Abstract
The biological functions of RNA are ultimately governed by the local environment at each nucleotide. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) chemistry is a powerful approach for measuring nucleotide structure and dynamics in diverse biological environments. SHAPE reagents acylate the 2′-hydroxyl group at flexible nucleotides because unconstrained nucleotides preferentially sample rare conformations that enhance the nucleophilicity of the 2′-hydroxyl. The critical corollary is that some constrained nucleotides must be poised for efficient reaction at the 2′-hydroxyl group. To identify such nucleotides, we performed SHAPE on intact crystals of the E. coli ribosome, monitored the reactivity of 1490 nucleotides in 16S ribosomal RNA, and examined those nucleotides that were hyper-reactive towards SHAPE and had well-defined crystallographic conformations. Analysis of these conformations revealed that 2′-hydroxyl reactivity is broadly facilitated by general base catalysis involving multiple RNA functional groups and by two specific orientations of the bridging 3′-phosphate group. Nucleotide analog studies confirmed the contributions of these mechanisms to SHAPE reactivity. These results provide a strong mechanistic explanation for the relationship between SHAPE reactivity and local RNA dynamics and will facilitate interpretation of SHAPE information in the many technologies that make use of this chemistry.
Introduction
The functions of almost all RNAs are governed or tuned by the base-paired secondary structure and higher-order tertiary interactions. Understanding the information expressed in RNA structure thus requires accurate and comprehensive knowledge about the local environment of most nucleotides in an RNA. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) chemistry provides comprehensive information on local nucleotide dynamics and has proven useful in probing structure-function interrelationships in RNA,1 RNA-protein interactions,2,3 RNA-small molecule interactions,4-6 and RNA folding dynamics.7,8 Because the nucleotide dynamics monitored by SHAPE correlate inversely with the likelihood that a nucleotide is constrained by base pairing, SHAPE data increase the accuracy of RNA secondary structure prediction.9-12 SHAPE has facilitated development of functional hypotheses in complex biological systems.13-17 Innovative refinements and applications of SHAPE include time-resolved7,8,18,19 and temperature-dependent analyses,5,20 examination of short RNAs21,22 and interfacing with new readout technologies,23 detection of long-range through-space interactions,2,24-27 and identification of optimal sites for attachment of fluorescent reporter probes.28
SHAPE reagents are small-molecule electrophiles like 1-methyl-7-nitroisatoic anhydride (1M7)29 and benzoyl cyanide (BzCN)18 that react preferentially with the 2′-hydroxyl group of flexible nucleotides to form a 2′-O-adduct (Fig. 1). The sites at which these adducts form are detected by either primer extension or by protection from exoribonuclease digestion. SHAPE provides a quantitative measure of local nucleotide order,30 interrogates all four nucleotides in a single experiment,31 and works well in diverse solution conditions and biological environments.14,32,33
Figure 1.
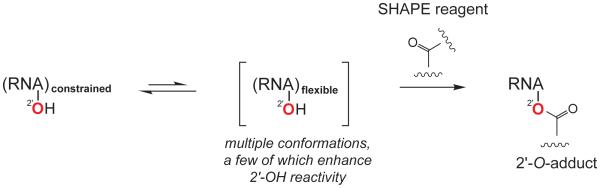
Scheme for SHAPE chemistry emphasizing that reactivity is governed primarily by the ability of a nucleotide to adopt a conformation that enhances the reactivity of the 2′-hydroxyl group towards SHAPE electrophiles.
The reactivity of the RNA 2'-ribose position is exquisitely sensitive to the nucleotide conformation.34,35 The mechanism that best explains the quantitative sensitivity of SHAPE to local nucleotide flexibility is that flexible nucleotides sample multiple conformations, and a small number of these conformations enhance the reactivity of the 2′-hydroxyl group towards SHAPE electrophiles (Fig. 1). Neither the nature of these reactive conformations nor mechanisms by which they enhance 2'-OH reactivity are fully understood.
If SHAPE is primarily governed by the ability of a given nucleotide to sample rare conformations that enhance 2'-OH reactivity, then there must exist a few nucleotides that are constrained in conformations poised for efficient reaction at the 2′-hydroxyl group. An important first step towards understanding the mechanism of SHAPE was the observation that cyclic adenosine monophosphate (cAMP) is 10-18 fold more reactive towards SHAPE reagents than are nucleotides in single-stranded RNA.35 In the fixed conformation of cAMP, the 3′-phosphodiester is constrained in a 3',5' cyclic linkage. The observed "hyper-reactivity" of cAMP is consistent with a model in which close proximity of the negatively charged internucleotide phosphodiester group destabilizes 2′-oxyanion formation during 2'-O-adduct synthesis. It is unlikely, however, that this is the only conformation that enhances the nucleophilicity of the 2′-hydroxyl.
To identify nucleotide conformations that are reactive towards SHAPE, we performed SHAPE on the 16S ribosomal RNA (rRNA) in intact crystals of the E. coli ribosome.36 We identified nucleotides that were hyper-reactive relative to the standard SHAPE reactivity scale and that also had well-defined experimental electron density. These nucleotides cluster into a small number of conformations that must facilitate the nucleophilic reactivity of the 2′-hydroxyl towards SHAPE reagents. We explored the mechanisms of these enhancements using a series of nucleotide analogues and showed that 2′-hydroxyl reactivity is enhanced by general base catalysis involving multiple RNA functional group partners and by two specific orientations of the 3′-phosphodiester backbone. These results support specific mechanisms for SHAPE reactivity and will facilitate interpretation of SHAPE reactivities in the many technologies that are now making use of this chemistry.
Results
In-crystal SHAPE reveals hyper-reactive nucleotides
The 1542-nucleotide 16S rRNA in crystals of E. coli 70S ribosomes represents an ideal system to discover the conformations of nucleotides reactive towards SHAPE. First, in the crystal, 16S rRNA is in a static, constrained state; second, individual nucleotides are in diverse conformations; and, third, structural information on nucleotide geometries exists at sufficiently high resolution (3.2 Å) to support development of concrete, testable hypotheses.
Crystals used in SHAPE analyses were essentially identical to those used in previous high-resolution structural studies.36 Before SHAPE probing, the crystals were washed extensively to remove uncrystallized ribosomes. The RNA in the crystals was modified by the addition of 1M7 at 4 °C. After washing to remove ribosomes that might have dissolved as a result of the modification reaction, the RNA was isolated by standard approaches and sites of SHAPE modification were identified by primer extension followed by capillary electrophoresis.10,37,38
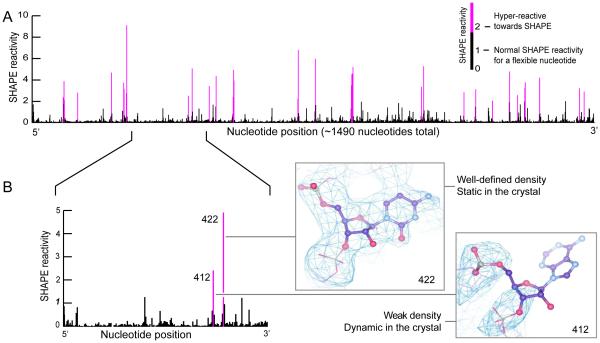
This analysis yielded a SHAPE reactivity profile for ~1490 of the 1542 nucleotides in the 16S rRNA (Fig. 2A). Nucleotide reactivities were normalized to a scale in which unreactive nucleotides had zero reactivity and the mean reactivity of the reactive nucleotides was defined as 1.0. On this scale, reactivities for the vast majority of the nucleotides analyzed fell in a range from zero to 1.5. However, 35 nucleotides had SHAPE reactivities in the range of 2 to 10 (in purple, Fig. 2A). These hyper-reactive nucleotides presumably adopt conformations that activate the 2'-OH group for the SHAPE reaction. The crystals contain two copies of the ribosome and we focused on the more highly ordered of these (see Methods for additional discussion). In the more ordered ribosome, 17 of the 35 hyper-reactive nucleotides had weak electron density in the crystal structure (Fig. 2B, nucleotide 412) and 18 were well defined by the experimental electron density (Fig. 2B, nucleotide 422).
Figure 2.
SHAPE chemistry performed on the 16S rRNA in 70S ribosome crystals identifies hyper-reactive nucleotides. (A) SHAPE reactivity histogram. Nucleotides that fall on the conventional SHAPE reactivity scale (normalized nucleotide reactivity <2) are black, nucleotides “hyper-reactive” towards SHAPE reagents are purple. (B) Section of data from part (A) showing two hyper-reactive nucleotides. One (nucleotide 412) has poor experimental electron density and is likely dynamic in the crystal, whereas the other (nucleotide 422) has well-defined density. Electron density is drawn at 1σ.
SHAPE reactivities are not governed by solvent or molecular accessibility
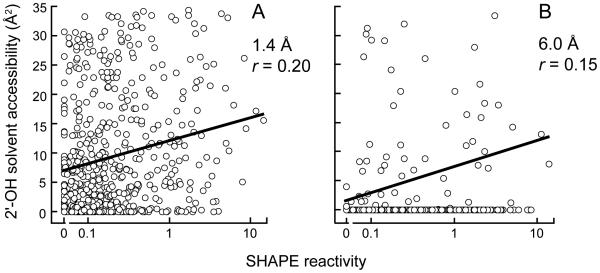
Although all evidence to date suggests that solvent accessibility does not strongly influence SHAPE reactivity,1 the in-crystal reactivity profile provided a unique opportunity to evaluate this relationship on a large data set. We compared SHAPE reactivities with the accessibility of the ribose 2′-hydroxyl group as a function of spherical probes ranging from 1.4 Å to 7.0 Å. The correlation between reactivity and solvent accessibility was low both for a probe size of 1.4 Å (r = 0.20) corresponding to water and for one of 6.0 Å (r = 0.15), which reflects the approximate molecular diameter of 1M7 (Fig. 3). In addition, one-half of the hyper-reactive nucleotides with well-defined conformations (9 of the 18) had low solvent accessibilities, less than 8 Å2, at the 2'-hydroxyl position. As many nucleotides with very low solvent and molecular accessibilities reacted readily with 1M7, SHAPE is essentially insensitive to solvent accessibility.
Figure 3.
Correlation between SHAPE reactivity and solvent accessibility at the O2′ position for single-stranded nucleotides in the 16S rRNA. Probe sizes of 1.4 Å and 6.0 Å correspond to the approximate molecular dimensions of water and 1M7, respectively. Pearson’s linear r-values are shown.
Structural families for hyper-reactive nucleotides
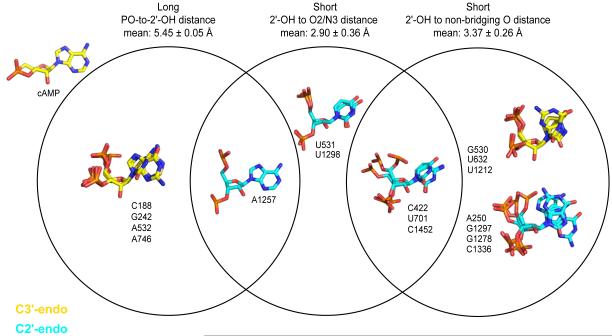
We grouped the 18 hyper-reactive nucleotides with well-defined conformations in the crystal by structural features, including ribose pucker, distance between 2'-OH and adjacent 3'-phosphodiester, and the presence of a nearby functional or catalytic group. Both the standard C3'-endo and the rarer C2'-endo conformation were represented with 7 and 11 examples, respectively (emphasized in yellow and cyan, Fig. 4). Given that nucleotides in the C3'-endo conformation occur roughly ten times more frequently in RNA than those in the C2'-endo conformation,39 the C2'-endo conformation is highly over-represented among hyper-reactive nucleotides, a feature noted previously.32
Figure 4.
Venn diagram of hyper-reactive nucleotide conformations. Nucleotides in the C3'- or C2'-endo ribose conformations are illustrated in yellow and cyan, respectively. Nucleotides are grouped according to the distance between the 2′-hydroxyl and a vector bisecting the non-bridging oxygen bonds in the 3′-phosphodiester group (see Fig. 5A) and proximity of the 2'-hydroxyl to a general base catalytic group. The conformation of hyper-reactive nucleotide A243 does not fall into any of these categories.
The orientation of the 3′-phosphodiester relative to the 2′-OH group was evaluated in two ways: (i) as the distance between the 2′-OH and a vector corresponding to the sum of the two phosphorus to non-bridging oxygen bond vectors (see PO-to-2'-OH distance, Fig. 5A, top) and (ii) as the distance between the 2′-OH and the closest non-bridging phosphate oxygen. Orientations of the 3'-phosphodiester group fell into two distinct conformations. Five nucleotides had orientations in which the non-bridging oxygen groups of the 3'-phosphodiester were oriented away from the 2'-OH group and thus the PO-to-2'-OH distance was relatively long at ~5.5 Å (Fig. 4, left-hand circle). Of these, four were in the C3'-endo conformation and closely resembled the conformation of cAMP (Fig. 4, upper left). This observation supports the model35 that electrostatic communication between the 3'-phosphodiester and the 2'-OH strongly modulates reactivity of the 2'-hydroxyl group. It is also possible for nucleotides in the C2'-endo conformation to have a long distance between the 3'-phosphodiester and the 2'-OH group; we observed one example in our study (Fig. 4, nucleotide A1257). There were ten examples of the conformation in which one of the non-bridging oxygen groups was within ~3.4 Å of the 2'-OH (Fig. 4, right-hand circle). In this case, the non-bridging oxygen likely facilitates SHAPE reactivity by functioning as a general base.
Figure 5.
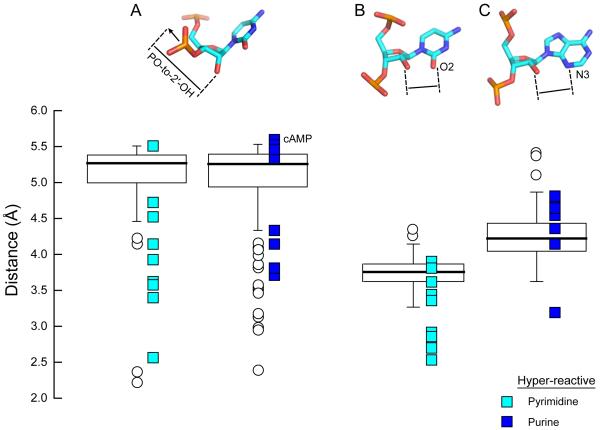
Geometric characteristics of nucleotides hyper-reactive by SHAPE, compared to single-stranded nucleotides with low B-factors in 16S rRNA. Rectangles span the central 50% of the data, the interquartile range, and the median is shown in a heavy line. Whiskers above and below each box give the largest or smallest non-outlier values; nucleotides with values >1.5 times the interquartile range are shown with circles. Distances for each hyper-reactive pyrimidine and purine nucleotide are shown by colored boxes. (A) Distance between the 2'-OH and the terminus of a vector bisecting the bonds to the non-bridging oxygens in the 3'-phosphodiester. (B) Distance between the 2'-OH and pyrimidine O2. (C) Distance between the 2'-OH and purine N3.
Six nucleotides adopt a conformation that brings the 2'-hydroxyl group within, or very close to, hydrogen bonding distance of oxygen or nitrogen functional groups with available lone pairs and that might also function as general bases (Fig. 4, center circle). In five of these nucleotides, hydrogen bonding between the 2'-OH and the base O2 group of cytosine or uridine was possible, whereas hydrogen bonding with the N3 group of adenosine was possible in one case (A1257). In three examples, the 2'-OH was within 4.2 Å of hydrogen bond acceptors involving non-adjacent nucleotides, suggesting that SHAPE reactivity might be facilitated by long-range, through-space interactions (Supporting Fig. 1). Of the 18 hyper-reactive nucleotides, only one, A243, did not fit into any of these conformational categories, and the reason for its hyper-reactivity is not understood.
Hyper-reactive nucleotides have unusual conformations
We next evaluated how common the identified conformations are by comparing the geometric parameters of the three classes of hyper-reactive nucleotides to all single-stranded nucleotides in the 16S rRNA with well-defined electron densities (182 nts total). Data are summarized using box plots (Fig. 5) in which the rectangle spans the central 50% of the data, or the interquartile range (IQR), and the median is shown in a heavy line. Extreme values, greater than 1.5 times the IQR, are shown explicitly as circle or square symbols. The distribution of PO-to-2'-OH distances (Fig. 5A) for the hyper-reactive nucleotides is notably different from that of all single-stranded nucleotides in the 16S rRNA and is distinct for pyrimidine and purine nucleotides. Of the 17 hyper-reactive nucleotides, only one fell into the IQR that characterized most single-stranded nucleotides in 16S rRNA. Five hyper-reactive nucleotides are characterized by a long PO-to-2'-OH distance, possible only when the 3'-phosphodiester points away from the 2'-OH group (Fig. 5A, top). The remaining 12 hyper-reactive nucleotides showed shorter than median PO-to-2'-OH distances, consistent with a hydrogen-bonding interaction.
The distribution of distances between the 2′-OH group and the pyrimidine O2 or purine N3 positions is also notably distinct from mean distances between the O2 or N3 position and all 2'-hydroxyl groups in single-stranded nucleotides in the 16S rRNA (Figs. 5B and C, respectively). Six of the hyper-reactive pyrimidine nucleotides had much shorter distances between the 2′-OH and the base O2 position than characterized most nucleotides in the16S rRNA. The distribution among the purine nucleotides was distinct both from single-stranded 16S rRNA nucleotides and from the hyper-reactive pyrimidines. One hyper-reactive purine nucleotide had a short distance between the 2′-hydroxyl and the N3 positions, whereas most of hyper-reactive purine nucleotides had longer than typical distances. These observations suggest that the most favorable conformations that facilitate 2'-OH reactivity may be different for pyrimidine and purine nucleotides but, in both cases, enhanced reactivity is strongly favored by an unusual nucleotide conformation.
Functional group substitution supports general base catalysis
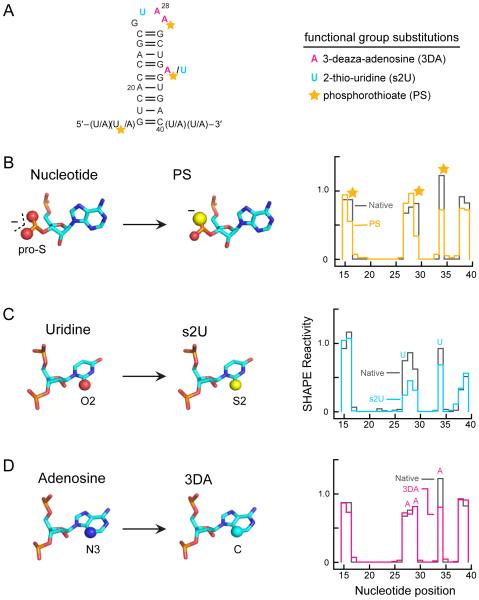
To test the hypothesis that specific RNA functional groups – especially the non-bridging phosphate oxygen and the pyrimidine O2 group – enhance the nucleophilic reactivity of the 2'-hydroxyl, we evaluated the effect of substituting individual functional groups in the context of a model RNA hairpin with a GNRA tetraloop and a single bulged nucleotide of either A or U. Two nucleotides flanking the 5' and 3' sides of the stem were designed to remain unpaired to permit normalization of SHAPE profiles (Fig. 6A). In-solution SHAPE indicated that none of the nucleotides in this RNA were hyper-reactive (Fig. 6, black lines at right).
Figure 6.
Effects of functional group substitutions on SHAPE reactivity. (A) Structure of the stem-loop RNA showing sites of nucleotide analogue incorporation during transcription. The bulged-A RNA contained flanking U sequences, and vice versa. (B-D) For each substituted transcript, modifications are illustrated on the left and SHAPE reactivities for the modified (colored lines) compared to the unmodified transcript (black lines) are shown on the right. (B) 3'-Phosphorothioate (PS), (C) 2-thio-uridine (s2U), and (D) 3-deaza-adenosine (3DA) functional group substitutions.
In the context of this model, we evaluated the impact of the non-bridging oxygen groups from the 3'-phosphodiester by analysis of a transcript containing phosphorothioate-substituted guanosine analogues. In this RNA, the pro-R oxygen was replaced with sulfur at the 3'-phosphodiester group for three single-stranded residues (Fig. 6A, yellow stars). In a standard nucleotide, the phosphate atom and non-bridging oxygens carry a net negative charge, whereas in the phosphorothioate analogue, the charge resides preferentially on the sulfur atom40 leaving the pro-S oxygen electron deficient and a weaker hydrogen bond acceptor and general base (Fig. 6B, left). SHAPE profiles for the native and phosphorothioate-substituted RNAs were similar, indicating that the stem-loop RNA folds into roughly the same structure in both RNAs. Strikingly, however, the SHAPE reactivities at each single-stranded nucleotide with a 3'-phosphorothioate substitution were 2-fold lower than the reactivities of these nucleotides in the unmodified transcript (Fig. 6B, graph). The electronic distribution at the 3'-phosphodiester group thus has a significant facilitating effect on 2'-OH reactivity.
The contribution of the pyrimidine O2 group to SHAPE reactivity was evaluated by substitution of the O2 atom with sulfur using 2-thio-uridine (Fig. 6C). Sulfur is a poor-hydrogen bond acceptor due to its size and diffuse electron cloud.41 The thiocarbonyl group in 2-thio-uridine thus disrupts the ability of this position to serve as a general base. SHAPE reactivities at U27 in the loop and at the bulged U34 in the 2-thio-uridine transcript decreased by 2-fold compared to reactivities of these nucleotides in the unsubstituted RNA (Fig. 6C). SHAPE reactivities for the two adenosine nucleotides in the loop, adjacent to U27, also decreased in the 2-thio-uridine-substituted RNA relative to those in the unmodified transcript suggesting that substitution of the O2 position affects the reactivity of neighboring nucleotides.
Finally, the contribution of the purine N3 position was tested using the nucleotide analogue 3-deaza-adenosine (Fig. 6D), which substitutes carbon for nitrogen and eliminates the ability of the substituted nucleotide to act as a general base. This substitution caused a 40% decrease in SHAPE reactivity at the bulged A relative to the position in the unmodified RNA but did not change the reactivity of the two adenosines in the loop. These data suggest the purine N3 group facilitates 2'-OH reactivity in some, but not all, structural contexts.
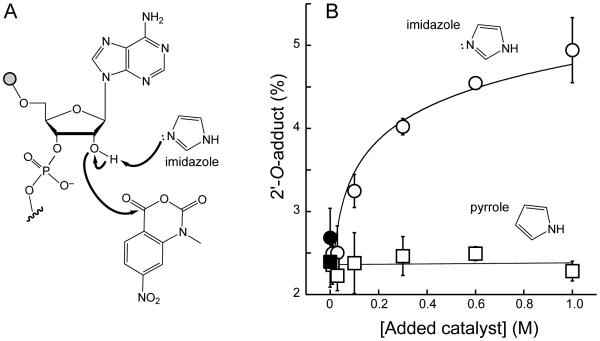
Catalysis by a solution-phase general base
SHAPE reactivity was strongly enhanced by general base catalysis involving functional groups in RNA (Figs. 4 and 6). An exogenous solution-phase base should therefore also enhance SHAPE reactivity. We monitored SHAPE reactivity of a 5'-labeled dinucleotide of adenosine and dideoxycytosine, which contains a single free 2′-hydroxyl that reacts with 1M7 to form a mono 2′-O-adduct (Fig. 7A). SHAPE reactivity was monitored at constant pH as a function of added imidazole, which contains lone pair electrons on nitrogen and is a good general base. Pyrrole, with a similar structure but without non-bonded lone pairs, was used as a control. Addition of 1 M imidazole increased 2′-O-adduct formation two fold, whereas pyrrole addition had no detectable effect on reactivity (Fig. 7B). These results strongly support the hypothesis that SHAPE reactivity is enhanced by general base catalysis involving both intra-nucleotide and through-space catalytic groups.
Figure 7.
Effect of a solution-phase catalyst on SHAPE adduct formation. (A) Mechanism for general base catalysis of the reaction of a 2′-hydroxyl towards SHAPE reagents mediated by imidazole. (B) Imidazole-versus pyrrole-mediated enhancement of 2′-O-adduct formation.
Discussion
RNA SHAPE chemistry detects local nucleotide flexibility in a way that correlates closely with independent measures of molecular order in RNA30,35 but that is largely independent of solvent or molecular accessibility (Fig. 3). SHAPE is best explained by a model in which reactive nucleotides preferentially sample relatively rare conformations that increase the nucleophilic reactivity of the 2'-OH group (Fig. 1). The corollary to this model, which provides a critical segue into understanding the mechanism of SHAPE, is that there must exist a few nucleotides that are constrained in reactive conformations.
Thirty-five nucleotides (~2% of those examined) in the 16S rRNA in crystals of E. coli ribosomes proved to have unusually high reactivities when interrogated by SHAPE in crystallo. This prevalence of hyper-reactive nucleotides corresponds closely to that observed in prior studies in our laboratory.9,20 The conformations of 18 of these nucleotides were well defined by the experimental electron density. Analysis of the conformations adopted by these nucleotides supports two overarching models for the underlying mechanism of SHAPE chemistry.
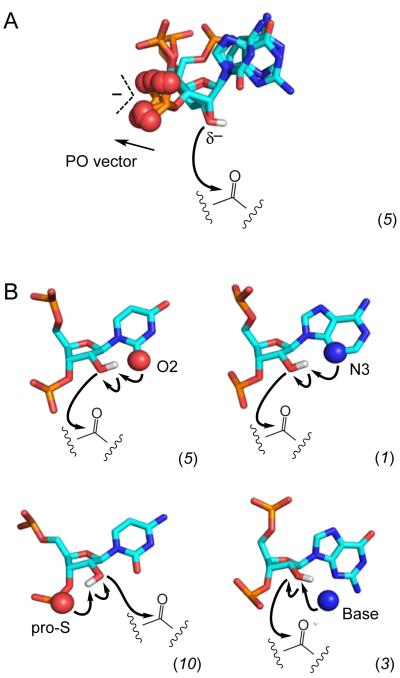
First, reactivity was enhanced by conformations in which the non-bridging oxygen atoms from the adjacent 3'-phosphodiester are directed away from the reactive 2'-OH group. This mechanism was initially characterized using cAMP,35 which remains one of the most hyper-reactive nucleotides identified to date: Its reactivity is ~15 on the scale used here for 16S rRNA (see Figs. 2A and 4). The phosphodiester-away mechanism is now supported by five examples of hyper-reactive nucleotides in the 16S rRNA (Fig. 4, left-hand circle). This conformation supports 2'-OH reactivity by reducing electrostatic destabilization of the reactive 2'-oxyanion (δ−), and perhaps other partial charges, in the transition state (Fig. 8A).
Figure 8.
Mechanisms of RNA SHAPE chemistry. (A) Stabilization of the intermediate reactive 2'-oxyanion. In the five superimposed hyper-reactive nucleotides, the non-bridging oxygen groups of the backbone (and the permanent charge) are directed away from the 2'-OH group. (B) General base catalysis. Examples of catalysis by the pyrimidine O2, purine N3, pro-S oxygen, and through-space groups are shown. For each panel, the number of occurrences observed in SHAPE analysis of 16S rRNA in crystallo is given in parentheses.
Second, SHAPE reactivity was enhanced by a proximal functional group capable of forming a hydrogen bond with the 2'-OH group (Fig. 4, middle and right-hand circles). These hyper-reactive conformations are remarkably diverse and include those that allow interactions with functional groups on the nucleobases, the pro-S non-bridging phosphate oxygen, and through-space hydrogen bond acceptors (Fig. 8B). In these cases, SHAPE reactivity is facilitated through deprotonation of the reactive 2'-OH via general base catalysis (equivalent to lowering the pKa of the 2'-OH group). This mechanism is strongly supported by functional group substitution studies showing that reducing the electron density on these atoms, which limits their ability to function as general bases, had a large effect on 2'-OH reactivity (Fig. 6).
The vast majority of nucleotides (85) in the 16S rRNA crystal that exhibited high (0.7–2.0) SHAPE reactivities had very little electron density and are likely dynamic. Only 18 nucleotides with high reactivities had well-defined positions in the crystal. Of these, 15 adopted conformations consistent with our reactivity-facilitating conformations (Supporting Table S1). Thus, SHAPE reactivity mechanisms defined by evaluating hyper-reactive positions appear to apply broadly to RNA nucleotides.
When general base catalysis predominates, pyrimidine and purine nucleotides may react via different preferred conformations. The interaction between the pyrimidine O2 and 2'-OH appears to be especially favorable and occurred in five examples in our set of hyper-reactive nucleotides (Fig. 4). For purine nucleotides, it appears to be more common for a non-bridging phosphodiester oxygen to function as the general base rather than the 2'-OH (of which we have a single example). Supporting this hypothesis, substitution of the pyrimidine O2 atom had a larger compromising effect on SHAPE reactivity than did substitution at the purine N3 group (Figs. 6C and D).
C2'-endo nucleotides were highly over-represented among the hyper-reactive nucleotides, a feature noted in studies of the Tetrahymena P546 domain.32 All examples of apparent intra-nucleotide general base catalysis involved nucleotides in the C2'-endo conformation (Fig. 4, center circle). Our data are consistent with a model in which the C2'-endo conformation facilitates 2'-OH reactivity primarily because it enhances the ability of the 2'-OH group to interact favorably with nearby general base groups, especially with the pyrimidine O2 or purine N3 atoms. However, the C2'-endo conformation is clearly not required for efficient SHAPE reactivity as evidenced by the many examples of hyper-reactive nucleotides in the C3'-endo conformation and by the high reactivity of cAMP, which is constrained in a C3'-endo-like conformation.
Finally, interactions with non-adjacent nucleotides can play a significant role in the nucleophilic reactivity of the 2'-OH group. First, phosphorothioate and 2-thio-uridine substitutions affected the reactivity of 2'-OH groups located one or two nucleotides away from the site of the substitution (Fig. 6B and C). Second, in addition to local interactions, three 2'-OH groups among the hyper-reactive nucleotides and 11 of 18 2'-OH groups among highly reactive nucleotides are close to a general base group located on a non-adjacent nucleotide in the 16S rRNA (Supporting Fig. S1 and Table S1). Third, the strong facilitating effect of imidazole indicates that through-space interactions can significantly enhance 2'-OH reactivity (Fig. 7). The net effect of all potential non-adjacent contributions provides a likely explanation for why SHAPE is uncorrelated with solvent accessibility (Fig. 3). Any accessibility penalty born by buried nucleotides is apparently compensated for by the density of functional groups able to function as general base catalysts in a folded RNA.
In sum, SHAPE reactivity at the 2'-OH group is facilitated by two major mechanisms (Fig. 8), each of which can be achieved by wide variety of nucleotide conformations. All hyper-reactive conformations identified through our analysis are incompatible with canonical Watson-Crick base pairing and fall predominantly into three well-defined structure classes. That diverse conformations facilitate reactivity strongly reinforces the basic biophysical model that SHAPE measures RNA dynamics. The data reported here explain the strong observed correlation between SHAPE reactivity and molecular motion30. In addition to understanding SHAPE-based analyses, the specific reactivity mechanisms established here emphasize the broad catalytic potential of functional groups in RNA and provide mechanistic insight into the general reactivity of distinct RNA conformations towards chemical probes, epigenetic modifying agents, and mutagens.
Methods
SHAPE analysis of 16S rRNA in 70S crystallized ribosomes
Crystals of the E. coli 70S ribosome were prepared as described.36 Ribosome crystals were washed three times in the same buffer used for cryo-protection and stabilization in the crystallographic studies, with the exception that the pH was increased from 4.8 to 6.5 [XB: 7% 2-methyl-2,4-penanediol, 7% PEG 8000, 24% PEG 400, 3.8 mM MgCl2, 380 mM NH4Cl, 5.5 mM putrescine, 5 mM spermidine, 22.5 mM MES (pH 6.5)] to remove free ribosomes prior to SHAPE analysis. Crystals were resuspended in 36 µL XB and SHAPE experiments were initiated by addition of 1/10 volume 1M7 (20 mM) in DMSO. Control reactions contained an equivalent volume of neat DMSO. Reactions were incubated for 2.5 hr at 4 °C (approximately 5 half lives for the hydrolysis rate of 1M7 in XB). Modified crystals were washed three times with XB to remove any free ribosomes, dissolved in 250 µL 1× TE and 50 µL 0.1 M dithiothreitol, and extracted three times with phenol:chloroform:isoamyl alcohol (25:24:1) and two times with chloroform:isoamyl alcohol (24:1). Total RNA was recovered by precipitation with ethanol, and 2′-O-adducts were detected by primer extension. Five primer pairs of 19-22 nucleotides were used with 3' positions complementary to 16S rRNA positions 323, 559, 947, 1112, and 1492.9 Primer extension products were resolved on an ABI 3130 capillary electrophoresis instrument. As judged from the long extensions obtained, RNA modification to form 2'-O-adducts was sparse, roughly 1 in ~300 nts were modified. Raw electropherograms were analyzed using ShapeFinder.37 Positions exhibiting high background were discarded and reactivities slightly less than zero were reset to zero. Three independent experiments were completed for each primer set and the resulting SHAPE data averaged (Fig. S2). Data sets were normalized by first excluding the top 2% of reactive nucleotides and then by dividing the reactivity for each nucleotide by the average of the next 8% most reactive nucleotides. The full dataset has been deposited in the SNRNASM database.42
Analysis of nucleotide geometries in 16S rRNA
All analyses started with the well-ordered E. coli ribosome visualized in a recent atomic-resolution structure, pdb id 3i1m.36 The crystals contain two ribosomes in each asymmetric unit. We focused our analysis on the better ordered (A) ribosome. For the nucleotides with well-defined conformations in both ribosome structures, nucleotide conformations were identical. Hyper-reactivities are likely to be underestimated as they reflect contributions from both highly ordered and less well-ordered ribosomes. The starting model was initially improved using MolProbity43 to identify all-atom clashes and suspect ribose sugar conformations. Corrections were carried out in Coot.44 Changes to the structure were only accepted if they survived refinement and did not compromise model geometry. The local conformations of nine of the well-ordered hyper-reactive nucleotides were ultimately adjusted (positions 250, 422, 531, 532, 701, 1278, 1297, 1336 and 1452). Nucleotides were initially aligned through the ribose moiety and then grouped according (i) to the distance between the 2′-OH and the vector bisecting the non-bridging oxygens in the adjacent 3′-phosphodiester and (ii) to the presence of a nearby functional group resulting in three categories (Fig. 4). One nucleotide (A243) did not fit into any of these three categories as determined by non-parametric box-plot analysis45 and Dixon’s Q-test.46 For the box-plot analysis (Fig. 5), well-ordered single-stranded nucleotides were defined as those not participating in conserved Watson-Crick base pairs, according to the secondary structures derived by comparative sequence analysis,47 and possessing an atomic displacement factor less than 60 in crystallographic refinement.
Nucleotide triphosphate analogues and RNA stem-loop synthesis
Phosphorothioate triphosphates and 2-thiouridine-5′-triphosphate were obtained from Glen Research and Trilink Biotechnologies, respectively. 3-Deaza-adenosine triphosphate was synthesized from 3-deaza-adenosine (Texas Biochemicals) as described.48 DNA templates directing synthesis of the stem-loop RNA [5′-(U/A)(U/A)GUC ACCAG CGUAA GCUG(A/U) GUGAC (U/A)(U/A)-3′] embedded in 5′ and 3′ structure cassette sequences35 were obtained by PCR. For analysis of phosphorothioate or 3-deaza-adenosine substitutions, the RNA contained a bulged adenosine and flanking uridine residues; for the 2-thiouridine substitution, the RNA contained a bulged uridine and flanking adenosine residues (Fig. 6). RNA constructs were synthesized by in vitro transcription [0.5 mL; 40 mM Tris (pH 8.0), 10 mM MgCl2, 10 mM dithiothreitol, 2 mM spermidine, 0.01% (v/v) Triton X-100, 4% (w/v) poly(ethylene) glycol 8000, 2 mM each NTP, PCR-generated template, T7 RNA polymerase; 37 °C; 4h]. Unsubstituted and nucleotide analogue-containing RNAs were synthesized using 0.1 and 0.2 mg/mL T7 RNA polymerase, respectively. RNAs were separated on a denaturing polyacrylamide gel (1× TBE, 8% polyacrylamide, 7 M urea, 29:1 acrylamide:bisacrylamide, 0.4 mm × 28.5 cm × 23 cm; 35 W, 1.5 h). Bands containing RNA were excised from the gel, and the RNA was recovered by overnight passive elution at 4 °C, followed by precipitation with ethanol. RNAs were resuspended in TE and stored at −20 °C.
SHAPE of the GUAA stem loop
RNAs (5 pmol) in 5 µL 1/2× TE were heated at 95 °C for 2 min, snap-cooled on ice, combined with 3 µL 3.3× folding buffer [333 mM Hepes (pH 8.0), 333 mM potassium chloride (pH 8.0), and 33 mM MgCl2], and incubated at 37 °C for 20 min. SHAPE experiments were initiated by adding 9 µL of the refolded RNA to 1 µL 100 mM 1M7 (in DMSO) or neat DMSO. Reactions were incubated at 37 °C for 2 min. SHAPE adducts were detected by primer extension and quantified using ShapeFinder.37 Data sets were normalized by dividing the value for each nucleotide by the average of the three most reactive nucleotides (10% of the total). SHAPE data sets for the native and analogue-containing RNAs were then re-scaled to the identical, single-stranded sequences in the flanking structure cassettes; SHAPE reactivities of these nucleotides were not affected by nucleotide analogue incorporation.
SHAPE reactivity as a function of added base
The [32P]-labeled adenosine-dideoxy cytosine (pAp-ddC; 1 µL, 100,000 cpm/µL) was incubated for 5 minutes at 37 °C in 8 µL 1.25× reaction buffer [62.5 mM HEPES (pH 8.0), 125 mM sodium chloride, 12.5 mM MgCl2] containing either imidazole or pyrrole at 0, 0.0375, 0.125, 0.375, 0.750, or 1.25 M (at pH 8.0). Adduct formation was initiated by adding 9 µL of pAp-ddC-containing solution to 1 µL 100 mM 1M7 (in DMSO) or neat DMSO. Reactions were incubated for 2 minutes at 37 °C. Reactions were diluted with 10 µL H2O and 20 µL formamide; 2 µL of the resulting solution was subjected to electrophoresis (1× TBE, 30% polyacrylamide; 29:1 acrylamide:bisacrylamide; 0.4 mm × 28.5 cm × 23 cm, 35 W, 1 hr); bands were quantified by phosphorimaging.
Supplementary Material
Acknowledgments
Acknowledgments. We thank Elizabeth O’Bryan for guidance in synthesizing 3-deaza-adenosine triphosphate. This work was supported by grants from the National Science Foundation (MCB-0919666 and MCB-1121024) to K.M.W.
Footnotes
Supporting Information Available: Figures illustrating hyper-reactive nucleotides for which the facilitating general base is located on a non-adjacent nucleotide and median and standard deviations for the complete in-crystal dataset. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).Weeks KM, Mauger DM. Acc. Chem. Res. 2011;44:1280–1291. doi: 10.1021/ar200051h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Duncan CD, Weeks KM. Biochemistry. 2008;47:8504–8513. doi: 10.1021/bi800207b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Mayerle M, Bellur DL, Woodson SA. J Mol Biol. 2011;412:453–465. doi: 10.1016/j.jmb.2011.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Dann CE, 3rd, Wakeman CA, Sieling CL, Baker SC, Irnov I, Winkler WC. Cell. 2007;130:878–892. doi: 10.1016/j.cell.2007.06.051. [DOI] [PubMed] [Google Scholar]
- (5).Stoddard CD, Gilbert SD, Batey RT. RNA. 2008;14:675–684. doi: 10.1261/rna.736908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Wang B, Wilkinson KA, Weeks KM. Biochemistry. 2008;47:3454–3461. doi: 10.1021/bi702372x. [DOI] [PubMed] [Google Scholar]
- (7).Mortimer SA, Weeks KM. Proc. Natl. Acad. Sci. USA. 2009;106:15622–15627. doi: 10.1073/pnas.0901319106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Lu C, Smith AM, Ding F, Chowdhury A, Henkin TM, Ke A. J Mol Biol. 2011;409:786–799. doi: 10.1016/j.jmb.2011.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Deigan KE, Li TW, Mathews DH, Weeks KM. Proc. Natl. Acad. Sci. USA. 2009;106:97–102. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Low JT, Weeks KM. Methods. 2010;52:150–158. doi: 10.1016/j.ymeth.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Bindewald E, Wendeler M, Legiewicz M, Bona MK, Wang Y, Pritt MJ, Le Grice SF, Shapiro BA. RNA. 2011;17:1688–1696. doi: 10.1261/rna.2640111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Washietl S, Hofacker IL, Stadler PF, Kellis M. Nucl. Acids Res. 2012 doi: 10.1093/nar/gks009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Costantino DA, Pfingsten JS, Rambo RP, Kieft JS. Nature Struct. Mol. Biol. 2008;15:57–64. doi: 10.1038/nsmb1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Jr., Swanstrom R, Burch CL, Weeks KM. Nature. 2009;460:711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Liang R, Kierzek E, Kierzek R, Turner DH. Biochemistry. 2010;49:8155–8168. doi: 10.1021/bi100286n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Gherghe C, Lombo T, Leonard CW, Datta SA, Bess JW, Jr., Gorelick RJ, Rein A, Weeks KM. Proc. Natl. Acad. Sci. USA. 2010;107:19248–19253. doi: 10.1073/pnas.1006897107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Grohman JK, Kottegoda S, Gorelick RJ, Allbritton NL, Weeks KM. J. Am. Chem. Soc. 2011;133:20326–20334. doi: 10.1021/ja2070945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Mortimer SA, Weeks KM. J. Am. Chem. Soc. 2008;130:16178–16180. doi: 10.1021/ja8061216. [DOI] [PubMed] [Google Scholar]
- (19).Mortimer SA, Weeks KM. Nature Protoc. 2009;4:1413–1421. doi: 10.1038/nprot.2009.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Wilkinson KA, Merino EJ, Weeks KM. J. Am. Chem. Soc. 2005;127:4659–4667. doi: 10.1021/ja0436749. [DOI] [PubMed] [Google Scholar]
- (21).Turner KB, Yi-Brunozzi HY, Brinson RG, Marino JP, Fabris D, Le Grice SF. RNA. 2009;15:1605–1613. doi: 10.1261/rna.1615409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Steen KA, Malhotra A, Weeks KM. J. Am. Chem. Soc. 2010;132:9940–9943. doi: 10.1021/ja103781u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Lucks JB, Mortimer SA, Trapnell C, Luo S, Aviran S, Schroth GP, Pachter L, Doudna JA, Arkin AP. Proc. Natl. Acad. Sci. USA. 2011;108:11063–11068. doi: 10.1073/pnas.1106501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Badorrek CS, Weeks KM. Nat Chem Biol. 2005;1:104–111. doi: 10.1038/nchembio712. [DOI] [PubMed] [Google Scholar]
- (25).Reymond C, Levesque D, Bisaillon M, Perreault JP. Structure. 2010;18:1608–1616. doi: 10.1016/j.str.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Legiewicz M, Zolotukhin AS, Pilkington GR, Purzycka KJ, Mitchell M, Uranishi H, Bear J, Pavlakis GN, Le Grice SF, Felber BK. J. Biol. Chem. 2010;285:42097–42104. doi: 10.1074/jbc.M110.182840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Kladwang W, Cordero P, Das R. RNA. 2011;17:522–534. doi: 10.1261/rna.2516311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Soulière MF, Haller A, Rieder R, Micura RJ. Am. Chem. Soc. 2011;133:16161–16167. doi: 10.1021/ja2063583. [DOI] [PubMed] [Google Scholar]
- (29).Mortimer SA, Weeks KM. J. Am. Chem. Soc. 2007;129:4144–4145. doi: 10.1021/ja0704028. [DOI] [PubMed] [Google Scholar]
- (30).Gherghe CM, Shajani Z, Wilkinson KA, Varani G, Weeks KM. J. Am. Chem. Soc. 2008;130:12244–12245. doi: 10.1021/ja804541s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Wilkinson KA, Vasa SM, Deigan KE, Mortimer SA, Giddings MC, Weeks KM. RNA. 2009;15:1314–1321. doi: 10.1261/rna.1536209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Vicens Q, Gooding AR, Laederach A, Cech TR. RNA. 2007;13:536–548. doi: 10.1261/rna.400207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, Giddings MC, Weeks KM. PLoS Biol. 2008;6:e96. doi: 10.1371/journal.pbio.0060096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Chamberlin SI, Merino EJ, Weeks KM. Proc. Natl. Acad. Sci. USA. 2002;99:14688–14693. doi: 10.1073/pnas.212527799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. J. Am. Chem. Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- (36).Zhang W, Dunkle JA, Cate JHD. Science. 2009;325:1014–1017. doi: 10.1126/science.1175275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Vasa SM, Guex N, Wilkinson KA, Weeks KM, Giddings MC. RNA. 2008;14:1979–1990. doi: 10.1261/rna.1166808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).McGinnis JL, Duncan CD, Weeks KM. Methods Enzymol. 2009;468:67–89. doi: 10.1016/S0076-6879(09)68004-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Murray LJ, Arendall W. B., 3rd, Richardson DC, Richardson JS. Proc. Natl. Acad. Sci. USA. 2003;100:13904–13909. doi: 10.1073/pnas.1835769100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Frey PA, Sammons RD. Science. 1985;228:541–545. doi: 10.1126/science.2984773. [DOI] [PubMed] [Google Scholar]
- (41).Donohue JJ. Mol. Biol. 1969;45:231–235. doi: 10.1016/0022-2836(69)90102-8. [DOI] [PubMed] [Google Scholar]
- (42).Rocca-Serra P, Bellaousov S, Birmingham A, Chen C, Cordero P, Das R, Davis-Neulander L, Duncan CD, Halvorsen M, Knight R, Leontis NB, Mathews DH, Ritz J, Stombaugh J, Weeks KM, Zirbel CL, Laederach A. RNA. 2011;17:1204–1212. doi: 10.1261/rna.2753211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Richardson JS, Schneider B, Murray LW, Kapral GJ, Immormino RM, Headd JJ, Richardson DC, Ham D, Hershkovits E, Williams LD, Keating KS, Pyle AM, Micallef D, Westbrook J, Berman HM. RNA. 2008;14:465–481. doi: 10.1261/rna.657708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Emsley P, Cowtan K. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- (45).Tukey JW. Exploratory data analysis. Addison-Wesley; Reading, MA: 1977. [Google Scholar]
- (46).Rorabacher DB. Anal. Chem. 1991;63:139–146. [Google Scholar]
- (47).Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Muller KM, Pande N, Shang Z, Yu N, Gutell RR. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Gillerman I, Fischer B. Nucleosides Nucleotides Nucleic Acids. 2010;29:245–256. doi: 10.1080/15257771003709569. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.