Abstract
Lysine 2,3-aminomutase (LAM) catalyzes the interconversion of L-lysine and L-β-lysine, a component of a number of antibiotics. The reaction requires the cofactors S-adenosyl-L-methionine (SAM), pyridoxal-5′-phosphate (PLP), and a [4Fe–4S] cluster. LAM is a founding member of the Radical SAM superfamily of enzymes. LAM is highly specific for L-lysine and will not accept most other amino acids as substrates. L-alanine and L-2-aminobutyrate at 0.2 M react as substrates for LAM at, respectively, 5 × 10–6 and 8 × 10–5 times the rate with saturating L-lysine. Saturating ethylamine accelerates the L-alanine reaction 70-fold, and saturating methylamine accelerates the L-2-aminobutyrate reaction 47-fold. The primary amines binding at the active site of LAM with L-alanine or L-2-aminobutyrate simulate L-lysine. The steady state kinetics of the reaction of L-alanine + ethylamine displays negative cooperativity with respect to L-alanine. The second order rate constant for production of β-alanine in the reaction of L-alanine and saturating ethylamine is 0.040 M–1 s–1, which is 2 × 10–5 times the value of kcat/Km for the reaction of L-lysine. When L-lysine is at a concentration 1/16th of Km, the lysyl-free radical intermediate is hardly detectable by EPR; however, the addition of L-alanine at high concentration (0.2 M) enhances free radical formation, and the addition of ethylamine further enhances radical formation. These facts complement the kinetic observations and support negative cooperativity in the reaction of L-alanine as a substrate for LAM. Present results and independent evidence support negative cooperativity in the reaction of L-lysine as well.
Introduction
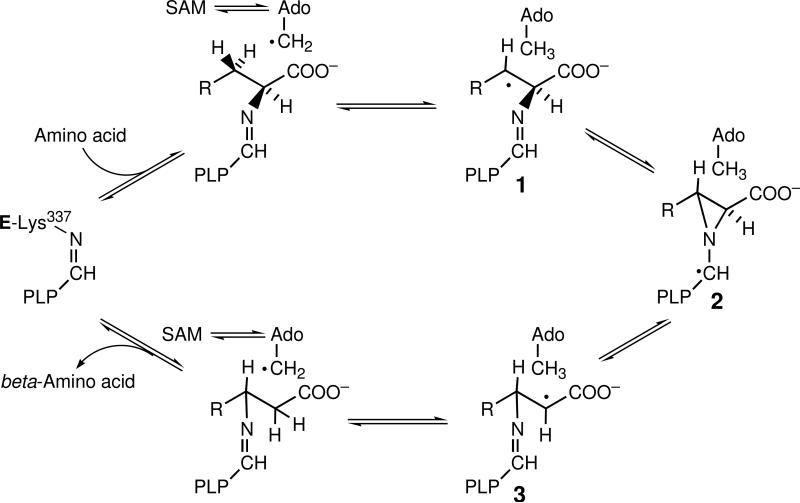
Lysine 2,3-aminomutase (LAM), the product of the gene kamA in Clostridium subterminale SB4, is a [4Fe–4S]-enzyme that catalyzes the pyridoxal-5′-phosphate (PLP)- and S-adenosyl-L-methionine (SAM)-dependent interconversion of L-lysine and L-β-lysine according to equation 1 by a radical mechanism.1-3 The radical intermediates are shown in Scheme 1,
 |
(1) |
where the 5′-deoxyadenosyl radical derived from SAM initiates the reaction by abstracting the 3-pro-R hydrogen from the side chain of L-lysine, which is bound through the α-amino group to PLP as an imine. Isotope-edited electron paramagnetic resonance (EPR)-spectroscopic and kinetic experiments by rapid-mix-freeze-quench EPR spectroscopy implicate radicals 1,3 and 5′-deoxyadenosine-5′-yl (Ado-CH2 •) as intermediates.4-10
Scheme 1.
LAM is a founding member of the Radical SAM superfamily of enzymes, characterized by the presence of the motif CxxxCxxC in the amino acid sequence.11 These enzymes carry out complex biochemical transformations by radical mechanisms. The superfamily includes more than 2845 proteins encoded by known genomes and are involved in more than 40 distinct biological processes.1
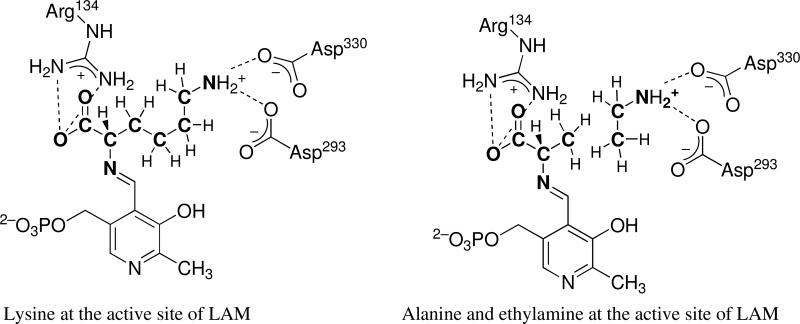
LAM is a homodimeric, [4Fe–4S]-enzyme that crystallizes as a homotetramer formed by association of two intercalated homodimers.12 The x-ray crystal structure shows that the α-amino group of L-lysine binds as an imine to the C4′-carboxaldehyde group of PLP, the carboxylate of lysine binds to the guanidinium group of Arg134, and the ε-aminium group of L-lysine binds to the β-carboxylate groups of both Asp293 and Asp330 in LAM. SAM binds through its amino and carboxylate groups to one iron of the [4Fe–4S].
An apparent fundamental chemical difficulty in catalysis by LAM is the reversible cleavage of SAM to the 5′-deoxyadenosyl radical and methionine. The [4Fe–4S] cluster plays the leading role in this process by ligating SAM to one of the four irons and mediating inner sphere electron transfer to the sulfonium center, leading to homolytic scission of the adenosyl-C5′—S bond according to equation 2. Ligation of SAM to the [4Fe–4S]-cluster elevates the reduction potential of SAM and lowers the barrier to electron transfer by 19 kcal/mol, thereby allowing the reaction to take place.13
 |
(2) |
LAM displays a very high degree of selectivity for L-lysine as a substrate. D-Lysine not only is rejected as a substrate, it is not even an inhibitor of LAM and will not bind to the active site at concentrations up to 100 mM. In this paper, we describe experiments designed to force the reaction of simple substrates. The results reveal aspects of the structural basis for the high degree of substrate selectivity in the actions of this enzyme.
The crystal structure of LAM shows that the dimeric unit includes extensive intercalation of subunit domains, described as domain swapping.12 Such intimate inter-subunit interactions open the way to subunit-subunit interactions; however, no deviation from traditional Michaelis-Menten kinetics can be detected in conventional initial rate analysis with L-lysine as the substrate.9 In this paper, we present kinetic and spectroscopic results with a very poor substrate, the results of which unmask an unusual form of negative cooperativity in the action of this enzyme.
Glutamate 2,3-aminomutase catalyzes the interconversion of L-glutamate and β-glutamate and is related to LAM in amino acid sequence, identical cofactor requirements, and a similar reaction mechanism.14 Efforts to elicit activity of this enzyme on L-alanine failed.
Experimental Procedures
Materials
The following were obtained from the named vendors: EPPS, PLP, sodium hydrosulfite, L-alanine, β-alanine, L-lysine, L-2-aminobutyrate, DL-3-aminobutyrate, L-cysteine, L-glutamate, L-glutamine, L-threonine, L-homoserine, L-tyrosine, L-phenylalanine, L-valine, L-leucine, L-isoleucine, L-histidine, L-tryptophan, L-ornithine, L-arginine, methylamine, ethylamine, n-propylamine, sodium formate, and sodium acetate from Sigma; DTT from Inalco; phenyl-Sepharose 6 Fast Flow (high substitution), Q-Sepharose Fast Flow, and Sephacryl S200 HR from Amersham Biosciences; E. coli BL21(DE3) CodonPlus RILP cells from Stratagene; L-[2,3,3,3-2H4]alanine from Cambridge Isotope; FeSO4 from JT Baker; PITC and amino acid standard solution #20088 from Pierce; Centricon YM10 from Millipore.. All other chemicals were obtained in highest available purity from commercial vendors and used as supplied. SAM was obtained from Sigma as the p-toluenesulfonate salt and purified over CM cellulose.8
Preparation of aminomutases
Expression of recombinant Clostridium subterminale SB4 LAM and Clostridium difficile glutamate 2,3-aminomutase in E. coli were as previously described.3,14 All purification steps of each enzyme were carried out in a Coy anaerobic chamber according to described procedures. Final purification of both proteins was by passage through a Sephacryl S200 HR column (2.5 x 25 cm) equilibrated with SB buffer (0.03 M sodium EPPS at pH 8.0, 20 μM FeSO4, 20 μM PLP, 0.25 mM of either L-lysine (LAM) or 1 mM L-glutamate (glutamate 2,3-aminomutase), and 1 mM DTT. Samples were frozen and stored in liquid N2.
Quantitative amino acid analysis
The molar concentrations of purified enzymes were determined by quantitative amino acid analysis based on the known amino acid composition as previously described.14
Assays of LAM with L-Alanine or β-Alanine
Prior to conducting assays, the activity of LAM was optimized by reductive incubation inside the Coy anaerobic chamber with 2 mM FeSO4, 2.4 mM PLP, and 10 mM L-cysteine in 0.2 M sodium EPPS buffer at pH 8.0 and 37 °C for 4 h. Activated LAM was separated from the reductive incubation components by gel filtration through a Sephacryl S200 Fast Flow column (2.5 x 15 cm) equilibrated with 0.03 M sodium EPPS buffer pH 8 buffer containing 1 mM DTT, 10 μM PLP, and 20 μM FeSO4. Colored fractions were collected and concentrated using Centricon YM10 centrifugation filtration columns. The concentrated enzyme (40-50 mg/ml) was frozen and stored in liquid N2.
Standard assays of activated LAM with L-alanine or β-alanine as substrates were conducted at 37 °C inside the anaerobic chamber. Product mixtures were assayed by derivatization of amino acids with PITC followed by HPLC.15 Stock solutions of reaction components were prepared at pH 8.0 and conditioned inside the anaerobic chamber. Standard assay solutions consisted of LAM (85 μM subunits), 80 mM Na-EPPS,at pH 8.0, 160 μM SAM, 0.7 mM sodium hydrosulfite, 200 mM L-alanine or β-alanine, and in the presence or absence of 0.2 M ethylamine. At timed intervals (0-20 min), 30 μL aliquots of reaction mixtures were quenched with 12 L of 2 N perchloric acid and centrifuged. Samples of the supernatant fluids were treated with PITC in the air as described.15 The PITC derivatives of L-alanine and β-alanine (10-50 μL) were separated using a C18 column (Phenomenex Gemini μ5, 4.6 mm x 25 cm, #G-4435-E0), flow rate 1 ml/min. Separation was attained by use of a linear gradient formed with 0.05 M ammonium acetate in water (A) and 0.1 M ammonium acetate in 44% water, 46% acetonitrile, and 10% methanol (B). The gradient was 0-10% B in 17.5 min, 10-35% B in 17.5 min, 35-100% B in 15 min. PITC-L-alanine emerged at 32 min and PITC-β-alanine at 28 min. The concentrations of product formed in quenched samples were determined from the fractions of the PITC-derivatives in the HPLC analysis. The peak area of either PITC-L-alanine or PITC-β-alanine was divided by the sum of the two and multiplied by the starting concentration of either L-alanine or β-alanine to obtain molar concentrations. The starting concentrations of L-alanine and β-alanine in quenched samples were measured by reference to amino acid standards.
In the kinetic analysis with L-alanine as the substrate or ethylamine as the activator, the standard procedure was followed with the following modifications. Initial rates were determined at varying substrate concentrations from 0.012 M to 0.265 M L-alanine, or at 0.2 M L-alanine with varying ethylamine concentrations from 0.012 to 0.20 M. The initial rates were employed in the calculation of kinetic parameters by the nonlinear least squares curve fitting programs (hypero and sigmoid) of Cleland.16
In assays components were substituted as appropriate: 0.2 M methylamine or propylamine for ethylamine; 0.2 M L-2-aminobutyrate for L-alanine; 0.2 M sodium formate, sodium acetate, or sodium bicarbonate with L-alanine in trials with glutamate 2,3-aminomutase. The PITC derivatives of L-2-aminobutyrate and DL-3 aminobutyrate (25-50 μL) were separated as described above except the gradient was modified to 0-10% B in 40 min, 10-35% B in 35 min, 35-100% B in 15 min. PITC-L-2-aminobutyrate emerged at 68 min and PITC-DL-3-aminobutyrate at 58 min.
Assays of the primary kinetic isotope effect were conducted at 190 mM of either L-alanine or L-[2,3,3,3-2H4]alanine with 200 mM ethylamine, other assay conditions as described above. The concentration of each amino acid was established prior to assay by quantitative amino acid analysis of PITC derivatives referenced to L-alanine standards.
EPR spectroscopy
Samples of LAM (450 μM subunits) reductively incubated with or without 0.25 mM L-Lysine were quickly mixed anaerobically at 24 °C with 240 mM Trissulfate, 1.4 mM SAM, 1.5 mM sodium hydrosulfite, and 120 mM L-alanine. In two experiments 0.2 M ethylamine or propylamine was also present as activators, and in one experiment the alkylamine-activator was excluded. A control sample containing 120 mM L-lysine without L-alanine was prepared as above. The samples were frozen at –150 °C as soon as possible after mixing (< 30 s). The EPR spectra of the frozen samples were recorded at 77 K using a Varian model E3 spectrometer with the following settings: field center 3250 Gauss,; scan width 200 Gauss; microwave frequency 9.1 GHz; microwave power 5 mW; modulation frequency 100 kHz; modulation amplitude 1.6 Gauss; time constant 0.3 sec; scan time 240 s; gain 125,000.
Results and Discussion
Activation of reaction of L-alanine by primary amines
LAM is highly selective for L-lysine as a substrate and does not display detectable activity toward L-glutamate, L-glutamine, L-threonine, L-homoserine, L-tyrosine, L-phenylalanine, L-valine, L-leucine, L-isoleucine, L-histidine, L-tryptophan, L-ornithine, or L-arginine. LAM displays very low but measurable activity toward 0.2 M L-alanine and 0.2 M L-2-aminobutyrate, producing β-alanine and L-3-aminobutyrate, respectively (Table 1). The rates in Table 1 for 0.2 M L-alanine or L-2-aminobutyrate are, respectively, 5 x 10–6 and 8 x 10–5 times the rate with saturating L-lysine.
Table 1.
2,3-Aminomutase Activities of Lysine 2,3-aminomutase and Glutamate 2,3-aminomutase.
| Enzyme | Substrate | Activator | Specific Activity |
|---|---|---|---|
| Lysine 2,3-aminomutase | L-Alanine | + ethylamine | 12 ± 0.34 |
| – ethylamine | 0.21 ± 0.02 | ||
| + methylamine | 5.7 ± 0.16 | ||
| + n-propylamine | 0.32 ± 0.03 | ||
| β-Alanine | + ethylamine | 1.2 ± 0.13 | |
| L-2-aminobutyrate | + methylamine | 140 ± 4.2 | |
| – methylamine | 3.7 ± 0.11 | ||
| Glutamate 2,3-aminomutase | L-Alanine | + formate | ndb |
| + acetate | nd | ||
| + bicarbonate | nd |
a Activities were measured at 37 °C at pH 8.0 (NaEPPS), with 0.16 mM SAM, 0.7 mM Na-hydrosulfite, and 85 μM LAM (subunits) in the anaerobic chamber. Substrate and amine or carboxylate concentrations were 0.2 M. Reactions were stopped at timed intervals and products derivatized as phenyisothiocyanates (PITC). The PITC derivatives were measured spectrophotometrically after separation by HPLC. Specific activities are expressed as nmol min−1 mg enzyme−1.
nd – not detectable (Activity <0.1 nmol min−1 mg enzyme−1)
The presence of ethylamine significantly enhances the action of LAM on L-alanine (Table 1). The kinetics of activation by ethylamine displays saturation with respect to ethylamine and obeys the Michaelis-Menten equation {v = V[EA]/([EA] + KEA}, where v is the measured velocity at a given concentration of ethylamine (EA), V is the maximum velocity at saturating ethylamime, and KEA is the dissociation constant of ethylamine. The value of KEA at 0.265 M L-alanine is 46 mM (Table 2). Ethylamine at 0.2 M accelerates the transformation of 0.2 M L-alanine into β-alanine 57-fold (Table 1). At 0.2 M, ethylamine is at 81% saturation, so that the acceleration would be 70-fold at saturating ethylamine. Methylamine at 0.2 M also accelerates the reaction of L-alanine 27-fold (Table 1). n-Propylamine displays very little stimulation in the reaction of L-alanine (Table 1).
Table 2.
Kinetic parameters in the action of lysine 2,3-aminomutase.a
| Parameter | Value | |
|---|---|---|
| L-Alanine (0.265 M) + varied C2H5NH3+ | KEA (mM) | 46 ± 6b |
| L-Alanine (varied) + 0.2 M C2H5NH3+ | KS (mM) | 29 ± 4c |
| L-Alanine (varied) + 0.2 M C2H5NH3+ | k (M–1 s–1) | 0.032 ± 0.002c |
| L-Alanine/[2,3-2H4]Alanine + 0.2 M C2H5NH3+ | kH/kD | 7.1 |
| L-Alanine (varied) + saturation M C2H5NH3+ | k (M–1 s–1) | 0.040 ± 0.002d |
| L-Lysine (varied) | kcat/Km (s–1 M–1)e | 8.0 × 103 |
pH 8.0, 37 °C, 0.2 M Na-EPPS buffer, 0.16 mM SAM, 85 μM LAM subunits.
Primary data in Figure S1 of Supplementary Information
Apparent value by fitting equation 5 to data in Figure 1.
Value at saturation of C2H5NH3+ obtained by correction for 81% saturation at 0.20 M
Literature value of apparent second order rate constant at pH 8.0 and 37 °C.9
By the principle of microscopic reversibility, LAM must catalyze the transformation of β-alanine into L-alanine. LAM does catalyze the reverse reaction at about one-tenth the rate of the forward reaction under comparable conditions (Table 1). L-2-Aminobutyrate at 0.2 M reacts 18-times more rapidly than 0.2 M L-alanine (Table 1). Methylamine at 0.2 M stimulates the reaction of L-2-aminobutyrate 38-fold.
The primary amines presumably stimulate reactions of L-alanine and L-2-aminobutyrate by binding to the active site in place of the propylamine moiety in the L-lysyl side chain, partially filling the subsite and stabilizing the transition state. Methylamine is less stimulatory than ethylamine, and propylamine is nearly ineffective. The related enzyme glutamate 2,3-aminomutase14 does not act on L-alanine and is not activated by formate, acetate, or bicarbonate.
The enhancement by primary amines in the reactivities of L-alanine and L-2-aminobutyrate as substrates can be rationalized by considering the effect of ethylamine on the reaction of L-alanine. The 70-fold enhancement is analogous to the chemical rescue of a mutated enzyme by a molecule in solution. For example, in GalT mutation of His166 to asparagine or glycine leads to inactive proteins because His166 is an essential nucleophilic catalyst in the reaction mechanism.17-19 The GalT activity absent in H166G-GalT can be rescued by addition of free imidazole, which can bind to the active site in place of the imidazolyl-methylene group vacated by substitution of glycine for His166.
Based on the structure of LAM with all cofactors in place and L-lysine bound to PLP at the active site12, the binding of ethylamine can be pictured as illustrated at the right in Scheme 2.
Scheme 2.
Replacement of L-lysine in the left image with L-alanine and ethylamine in the right image allows a reasonable fit of atoms permitting L-alanine and ethylamine to simulate L-lysine. In this picture the carbon backbone of L-lysine includes 4-carbons and 8-hydrogens and the simulation with L-alanine and ethylamine includes 3-carbons and 8-hydrogens. With one carbon less, the simulation succeeds at a less efficient rate, presumably because the van der Waals contacts are permissive. Methylamine in a similar picture would have 2-carbons and 8-hydrogens and is less efficient. Considering n-propylamine in this formulation, there would be parity with lysine in 4-carbons; however, the site would be burdened with 10-hydrogens, two more than in the L-lysyl side chain. The fact that n-propylamine is hardly effective can be attributed to unfavorable van der Waals contacts owing to the bulk of two additional hydrogens. Interiors of proteins are tightly packed,20 and they cannot accommodate new steric demands without suffering global effects on structure. An analogous situation exists in the chemical rescue of GalT, where rescue of H166A-GalT by imidazole is 1/630th as effective as rescue of H166G-GalT.19 The complex of H166A-GalT with imidazole entails two additional hydrogens in place of the His166 side chain.
Enhancements by primary amines in the reactions of L-alanine and L-2-aminobutyrate as substrates for LAM are strictly analogous to stimulations by methylguanidine and ethylamine of trypsin acting on N-acetylglycine ethyl ester, on a somewhat lower scale. Trypsin selectively catalyzes the hydrolysis of N-acyl esters and amides of L-arginine and L-lysine, but also acts at a very slow rate on N-acetylglycine ethyl ester, a substrate lacking a basic side chain. Methylguanidine and ethylamine stimulate the trypsin-catalyzed hydrolysis of N-acetylglycine ethyl ester by factors of 4 and 12, respectively.21
Hydrogen transfer and rate limitation
The observed rate for the reaction of L-[2,3-2H4]alanine is less than one-seventh the rate for the reaction of L-alanine (Table 2). The ratio of rates at 0.19 M substrates and 0.2 M ethylamine is 7.1 favoring unlabeled L-alanine. The result clearly shows that hydrogen transfer is rate limiting in the reaction of L-alanine. The kinetic isotope effect is similar to the value 5.4 in the reaction of L-lysine.10
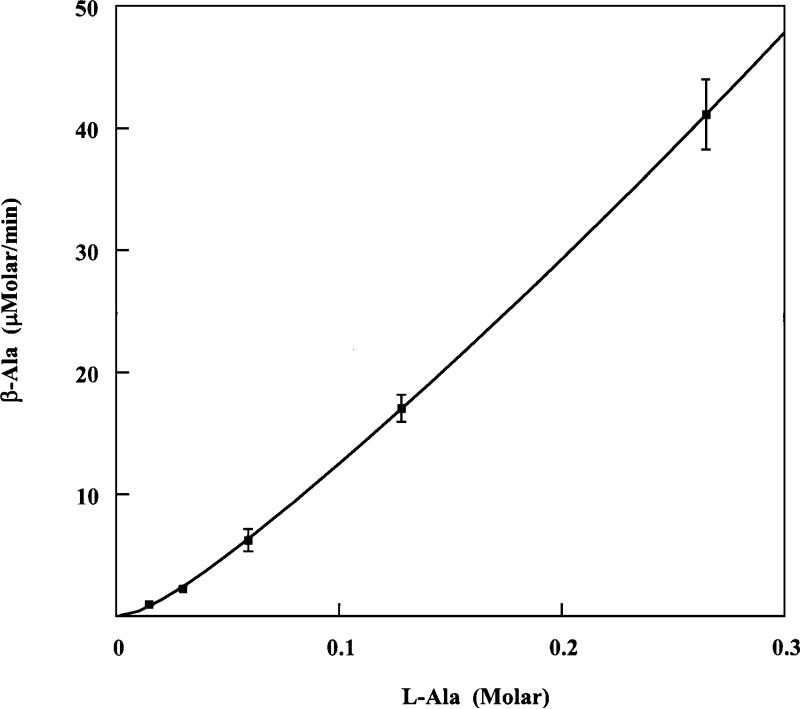
Kinetics of the reaction of L-alanine
The concentration dependence of the rate in the ethylamine activated reaction of L-alanine is complex. As shown in Figure 1, the initial rates increase with increasing concentrations of L-alanine and reach a linear phase. The unusual features are the concavity of the curve at low concentrations of L-alanine and the absence of a saturation effect at high concentrations. In contrast, the reaction of lysine follows classical Michaelis-Menten saturation kinetics.9 Complexity of the type in Figure 1 might be accounted for by any of a variety of mechanisms. The simplest mechanism is the cooperative substrate sequence in equations 3 and 4, in which the free enzyme is essentially inactive and the binding of
| (3) |
| (4) |
one molecule of L-alanine to a molecule of enzyme does not lead to production of β-alanine at a significant rate (equation 3). Then the mono-ligated enzyme E.Ala reacts in a second order process with free L-alanine to produce β-alanine. The initial nonlinear part of the curve represents binding the first molecule of L-alanine to a molecule of enzyme to form the active species E.Ala. The observed activity increases exponentially as the enzyme becomes saturated to its catalytically active state (E.Ala). The linear portion represents the second order reaction of the active enzyme with free L-alanine to produce β-alanine. The initial rate equation for the kinetic model is equation 5, which is fitted to the data in Figure 1 to obtain the kinetic parameters in Table 2.
| (5) |
Figure 1.
Initial rates of 2,3-aminomutase activity of LAM, with L-alanine + ethylamine simulating the substrate L-lysine.
Initial rates of β-alanine formation are plotted versus L-alanine concentrations. Rates were measured by the procedure in Experimental Procedure at pH 8.0 and 37 °C in the presence of 0.2 M ethylamine. Initial rates were measured in quadruplicate. Equation 5 was recast into the form v/[Ala] = k[Eo][Ala]/(KS + [Ala]) and fitted to the data using the program Hypero of Cleland.16 Values of k and KS are given in Table 2.
In an enzymatic reaction, expectations of saturation kinetics are often not realized in the reactions of very poor substrates, which might bind too weakly to display saturation behavior under physicochemically reasonable or accessible conditions. In Figure 1 the only observed saturation is the initial binding of L-alanine. The catalytically active enzyme E.Ala is expected to bind L-alanine in the process of transforming it into β-alanine, but this is not observed at concentrations as high as 0.265 M. It is not practical to study the reaction at higher concentrations, so we choose to fit the data to equation 5 and interpret the results in terms of the mechanism in equations 3 and 4. The parameter KS is the dissociation constant for E.Ala. The parameter k is the second order rate constant for the transformation of L-alanine into β-alanine by E.Ala. This rate constant in Table 2 can be compared with the apparent second order rate constant in the LAM-catalyzed reaction of L-lysine, which is kcat/Km = 8.0 x 103 M–1 s–1 at the pH and temperature of Table 2.9 Comparison with the value in Table 2 of k = 0.040 M–1 s–1 for the reaction of L-alanine shows that LAM catalyzes the reaction of free L-lysine at 2 x 105-times the rate of L-alanine.
The simplest and most likely molecular basis for the kinetic mechanism (equations 3 and 4) is that L-alanine bound in E.Ala occupies the active site in one subunit of the dimeric unit in LAM. This species is either unreactive or reacts very slowly. However, it is conformationally poised to catalyze the reaction of a second molecule of L-alanine at the active site of the second subunit. This is a version of negative cooperativity in the action of LAM on L-alanine, a poor substrate.
Negative cooperativity is increasingly recognized in the actions of a number of enzymes.22 It was first described quantitatively for skeletal muscle glyceraldehyde-3-phosphate dehydrogenase (GAPDH), a homotetrameric enzyme that catalyzes the phosphate-dependent dehydrogenation of glyceraldehyde-3-phosphate by NAD to form NADH and 1,3-diphosphoglycerate.23 The subunits of GADPH bind NAD with decreasing affinity as the binding sites in each subunit become occupied. The dimeric pyruvate dehydrogenase component (E1) of the pyruvate dehydrogenase complex binds thiamine diphosphate to each subunit, but inter-subunit communications allow only one of the coenzyme molecules to function chemically at a time in an alternating sites mechanism.24 Negative cooperativity is described as a general property of thiamine diphosphate-dependent enzymes, Type 1 fatty acid synthases, and F1-ATPases.25-27
Extreme forms of negative cooperativity lead to behavior in which it appears that just half of the available sites in an enzyme function in a physicochemical experiment. In such cases, all sites are nominally identical, but reaction at one site in an oligomer such as a dimer leads to a large decrease in function at the other site. This behavior is observed in many enzymes, among them the small subunit of ribonucleotide reductase from Escherichia coli, tryptophanyl tRNA synthetase from Deinococcus radiodurans, and human 6-phosphogluconate dehydrogenase.28-30
Half-site phenomena have often been found to be illusory because of the difficulties intrinsic to bioanalytical methods, and many reported instances have been found to be incorrect. Among the difficulties are unreliable measurements of molar protein concentrations, uncertainties about the purity or chemical homogeneity of biological samples, and uncertainty about association/dissociation behavior in multisubunit enzymes. Kinetics alone is often insufficient evidence for assigning negative cooperativity or half-sites reactivity. In the present case, EPR spectroscopy in the next section provides complementary evidence supporting negative cooperativity in the reaction of L-alanine with LAM
EPR spectroscopy of activation by L-alanine
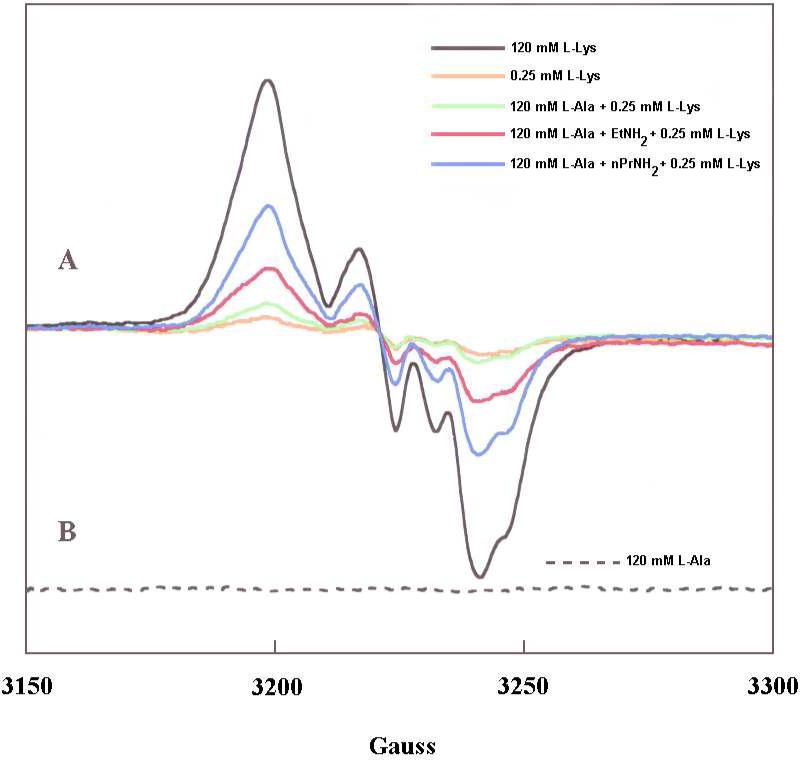
Unlike the reaction of L-lysine with LAM, the reaction of L-alanine does not elicit an EPR signal in the steady state. Freeze-quenching of the reaction of the complex of LAM.PLP.SAM.[4Fe–4S] with 0.12 M L-lysine in the steady state reveals the prominent EPR signal of radical intermediate 3 in Scheme 1.4,5 This spectrum is shown as the black trace in part A of Figure 2. The dashed trace in part B of Figure 2 is the EPR trace in a comparable reaction of L-alanine, showing no EPR signal. The absence of an EPR signal with L-alanine can be explained on the basis that it is such a poor substrate that reaction intermediates exist at concentrations too low to be detected by EPR.
Figure 2.
EPR spectra of product radical formation at the active site in LAM.
Procedure is described in the Experimental section. The black spectrum is that of the product-related radical 3 in Scheme 1 elicited by the natural substrate L-lysine at 120 mM in the steady state. The dashed spectrum is that obtained upon substitution of L-alanine for L-lysine. The weak orange signal was obtained with L-lysine at a concentration 1/16 the Km for L-lysine, the green spectrum upon addition of 0.2 M L-alanine, and the red spectrum upon further addition of 0.2 M ethylamine. The blue spectrum was obtained upon substitution of n-propylamine for ethylamine in the preceding experiment.
However, L-alanine proves to be a useful probe in the reaction of L-lysine, as monitored by EPR detection of radical 3 in Scheme 1. The presence of 0.2 M L-alanine enhances the EPR spectrum of radical 3 generated at low concentrations of L-lysine. The value of Km in the reaction of L-lysine is 4 mM,3,9 so that very little radical 3 can be detected at 0.25 mM L-lysine (orange trace in Figure 2). However, the presence of L-alanine (0.2 M) enhances this spectrum (green trace in Figure 2). Moreover, the additional presence of ethylamine (EtNH2) dramatically enhances the trace for radical 3 (red trace in Figure 2), and n-propylamine (blue trace) further enhances the signal.
Enhancement of the EPR signal in the reaction of L-lysine by L-alanine and further enhancement by primary amines shows that “simulated L-lysine” in a second binding site stimulates radical formation by reaction of L-lysine in the active site. The effect at a low concentration of L-lysine (0.25 mM) requires high concentrations (0.2 M) of L-alanine and ethylamine, much higher concentrations than the values of dissociation constants for these molecules (KS and KEA, Table 2) binding to free LAM. This result is consistent with strongly negative cooperativity in binding at the active site.
Independent evidence implicates negative cooperativity in binding L-lysine to the two active sites in dimeric LAM. When purified through several steps, including chromatography, in the complete absence of L-lysine, LAM contains significant amounts of L-lysine and L-β-lysine.31 The purified enzyme is very unstable, especially to chromatography, but it is stabilized by the inclusion of 0.1 mM L-lysine in the purification buffers.32 Tight binding of L-lysine and stabilization by 0.1 mM L-lysine indicates a high affinity binding site, and the value of Km (4 mM) in kinetic experiments and the requirement for high concentrations to elicit the EPR signal of radical 3 (Scheme 1) is indicative of weaker affinity in the catalytic process. Moreover, 4-thia-L-lysine is a substrate that exhibits a Km-value of 1.4 mM, but as an inhibitor of the reaction of L-lysine the competitive inhibition constant is 0.1 mM.9
Some of the results reported herein and elsewhere could be accounted for by invoking an alternative binding site for amino acids like L-alanine, such as an allosteric site. However, no alternative sites appear in the x-ray crystal structure from crystals developed in high enough concentrations of L-lysine to saturate the two binding sites in the functional dimer of LAM.12 Moreover, it is doubtful that an allosteric site would display such a low affinity as observed in the present experiments with L-alanine: such a low affinity site would certainly have no biological significance.
All things considered, the simplest interpretation of the results mandates high affinity binding of L-lysine at one active site of the dimeric LAM to form a mono-ligated species that displays little or no activity. The mono-ligated LAM binds a second molecule of L-lysine with lower affinity, and the bis-ligated LAM is fully active. L-Alanine can occupy one binding site in the dimer with low affinity (KS = 29 mM, Table 2), and binding to the second subunit is so weak that it cannot be detected in kinetic experiments. In the present EPR experiments with L-lysine bound to one subunit, L-alanine and ethylamine simulate the second molecule of L-lysine. In the kinetic experiments, both L-alanine + ethylamine and L-2-aminobutyrate + methylamine simulate L-lysine as substrates.
The steady state kinetics of the reaction of L-lysine does not reveal cooperativity, which must be explained in the light of the present results with L-alanine + ethylamine as a simulation of L-lysine. In view of the fact that LAM binds L-lysine and analogs such as 4-thia-L-lysine very tightly, while displaying much higher values of Km, and fast turnover, cooperativity likely would not be observed or would be very difficult to observe in conventional steady state kinetic experiments.
Equilibria in aminomutase reactions
Experiments indicate that under comparable conditions the rate at which LAM catalyzes the reverse reaction of β-alanine to form L-alanine is about one-tenth the rate of the forward process. The rate difference is consistent with the expectation that the equilibrium constant is likely to be in the range of 7-15 in favor of β-alanine. An experimental value of the equilibrium constant for the reaction of L-alanine is not available. However, the equilibrium constants for the formation of β-L-lysine from L-lysine range from 7.2 to 10.8 depending on temperature between 21 and 37 °C,33 and for the reaction of glutamate 2,3-aminomutase it is 15.7 at 37 °C.14 The values of these equilibrium constants are attributed the greater enthalpic strength of the C—N bonds in β-L-lysine and β-glutamate than in L-lysine and L-glutamate, respectively.33 The greater strength of the β-C—N bonds is explained by the fact that these groups are flanked by sp3-carbons on both sides, C2 and C4. In L-lysine and L-glutamate the α-C—N groups are flanked by sp2-C1 and sp3-C3. The β-carbon in the β-C–N bond is less electropositive than in the α-C–N bond owing to the higher electronegativity of sp2-C1 in the α-amino acid. The higher electropositivity of the α-carbon, relative to the β-carbon makes the electronegativity difference between bonding atoms in the α-C—N bond smaller than in the β-C–N bond. Because electronegativity difference between bonding atoms contributes to enthalpic bond strength, the β-C–N bond is stronger in β-L-lysine and β-glutamate than in L-lysine and L-glutamate, respectively. This effect makes the β-amino acid more stable than the α-amino acid and leads to equilibrium constants of 7 to 15 for LAM and glutamate 2,3-aminomutase. The same should be true of β-alanine and L-alanine, leading to an equilibrium constant of 7-15.
Not all aminomutase reactions display equilibrium preferences for one isomer. The equilibrium constant for the reaction of lysine 5,6-aminomutase is near unity.34 This can be attributed to the fact that in both the substrate and the product the C–N bonds are flanked by sp3-carbons. Thus, the carbon in the C—N bond is electronically similar in the substrate and product, so that the C—N bonds are nearly isoenergetic.
Conclusions
L-Alanine and ethylamine binding to LAM simulates to a degree the binding of L-lysine, as does the binding of L-2-aminobutyrate and methylamine. The resultant complexes react as substrates to form β-alanine and 3-aminobutyrate, respectively, albeit at very slow rates. The aminomutation of L-alanine + ethylamine simulating L-lysine displays negative cooperativity with respect to L-alanine, as indicated by kinetic experiments. Negatively cooperative binding of L-alanine + ethylamine in simulation of L-lysine is complemented and supported by the enhancement of lysyl-radical formation in the presence of L-alanine and further enhancement by L-alanine in combination with ethylamine. Available evidence suggests that L-lysine also reacts with negative cooperativity.
Supplementary Material
Acknowledgements
The authors thank Dr. Laurie A. Reinhardt for fitting the kinetic data and Professor George H. Reed for the use of the EPR spectrometer. This research was supported by Grant No. DK 28607 from the National Institute of Diabetes and Digestive and Kidney Diseases.
Footnotes
List of Abbreviations: Ala, L-alanine; DTT, dithiothreitol; EPPS, N-[2-hydroxyethyl]piperazine-N′-3-propanesulfonic acid; EPR, electron paramagnetic resonance; HPLC, high performance liquid chromatography; LAM, lysine 2,3-aminomutase; PLP, pyridoxal-5′-phosphate; PITC, phenylisothiocyanate; SAM, S-adenosyl-L-methionine; Tris, tris-(hydroxymethyl)methylamine.
References and Notes
- 1.Frey P,A, Hegeman AD, Ruzicka FJ. Crit. Rev. Biochem. Mol. Biol. 2008;43:63–88. doi: 10.1080/10409230701829169. [DOI] [PubMed] [Google Scholar]
- 2.Frey PA, Th O. Magnusson, Chem. Rev. 2003;103:2129–2148. doi: 10.1021/cr020422m. [DOI] [PubMed] [Google Scholar]
- 3.Ruzicka FJ, Lieder KW, Frey PA. J. Bacteriol. 2000;182:469–476. doi: 10.1128/jb.182.2.469-476.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ballinger MD, Reed GH, Frey PA. Biochemistry. 1992;31:949–953. doi: 10.1021/bi00119a001. [DOI] [PubMed] [Google Scholar]
- 5.Ballinger MD, Frey PA, Reed GH. Biochemistry. 1992;31:10782–10789. doi: 10.1021/bi00159a020. [DOI] [PubMed] [Google Scholar]
- 6.Ballinger MD, Frey PA, Reed GH, LoBrutto R. Biochemistry. 1995;34:10086–10093. doi: 10.1021/bi00031a033. [DOI] [PubMed] [Google Scholar]
- 7.Chang CH, Ballinger MD, Reed GH, Frey PA. Biochemistry. 1996;35:11081–11084. doi: 10.1021/bi960850k. [DOI] [PubMed] [Google Scholar]
- 8.Wu W, Lieder KW, Reed GH, Frey PA. Biochemistry. 1995;34:10532–10537. doi: 10.1021/bi00033a027. [DOI] [PubMed] [Google Scholar]
- 9.Miller J, Bandarian V, Reed GH, Frey PA. Arch. Biochem. Biophys. 2001;387:281–288. doi: 10.1006/abbi.2001.2261. [DOI] [PubMed] [Google Scholar]
- 10.Magnusson O. Th., Reed GH, Frey PA. Biochemistry. 2001;40:7773–7782. doi: 10.1021/bi0104569. [DOI] [PubMed] [Google Scholar]
- 11.Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. Nucleic Acids Res. 2001;29:1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lepore BW, Ruzicka FJ, Frey PA, Ringe D. Proc. Natl. Acad. Sci. U.S.A. 2005;102:13819–13824. doi: 10.1073/pnas.0505726102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang SC, Frey PA. Biochemistry. 2007;46:12889–12895. doi: 10.1021/bi701745h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruzicka FJ, Frey PA. Biochim. Biophys. Acta. 2007;1774:286–296. doi: 10.1016/j.bbapap.2006.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heinrikson RL, Meredith SC. Anal. Biochem. 1984;136:65–74. doi: 10.1016/0003-2697(84)90307-5. [DOI] [PubMed] [Google Scholar]
- 16.Cleland WW. Methods Enzymol. 1979;63:103–138. doi: 10.1016/0076-6879(79)63008-2. [DOI] [PubMed] [Google Scholar]
- 17.Field TL, Reznikoff WS, Frey PA. Biochemistry. 1989;28:2094–2099. doi: 10.1021/bi00431a019. [DOI] [PubMed] [Google Scholar]
- 18.Kim J, Ruzicka F, Frey PA. Biochemistry. 1990;29:10590–10593. doi: 10.1021/bi00499a003. [DOI] [PubMed] [Google Scholar]
- 19.Ruzicka FJ, Geeganage S, Frey PA. Biochemistry. 1998;37:11385–11392. doi: 10.1021/bi980877z. [DOI] [PubMed] [Google Scholar]
- 20.Richards FM. J. Mol. Biol. 1974;82:1–14. doi: 10.1016/0022-2836(74)90570-1. [DOI] [PubMed] [Google Scholar]
- 21.Inagami T, York SS. Biochemistry. 1968;7:4045–4052. doi: 10.1021/bi00851a036. [DOI] [PubMed] [Google Scholar]
- 22.Koshland DE, Jr., Hamadani K. J. Biol. Chem. 2002;277:46841–46844. doi: 10.1074/jbc.R200014200. [DOI] [PubMed] [Google Scholar]
- 23.Conway A, Koshland DE. Biochemistry. 1968;7:4011–4023. doi: 10.1021/bi00851a031. [DOI] [PubMed] [Google Scholar]
- 24.Brahma A, Banerjee N, Bhattacharyya D. FEBS Journal. 2009:6725–6740. doi: 10.1111/j.1742-4658.2009.07386.x. [DOI] [PubMed] [Google Scholar]
- 25.Frank RA, Leeper FJ, Luisi BF. Cell. Mol. Life Sci. 2007;64:892–905. doi: 10.1007/s00018-007-6423-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schweizer E, Hofmann J. Microbiol. Mol. Biol. Rev. 2004;68:501–517. doi: 10.1128/MMBR.68.3.501-517.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Falson P, Goffeau A, Boutry M, Jault JM. Biochim. Biophys. Acta. 2004;1658:133–140. doi: 10.1016/j.bbabio.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 28.Sjöberg BM, Karlsson M, Jörnvall H. J. Biol. Chem. 1987;262:9736–9743. [PubMed] [Google Scholar]
- 29.Buddha MR, Crane BR. J. Biol. Chem. 2005;280:31965–31973. doi: 10.1074/jbc.M501568200. [DOI] [PubMed] [Google Scholar]
- 30.Dallocchio F, Matteuzzi M, Bellini T. Biochem. J. 1985;227:305–310. doi: 10.1042/bj2270305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moss M, Frey PA. J. Biol. Chem. 1987;262:14859–14862. [PubMed] [Google Scholar]
- 32.Moss ML, Frey PA. J. Biol. Chem. 1990;265:18112–18115. [PubMed] [Google Scholar]
- 33.Chen D, Tanem J, Frey PA. Biochim. Biophys. Acta. 2007;1774:297–302. doi: 10.1016/j.bbapap.2006.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang K-H, Chang CH, Frey PA. Biochemistry. 2001;40:5190–5199. doi: 10.1021/bi010157j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.