Abstract
The perpetual arms race between bacteria and phage has resulted in the evolution of efficient resistance systems that protect bacteria from phage infection. Such systems, which include the CRISPR-Cas and restriction-modification systems, have proven to be invaluable in the biotechnology and dairy industries. Here, we report on a six-gene cassette in Bacillus cereus which, when integrated into the Bacillus subtilis genome, confers resistance to a broad range of phages, including both virulent and temperate ones. This cassette includes a putative Lon-like protease, an alkaline phosphatase domain protein, a putative RNA-binding protein, a DNA methylase, an ATPase-domain protein, and a protein of unknown function. We denote this novel defense system BREX (Bacteriophage Exclusion) and show that it allows phage adsorption but blocks phage DNA replication. Furthermore, our results suggest that methylation on non-palindromic TAGGAG motifs in the bacterial genome guides self/non-self discrimination and is essential for the defensive function of the BREX system. However, unlike restriction-modification systems, phage DNA does not appear to be cleaved or degraded by BREX, suggesting a novel mechanism of defense. Pan genomic analysis revealed that BREX and BREX-like systems, including the distantly related Pgl system described in Streptomyces coelicolor, are widely distributed in ∼10% of all sequenced microbial genomes and can be divided into six coherent subtypes in which the gene composition and order is conserved. Finally, we detected a phage family that evades the BREX defense, implying that anti-BREX mechanisms may have evolved in some phages as part of their arms race with bacteria.
Keywords: CRISPR, PGL, pglZ, phage defense
See also: R Barrangou & J van der Oost (January 2015)
Introduction
The ongoing arms race between bacteria and bacteriophages (phages) has led to the rapid evolution of extensive mechanisms to combat phage infection (Labrie et al, 2010; Stern & Sorek, 2011). Among these are restriction-modification systems (Tock & Dryden, 2005), abortive infection (Abi) mechanisms (Chopin et al, 2005), and the CRISPR-Cas adaptive defense system (Sorek et al, 2008; van der Oost et al, 2009; Deveau et al, 2010; Horvath & Barrangou, 2010). The relatively recent discovery of the complex and abundant CRISPR-Cas system highlights the fact that our knowledge of the arsenal of phage-defense mechanisms encoded in bacterial and archaeal genomes is incomplete. Indeed, accumulating evidence suggest that many additional phage resistance systems present in microbial genomes have yet to be discovered (Stern & Sorek, 2011; Makarova et al, 2013; Swarts et al, 2014).
A recent study has reported that genes involved in phage resistance, such as restriction-modification enzymes and toxin–antitoxin systems, are non-randomly clustered to specific genomic locations in bacterial and archaeal genomes, forming genomic ‘defense islands’ (Makarova et al, 2011). One of the genes found enriched within defense islands is pglZ, a putative member of the alkaline phosphatase superfamily. This gene was previously reported as essential for a unique phage resistance phenotype in Streptomyces coelicolor A3(2), denoted phage growth limitation (Pgl) (Chinenova et al, 1982). Streptomyces coelicolor strains carrying the Pgl system are sensitive to the first cycle of infection by phage ΦC31, but are resistant to phages emerging from this first cycle. Further studies mapped the Pgl phenotype to a cluster of four genes that were able to reconstitute the phenotype upon transfer to a new host (Sumby & Smith, 2002). These genes include pglZ, a putative phosphatase; pglW, a serine/threonine kinase domain-containing protein; pglX, a protein containing an adenine-specific DNA methyltransferase motif; and pglY, a protein containing a P-loop domain (Sumby & Smith, 2002). The Pgl system was active against ΦC31 and its homoimmune relatives, but not to any of the other phage that were tested (Laity et al, 1993). A molecular mechanism to explain the activity of the Pgl system was never deciphered.
Based on the enrichment of pglZ-domain genes in genomic defense islands, it was suggested that genes containing this domain are involved in phage defense in multiple species (Makarova et al, 2011). In this work, we analyzed ∼1,500 bacterial and archaeal genomes and found that pglZ-domain genes are present in about 10% of these genomes. Moreover, in more than half of the cases, the pglZ-domain gene was found embedded in a gene cluster composed of six genes, two of which (pglZ and pglX) display considerable sequence homology to genes in the Pgl system, and four additional genes that encode a putative protease, a protein with an ATPase domain, a predicted RNA-binding protein, and a gene of unknown function. We hypothesized that this gene cluster forms a novel phage-defense system, which we denote BREX (Bacteriophage Exclusion). Indeed, we show that transfer of this new system from Bacillus cereus into Bacillus subtilis provides B. subtilis with resistance to a broad range of Bacillus phages, both virulent and temperate phage. Our data show that phage adsorption occurs in BREX-containing strains, but that neither phage DNA replication nor lysogeny occurs, and that this system does not act via an abortive infection mechanism. This system does not display the Pgl phenotype and hence probably functions through a novel mode of action, different than that of the Pgl system. We provide further evidence that the system methylates chromosomal DNA at a specific motif and that this methylation is likely to be essential for the system's activity. Pan genomic and phylogenetic analyses further show that BREX undergoes extensive horizontal gene transfer and that pglZ-containing gene clusters can be divided into six coherent BREX subtypes in which the gene order and composition are conserved. Each BREX subtype contains 4–8 genes, some of which are core genes while others are subtype-specific. Finally, we found that a minority of phages escaped BREX defense, implying that these phages may have evolved anti-BREX mechanisms.
Results
BREX is abundant in bacteria and archaea
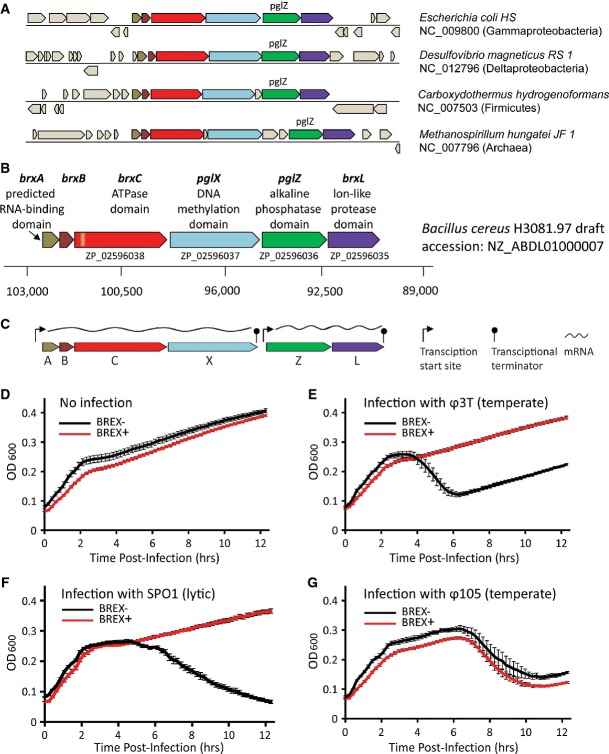
Previous analyses of pglZ-domain genes demonstrated that this domain is enriched in defense islands of bacteria and archaea (Makarova et al, 2011), and documented a number of genes commonly associated with pglZ-domain genes (Makarova et al, 2011, 2013). To understand whether there is higher order organization among pglZ and its associated genes, we performed homology searches and found 144 occurrences of pglZ-domain genes in the 1,447 finished bacterial and archaeal genomes analyzed (Supplementary Table S1, Materials and Methods). Remarkably, in 55% of the cases (79 of 144), the pglZ gene was embedded within a 6-gene cluster arranged in a highly conserved order in a diverse array of bacteria and archaea (Fig1A; Supplementary Table S2). Subsequent searches conducted on 5,493 draft genome sequences deposited in NCBI showed that this 6-gene cluster is present in an additional 290 genomes (Supplementary Table S3).
Figure 1. BREX confers resistance to phage.
- A The pglZ gene reproducibly appears within a six-gene cluster in various genomes. Representative appearances are shown.
- B The BREX locus in B. cereus H3081.97. Coordinates below the genes denote the position along the NZ_ABDL01000007 contig in the draft genome of B. cereus H3081.97. Orange box within brxC represents the position of the ATPase P-loop motif.
- C Operon organization of the B. cereusBREX system integrated in the B. subtilis genome. Expression throughout the operons was validated by RNA-seq. Positions of promoters and terminators were inferred by 5′ and 3′ RACE, respectively.
- D–G Culture dynamics of phage-infected wild-type (black) versus BREX-containing (red) strains of B. subtilisBEST7003. Bacterial strains were exposed to phage at time = 0 h, and optical density measurements were read in a 96-well plate format. Each experiment was performed three times with three technical triplicates for each biological replicate. Error bars represent SEM. Shown are representative results for three of the ten phages tested; data for the remaining seven phages are found in Supplementary Fig S2.
Two of the six genes found in this conserved cluster share homology with genes from the previously reported Pgl system (Sumby & Smith, 2002): pglZ, coding for a protein with a predicted alkaline phosphatase domain, and pglX, coding for a protein with a putative methylase domain. The four additional genes include (i) a Lon-like protease-domain gene, denoted here brxL; (ii) a gene coding for a protein with significant structural homology to the RNA-binding antitermination protein NusB (brxA, see Supplementary Fig S1); (iii) a gene of unknown function (brxB); and (iv) a large, ∼1,200 amino acid protein with an ATP-binding motif (GXXXXGK[T/S]), which we denote brxC. Although this does not resemble any classical combination of genes currently known to be involved in phage defense, the preferential localization of this conserved gene cluster in the genomic vicinity of other defense genes suggests that it could form a novel phage-defense system. We denote this putative defense system the BREX (Bacteriophage Exclusion) system.
BREX confers resistance to phage infection in Bacillus subtilis
To determine whether the BREX system provides protection against phage infection, the complete BREX system from Bacillus cereus H3081.97 (Fig1B) was cloned into a Bacillus subtilis BEST7003 strain lacking an endogenous BREX system. Proper integration of the intact system into the B. subtilis genome was verified by complete genome sequencing. We then verified, using RNA-seq, that the genes of the integrated system are transcribed in Bacillus subtilis when grown in exponential phase in rich medium. Furthermore, using 5′ and 3′ RACE, we determined that the system is transcribed as two operons with the first four genes, brxA-brxB-brxC-pglX, forming a single transcriptional unit, while the last two genes, pglZ-brxL, are co-expressed as a second transcriptional unit (Fig1C, Materials and Methods). The observation that the genes in the putative BREX system are co-transcribed as two long polycistronic mRNAs further supports that they work together as components of a functional system.
Ten B. subtilis phages were selected for phage infection experiments, spanning a wide range of phage phylogeny, including Myoviridae (phages SPO1 and SP82G), lambda-like Siphoviridae phages (Φ105,rho10, rho14,and SPO2), and SPβ-like Siphoviridae phages (SPβ,Φ3T, SP16, and Zeta). Two of the phages are obligatory lytic (SPO1 and SP82G), while the remaining are temperate (Table1). The phage sensitivity of B. subtilis strains either containing or lacking the BREX system was evaluated using both optical density measurements in a 96-well plate format, and double agar overlay plaque assays.
Table 1.
BREX protection against the phages used in this study
| Phage | Genus | Family | Life cycle | Infection blocked by BREX? | Efficiency of BREX protectiona |
|---|---|---|---|---|---|
| SPβ | SPβ-like | Siphoviridae | Temperate | Yes | > 105 |
| SP16 | SPβ-like | Siphoviridae | Temperate | Yes | > 105 |
| Zeta | SPβ-like | Siphoviridae | Temperate | Yes | > 105 |
| Φ3T | SPβ-like | Siphoviridae | Temperate | Yes | > 105 |
| SPO2 | Lambda-like | Siphoviridae | Temperate | Yes | > 105 |
| Φ105 | Lambda-like | Siphoviridae | Temperate | No | 1 |
| rho10 | Lambda-like | Siphoviridae | Temperate | No | 1 |
| rho14 | Lambda-like | Siphoviridae | Temperate | No | 1 |
| SPO1 | SPO1-like | Myoviridae | Obligatory lytic | Yes | 8 × 102 ± 0.02 |
| SP82G | SPO1-like | Myoviridae | Obligatory lytic | Yes | 1.8 × 101 ± 0.08 |
Protection efficiency was calculated as the ratio between the number of plaques formed on the BREX-lacking strain divided by the number of plaques formed on the BREX-containing strain with the same phage titer, using increasing titers. Standard deviation is calculated from a biological triplicate of the plaque experiment.
Upon phage infection, the B. subtilis strain containing the BREX system showed resistance to seven of the ten phages tested (Table1). Growth curves of BREX-containing bacteria infected with these seven phages at a multiplicity of infection (MOI) of 10−3–10−4 were similar to the uninfected bacteria, while declines in optical density measurements were observed for the control strain lacking the BREX system, indicating lysis of the infected cells (Fig1D–F; Supplementary Fig S2). These results confirm that BREX is a phage-defense system that provides protection against a wide array of phages, including both virulent and temperate ones.
In contrast to the protection from phage infection observed with the first seven phages tested, phage resistance was not observed upon infection with phage Φ105 and its close relatives, rho10 and rho14. Similar kinetics of cell lysis were observed for strains either containing or lacking the BREX system (Fig1G; Supplementary Fig S2). Considering that phage Φ105 is estimated to share high (83–97%) genome homology with rho10 and rho14 (Rudinski & Dean, 1979), the inability of the BREX system to protect against these three phages could indicate that this phage family has evolved strategies to counteract the BREX defense, as has been observed with other bacterial defense systems (Bondy-Denomy et al, 2013). If such strategies exist, their identification could provide insight into the mechanism of action of the BREX system. Alternatively, the resistance of phage Φ105 and its relatives to the BREX system could also stem from intrinsic differences in the infection cycle of these phages making them immune to BREX-mediated defense, or because they do not encode a target for the BREX activity.
To further evaluate the level of protection provided by the BREX system against the phages that were tested, plaque assays were performed using increasing phage concentrations. For five of the phages, no plaques were observed when the BREX-containing strain was challenged even with the highest phage concentrations, indicating that the BREX system provides at least 105-fold protection against cell lysis upon infection (Table1). Plaque assays also confirmed that phage Φ105 and its relatives evade BREX defense, because similar efficiencies of plating and plaque morphology were observed in both BREX-containing and wild-type control strains (Table1).
Interestingly, for two of the phages tested, SPO1 and SP82G, plaque assays showed only a 101-fold reduction in plaque numbers in BREX-containing strains (Table1). These results were consistent with the observation that incubation of the BREX-containing strain with these two phages for extended periods of time (> 20 h) often resulted in an eventual culture decline occurring at apparently stochastic points in time (Fig2A). To gain further insight into the nature of the incomplete BREX defense against these phages, we performed a one-step phage growth curve assay (Carlson, 2005) with SPO1. Briefly, this experiment involves mixing SPO1-infected cells with a SPO1-sensitive B. subtilis cells and plating them together using an agar overlay method. Phage bursts from successful infections are visualized as a single plaque on a lawn from the SPO1-sensitive B. subtilis strain, enabling an evaluation of the number of phages that have adsorbed and completed a successful infection cycle (Materials and Methods). Enumeration of plaques during the first 45 min of the time course infection indicated that the SPO1 phage was able to complete the lytic cycle only in 9 ± 4% of the initially infected cells (Fig2B). A delay in the kinetics of the phage cycle was also observed, with phage bursts observed 75 and 105 min following infection of BREX-lacking and BREX-containing cells, respectively (Fig2B). Together, these results suggest that the BREX system provides significant, but not complete, protection from infection by phages SPO1 and SP82G.
Figure 2. Phage infection dynamics in BREX-containing cells.
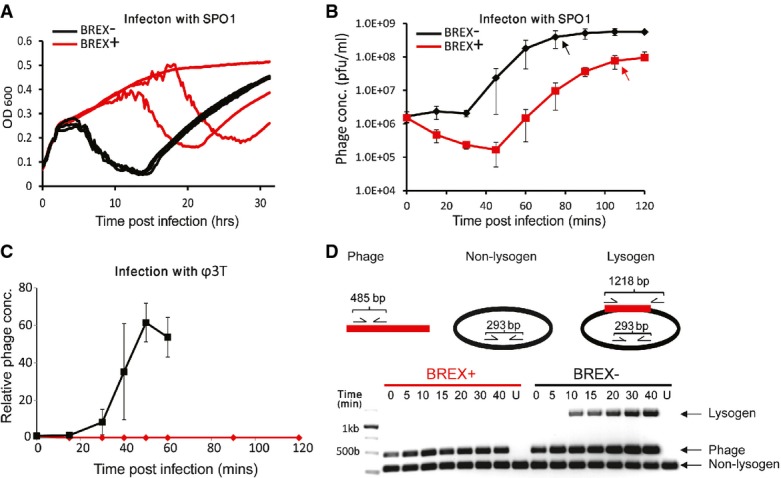
- Culture dynamics over an extended period (> 30 h) for SPO1-infected wild-type (black) versus BREX-containing (red) strains of B. subtilisBEST7003. Each curve represents a single technical replicate grown in a single well on a 96-well plate. Culture decline is temporally reproducible for the BREX-lacking strain but occurs later, at apparently stochastic time points, for the BREX-containing strain. Re-growth following culture crash represents phage-resistant mutants.
- Phage production during a one-step phage growth curve experiment with wild-type (black) and BREX-containing (red) strains of B. subtilisBEST7003 infected with SPO1. Error bars represent SD. Y-axis represents absolute phage concentrations. Black and red arrows point to the time point of maximal burst for BREX-lacking and BREX-containing strains, respectively.
- Phage production during a one-step phage growth curve experiment with wild-type (black) and BREX-containing (red) strains of B. subtilis BEST7003 infected with Φ3T. Error bars represent SD. Y-axis represents relative phage concentrations normalized to the value at the beginning of the infection.
- Multiplex PCR assay showing lysogeny during a phage infection time course in the strain lacking BREX (black), but not in BREX-containing strain (red), or uninfected (U) strains. Amplicons for the bacterial DNA, phage DNA, and lysogen-specific DNA are 293, 485, and 1,218 bp, respectively.
The mode of action of BREX is different than that of the Pgl system
BREX is an apparently complex system with proteins that are predicted to have multiple biochemical activities (e.g., protease, phosphatase, methylase). Fully deciphering its mechanism of action and understanding the role of each of its six proteins in phage defense is expected to be non-trivial and would probably require multiple genetic, biochemical, and structural studies. Here, we set out to provide initial insights into the function of the BREX system.
Due to the homology of a subset of the genes in the BREX system to genes in the Pgl system, we first examined whether BREX also functions through the Pgl mode of action. The Pgl phenotype observed in S. coelicolor A3 predicts that the first infection cycle by the phage would be successful, producing viable phage progeny. We used one-step phage growth curve assays to examine the first infection cycle of phage Φ3T in BREX-containing cells. While wild-type control strains displayed phage burst sizes of 61.5 ± 10.2 particles per infected cell (Fig2C), there was no production of Φ3T phage during infections of BREX-containing cells under similar conditions. To exclude the possibility that productive phage infection could occur, but at later times, experiments were extended to 120 min (three infection cycles in wild-type strains) in BREX-containing cells. Plaques were not observed, even at later times (Fig2C). These results demonstrate that unlike the S. coelicolor Pgl system, the BREX system halts Φ3T production prior to the first round of infection.
Previous experiments with the S. coelicolor Pgl system demonstrated that although the Pgl defense system prevents continued propagation of the temperate phage ΦC31, it does not block lysogeny of the phage (Chinenova et al, 1982). To determine whether BREX also permits lysogeny, we examined phage Φ3T integration into the B. subtilis genome during infection using a PCR assay. In wild-type control strains, lysogeny was first detected 10 min following phage infection (Fig2D). However, no evidence for phage integration into the host genome was found in BREX-containing cells. Evaluation of lysogeny in bacterial colonies that survived the phage infection also indicated that none of the surviving BREX-containing colonies were lysogens, while all surviving colonies tested in strains lacking the BREX system were lysogenic for phage Φ3T. Together, these results suggest that although BREX and Pgl share a subset of genes, the two systems probably exert their defense through different modes of action.
BREX is not an abortive infection system
One of the common forms of phage defense is abortive infection (Abi), where infected cells commit ‘suicide’ before phage progeny are produced, thus protecting the culture from phage propagation (Chopin et al, 2005). Such a phenotype predicts that with a high MOI, where nearly all bacteria are infected in the first cycle, massive cell death will be observed in the culture. To test whether the BREX system works through an Abi mechanism, we infected the BREX-containing B. subtilis strain with increasing concentrations of the Φ3T phage against which the BREX was shown to provide resistance. Even at an MOI > 1, no significant growth arrest or culture decline was found in the liquid culture, suggesting that the BREX is not an Abi system (Fig3A).
Figure 3. Initial characterization of BREX activity.
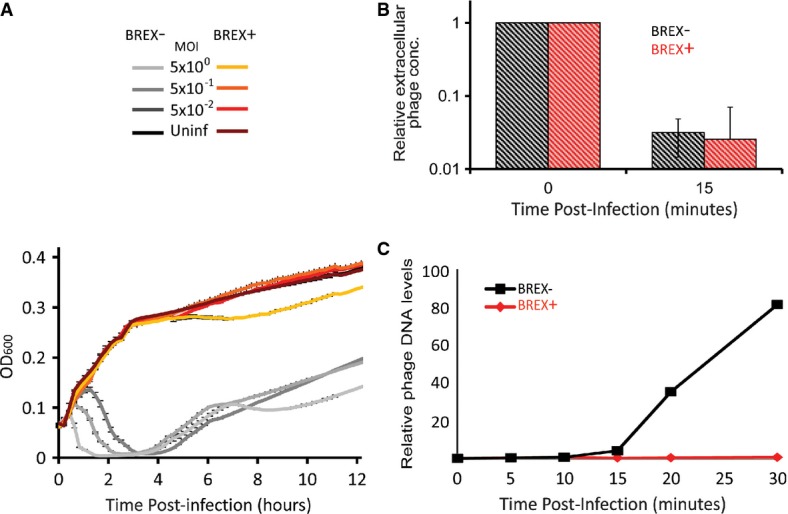
- BREX does not work via an abortive infection mechanism. Increasing the MOI of the Φ3T infection from 0.05 to 5 shortens the time to culture crash for the BREX-lacking strain, but does not result in culture decline for BREX-containing strain. Error bars represent SD of technical triplicates.
- The system does not interfere with phage adsorption to bacterial cells. Strains either containing (red) or lacking (black) the BREX system were infected with phage Φ3T and then chloroform-treated 15 min following infection. The culture was plated on Φ3T-sensitive B. subtilis cells and plaques, representing extracellular, unadsorbed phages, were counted.
- Phage DNA replication does not occur in BREX-containing cells (red), but is observed in the BREX-lacking strain (black). Y-axis represents relative phage concentrations normalized to the value at the beginning of the infection, as measured by Illumina sequencing.
BREX allows phage adsorption but blocks phage DNA replication
To gain further insight into the stage at which the infection cycle is blocked by BREX, we asked whether phage adsorption is prevented. Adsorption assays showed that Φ3T efficiently adsorbs to both BREX-containing and BREX-lacking cells, indicating that BREX does not block adsorption (Fig3B). We then assayed whether BREX allows phage DNA replication within infected cells. For this, we extracted total cellular DNA (including chromosomal DNA and intracellular phage DNA) at successive time points following a high-MOI infection by Φ3T and submitted the extracted DNA to Illumina sequencing. Since host DNA is not degraded following Φ3T infection (Supplementary Fig S3), mapping the sequenced reads to the reference B. subtilis and Φ3T genomes allowed quantification of the number of Φ3T genome equivalents per infected cell at each time point. In wild-type control cells, phage DNA replication began between 10 and 15 min following infection, and after 30 min, phage DNA levels were elevated 81-fold relative to that observed at the 10-min time point (Fig3C). In contrast, no increase in phage DNA levels was observed in BREX-containing cells (Fig3C). These results indicate that phage DNA replication does not occur in BREX-containing cells and that this system exerts its function at the early stages of the infection cycle.
BREX methylates bacterial DNA but does not degrade phage DNA
The presence of a predicted m6A DNA adenine methylase (the pglX gene) in the BREX system prompted us to examine whether either bacterial or phage DNA are methylated in a BREX-dependent manner. To test this, we used the PacBio sequencing platform that directly detects m6A modifications in sequenced DNA (Murray et al, 2012). In DNA extracted from BREX-containing cells, the PacBio platform clearly detected m6A methylation on the 5th position of the non-palindromic hexamer TAGGAG (Fig4A). While nearly all TAGGAG motifs were methylated in BREX-containing B. subtilis cells (Fig4B), no methylation on this motif was observed in the strain lacking the BREX system. These results suggest that BREX drives motif-specific methylation on the genomic DNA of the bacteria in which it resides.
Figure 4. Methylation activity of BREX.
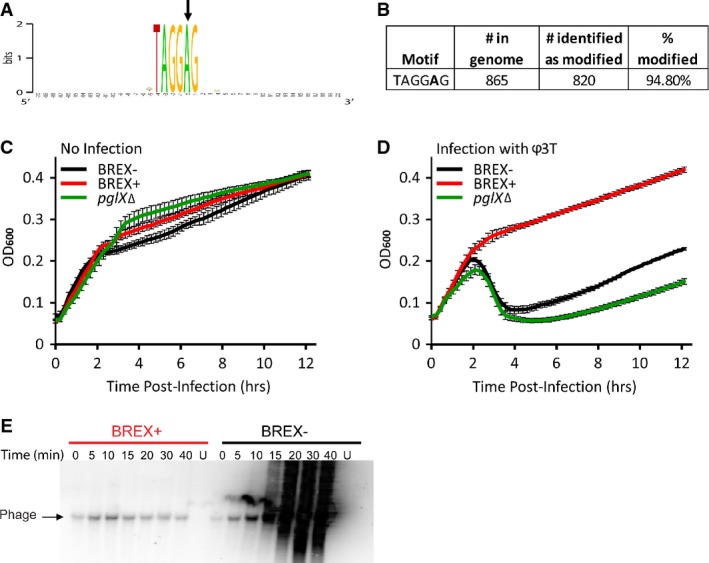
- A Consensus sequence around m6A modified bases in the BREX-containing B. subtilis genome. The modified base is marked by an arrow. Modifications were directly detected by DNA sequencing using the PacBio platform.
- B Statistics of modified motifs in the BREX-containing B. subtilis genome.
- C, D Culture dynamics of non-infected (C) or phage-infected (D) cultures. Curves depict culture dynamics of strains lacking BREX (black) and BREX-containing (red) strains of B. subtilisBEST7003, as well as a BREX-containing strain where the pglX methylase was deleted (green). Axes and error bars are as in Fig1.
- E Southern blot analysis of phage Φ3T genome during infection. Numbers indicate time (in min) following infection; U, uninfected. Probe was designed to match positions 94,645–95,416 in the phage genome. Each lane contains 200 ng total DNA.
To examine whether BREX also methylates the invading phage DNA, we extracted total cellular DNA (including chromosomal DNA and intracellular phage DNA) at 10 and 15 min following a high-MOI infection by Φ3T and subjected the extracted DNA to PacBio sequencing. As in the previous assay, we found that TAGGAG motifs in the bacterial genome were methylated throughout the infection. However, none of the 43 TAGGAG motifs present on the phage genome was found to be methylated at any of the time points sampled during infection.
The presence of bacterial-specific methylation could suggest that the BREX system encodes some kind of restriction-modification activity and that the methylation of TAGGAG motifs in the bacterial genome may serve to differentiate between self and non-self DNA. This hypothesis would predict that deletion of the methylase gene, pglX, would be detrimental to the cell, as the genomic TAGGAG motifs will no longer be protected from the putative restriction activity of BREX. However, deletion of the pglX from the BREX system that we integrated into B. subtilis was not toxic to the cells (Fig4C). Moreover, BREX-containing strains having a deletion of pglX were sensitive to all phage tested (Fig4D). These results show that pglX is essential for BREX-mediated phage resistance and also suggest that the BREX mechanism of action is not consistent with a simple restriction-modification activity.
To further test whether BREX leads to cleavage or degradation of phage DNA, we examined the integrity of phage DNA using Southern blot analyses on total cellular DNA extracted from phage-infected cells at increasing time points following infection. The Southern blot demonstrated extensive replication of phage DNA in BREX-lacking cells and confirmed that no phage DNA replication occurs in BREX-containing cells (Fig4E). However, in BREX-containing cells, the phage DNA appeared intact, with no signs of phage DNA cleavage or processive degradation (Fig4E). These results further imply that BREX inhibits phage propagation in a mechanism other than direct degradation of phage DNA.
BREX can confer mild protection against plasmid transformation
Many defense systems, including restriction enzymes and CRISPR, can confer resistance against both invading phages and plasmids. To examine whether BREX can also block plasmids, we compared plasmid transformation efficiency between BREX-containing and control cells, using three different plasmids (two integrative and one episomal, low copy plasmid, with sizes ranging between 6.7 and 8.8 kb). No considerable reduction in transformation efficiency was observed for the two integrative plasmids, whereas a mild effect (∼1 order of magnitude) was observed for the episomal plasmid (Supplementary Fig S4). These results show that plasmids can also be targeted, to a certain extent, by the BREX system. None of the plasmids, however, was blocked as efficiently as many of the phages we tested (> 5 orders of magnitude). This may suggest that the plasmids we used do not contain strong targets for BREX or that BREX specifically targets other characteristics of foreign DNA that are inherent to phage infection.
BREX belongs to a superfamily of defense systems divided into six major subtypes
The experimental results presented above show that BREX is a phage-defense system that contains significant mechanistic complexity. To gain a deeper understanding of the evolution of this system and its relatives, we set out to perform a phylogenetic analysis of its main components.
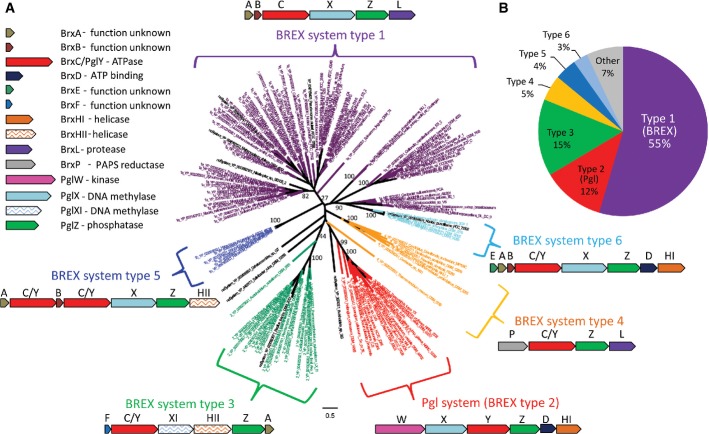
As indicated above, genes with pglZ domains were found in ∼10% of all the completely sequenced (finished) genomes analyzed in this study. This domain is present in at least two characterized phage-defense systems: the BREX system described here and the Pgl system described in S. coelicolor. To gain a more global view of potential defense systems containing pglZ-domain genes, we reconstructed the phylogenetic tree of all the PglZ-domain proteins collected. This tree shows clear clustering of PglZ into several distinct phyletic groups (Fig5A). The largest group of pglZ-domain genes (colored purple in Fig5A; Fig5B) corresponds to the BREX system, and PglZ genes belonging to this group were found to be embedded in the typical BREX six-gene cluster we described here. A second, distinct clade of the PglZ tree (red clade in Fig5A) contains the S. coelicolor pglZ, previously shown to be part of the Pgl system. An examination of the genes in the near vicinity of this Pgl clade showed that all pglZ genes were embedded in a pglWXYZ gene cluster typical of the Pgl system (Sumby & Smith, 2002), and this clade therefore encompasses the Pgl systems.
Figure 5. PglZ phylogeny and classification of BREX subtypes.
- Shown is a phylogenetic tree of the PglZ protein instances detected in this study. The tree is color-coded according to the operon organization of the neighboring genes (BREX types). The gene order and genomic organization of each type is illustrated next to its relevant branch on the PglZ tree. Numbers depict bootstrap values.
- Prevalence of the different BREX subtypes within the BREX superfamily of defense systems among the sequenced genomes that were analyzed.
In a similar manner, for almost every major clade of the PglZ tree, we were able to detect a coherent set of 4–8 associated genes appearing in a highly conserved order, defining clear organizational multi-gene systems (Fig5A). The composition and order of these genes was highly coherent within each phyletic group but differed between the clades. We hypothesize that all these systems represent a superfamily of phage-defense systems that includes BREX, Pgl, and four additional related defense systems. Moreover, since the individual clades separate close to the root of the PglZ tree (Fig5A), and since the six systems we defined are widespread across the entire bacterial and archaeal tree of life (Fig6), our results suggest that the separation between the systems occurred at an ancient point in the evolutionary history of bacteria and archaea. The BREX system is by far the most common system in this superfamily among the genomes sequenced thus far (Fig5B). For this reason, we suggest to name the superfamily after this system, with type 1 BREX representing the system described above in this paper, type 2 BREX corresponding to the Pgl system, and types 3–6 representing the additional putative defense systems belonging to the BREX superfamily.
Figure 6. Distribution of BREX systems across the phylogenetic tree of bacteria and archaea.
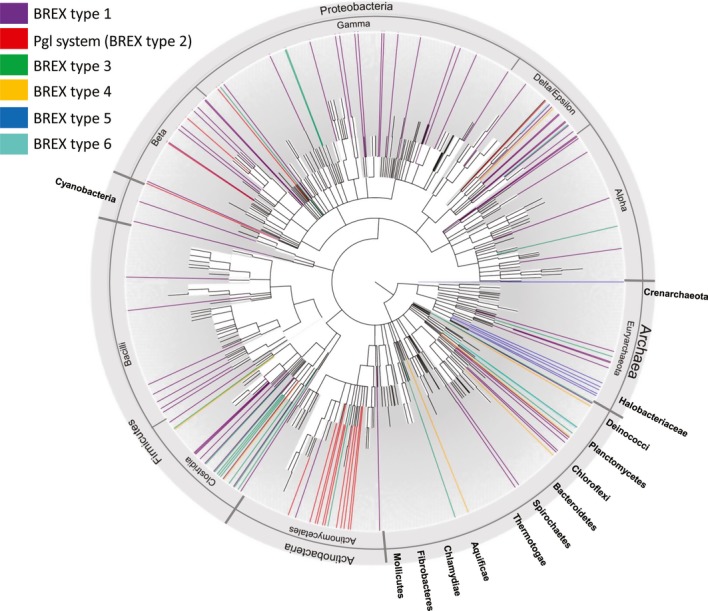
Shown is the common tree of bacteria and archaea as represented in the NCBI Taxonomy resource (Materials and Methods). Organisms in which a BREX system exists are colored; color code follows the BREX subtype coloring from Fig5. Extensive horizontal transfer is observed by the lack of coherence between the species tree and the PglZ phylogeny.
Overall, 13 gene families were found to be associated with BREX systems (Table2), largely consistent with previous reports on genes enriched in the vicinity of pglZ in defense islands (Makarova et al, 2011). By definition, all systems contain a pglZ-domain gene. In addition, all of them harbor a large protein (∼1,200 aa) with a P-loop motif. The P-loop motif (GXXXXGK[T/S]) is a conserved ATP/GTP-binding motif that is ubiquitously found in many ATP-utilizing proteins such as kinases, helicases, motor proteins, and proteins with multiple other functions (Thomsen & Berger, 2008). In general, the P-loop-containing genes in the various BREX subtypes share little homology: For example, the brxC gene of BREX type 1 and pglY gene of Pgl system share homology only across 4% of their protein sequence, and this homology is concentrated around the P-loop motif (Supplementary Fig S5). Despite the low homology, distant homology analysis with HHpred (Soding, 2005) showed that they share a domain of unknown function denoted DUF499 (Table2). We therefore hypothesize that the P-loop-containing genes in all six BREX subtypes share a similar role in the system and hence refer to these genes (brxC/pglY) as having a common function (Table2 and Fig5). Apart from the two core genes pglZ and brxC/pglY that appear in each of the six BREX subtypes, the remaining genes are subtype specific or are restricted to only a subset of the BREX subtypes.
Table 2.
Genes composing the BREX systems
| Gene | Subsystems in which gene appears | Associated domains | Domain annotation | Median gene size (aa) |
|---|---|---|---|---|
| pglZ | Core gene | pfam08665 | Alkaline phosphatase | 835 |
| brxC/pglY | Core gene | DUF499, DUF2791 (pfam10923) | ATP binding | 1,208 |
| brxA | 1,3,5,6 | DUF1819 (pfam08849) | Unknown function | 232 |
| pglX | 1,2,5,6 | pfam13659 (COG1002/COG0286) | Adenine-specific methylase | 1,175 |
| brxL | 1,4 | COG4930 | Lon-like protease | 682 |
| brxHI | 2,6 | COG1201 | Lhr-like helicase | 712 |
| brxHII | 3,5 | COG0553 | DNA/RNA helicases | 965 |
| brxD | 2,6 | DUF2791 (pfam10923) | ATP binding | 442 |
| brxE | 6 | Unknown function | 201 | |
| brxB | 1,5,6 | DUF1788 (pfam08747) | Unknown function | 193 |
| pglXI | 3 | COG0863/COG1743 (pfam01555) | Adenine-specific methylase | 920 |
| pglF | 3 | ATPase | 158 | |
| pglW | 2 | COG0515 | Serine/threonine protein kinase | 1,413 |
| brxP | 4 | COG0175 (pfam01507), pfam13182 | Phosphoadenosine phosphosulfate reductase | 774 |
Although the Pgl system (type 2 BREX) was previously described as being comprised of only four genes (pglW, X, Y, and Z)(Sumby & Smith, 2002), in 89% of the occurrences of this system (16/18 instances), we found that two additional genes, denoted here brxD and brxHI, were also associated with the system. Given that both these genes appear in the same order in the type 6 BREX system (Fig5A), one may speculate that these genes play an integral part of the type 2 BREX (Supplementary Table S4). The first gene, brxD, encodes a small protein predicted to bind ATP, while the second gene, brxHI, encodes a predicted helicase.
The type 3 BREX system was found in 20 genomes and is similar, in terms of gene composition, to the common BREX type 1 (Supplementary Table S5). Both systems contain the short protein BrxA, which shares structural similarity with the RNA-binding protein, NusB (Supplementary Fig S1). In addition, both type 1 and 3 systems contain a gene encoding a predicted adenine-specific DNA methylase (pglX and pglXI for subtypes 1 and 3, respectively), although the methylase domain differs between the subtypes (pfam13659 and pfam01555 in pglX and pglXI, respectively) (Fig5A and Table2). It is therefore likely that PglX and PglXI perform the same host DNA methylation function although they do not share sequence homology. BREX type 3 systems do not contain a protein with a Lon-like protease domain, but instead contain a predicted helicase protein, brxHII (Fig5A). In addition, the gene of unknown function brxB is replaced with another gene of unknown function, denoted here as brxF.
The less abundant type 4 BREX system is composed of four genes, two of which are the core brxC/pglY and pglZ genes, and the third, brxL, contains a Lon-like protease domain (Fig1; Supplementary Table S6). The fourth gene, which we denote brxP, is specific to type 4 BREX and contains a phosphoadenylyl-sulfate reductase domain (COG0175/pfam01507). Interestingly, this domain is associated with the phage resistance DND system that performs sulfur modifications on the DNA backbone, providing an additional link between BREX systems and phage resistance (Wang et al, 2007, 2011; You et al, 2007).
The two least common BREX subtypes, type 5 and type 6, are similar to the type 1 BREX system, but contain some additional variations (Fig5; Supplementary Tables S7 and S8). In type 5 BREX, the gene containing the Lon-like protease domain is replaced by a helicase-domain gene (which we denote brxHII), and there is a duplication of brxC/pglY. In subfamily 6, the protease is replaced by brxD and brxHI, a pair which also appears in type 2 BREX (Fig5A), and an additional gene of unknown function, which we denote brxE, is found as the first gene in the cluster.
Overall, 135 of the 144 pglZ genes we detected in microbial genomes (94%) were found to be embedded as part of one of the six BREX systems described (Supplementary Table S9), and seven of the remaining pglZ genes were clearly part of degraded (probably pseudogenized) systems. In most cases, we found a single BREX system per organism, with only 8 (6.3%) of genomes harboring more than one subtype (Supplementary Table S9). In addition, in 14% (19/135) of the identified systems, one of the genes was missing (Supplementary Tables S2, S4, S5, S6, S7, and S8), possibly representing inactivated systems. Phage-defense systems often encode toxic genes (Makarova et al, 2012) or impose a fitness cost (Gomez & Buckling, 2011; Hall et al, 2011; Stern & Sorek, 2011), and it is possible that BREX systems also inflict a fitness cost, leading to gene loss in the absence of phage pressure.
Extensive horizontal transfer of BREX systems
An examination of the distribution of BREX systems across microbial species shows that these systems undergo extensive horizontal transfer (Fig6). First, the distribution of systems across the species tree is patchy; and second, the PglZ tree is not consistent with the species tree, with closely related species accommodating distantly related PglZ and vice versa. Nevertheless, phylogenetic trees reconstructed from additional BREX genes generally recapitulate the structure of the PglZ tree, suggesting that genes within specific BREX systems co-evolve and are co-horizontally transferred (Supplementary Fig S6).
Despite the extensive horizontal transfer observed for the BREX systems, some clades show enrichment in specific subtypes: type 1 BREX is enriched in Deltaproteobacteria (P = 0.001); type 2 (the Pgl system) appears almost solely in Actinobacteria (P = 4.8 × 10−9); and type 5 is exclusive to the archaeal class Halobacteria. The enrichment of specific subtypes within specific phyla might link the ancestry of these subtypes to the phyla in which they are enriched; alternatively, phylum-specific BREX subtypes might rely on additional, phylum-specific cellular mechanisms that are not directly encoded in the BREX genes, or provide defense against phages that predominantly attack the specific phyla.
The relative frequency of BREX in archaea (10%) is similar to that observed in bacteria. Only subtypes 1, 3, and 5 are represented in the 111 archaeal genomes analyzed in this study. However, the absence of subtypes 2, 4, and 6 from archaeal genomes could be the result of their rarity and the relative paucity of sequenced archaeal genomes, comprising only 111 out of the 1,447 genomes analyzed.
Frequent interruptions in the adenine-specific methylase pglX
One of the strains we obtained when engineering the B. cereus type 1 BREX system into B. subtilis was not active against any of the tested phages although PCR analysis showed that it contained the complete BREX system. Upon Illumina whole-genome re-sequencing of the engineered strain, we observed a frameshift mutation in the adenine-specific methylase gene pglX, resulting from a single nucleotide deletion occurring in a stretch of seven guanine (G) residues at position 2,128 (out of 3,539 bp) of this gene. These results further support the finding that the pglX gene is essential for the function of the type 1 BREX system. Moreover, they resemble the results described for the Pgl system in S. coelicolor, where the sequence of pglX was prone to single nucleotide deletions or insertions, leading to phase variation in the activity of the Pgl system in a subpopulation of the bacteria (Laity et al, 1993; Sumby & Smith, 2003). We therefore examined more broadly additional evidence for genetic variability of pglX in nature.
In 11% of the BREX systems we documented (15/135), the pglX gene presented irregularities with respect to the common BREX organization (Fig7A; Supplementary Tables S2, S4, S5, S6, S7, and S8). These included seven instances of premature stop codons in the middle of the gene, two instances of gene duplication and six occurrences where a full-length pglX gene was adjacent to one or more partial forms of pglX (with an extreme example in Methanobrevibacter smithii, where five truncated forms of the pglX are found near the full-length gene (Fig7B)). The complete and truncated forms of the methylase usually reside on opposite strands and are accompanied by a gene annotated as a recombinase, possibly involved in switching between the two versions of pglX. Indeed, when analyzing the genomes of two strains of Lactobacillus rhamnosus GG that were sequenced independently (NCBI accessions FM179322 and AP011548), we found that the pglX sequence is identical between the strains except for a cassette of 313 bp that was switched between the full-length and truncated pglX genes (Fig7C). The interchanged cassette was flanked by two inverted repeats suggesting a recombination-based cassette switching possibly mediated by the accompanying recombinase. DNA shuffling via recombination events was previously shown to control phase variation in bacterial defense-related genes to alter the specificity or to mitigate toxic effects of specific genes in the absence of phage pressure (Hallet, 2001; Cerdeno-Tarraga et al, 2005; Bikard & Marraffini, 2012). Since no other gene except for pglX presented such high rates of irregularities, our results mark PglX as possibly undergoing frequent phase variation, further implicating this gene as the specificity-conferring element in the BREX system or, alternatively, marking it as particularly toxic.
Figure 7. Frequent irregularities in the adenine-specific methylase pglX in BREX type 1.
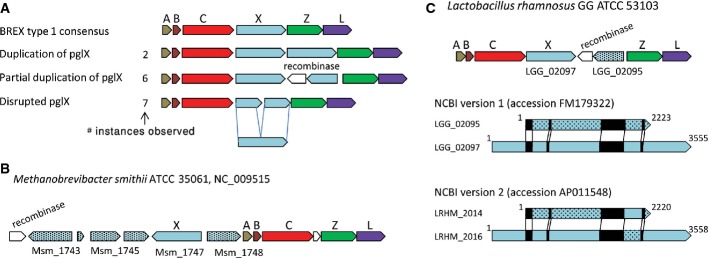
- Irregular genotypes (duplication, inversion, and premature stop codon) associated with pglX.
- Genomic organization of BREX system type 1 in Methanobrevibacter smithii ATCC 35061.
- Genomic organization of BREX system type 1 in Lactobacillus rhamnosusGG. A cassette switch between the short and the long forms of pglX is observed when the sequences of two isolates of Lactobacillus rhamnosusGG (accessions FM179322 and AP011548, respectively) are compared. Repeat sequences between the short and long forms are shown in black.
Discussion
In this study, we describe a phage resistance system that is widespread in bacteria and archaea. The most abundant subtype of this system, when transferred from B. cereus to the model organism B. subtilis, confers complete or partial resistance against phages spanning a wide phylogeny of phage types, including virulent and temperate phages. The abundance of this system and the efficiency with which it protects against phages imply that it plays an important role as a major line of innate defense encoded by bacteria against phages. Nevertheless, our identification of a family of phages that can completely overcome this system suggests that as in the case of CRISPRs (Bondy-Denomy et al, 2013), phages may have evolved molecular mechanisms to shut down or circumvent BREX defense.
The major phage resistance systems that were characterized to date, including the restriction-modification and CRISPR-Cas systems, encode mostly for proteins that interact with and manipulate DNA and RNA molecules. Indeed, the BREX system contains such proteins including methylases and putative helicases. However, BREX systems also contain genes coding for proteins predicted to be involved in the manipulation of other proteins, such as the Lon-like protease, BrxL, and possibly also the predicted alkaline phosphatase, PglZ, and the serine/threonine kinase, PglW. This could imply that the defense mechanism employed by the BREX system takes place later in the infection where phage proteins are already produced and can be manipulated by PglZ and/or BrxL. Alternatively, these BREX proteins might target phage proteins co-injected with the phage DNA early in the infection cycle. Our data suggest that the BREX system acts sometime before phage DNA replication (Fig3C). Finally, these proteins might also interact with other bacterial-encoded proteins, or with other components of the BREX system itself, to regulate the BREX activity.
Our results show that the B. cereus BREX system methylates adenine residues on the 5th position of TAGGAG motifs in the bacterial genome, a function that is probably mediated by PglX. It is therefore likely that this methylation serves as part of the self/non-self recognition machinery of BREX. One may hypothesize that BREX targets non-modified TAGGAG motifs in a way akin to restriction-modification (R-M) systems. However, a few lines of evidence suggest that the BREX is not a simple restriction-modification (R-M) system. First, we failed to detect cleavage or processive degradation of phage DNA in infected BREX-containing cells (Fig4E). Second, in classical R-M systems, methylation is usually performed on palindromic sites, meaning that both strands of the DNA are modified at the same motif. This allows the R-M methylase to identify hemi-methylated sites during bacterial DNA replication, such that the non-methylated, newly synthesized strand duplexed with a methylated site on the old strand becomes methylated as well. The BREX methylation site, however, is non-palindromic, so that only one strand is being methylated. During DNA replication, therefore, one duplex will carry the methylated site while the other will not; it is not clear how the BREX system would differentiate the non-methylated newly synthesized bacterial DNA from non-methylated DNA of invading phage, and why the bacterial DNA is methylated while the invading phage is targeted. A third inconsistency with the hypothesis that BREX is an R-M system stems from our results with the ΔpglX BREX strain. In many R-M systems, deletion of the methylase would be detrimental to cellular growth, because in the absence of DNA modification, the restriction enzyme would cleave the bacterial DNA. However, the ΔpglX BREX strain showed no observable growth defect compared to the wild-type BREX strain, but did lose its defensive activity against phage.
The resemblance of the methylated TAGGAG motif to the consensus ribosomal binding site (RBS) of Bacillus, AAAGGAGG, may lead to a hypothesis on a putative linkage between the BREX functionality and translation initiation. It remains to be seen, however, whether the same motif is being methylated in all instances of the BREX system or whether each system carries a different motif specificity as in the case of R-M systems.
Some preliminary hints that might help in future elucidation of the BREX mechanism of action can be found in the interchangeability of protein domains between the different BREX subtypes. For example, the DNA methylase common to BREX types 1, 2, 5, and 6 is replaced by BrxP, a protein containing a phosphoadenylyl-sulfate reductase domain. This domain was previously found in the DND phage resistance system where it is involved in performing sulfur modifications on the DNA backbone to differentiate between phage and host DNA (Wang et al, 2007, 2011; You et al, 2007). It is therefore possible that the PglX methylase and BrxP reductase perform analogous DNA-modifying functions in type 1 and type 4 systems, respectively, which enables BREX to differentiate between phage and host DNA. Similarly, the Lon-like protease in the type 1 and 4 systems is replaced by a helicase in subtypes 2, 5, and 6. It will be of interest to determine the function of the protease and helicase in these systems.
The family of systems we identified here also includes the Pgl anti-phage system of the S. coelicolor A3, which is a less common BREX variant denoted here as a type 2 BREX. Four genes, pglW, pglX, pglY, and pglZ, were shown to be necessary and sufficient for the Pgl phenotype (Sumby & Smith, 2002). However, in this study, we identified two additional genes, encoding an ATP/GTP-binding protein (pglD) and a helicase (pglHI) that are almost always associated with the four Pgl genes in type 3 systems (Fig5). It is possible that the unique Pgl phenotype observed for S. coelicolor A3 and its phage ΦC31 stems from an incomplete BREX system found in that strain, perhaps due to mutational inactivation of pglD or pglHI. This hypothesis is also in line with the lack of Pgl defense against any other phage except for ΦC31 and its homoimmune relatives. Consistently, the type 1 system we tested in B. subtilis seems to confer resistance also against the first cycle of phage infection and hence could not be defined as reproducing the Pgl phenotype. An alternative explanation might be that the pglD and pglHI genes are an integral part of the system but do not participate in the act of defense itself, as in the case of the cas1 and cas2 genes in the CRISPR-Cas system (Terns & Terns, 2011).
The abundance of pglZ in microbial genomes and its preferred localization to defense islands has previously been reported by Makarova et al (2011, 2013). They observed that pglZ is frequently found associated with additional genes, and suggested its involvement in a complex PGL-like anti-phage system. Our results now divide pglZ-containing gene clusters into several coherent types in terms of gene composition, gene order, and genomic organization, suggesting that these subtypes act as discrete functional units. Furthermore, the genes composing the subtypes of these systems are co-evolving and horizontally transferred together, as observed from the similar phylogeny of the two core proteins, PglZ and BrxC/PglY (Fig5; Supplementary Fig S5).
It is now well appreciated that the arms race between bacteria and phage is playing a major role in shaping the evolution of bacterial and phage genomes (Stern & Sorek, 2011). The discovery of a major additional line of defense in bacteria may, therefore, shed more light on this complex arms race. Previously, deep understanding of the molecular mechanism of complex phage resistance systems such as restriction-modification and CRISPR-Cas has led to revolutions in molecular engineering applications (Cong et al, 2013; DiCarlo et al, 2013), as such systems provide both specificity and targeting of nucleic acids. With further mechanistic studies into the potentially functional sophistication encoded by the multi-gene BREX system, this system might in the future prove to be yet another powerful molecular tool.
Materials and Methods
Genomic data and molecular phylogeny of the PglZ protein
A set of 1,447 completely sequenced prokaryotic genomes (1,336 bacterial and 111 archaeal genomes, Supplementary Table S1) were downloaded from the NCBI FTP site (http://ftp.ncbi.nih.gov/genomes/Bacteria/) in September 2011 and used for subsequent analyses. Several PglZ protein sequences were used as a query in a PSI-BLAST search against the 1,447 prokaryotic genomes with an inclusion threshold e-value of 0.001. Proteins that did not contain the pglZ domain or that were shorter than 600 aa were filtered out. The remaining protein sequences were used to build a PglZ tree as follows. Amino acid sequences were aligned using the MAFFT algorithm (Katoh et al, 2002). The Fourier transform approximation was disabled, and substitution rates were modeled with JTT (Jones et al, 1992) and BLOSUM45 matrix, which is suitable for diverged sequences. The gene tree was reconstructed using the probabilistic RAxML algorithm, with 100 bootstrap replicates, substitutions modeled with JTT (Jones et al, 1992), while allowing for rate variability among sites.
Identification of BREX types 1–6
System types were characterized based on manual observation of phyletic clusters in the PglZ tree. The specific genes associated with each PglZ phyletic type were defined using the IMG genome browser (https://img.jgi.doe.gov/cgi-bin/w/main.cgi). A representative protein sequence of each of the individual genes (Table2) was then used as query in a PSI-BLAST search with an inclusion threshold e-value of 0.05. Only gene clusters containing the two core genes and at least two additional genes were considered, under the added constraint that the genomic distance between the first and last genes in the system should be under 30 kb. In the case of PglY, homology was based on the shared motifs (the P-loop motif GXXXXGK(T/S) (DUF2791)) and DUF499 combined with the conserved size of the gene in the different subtypes (∼1,200 aa). The filtered clusters were manually assigned to systems according to gene content. Only clusters containing the complete set of genes or missing one non-core gene were included in the final set (Supplementary Tables S2, S3, S4, S5, S6, S7, and S8). In the case of BREX 2, systems missing both pglD and pglHI were also included. The blastx program was used to scan intergenic regions in the clusters for unannotated genes.
Protein domains were annotated using the conserved domain database (CDD) (Marchler-Bauer & Bryant, 2004) and HHpred (Soding, 2005). In the latter case, queries were carried out using representative sequences against the PDB, SCOP, interpro, pfam, smart, tigrfam, and COG databases using default search parameters.
The consensus organisms tree for Fig6 was derived from the NCBI ‘common tree’ downloaded from the NCBI Taxonomy portal at http://www.ncbi.nlm.nih.gov/Taxonomy/CommonTree/wwwcmt.cgi. In order to check whether subtypes are overrepresented in specific bacterial phyla, the two following ratios were compared, using a hypergeometric statistical test: (i) number of instances of a specific subtype in a specific phylum/total number of the specific subtype in bacteria; (ii) total number of genomes of the specific phylum analyzed/total number of bacterial genomes analyzed. P-value ≤ 0.05 was considered statistically significant following Benjamini and Hochberg correction for multiple testing.
Strain construction
The type 1 BREX system was amplified in fragments from the B. cereus H3081.97 genome (kindly provided by A. Hoffmaster and C. Beesley) from position 89,288–103,514 (GenBank ABDL02000007.1). The PCR-amplified fragments were transformed into S. cerevisiae together with the pYES1L vector (Invitrogen), where they were assembled in vivo into a circular plasmid. Genomic DNA from the transformed S. cerevisiae strain was then transformed into E. coli BL21-AI where the plasmid was amplified, and then integrated into the proB gene in B. subtilis BEST7003, along with a chloramphenicol resistance cassette. PCR and Illumina-based whole-genome sequencing were performed to confirm the presence of the intact BREX system within B. subtilis BEST7003. Control strains contain only the chloramphenicol resistance cassette integrated at the proB locus. The pglX deletion strain was constructed in a manner similar to that described above, but with PCR fragments that created a deletion from position 94,655–98,163 (GenBank ABDL02000007.1), leaving only 31 nucleotides of the pglX gene.
Growth dynamics of phage-infected cultures
Overnight cultures were diluted 1:100 in LB media supplemented with 0.1 mM MnCl2 and 5 mM MgCl2 and then grown to logarithmic phase. Standard phage infections were performed at multiplicities of infection ranging from 10−3 to 10−4. High concentration phage infections were performed at multiplicities of infection ranging from 0.05 to 5. Optical density measurements at a wavelength of 600 nm were taken every 13 min using a TECAN Infinite 200 plate reader in a 96-well plate.
Plaque assays
Small drop plaque assays were initially performed using 0.75% agar plates containing bacterial cultures that were diluted 1:13 in LB media supplemented with 0.1 mM MnCl2 and 5 mM MgCl2. Serial dilutions of the phage between 2 × 100 and 2 × 105 pfu were spotted on these plates, and plaques were counted after overnight growth at room temperature. Further confirmation of plaque numbers was performed by an agar overlay assay. The bottom agar was composed of LB media supplemented with 0.1 mM MnCl2 and 5 mM MgCl2 and 1.5% agar. The top agar was prepared by diluting logarithmic phase bacterial cultures 1:30 in LB media supplemented with 0.1 mM MnCl2 and 5 mM MgCl2 and 0.5% agar with the addition of serial dilutions of the phage. Plaques were counted after overnight growth at room temperature.
One-step phage growth curve assays
One-step phage growth curve experiments were performed as described by Carlson (2005). Logarithmic phase cultures were infected with either phage SPO1 or Φ3T at an MOI of 0.05. The infection culture was diluted 1:10,000 after 18 min of growth at 37°C, to reduce the likelihood of phage infection following cell lysis. To evaluate the number of infective centers and extracellular phage present in the infection mixture, samples were taken throughout the phage infection time course, were mixed with a phage-sensitive B. subtilis strain, and were plated using the agar overlay method together, as described above. Phage adsorption was inferred by evaluating the number of extracellular phage present in the mixture at early time points. This was assayed by mixing the infection mixture with chloroform, incubating it at 37°C for 4 min, followed by a 4-min incubation on ice, and 30 min at room temperature. The aqueous phase was then mixed with a phage-sensitive strain and plated using the agar overlay method described above. The derived results allow evaluation of the extracellular phage levels, since the chloroform will kill all bacteria, including those with phage adsorbed, and at early time points, phage have not yet assembled inside the cell and are therefore unable to form plaques. A drop in extracellular phage levels indicates that adsorption has occurred.
Phage infection time courses, genomic DNA sequencing, and methylation analysis
Phage infection time courses for methylome analysis, detection of lysogeny and relative phage abundance were performed at an MOI of 4. Cell pellets were washed three times in 10 mM Tris pH 7.4 to remove unadsorbed phage, followed by DNA extraction. DNA library preparations and sequencing for methylome analysis were performed at the Yale Center for Genome Analysis.
To determine the relative abundance of bacterial and phage Φ3T DNA levels, DNA was first fragmented using NEBNext dsDNA Fragmentase (NEB) as per manufacturer's instructions. The relative abundance of bacterial and phage Φ3T DNA levels was determined by Illumina sequencing of the DNA libraries of phage Φ3T-infected time course cultures. Resulting sequence reads were mapped to the phage and host genomes as previously described (Wurtzel et al, 2010). Reads from DNA sequences shared by both B. subtilis BEST7003 and phage Φ3T DNA were discarded from the dataset. The remaining mapped reads were enumerated to compare the number of reads mapping to the B. subtilis BEST7003 DNA sequence relative to phage Φ3T DNA at each time point, normalized to the genome size.
Detection of phage lysogeny
Genomic DNA sequencing of a lysogen containing phage Φ3T was performed using Illumina sequencing to determine the DNA sequence of the Φ3T phage, and the site of phage integration in the genome. Integration of the Φ3T phage was determined to be at a GTAGG site on the B. subtilis BEST7003 bacterial genome at position 2,106,060–2,106,064. Multiplex PCR assays were used to detect phage Φ3T DNA, B. subtilis BEST7003 DNA, and the novel junction created in the lysogenized strain. Primers used to detect phage Φ3T were GAGGTTCGCTAGGGCGAAAT and TCTCTGCTTTGATTCGTCCATGA. Primers for detection of B. subtilis BEST7003 and the unique junction found in the lysogen were TGCCTGCATGAGCTGATTTG and GGCAGGAATGAATGGTGGATATTG, and TCATGCTCCGGATTTGCGAT and TGCCTCCTTTCGATTTTGTTACC, respectively.
Plasmid transformation assays
Competent cell preparation and transformation for BREX-containing and control B. subtilis BEST7003 bacteria were performed as described in Itaya and Tsuge (2011). The integrative plasmids pAX01 (7.8 kb) and pDG1731 (6.7 kb) and the episomal plasmid pHCMC05 (8.8 kb) were obtained from the Bacillus Genetic Stock Center (BGSC). pHCMC05 was further engineered to replace the chloramphenicol resistance gene by a spectinomycin-resistance gene. The spectinomycin-resistance gene was amplified from the pDG1731 plasmid using primers AAAAATTTAGAAGCCAATGAAATC and CCCCCTATGCAAGGGTTTAT and integrated between the MfeI and BsaBI restriction sites of pHCMC05, thus replacing the chloramphenicol resistance gene. 100 fmol of plasmid was used per transformation. Before plating the competent bacteria on selection plates, a live count was performed using a series of serial dilutions plated on LB plates. Transformation efficiency was calculated as the number of colonies that grew on selection plates divided by the live count on LB plates.
Acknowledgments
We thank Udi Qimron, Debbie Lindell, Ilana Kolodkin-Gal, Uri Gophna, Daniel Dar, Azita Leavitt, Sarit Edelheit, Asaf Levy, and Eran Mick for comments and stimulating discussions. We also thank M. Itaya for kindly providing the BEST7003 strain, the BGSC for phage isolates and plasmids, Yana Gofman for assistance and advice in structural analyses, and A. Hoffmaster and C. Beesley for kindly sharing the B. cereus lysate. R.S. was supported, in part, by ISF (personal grant 1303/12 and I-CORE grant 1796), ERC-StG Program (grant 260432), HFSP (grant RGP0011/2013), the Abisch-Frenkel Foundation, and a DIP grant from the Deutsche Forschungsgemeinschaft. O.C. was the beneficiary of a postdoctoral grant from the AXA Research Fund.
Author contributions
HS conceived the project, identified the systems, and performed most of the computational analyses. TG designed the experiments, performed them, and analyzed the results. EW established the phage infection platform with the B. subtilis BEST7003 host. OC performed the phylogenetic analyses. SD analyzed the transcriptome and genome data. YCA performed infection experiments. SA performed some of the computational analyses. GO designed and performed the plasmid transformation efficiency experiments, and assisted with the pglX deletion strain. RS analyzed the results, overviewed the project, and wrote the paper together with TG and HS.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary Information
Supplementary Table S1
Supplementary Table S2
Supplementary Table S3
Supplementary Table S4
Supplementary Table S5
Supplementary Table S6
Supplementary Table S7
Supplementary Table S8
Supplementary Table S9
Review Process File
References
- Bikard D, Marraffini LA. Innate and adaptive immunity in bacteria: mechanisms of programmed genetic variation to fight bacteriophages. Curr Opin Immunol. 2012;24:15–20. doi: 10.1016/j.coi.2011.10.005. [DOI] [PubMed] [Google Scholar]
- Bondy-Denomy J, Pawluk A, Maxwell KL, Davidson AR. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature. 2013;493:429–432. doi: 10.1038/nature11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson K. Working with Bacteriophages: common techniques and methodological approaches. In: Kutter E, Sulakvelidze A, et al., editors. Bacteriophages, Biology and Applications. Florida: CRC Press; 2005. pp. 437–494. [Google Scholar]
- Cerdeño-Tárraga AM, Patrick S, Crossman LC, Blakely G, Abratt V, Lennard N, Poxton I, Duerden B, Harris B, Quail MA, Barron A, Clark L, Corton C, Doggett J, Holden MT, Larke N, Line A, Lord A, Norbertczak H, Ormond D, et al. Extensive DNA inversions in the B. fragilis genome control variable gene expression. Science. 2005;307:1463–1465. doi: 10.1126/science.1107008. [DOI] [PubMed] [Google Scholar]
- Chinenova TA, Mkrtumian NM, Lomovskaia ND. Genetic characteristics of a new phage resistance trait in Streptomyces coelicolor A3(2) Genetika. 1982;18:1945–1952. [PubMed] [Google Scholar]
- Chopin MC, Chopin A, Bidnenko E. Phage abortive infection in lactococci: variations on a theme. Curr Opin Microbiol. 2005;8:473–479. doi: 10.1016/j.mib.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveau H, Garneau JE, Moineau S. CRISPR/Cas system and its role in phage-bacteria interactions. Annu Rev Microbiol. 2010;64:475–493. doi: 10.1146/annurev.micro.112408.134123. [DOI] [PubMed] [Google Scholar]
- DiCarlo JE, Norville JE, Mali P, Rios X, Aach J, Church GM. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013;41:4336–4343. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez P, Buckling A. Bacteria-phage antagonistic coevolution in soil. Science. 2011;332:106–109. doi: 10.1126/science.1198767. [DOI] [PubMed] [Google Scholar]
- Hall AR, Scanlan PD, Morgan AD, Buckling A. Host-parasite coevolutionary arms races give way to fluctuating selection. Ecol Lett. 2011;14:635–642. doi: 10.1111/j.1461-0248.2011.01624.x. [DOI] [PubMed] [Google Scholar]
- Hallet B. Playing Dr Jekyll and Mr Hyde: combined mechanisms of phase variation in bacteria. Curr Opin Microbiol. 2001;4:570–581. doi: 10.1016/s1369-5274(00)00253-8. [DOI] [PubMed] [Google Scholar]
- Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- Itaya M, Tsuge K. Construction and manipulation of giant DNA by a genome vector. Methods Enzymol. 2011;498:427–447. doi: 10.1016/B978-0-12-385120-8.00019-X. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat Rev Microbiol. 2010;8:317–327. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- Laity C, Chater KF, Lewis CG, Buttner MJ. Genetic analysis of the phi C31-specific phage growth limitation (Pgl) system of Streptomyces coelicolor A3(2) Mol Microbiol. 1993;7:329–336. doi: 10.1111/j.1365-2958.1993.tb01124.x. [DOI] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Snir S, Koonin EV. Defense islands in bacterial and archaeal genomes and prediction of novel defense systems. J Bacteriol. 2011;193:6039–6056. doi: 10.1128/JB.05535-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Anantharaman V, Aravind L, Koonin EV. Live virus-free or die: coupling of antivirus immunity and programmed suicide or dormancy in prokaryotes. Biol Direct. 2012;7:40. doi: 10.1186/1745-6150-7-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Koonin EV. Comparative genomics of defense systems in archaea and bacteria. Nucleic Acids Res. 2013;41:4360–4377. doi: 10.1093/nar/gkt157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32:W327–W331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray IA, Clark TA, Morgan RD, Boitano M, Anton BP, Luong K, Fomenkov A, Turner SW, Korlach J, Roberts RJ. The methylomes of six bacteria. Nucleic Acids Res. 2012;40:11450–11462. doi: 10.1093/nar/gks891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJ. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem Sci. 2009;34:401–407. doi: 10.1016/j.tibs.2009.05.002. [DOI] [PubMed] [Google Scholar]
- Rudinski MS, Dean DH. Evolutionary considerations of related Bacillus subtilis temperate phages phi 105, rho 14, rho 10, and rho 6 as revealed by heteroduplex analysis. Virology. 1979;99:57–65. doi: 10.1016/0042-6822(79)90036-9. [DOI] [PubMed] [Google Scholar]
- Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- Sorek R, Kunin V, Hugenholtz P. CRISPR–a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol. 2008;6:181–186. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- Stern A, Sorek R. The phage-host arms race: shaping the evolution of microbes. BioEssays. 2011;33:43–51. doi: 10.1002/bies.201000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumby P, Smith MC. Genetics of the phage growth limitation (Pgl) system of Streptomyces coelicolor A3(2) Mol Microbiol. 2002;44:489–500. doi: 10.1046/j.1365-2958.2002.02896.x. [DOI] [PubMed] [Google Scholar]
- Sumby P, Smith MC. Phase variation in the phage growth limitation system of Streptomyces coelicolor A3(2) J Bacteriol. 2003;185:4558–4563. doi: 10.1128/JB.185.15.4558-4563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarts DC, Jore MM, Westra ER, Zhu Y, Janssen JH, Snijders AP, Wang Y, Patel DJ, Berenguer J, Brouns SJ, van der Oost J. DNA-guided DNA interference by a prokaryotic Argonaute. Nature. 2014;507:258–261. doi: 10.1038/nature12971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terns MP, Terns RM. CRISPR-based adaptive immune systems. Curr Opin Microbiol. 2011;14:321–327. doi: 10.1016/j.mib.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen ND, Berger JM. Structural frameworks for considering microbial protein- and nucleic acid-dependent motor ATPases. Mol Microbiol. 2008;69:1071–1090. doi: 10.1111/j.1365-2958.2008.06364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tock MR, Dryden DT. The biology of restriction and anti-restriction. Curr Opin Microbiol. 2005;8:466–472. doi: 10.1016/j.mib.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Wang L, Chen S, Xu T, Taghizadeh K, Wishnok JS, Zhou X, You D, Deng Z, Dedon PC. Phosphorothioation of DNA in bacteria by dnd genes. Nat Chem Biol. 2007;3:709–710. doi: 10.1038/nchembio.2007.39. [DOI] [PubMed] [Google Scholar]
- Wang L, Chen S, Vergin KL, Giovannoni SJ, Chan SW, DeMott MS, Taghizadeh K, Cordero OX, Cutler M, Timberlake S, Alm EJ, Polz MF, Pinhassi J, Deng Z, Dedon PC. DNA phosphorothioation is widespread and quantized in bacterial genomes. Proc Natl Acad Sci USA. 2011;108:2963–2968. doi: 10.1073/pnas.1017261108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzel O, Dori-Bachash M, Pietrokovski S, Jurkevitch E, Sorek R. Mutation detection with next-generation resequencing through a mediator genome. PLoS ONE. 2010;5:e15628. doi: 10.1371/journal.pone.0015628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You D, Wang L, Yao F, Zhou X, Deng Z. A novel DNA modification by sulfur: DndA is a NifS-like cysteine desulfurase capable of assembling DndC as an iron-sulfur cluster protein in Streptomyces lividans. Biochemistry. 2007;46:6126–6133. doi: 10.1021/bi602615k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Table S1
Supplementary Table S2
Supplementary Table S3
Supplementary Table S4
Supplementary Table S5
Supplementary Table S6
Supplementary Table S7
Supplementary Table S8
Supplementary Table S9
Review Process File