10.1111/j.1755-148X.2010.00697.x. [DOI] [PubMed] [Google Scholar]
- 31.Hoek KS, et al. In vivo switching of human melanoma cells between proliferative and invasive states. Cancer Research. 2008;68:650–656. doi: 10.1158/0008-5472.CAN-07-2491. [DOI] [PubMed] [Google Scholar]
- 32.Giuliano S, et al. Microphthalmia-associated transcription factor controls the DNA damage response and a lineage-specific senescence program in melanomas. Cancer Research. 2010;70:3813–3822. doi: 10.1158/0008-5472.CAN-09-2913. [DOI] [PubMed] [Google Scholar]
- 33.Huber WE, et al. A tissue-restricted cAMP transcriptional response: SOX10 modulates alpha-melanocyte-stimulating hormone-triggered expression of microphthalmia-associated transcription factor in melanocytes. Journal of Biological Chemistry. 2003;278:45224–45230. doi: 10.1074/jbc.M309036200. [DOI] [PubMed] [Google Scholar]
- 34.Lang D, et al. Pax3 functions at a nodal point in melanocyte stem cell differentiation. Nature. 2005;433:884–887. doi: 10.1038/nature03292. [DOI] [PubMed] [Google Scholar]
- 35.Dorsky RI, Raible DW, Moon RT. Direct regulation of nacre, a zebrafish MITF homolog required for pigment cell formation, by the Wnt pathway. Genes & Development. 2000;14:158–162. [PMC free article] [PubMed] [Google Scholar]
- 36.Takeda K, et al. Induction of melanocyte-specific microphthalmia-associated transcription factor by Wnt-3a. Journal of Biological Chemistry. 2000;275:14013–14016. doi: 10.1074/jbc.c000113200. [DOI] [PubMed] [Google Scholar]
- 37.Widlund HR, et al. Beta-catenin-induced melanoma growth requires the downstream target Microphthalmia-associated transcription factor. The Journal of Cell Biology. 2002;158:1079–1087. doi: 10.1083/jcb.200202049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hemesath TJ, Price ER, Takemoto C, Badalian T, Fisher DE. MAP kinase links the transcription factor Microphthalmia to c-Kit signalling in melanocytes. Nature. 1998;391:298–301. doi: 10.1038/34681. [DOI] [PubMed] [Google Scholar]
- 39.Wu M, et al. c-Kit triggers dual phosphorylations, which couple activation and degradation of the essential melanocyte factor Mi. Genes & Development. 2000;14:301–312. [PMC free article] [PubMed] [Google Scholar]
- 40.Wellbrock C, Marais R. Elevated expression of MITF counteracts B-RAF-stimulated melanocyte and melanoma cell proliferation. The Journal of Cell Biology. 2005;170:703–708. doi: 10.1083/jcb.200505059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boni A, et al. Selective BRAFV600E inhibition enhances T-cell recognition of melanoma without affecting lymphocyte function. Cancer Research. 2010;70:5213–5219. doi: 10.1158/0008-5472.CAN-10-0118. [DOI] [PubMed] [Google Scholar]
- 42.Wellbrock C, et al. Oncogenic BRAF Regulates Melanoma Proliferation through the Lineage Specific Factor MITF. PLoS ONE. 2008;3:e2734. doi: 10.1371/journal.pone.0002734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mansky KC, Sankar U, Han J, Ostrowski MC. Microphthalmia Transcription Factor Is a Target of the p38 MAPK Pathway in Response to Receptor Activator of NF- B Ligand Signaling. Journal of Biological Chemistry. 2002;277:11077–11083. doi: 10.1074/jbc.M111696200. [DOI] [PubMed] [Google Scholar]
- 44.Miller AJ, Levy C, Davis IJ, Razin E, Fisher DE. Sumoylation of MITF and its related family members TFE3 and TFEB. Journal of Biological Chemistry. 2005;280:146–155. doi: 10.1074/jbc.M411757200. [DOI] [PubMed] [Google Scholar]
- 45.Murakami H, Arnheiter H. Sumoylation modulates transcriptional activity of MITF in a promoter-specific manner. Pigment Cell Res. 2005;18:265–277. doi: 10.1111/j.1600-0749.2005.00234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haq R, et al. Oncogenic BRAF regulates oxidative metabolism via PGC1α and MITF. Cancer Cell. 2013;23:302–315. doi: 10.1016/j.ccr.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johannessen CM, et al. A melanocyte lineage program confers resistance to MAP kinase pathway inhibition. Nature. 2014;504:138–142. doi: 10.1038/nature12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yokoyama S, et al. A novel recurrent mutation in MITF predisposes to familial and sporadic melanoma. Nature. 2011;480:1–7. doi: 10.1038/nature10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bertolotto C, et al. A SUMOylation-defective MITF germline mutation predisposes to melanoma and renal carcinoma. Nature. 2011;480:94–98. doi: 10.1038/nature10539. [DOI] [PubMed] [Google Scholar]
- 50.Shoag J, et al. PGC-1 Coactivators Regulate MITF and the Tanning Response. Molecular Cell. 2013;49:145–157. doi: 10.1016/j.molcel.2012.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Feige EE, et al. Hypoxia-induced transcriptional repression of the melanoma-associated oncogene MITF. Proc. Natl. Acad. Sci. U.S.A. 2011;108:E924–E933. doi: 10.1073/pnas.1106351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoek KS, et al. Novel MITF targets identified using a two-step DNA microarray strategy. Pigment Cell & Melanoma Research. 2008;21:665–676. doi: 10.1111/j.1755-148X.2008.00505.x. [DOI] [PubMed] [Google Scholar]
- 53.Strub T, et al. Essential role of microphthalmia transcription factor for DNA replication, mitosis and genomic stability in melanoma. Oncogene. 2011;30:2319–2332. doi: 10.1038/onc.2010.612. [DOI] [PubMed] [Google Scholar]
- 54.McGill GG, et al. Bcl2 regulation by the melanocyte master regulator Mitf modulates lineage survival and melanoma cell viability. Cell. 2002;109:707–718. doi: 10.1016/s0092-8674(02)00762-6. [DOI] [PubMed] [Google Scholar]
- 55.Levy C, et al. Lineage-specific transcriptional regulation of DICER by MITF in melanocytes. Cell. 2010;141:994–1005. doi: 10.1016/j.cell.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Khaled M, Levy C, Fisher DE. Control of melanocyte differentiation by a MITF-PDE4D3 homeostatic circuit. Genes & Development. 2010;24:2276–2281. doi: 10.1101/gad.1937710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Du J, et al. Critical role of CDK2 for melanoma growth linked to its melanocyte-specific transcriptional regulation by MITF. Cancer Cell. 2004;6:565–576. doi: 10.1016/j.ccr.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 58.Jacobs AL, Schär P. DNA glycosylases: in DNA repair and beyond. Chromosoma. 2011;121:1–20. doi: 10.1007/s00412-011-0347-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carreira S, Liu B, Goding CR. The gene encoding the T-box factor Tbx2 is a target for the microphthalmia-associated transcription factor in melanocytes. Journal of Biological Chemistry. 2000;275:21920–21927. doi: 10.1074/jbc.M000035200. [DOI] [PubMed] [Google Scholar]
- 60.Vance KW, Carreira S, Brosch G, Goding CR. Tbx2 is overexpressed and plays an important role in maintaining proliferation and suppression of senescence in melanomas. Cancer Research. 2005;65:2260–2268. doi: 10.1158/0008-5472.CAN-04-3045. [DOI] [PubMed] [Google Scholar]
- 61.Carreira S, et al. Mitf cooperates with Rb1 and activates p21Cip1 expression to regulate cell cycle progression. Nature. 2005;433:764–769. doi: 10.1038/nature03269. [DOI] [PubMed] [Google Scholar]
- 62.Loercher AE, Tank EMH, Delston RB, Harbour JW. MITF links differentiation with cell cycle arrest in melanocytes by transcriptional activation of INK4A. The Journal of Cell Biology. 2005;168:35–40. doi: 10.1083/jcb.200410115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Haq R, et al. BCL2A1 is a lineage-specific antiapoptotic melanoma oncogene that confers resistance to BRAF inhibition. Proc. Natl. Acad. Sci. U.S.A. 2013;110:4321–4326. doi: 10.1073/pnas.1205575110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dynek JN, et al. Microphthalmia-associated transcription factor is a critical transcriptional regulator of melanoma inhibitor of apoptosis in melanomas. Cancer Research. 2008;68:3124–3132. doi: 10.1158/0008-5472.CAN-07-6622. [DOI] [PubMed] [Google Scholar]
- 65.McGill GG, Haq R, Nishimura EK, Fisher DE. c-Met expression is regulated by Mitf in the melanocyte lineage. Journal of Biological Chemistry. 2006;281:10365–10373. doi: 10.1074/jbc.M513094200. [DOI] [PubMed] [Google Scholar]
- 66.Straussman R, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487:500–504. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sánchez-Martín M, et al. SLUG (SNAI2) deletions in patients with Waardenburg disease. Human Molecular Genetics. 2002;11:3231–3236. doi: 10.1093/hmg/11.25.3231. [DOI] [PubMed] [Google Scholar]
- 68.Liu F, Fu Y, Meyskens FL. MiTF regulates cellular response to reactive oxygen species through transcriptional regulation of APE-1/Ref-1. Journal of Investigative Dermatology. 2009;129:422–431. doi: 10.1038/jid.2008.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Busca R. Hypoxia-inducible factor 1 is a new target of microphthalmia-associated transcription factor (MITF) in melanoma cells. The Journal of Cell Biology. 2005;170:49–59. doi: 10.1083/jcb.200501067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ito S, Wakamatsu K. Chemistry of mixed melanogenesis--pivotal roles of dopaquinone. Photochemistry and Photobiology. 2008;84:582–592. doi: 10.1111/j.1751-1097.2007.00238.x. [DOI] [PubMed] [Google Scholar]
- 71.Samokhvalov A, et al. Oxidation potentials of human eumelanosomes and pheomelanosomes. Photochemistry and Photobiology. 2006;81:145–148. doi: 10.1562/2004-07-23-RC-245. [DOI] [PubMed] [Google Scholar]
- 72.Meredith P, Sarna T. The physical and chemical properties of eumelanin. Pigment Cell Res. 2006;19:572–594. doi: 10.1111/j.1600-0749.2006.00345.x. [DOI] [PubMed] [Google Scholar]
- 73.Chen KG, Valencia JC, Gillet J-P, Hearing VJ, Gottesman MM. Involvement of ABC transporters in melanogenesis and the development of multidrug resistance of melanoma. Pigment Cell & Melanoma Research. 2009;22:740–749. doi: 10.1111/j.1755-148X.2009.00630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mitra D, et al. An ultraviolet-radiation-independent pathway to melanoma carcinogenesis in the red hair/fair skin background. Nature. 2012;491:413–417. doi: 10.1038/nature11624. [DOI] [PMC free article] [PubMed] [Google Scholar]