Abstract
Sterol 14α-demethylase (cytochrome P450 family 51 (CYP51)) is an essential enzyme occurring in all biological kingdoms. In eukaryotes, it is located in the membrane of the endoplasmic reticulum. Selective inhibitors of trypanosomal CYP51s that do not affect the human CYP51 have been discovered in vitro and found to cure acute and chronic mouse Chagas disease without severe side effects in vivo. Crystal structures indicate that CYP51 may be more rigid than most CYPs, and it has been proposed that this property may facilitate antiparasitic drug design. Therefore, to investigate the dynamics of trypanosomal CYP51, we built a model of membrane-bound Trypanosoma brucei CYP51 and then performed molecular dynamics simulations of T. brucei CYP51 in membrane-bound and soluble forms. We compared the dynamics of T. brucei CYP51 with those of human CYP51, CYP2C9, and CYP2E1. In the simulations, the CYP51s display low mobility in the buried active site although overall mobility is similar in all the CYPs studied. The simulations suggest that in CYP51, pathway 2f serves as the major ligand access tunnel, and both pathways 2f (leading to membrane) and S (leading to solvent) can serve as ligand egress tunnels. Compared with the other CYPs, the residues at the entrance of the ligand access tunnels in CYP51 have higher mobility that may be necessary to facilitate the passage of its large sterol ligands. The water (W) tunnel is accessible to solvent during most of the simulations of CYP51, but its width is affected by the conformations of the heme's two propionate groups. These differ from those observed in the other CYPs studied because of differences in their hydrogen-bonding network. Our simulations give insights into the dynamics of CYP51 that complement the available experimental data and have implications for drug design against CYP51 enzymes.
Keywords: sterol 14α-demethylase, CYP51, cytochrome P450, molecular dynamics simulation, membrane-bound protein, protein dynamics, ligand tunnel, heme propionate groups
INTRODUCTION
Sterol 14α-demethylase (14DM or cytochrome P450 family 51 (CYP51)) enzymes form a cytochrome P450 (CYP, P450) family essential in sterol biosynthesis (Lepesheva and Waterman, 2004; Lepesheva et al., 2010a). The catalytic function of CYP51 is to remove the 14α-methyl group from cyclized sterol precursors such as lanosterol (Lepesheva and Waterman, 2004). CYP51 is the most ancient CYP family and has been found in all the biological kingdoms (Aoyama et al., 1996; Nelson, 1999; Yoshida et al., 2000; Lepesheva and Waterman, 2004). Drugs (azoles such as itraconazole) that inhibit CYP51 in fungi have been used to treat human fungal infection for more than 20 years (Yoshida, 1988; Lepesheva et al., 2008). CYP51 is also considered a good target for antiprotozoan drugs (Lepesheva and Waterman, 2011). The inhibitor N-[(1R)-1-(2,4-dichlorophenyl)-2-(11H-imidazol-1-yl)ethyl]-4-(5-phenyl-1,3,4-oxadiazol-2-yl)benzamide (VNI) has been shown to inhibit CYP51 in protozoan pathogens, such as Trypanosoma brucei and Trypanosoma cruzi, specifically but not to affect the human CYP51 (Lepesheva et al., 2010a). VNI was tested in mouse models of the acute and chronic forms of Chagas disease, and the VNI-treated mice were all able to survive without severe observable side effects (Villalta et al., 2013). Other inhibitors have also been found that target parasitic CYP51s such as 14α-methylenecyclopropyl-Δ7-24,25-dihydrolanosterol (MCP, LNP) (Hargrove et al., 2012a) and (R)-4′-chloro-N-(1-(2,4-dichlorophenyl)-2-(11H-imid-azol-1-yl)ethyl)biphenyl-4-carboxamide (Lepesheva et al., 2010b; Hargrove et al., 2012b; Andriani et al., 2013).
There are differences in many of the properties of CYP51 compared with other P450s. The very low root-mean-square deviation (RMSD) (0.75 Å) between the Cα atoms of the ligand-free and VNI-bound crystal structures of T. brucei CYP51 suggests high structural rigidity of the enzyme (Lepesheva et al., 2010a). This notion has been supported by the analysis of crystal structures of other eukaryotic CYP51 orthologs, including T. cruzi, Leishmania infantum, and human, in ligand-free forms and co-crystallized in complexes with different heme-binding inhibitors and with the substrate analog MCP (Lepesheva et al., 2010b; Hargrove et al., 2011; Hargrove et al., 2012a).
In crystal structures of eukaryotic CYP51 enzymes, there is only one wide open ligand access tunnel (Lepesheva et al., 2010a). This corresponds to tunnel 2f in Wade's nomenclature (Cojocaru et al., 2007) and is lined by helices A′ and F″ and the tip of the β4 hairpin (Lepesheva et al., 2010b; Lepesheva et al., 2010a; Hargrove et al., 2011; Hargrove et al., 2012a). This tunnel is open (to a probe with a radius of 1.4 Å, corresponding to the size of a water molecule) in all crystal structures and often accommodates the longest portion of the ligand molecule that, in the case of VNI, approaches the tunnel's outer entrance (Protein Data Bank (PDB) ID: 3GW9) and, in the case of posaconazole, extends from the active site to the protein surface (PDB ID: 3K1O) (Lepesheva et al., 2010b).
Little is known about the influence of the membrane on CYP51, the dynamics of CYP51, and the opening and closing of ligand tunnels in CYP51. Here, we address the questions: (i) In which orientation is the T. brucei CYP51 embedded in the membrane, and how is the flexibility of the protein influenced when it is in the membrane? (ii) Is CYP51 more rigid compared with other membrane-bound CYPs as suggested by the crystal structures? (iii) Why do inhibitors of parasitic CYP51s not inhibit the human ortholog? Is this because of different flexibility or different interactions of the binding cavity residues? (iv) How do the ligand tunnels open and close? Which tunnels can be used for ligand (including substrate, product, inhibitor, and water) access and egress?
CYP51 is anchored in the membrane by a single N-terminal transmembrane helix and has part of its globular domain embedded in the membrane. The positioning of CYPs in the membrane has been studied experimentally and by computer simulation (Frey and David, 1987; Ohta et al., 1992; Black et al., 1994; Bayburt and Sligar, 2002; Headlam, 2003; Pikuleva, 2004; Ozalp et al., 2006; Berka et al., 2011; Cojocaru et al., 2011; Denisov et al., 2012; Yamamoto et al., 2013; Monk et al., 2014). A few CYPs, including CYP1A2, CYP2A6, CYP2C9, CYP2D6, CYP2E1, CYP3A4, and CYP19, have been modeled and simulated in phospholipid bilayers using different approaches (Berka et al., 2011; Cojocaru et al., 2011; Denisov et al., 2012; Sgrignani and Magistrato, 2012; Baylon et al., 2013; Berka et al., 2013; Haider et al., 2013). The protocol described in Cojocaru et al. (2011), which involves a combination of coarse-grained and all-atom molecular dynamics (MD) simulations, does not use prior knowledge about the protein–membrane interactions and can be applied to predict the orientation of CYPs in the membrane using solely the crystal structure of the globular domain.
To study the influence of the membrane on T. brucei CYP51, we built a model of T. brucei CYP51 in a 1-palmitoyl-2-oleoylsn-glycero-3-phosphocholine (POPC) bilayer using a procedure similar to that in Cojocaru et al. (2011). The positioning of CYP51 in the membrane in this model is in good agreement with available experimental data (Frey and David, 1987; Ohta et al., 1992; Black et al., 1994; Bayburt and Sligar, 2002; Baylon et al., 2013). We refer to this model as the “membrane-bound” T. brucei CYP51. We also performed MD simulations of CYPs immersed in aqueous solvent without a bilayer. We refer to these models as “soluble” CYPs. Here, we describe these models and the analysis of the protein dynamics during the simulations. Simulations of several hundreds of nanosecond duration have been performed for some soluble CYPs, including P450cam (Miao and Baudry, 2011; Miao et al., 2012) and the major human–drug metabolizing CYPs (Denisov et al., 2012; Hendrychova et al., 2012; Berka et al., 2013). Here, we instead carried out a number of shorter simulations of different CYPs in soluble and membrane-bound forms in order to perform a comparative analysis of the dynamics of the active site and ligand tunnel regions. We focused on comparing simulations of the soluble models of T. brucei CYP51 and human CYP51 with human CYP2C9 and human CYP2E1 and simulations of the membrane-bound model of T. brucei CYP51 with membrane-bound models of human CYP2C9. The analysis reveals features specific to CYP51 relating to the flexibility of the active site and the heme propionate groups, ligand access and egress tunnels, the opening of the water tunnel, and hydrogen-bonding networks. These have implications for the recognition of substrates and inhibitors by CYP51.
MATERIALS AND METHODS
Models of soluble T. brucei and human CYP51
The crystal structure of the ligand-free form of T. brucei CYP51 (PDB id: 3G1Q solved at 1.89-Å resolution) was used to build models for the simulations (Lepesheva et al., 2010a). The first three residues of the N-terminus were mutated from GKL to the wild-type PTD sequence with the mutation tool in PyMOL (Schrodinger, 2010). The PDB2PQR server (Dolinsky et al., 2004) was used to assign the protonation states of the ligand-free CYP51 at pH 7. HIS420 was doubly protonated. For the ligand-bound model, MCP (14α-methylenecyclopropyl-Δ7-24,25-dihydrolanosterol) was positioned in the active site of the ligand-free model with the Visual Molecular Dynamics (VMD) software (Humphrey et al., 1996) by using the transformation matrix for superimposing the protonated ligand-free model and the MCP-bound crystal structure (PDB id: 3P99). This procedure could be applied because the cavity lining residues were in almost identical positions in the two crystal structures. This model was used rather than the crystal structure of the complex to permit direct comparison of the ligand-free and ligand-bound systems.
For the simulation of human ligand-free CYP51, the crystal structure of human CYP51 (PDB ID: 3LD6 solved at 2.80-Å resolution) with ketoconazole removed was used. This structure was chosen because it has the best resolution. The same procedure to that for T. brucei CYP51 was used to protonate the structure. HIS167 and HIS447 (close to the heme center) were doubly protonated.
MD simulations of soluble CYP51
The modeled protein structures were further processed with the tleap program of AmberTools 1.5 (Case et al., 2010). The Amber ff99SB force field was used for the protein and the generalized Amber force field (GAFF) for the POPC phospholipids (Wang et al., 2004; Hornak et al., 2006). The parameters for the heme center were assigned as in Harris et al. (2004) and Cojocaru et al. (2011). The partial charges of MCP were derived using a two-stage Restrained Electrostatic Potential (RESP) fit method: RESP-A1A (HF/6-31G*) of the R.E.D server (Bayly et al., 1993; Dupradeau et al., 2010; Vanquelef et al., 2011). The parameter files of the two ligands were generated using the Antechamber program of the AmberTools 1.5 with the GAFF (Wang et al., 2004; Wang et al., 2006; Case et al., 2010). The systems were solvated in an octahedral box of three-site transferrable intermolecular potential (TIP3P) water molecules that extended at least 12 Å from the protein and were neutralized with counter ions (Jorgensen and Madura, 1983). The Amber (version 11) program was used to minimize and equilibrate the systems. Periodic boundary conditions with the Particle Mesh Ewald (PME) model for electrostatics were used. The energy minimization was performed in 14 stages, each with a harmonic restraint (decreasing from 1000 to 0 kcal/mol Å2) to the crystallographic positions on the non-hydrogen atoms. For each restraint value, 1000 minimization cycles were performed consisting of 100 cycles of steepest descent and 900 cycles of conjugate gradient. Heating to 300 K was then performed for 25 ps under constant temperature, constant volume (NVT) conditions with a restraint of 50 kcal/mol Å2 on all non-hydrogen atoms. A time step of 1 fs was used. Langevin dynamics with a collision frequency γ = 1/ps was performed. After heating, the system was further equilibrated in eight stages each with a decreasing restraint (50 to 0 kcal/mol Å2) on non-hydrogen atoms. For each restraint force constant, 25 ps of equilibration was performed. The temperature was maintained at 300 K by Langevin dynamics with a collision frequency γ = 1/ps, and the pressure was maintained at 1.0 bar by isotropic position scaling with a relaxation time of 1 ps. The same setup was applied to a final equilibration under constant pressure, constant temperature (NPT) conditions with no positional restraints for 1 ns.
The production runs for the ligand-free T. brucei CYP51, MCP-bound T. brucei CYP51, and ligand-free human CYP51 models were performed with NAMD 2.9 (Phillips et al., 2005). For the MCP-bound T. brucei CYP51 simulation, the same force field as the simulations of the ligand-free T. brucei CYP51 was used together with the same minimization and equilibration protocol. The switching function was set to “on”, and the non-bonded cutoff was set to 10 Å. All the bonds to hydrogen atoms were constrained with the SHAKE algorithm. Temperature was controlled by Langevin dynamics with a damping coefficient of 0.5/ps at 300 K. Pressure was controlled by the Nosé–Hoover– Langevin piston method with an oscillation period of 1 ps and a damping timescale of 1 ps at 1.011325 bar. The production runs were 30 ns for the two models of ligand-free and MCP-bound T. brucei CYP51 and 15 ns for the model of ligand-free human CYP51 with a time step of 1.5 fs.
MD simulations of the models of soluble CYP2C9 and CYP2E1
The MD simulation trajectories of the models of soluble CYP2C9 and CYP2E1 at 300 K from previous studies were analyzed (Table 1; Cojocaru et al., 2011 and unpublished data). The simulations of human CYP2C9 were performed using two models built from the crystal structure (PDB id: 1R9O) with different conformations of the F–G loop region (one model with F′ and G′ helices and the other model with an F–G loop only; Cojocaru et al., 2011). The simulations of human CYP2E1 were performed using two CYP2E1 models based on two crystal structures (PDB ID: 3KOH, 3T3Z; unpublished data). The trajectories were compared with those of the soluble CYP51s.
Table 1.
Main features of the all-atom molecular dynamics production simulations of the different CYPs
| CYP | PDB ID | Environmentb | Ligandc | Ensembled | Time (ns) |
|---|---|---|---|---|---|
| CYP51 (T. brucei) | 3G1Q | SOL | ------ | NPT | 30 |
| 3G1Q | MEM | ------ | NPγT | 60 | |
| 3P99e | SOL | MCP | NPT | 30 | |
| 3P99e | MEM | MCP | NPγT | 15 | |
| CYP51 (human) | 3LD6 | SOL | ------ | NPT | 15 |
| CYP2C9_M1 (human)a | 1R9O | SOL | ------ | NPT | 28.28 |
| 1R9O | MEM | ------ | NPγT | 18.75 | |
| 1R9O | SOL | FLO | NPT | 27.84 | |
| 1R9O | MEM | FLO | NPγT | 11.25 | |
| CYP2C9_M2 (human)a | 1R9O | SOL | ------ | NPT | 27.78 |
| 1R9O | MEM | ------ | NPγT | 18.75 | |
| 1R9O | SOL | FLO | NPT | 27.4 | |
| 1R9O | MEM | FLO | NPγT | 11.25 | |
| CYP2E1 (human) | 3KOH | SOL | ------ | NPT | 10 |
| 3T3Z | SOL | ------ | NPT | 10 |
The F-G loop region in the crystal structure of human CYP2C9 (PDB id: 1R9O) is missing. Two models of the F-G loop were generated for the simulations (Cojocaru et al., 2011). Model 1 has F′ and G′ helices, and model 2 has an unstructured F-G loop.
SOL, the soluble CYPs (no membrane); MEM, the membrane-bound CYPs in a lipid bilayer.
The ligands used in the simulation. FLO, 4′-hydroxy flurbiprofen; MCP, 14α-methylenecyclopropyl-Δ7-24,25-dihydrolanosterol; “----”, ligand-free structures.
In the NPγT simulations of the membrane-bound CYPs, a surface tension, γ = 60 dyn/cm, was applied to the plane of the membrane together with the pressure control to maintain the bilayer dynamics (Cojocaru et al., 2011). A comparison of the area per lipid, the bilayer thickness, and the order parameters of the membrane in the 60-ns simulation of the membrane-bound CYP51 with experimental data is plotted in Figures S6 and S7.
Only the ligand MCP of 3P99 was transferred into the active site of the equilibrated ligand-free CYP51 structure (originally derived from the crystal structure 3G1Q). The simulations were then performed starting with this structure (see Methods).
Coarse-grained model of membrane-bound T. brucei CYP51
A model of the membrane-bound ligand-free T. brucei CYP51 was built using an improved version of the protocol described in Cojocaru et al. (2011). MODELLER_9.10 (Sali and Blundell, 1993) was used to model the missing residues. The missing residues 1 to 21 were predicted to form the transmembrane helix and modeled as a helix. Residues 22 to 34 were modeled as a loop (residues 22–28 were missing in the crystal structure; more residues were modeled for the superimposed of the globular domain). The modeled transmembrane helix and loop structure were then coarse grained using MARTINI2.1 with an elastic network between the non-bonded beads (Marrink et al., 2004; Marrink et al., 2007; Monticelli et al., 2008; Periole et al., 2009). The elastic bond force constant was 1.19 kcal/mol Å2 (500 kJ/mol nm2), and the upper bond length cutoff was 9 Å. The coarse-grained structure (modeled residues 1 to 34) was inserted into a pre-equilibrated bilayer of 602 coarse-grained POPC molecules (Marrink et al., 2004). The overlapping lipids within 3 Å of the transmembrane helix were removed. A 5-μs simulation was performed to find the preferred orientation of the transmembrane helix alone in the membrane. The protocol is the same as for the whole membrane-bound protein system (refer to succeeding texts).
A coarse-grained model of the T. brucei CYP51 globular domain (PDB id: 3G1Q) was also generated with MARTINI and combined with the preferred orientation of the transmembrane helix in the 5-μs simulation. The elastic network of the linker region (residues 22–36) connecting the transmembrane helix and the globular domain was removed. Ten different initial orientations of the globular domain with different angles and height above the membrane were generated by changing the dihedral angles of this flexible loop region randomly as described in Cojocaru et al. (2011).
Each complex was then solvated in a box of coarse-grained water molecules with the same x and y dimensions as the POPC bilayer taken from Marrink et al. (2004) and extending 26 nm along the z axis. The system was minimized with the steepest descent method using GROMACS_4.5.5 (Berendsen et al., 1995; Lindahl et al., 2001; Van der Spoel et al., 2005; Hess et al., 2008). The system was minimized until the maximum force was less than 10 kJ/mol nm (23.8 cal/mol Å) for 10 000 steps. Then, the system was relaxed at 200 K for 50 000 steps. After that, a 3-μs simulation with a time step of 20 fs was performed. Periodic boundary conditions were applied. The electrostatic interactions and van der Waals interactions were calculated with the “Shift” algorithm. The lower and upper cutoffs for electrostatic interactions were 0 and 12 Å, whereas those of the van der Waals interactions were 9 and 12 Å. Temperature coupling using velocity rescaling with a stochastic term was applied with a 1-ps time constant for coupling at 310 K. Semi-isotropic Berendsen pressure coupling was used with a 2-ps coupling time constant at 1 bar.
To check for possible artifacts from the definition of the elastic network, four more simulations of the coarse-grained T. brucei CYP51 in the POPC bilayer were performed after the removal of the elastic network for residues 22–23, 22–31, 22–33, and 22–35. Despite the differences in the definition of the elastic network, three of these four simulations converged to the same orientation as the previous simulations except the simulation with the elastic network removed for residues 22–31, in which the protein did not embed in the membrane, and the F–G loop region was out of the membrane. Although the extent to which the elastic network was different, the other three simulations converged to the same final orientation as the nine out of 10 previous simulations. To check whether the flexibility of the F–G loop influences the final orientation, one additional simulation was performed with the elastic network of the F–G loop (residues 22–36 and 207–224) removed. This simulation converged to the same final orientation as well.
Reverse conversion from the coarse-grained to the all-atom model
The conversion from the coarse-grained model to the all-atom model was performed separately for the membrane, the transmembrane helix (residues 1–21), the linker region (residues 22–36), and the globular domain (residues 37–476). The trans-membrane helix and the globular domain were converted by superimposing the modeled structure of the transmembrane helix and the equilibrated all-atom models from the soluble ligand-free T. brucei CYP51 simulation, respectively, onto the coarse-grained models. The same protocol as in Cojocaru et al. (2011) was used to convert the lipids.
Because of the large conformational changes of the linker region, the MARTINI reverse conversion tool was used to convert the linker region (Rzepiela et al., 2010). Firstly, the MARTINI mapping information of the linker regions, which correlates the all-atom amino acids with the coarse-grained beads of the amino acids, was used to position the all-atom amino acids onto the coarse-grained beads after the coarse-grained simulations. Then, a subsequent simulated annealing was performed for 60 000 steps with a time step of 2 fs. The linker was coupled at 1300 K at the beginning of the annealing; the temperature was decreased linearly to 300 K. The four parts (transmembrane helix, the linker, the globular domain, and the POPC bilayer) of the system were then combined, and the system was neutralized in a NaCl concentration of 150 mM and solvated in a box of TIP3P water molecules (same dimension as the membrane in the x and y directions and extending 16 nm in the z direction). For the MCP-bound model, MCP was superimposed onto the active site of the membrane-bound ligand-free T. brucei CYP51 by using the transformation matrix calculated by superimposing it with the crystal structure (PDB id: 3P99).
All-atom MD simulations of membrane-bound T. brucei CYP51
The ff99SB force field was used for the protein, and GAFF was used for the POPC bilayer (Wang et al., 2004; Hornak et al., 2006; Jójárt and Martinek, 2007). Energy minimization of the all-atom system was performed in Amber (version 12; Case et al., 2012) with decreasing force constants from 1000 to 0 kcal/mol Å2. One hundred cycles of steepest descent and 900 cycles of conjugate gradient were used for each restraint value. The system was then equilibrated in NAMD2.9 as described previously (Cojocaru et al., 2011).
For the production runs, periodic boundary conditions were used, and the electrostatic interactions were calculated using the PME method. The NPT ensemble was applied with a target surface tension of 60 dyn/cm. All bonds to hydrogen atoms were constrained with the SHAKE algorithm. Temperature was controlled by Langevin dynamics with a damping coefficient of 0.5/ps at 310 K on non-hydrogen atoms. Pressure was controlled by the Nosé–Hoover–Langevin piston method with an oscillation time of 1000 fs and a damping time of 1000 fs. A production run of 60 ns was performed for the ligand-free model without restraints with a time step of 1.5 fs. A production run of 15 ns was performed for the MCP-bound model with the same setup.
Data analysis
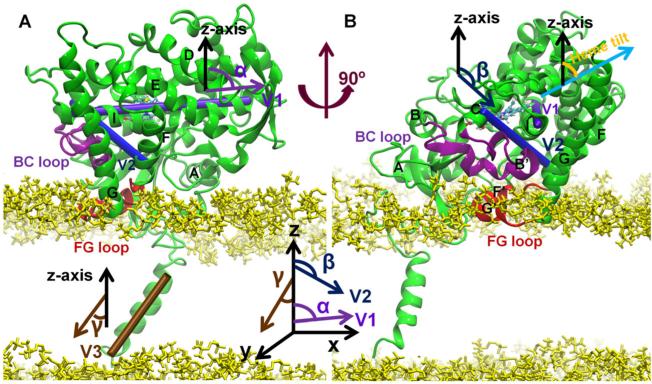
Three parameters (Cojocaru et al., 2011) were used to define the position and orientation of the globular domain on the membrane (Figure 1): (i) d—the distance of CYP51 from the membrane was calculated by subtracting the z-coordinate of the center of mass of the protein from the z-coordinate of the center of mass of the membrane; (ii) α—angle between the z-axis and vector V1; V1 is the vector defined by the center of mass of the first (residues 278–282) and last (residues 304–308) helical turns of the I helix; (iii) β—angle between the z-axis and vector V2; V2 is the vector de-fined by the center of mass of one helical turn of the C helix (residues 122–126) and one turn of the F helix (residues 202–206). For the coarse-grained simulations, the system was analyzed only after the system was equilibrated and the parameters fluctuated stably.
Figure 1.
Two views of CYP51 in the bilayer at the last snapshot of the 60-ns simulation of ligand-free T. brucei CYP51. Important secondary structure elements are labeled. The B–C loop is shown in purple and the F–G loop in red. The head groups of the POPC lipids are shown in yellow and the membrane plane is the x–y plane. Two vectors (V1 and V2) that define the orientation of the protein with respect to the membrane are represented by cylinders. V1 follows the I helix. V2 points from the first helical turn of the C helix to the last helical turn of the F helix. Vector V3 points along the transmembrane helix. The “heme tilt” (orange) is the angle between the heme plane (cyan arrow) and the normal to the membrane plane (black arrow). Detailed information of the three angles is shown next to the cylinder representation of the three vectors. For more details, refer to Materials and Methods: Data Analysis.
The angle γ between the z-axis and vector V3 was used to de-fine the orientation of the transmembrane helix in the membrane. V3 is the vector defined by the center of mass of the first (residues 1–5) and last (residues 17–21) helical turns of the transmembrane helix. The heme tilt angle is the angle between the z-axis and the average heme plane defined by the “NA,” “NB,” “NC,” and “ND” atoms of the heme. The “height” of the proteins was calculated by taking the z-coordinate of the highest non-hydrogen atom of the protein above the membrane and subtracting the average z-coordinate of the phosphate groups (atoms: “P,” “O1P,” “O2P,” “O3P,” and “O4P”) of the upper leaflet of the lipid bilayer.
All the previously mentioned parameters were calculated in VMD (Humphrey et al., 1996). The RMSD and root-mean-square fluctuation calculations were performed using the cpptraj program of the AmberTools 12 (Case et al., 2012). For the comparison of the computed B-factors, the four CYPs (human CYP51, T. brucei CYP51, human CYP2C9, and human CYP2E1) were aligned with the sequence and structure alignment tool Profile Multiple Alignment with Predicted Local Structures and 3D constraints (PROMALS3D; Pei et al., 2008).
The area per lipid and the thickness of the membrane were analyzed with the grid-based tool GridMAT-MD (Allen et al., 2009). Two hundred grids were assigned to the x–y plane, and the x and y ratio was then modified according to the dimensions of the membrane of the CYP system. Lipids that were within 10 Å of the protein were removed for the calculation. The order parameters were calculated with the g_order tool of the GROMACS_4.6 package (Pronk et al., 2013).
The opening and closing of the ligand tunnels was analyzed using CAVER3.0 (Chovancova et al., 2012). PDB format coordinate files (100–200) were generated along each trajectory and were used as the input for the CAVER analysis. For example, in the membrane-bound ligand-free model of T. brucei CYP51, 200 PDB files were generated from the 60-ns simulation with a 0.3-ns sampling interval. The probe radius was assigned a value of 1.4 Å, corresponding to the size of a water molecule. A tunnel was considered to be open when the radius of the bottleneck was larger than 1.4 Å.
RESULTS AND DISCUSSION
Model of T. brucei CYP51 in the POPC bilayer
One predominant orientation of the membrane-bound T. brucei CYP51
An improved version of the protocol in Cojocaru et al. (2011) as described in the Methods section was used to build the membrane-bound ligand-free model of T. brucei CYP51. In the 5-μs coarse-grained simulation of the transmembrane helix alone in the POPC bilayer, the transmembrane helix adopted an orientation of 17 ± 8° to the normal of the membrane (angle γ in Figure 1). In the coarse-grained simulations of the whole protein in the POPC bilayer, nine out of the 10 simulations converged to one final orientation after the system was equilibrated (Figure S1). The three parameters defining the position of the protein after the equilibration of the nine converged coarse-grained simulations were d = 41.3 ± 1.8 Å, α = 82.2 ± 6.3°, and β = 126.5 ± 8.4° (Figure S2). One snapshot from the coarse-grained simulations having values of all three parameters within 1% of the mean values of the parameters was taken as the starting structure for the all-atom simulations. In the all-atom simulations of the ligand-free and MCP-bound T. brucei CYP51, the globular domain stayed in about the same orientation above the membrane. The parameters were d = 38.1 ± 0.9 Å, α = 82.8 ± 5.2°, and β = 122.9 ± 6.0° (Figure S2).
Comparison with experimental data and with other CYP models supports the atomic detail models of the membrane-bound T. brucei CYP51
In the simulation of the membrane-bound ligand-free T. brucei CYP51, the tilt angle of the transmembrane helix to the normal of the bilayer was 20.1 ± 6.5°, which is similar to the transmembrane helix tilt angle of ~17° measured for this helix from CYP2B4 by solid-state nuclear magnetic resonance spectroscopy (Yamamoto et al., 2013). In the model of membrane-bound T. brucei CYP51, the F–G loop region (residues 206–224) is embedded in the membrane, which is consistent with experiments on other CYPs (Headlam, 2003; Pikuleva, 2004; Ozalp et al., 2006). Table 2 shows a comparison of the model of the membrane-bound T. brucei CYP51 with experimental data and the models of CYP2C9 (Cojocaru et al., 2011) and CYP3A4 (Baylon et al., 2013). The CYP51 model is mostly in good agreement with these experiments and shows about the same degree of agreement with experiments as the CYP2C9 and CYP3A4 models. The measured heme tilt angle (between the heme plane and the normal to membrane plane, as shown in Figure 1B) of CYP3A4 was 59.7 ± 4.1° in experiments (Baylon et al., 2013). This angle was 63.1 ± 4.5° in our simulation of the ligand-free membrane-bound T. brucei CYP51. This is close to the experimental value (Table 2) and that in models of six human drug-metabolizing CYPs (CYP1A2, 67 ± 6°; CYP2A6, 69 ± 5°; CYP2C9, 61 ± 4°; CYP2D6, 72 ± 6°; CYP2E1, 60 ± 5°; and CYP3A4, 56 ± 5°; Berka et al., 2013). The membrane-bound model of T. brucei CYP51 also agrees quite well with the predicted orientation (d, ~41.6 Å; α, 83.7°; β, 131.8°) in the Orientations of Proteins in Membranes (OPM) database for CYP51 although OPM predicted a different orientation for CYP2C9 with the A helix region (residues 45–66) not embedded in the membrane (Lomize et al., 2006; Cojocaru et al., 2011). After we built our models of T. brucei CYP51, the structure of the full-length Saccharomyces cerevisiae CYP51 was determined by crystallography (Monk et al., 2014). The orientation of our model in lipid bilayer is consistent with this structure of a fungal CYP51. The computed B-factors of the membrane-bound CYP51 in the simulation were compared with those of the crystal structure and the simulated soluble form (Figure S3). The trends are consistent for the three systems although, as expected, the computed B-factors vary over a wider range in the simulations. The F–G loop region has lower mobility in the membrane-bound simulation because this region is embedded in the membrane.
Table 2.
Comparison of membrane-bound T. brucei CYP51 model with the experiments, two human CYP2C9 models with different F-G loop structures (Cojocaru et al., 2011), and a CYP3A4 model (Baylon et al., 2013)
| CYP51 |
CYP2C9_M1 |
CYP2C9_M2 |
CYP3A4 |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Residue numbera | Exp. data | Exp. CYP | Locationb | Agreementc | Location | Agreement | Location | Agreement | Location | Agreement |
| 1-27 | Mh | 2B2 | M | + | M | + | M | + | M | + |
| 28 | Mh/Ci | 2B2, 2B4 | C | + | C | + | C | + | M | + |
| 29-40 | Mh | 2B2 | M | + | M & C | +− | M | + | M & C | +− |
| 42-49 | Mh | 2B2 | M & C | +− | M | + | M & C | +− | M & C | +− |
| 63-75 | Ch | 2B2 | C | + | M | - | M | - | M & C | +− |
| 95-100 | Mh | 2B2 | C | - | C | - | C | - | C | - |
| 110-118 | Ch | 2B2 | C | + | C | + | C | + | C | + |
| 124-133 | Ch | 2B2 | C | + | C | + | C | + | C | + |
| 186-193 | Ch | 2B2 | C | + | C | + | C | + | C | + |
| 209-218 | Mh | 2B2 | M & C | +− | M | + | M | + | M | + |
| 220-227 | Ch | 2B2 | M | - | M & C | +− | C | + | M & C | +− |
| 310-318 | Ch | 2B2 | C | + | C | + | C | + | C | + |
| 390-406 | Ch | 2B2 | C | + | C | + | C | + | C | + |
| 39,72 | Mj | 21A1 | M | + | M | + | M | + | M | + |
| 221 | Cj | 21A1 | M | - | M | +− | C | + | M | - |
| 83, 123,190, 235, 341 | Cj | 21A1 | C | + | C | + | C | + | C | + |
| 373d | Mj | 21A1 | C | - | M | + | M | +− | C | - |
| Heme tilt angle (°)e | 59.7 ± 4.1k | 3A4 | 63.1 ±4.5 | + | 43.2 ± 3.4 | - | 38.5 ± 5.4 | - | 61.5 | + |
| 38-78j | 17A1, 21A1 | + | + | + | + | |||||
| Height_1 (Å)f | 35 ± 9l | 2B4 | 50.5 ± 2.2 | - | 47.9 ± 1.7 | - | 47.1 ± 1.9 | - | 58.0 | - |
| Height_2 (Å)g | 38.1 ± 0.9 | + | 33.6 ± 0.7 | + | 41.0 ± 1.0 | + | 39.6 | + | ||
The residue numbers are adjusted from the CYP2C9 residue numbers according to the alignment of CYP51 with CYP2C9 (Figure S2) and the alignment of CYP51 with CYP3A4 using the “align” function of PyMOL (Schrodinger, 2010).
The location of the residues with respect to the membrane: “M” indicates that the residues are either in the membrane or in the head group region of the membrane. “C” indicates that the residues are in the cytosol. “M & C” indicates that the fragment is partly in the membrane and partly in the cytosol. “M/C” indicates that there are experiments showing that a particular residue is in the membrane, while other experiments showed that this residue is in the cytosol.
These columns show whether the model is in agreement with the experiments. “+”—the model is in good agreement with the experiments. “+-”—the model is partly in agreement with the experiments.“-”—the model is not in agreement with the experiments.
Residue 373 was shown to be deeper in the membrane than the residues 39 and 72.
“Heme tilt angle” is the angle between the heme plane and the normal of the membrane plane. For CYP51 and the two models of CYP2C9, the heme tilt angle is computed as an average over snapshots from the production runs of the simulations. For CYP3A4, the angle is computed for the final model.
“Height_1” is the height of the CYP above the lipid bilayer measured by atomic force microscopy in the experiments and the distance between thez-coordinate of the highest non-hydrogen atom of the protein and the average z-coordinate of the phosphate group of the upper leaflet of the lipid bilayer.
“Height_2” is the distance between the center of mass of the protein and the center of mass of the membrane.
References are as follows:
Bayburt and Sligar (2002) and experiments were carried out for the CYP listed in the third column.
Dynamics of CYP51 and implications
Lower mobility in the active site of CYP51 but not higher mobility at the ligand access tunnel entrance
Comparison of the crystal structures suggested that CYP51 is less mobile than other CYPs (Lepesheva et al., 2010a; Hargrove et al., 2012a). Therefore, we compared the computed B-factors in the ligand-free simulations of the four CYPs studied.
The simulations of soluble models of CYPs are converged shortly after equilibration, for example, in the simulation of the soluble model of ligand-free T. brucei CYP51 (Figure S4). In the simulations of the soluble CYPs, the average computed B-factor of T. brucei CYP51 (30.0 ± 32.4 Å2), averaged over all non-hydrogen atoms and the last 9 ns of the simulations, and of human CYP51 (24.1 ± 22.7 Å2) is similar to that of CYP2C9 (29.4 ± 39.8 and 31.5 ± 39.4 Å2) and of CYP2E1 (28.8 ± 33.2 and 27.9 ± 39.0 Å2) (Figure 2A). In the membrane-bound simulations, the average B-factor (over the last 12 ns) of the globular domain of T. brucei CYP51 (35.7 ± 33.2 Å2) is also similar to that of CYP2C9 (32.1 ± 33.2 and 32.5 ± 33.2 Å2; Figure 2B).
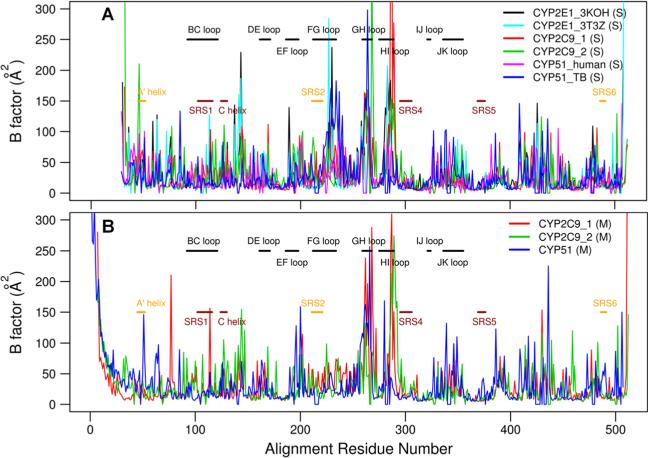
Figure 2.
Comparison of the B-factors of the non-hydrogen atoms of different CYPs with the loop regions (black), the ligand binding cavity residues (dark red), and ligand tunnel opening residues (orange) labeled. The residue numbers of the CYPs correspond to the PROMALS3D alignment (Figure S2). (A) Comparison of B-factors averaged over the last 9 ns of the two simulations with soluble ligand-free forms of the two human CYP2C9 models, the two simulations with the two human CYP2E1 models, and the simulations of T. brucei CYP51 and human CYP51. (B) Comparison of the B-factors averaged over the last 12 ns of the simulations with the membrane-bound ligand-free forms of CYP2C9 and CYP51.
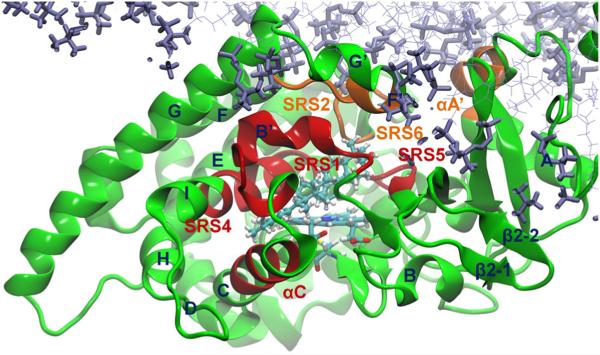
As the computed B-factors vary widely along the sequence, we next compared the B-factors of two groups of residues, the substrate binding cavity residues and the tunnel 2f entrance residues (Hargrove et al., 2011). Residues of these two groups on T. brucei CYP51 are shown in Figure 3, and the corresponding residues of human CYP2C9 and human CYP2E1 were taken from the alignment (Figure S5). The substrate binding cavity consists of the substrate recognition site 1 (SRS1, αB′–B′/C loop, residues 102–116), the C helix (residues 124–130), SRS4 (residues 284–295), and SRS5 (residues 355–361) (Gotoh, 1992; Hargrove et al., 2011). The ligand access tunnel 2f has entrance residues including αA′ (residues 45–52), SRS2 (αF″, residues 208–214), and SRS6 (β4 hairpin, residues 457–462). The average B-factors of the Cα atoms of these regions were computed and are given in Table 3. If the average of the computed B-factors of the Cα atoms was relatively low, the region was considered to have low mobility.
Figure 3.
The substrate binding cavity residues (dark red) and the ligand tunnel entrance residues (orange) shown in the last snapshot of the simulation of the membrane-bound ligand-free T. brucei CYP51. The substrate binding cavity residues are residues 102–116 (SRS1, αB′–B′/C loop), 124–130 (αC), 284–295 (SRS4, αI), and 355–361 (SRS5). The residues at the entrance of ligand tunnel 2f include αA′ (residues 45–52), SRS2 (αF″, residues 208–214), and SRS6 (β4 hairpin, residues 457–462). The ligand MCP is shown in cyan. The bilayer is shown in violet.
Table 3.
Average computed B-factors (Å2)of Cα atoms in specific regions of the simulated CYPs
| CYP | Environmenta | SRS1 | C helix | SRS4 | SRS5 | A′ helix | SRS2 | SRS6 |
|---|---|---|---|---|---|---|---|---|
| T. brucei CYP51 | S | 11.9 ± 2.0 | 8.7 ± 1.4 | 7.1 ± 1.1 | 6.8 ± 2.2 | 18.1 ± 4.9 | 39.9 ± 18.6 | 18.2 ± 9.2 |
| Human CYP51 | S | 8.7 ± 2.6 | 7.4 ± 1.5 | 9.1 ± 4.0 | 5.8 ± 1.3 | 40.7 ± 11.9 | 19.1 ± 8.0 | 10.4 ± 3.6 |
| Human CYP2C9_1 | S | 23.2 ± 8.6 | 10.1 ± 2.7 | 5.6 ± 1.2 | 5.9 ± 1.1 | 19.6 ± 13.0 | 17.0 ± 3.9 | 11.3 ± 4.8 |
| Human CYP2C9_2 | S | 19.8 ± 9.3 | 13.7 ± 4.8 | 13.8 ± 7.9 | 4.8 ± 0.9 | 11.2 ± 5.7 | 20.4 ± 15.1 | 6.3 ± 0.7 |
| Human CYP2E1_1 | S | 8.7 ± 2.1 | 9.7 ± 2.5 | 5.8 ± 2.2 | 8.5 ± 5.0 | 20.7 ± 7.2 | 9.0 ± 2.1 | 8.0 ± 1.4 |
| Human CYP2E1_2 | S | 12.5 ± 5.5 | 11.3 ± 2.3 | 5.3 ± 1.9 | 7.8 ± 4.5 | 12.5 ± 7.3 | 8.3 ± 3.5 | 9.0 ± 3.5 |
| T. brucei CYP51 | M | 11.5 ± 2.6 | 8.3 ± 0.8 | 8.8 ± 3.7 | 9.8 ± 6.2 | 37.5 ± 14.3 | 16.9 ± 2.1 | 32.7 ± 15.1 |
| Human CYP2C9_1 | M | 15.8 ± 5.0 | 7.4 ± 0.8 | 13.0 ± 6.9 | 6.1 ± 1.2 | 10.4 ± 4.6 | 20.0 ± 5.3 | 7.5 ± 2.0 |
| Human CYP2C9_2 | M | 16.4 ± 9.1 | 16.8 ± 8.6 | 15.1 ± 8.2 | 5.5 ± 0.7 | 10.5 ± 2.4 | 18.8 ± 15.9 | 7.6 ± 2.2 |
The B-factors for the soluble simulations were computed from the last 9 ns of the two simulations with soluble ligand-free forms of the two human CYP2C9 models, the two simulations with the two human CYP2E1 models, and the simulations of T. brucei CYP51 and human CYP51. The B-factors of the membrane-bound simulations were computed from the last 12 ns of the simulations with the membrane-bound ligand-free forms of CYP2C9 and CYP51.
Simulations were performed without a lipid bilayer (S, soluble model) or with a lipid bilayer (M, membrane-bound model).
In the simulations of the soluble proteins, the average computed B-factor of the Cα atoms of the binding cavity residues of T. brucei CYP51 (9.1 ± 2.8 Å2) and human CYP51 (8.1 ± 3.0 Å2) is similar to that in the simulations of the two human CYP2E1 models (8.0 ± 3.2 and 9.3 ± 5.0 Å2) but lower than that in the simulations of the two human CYP2C9 models (12.7 ± 9.6 and 14.2 ± 8.9 Å2) as shown in Figure 2A. In the membrane-bound simulations, the substrate binding cavity also has lower mobility in the simulation of the membrane-bound model of T. brucei CYP51 (9.8 ± 3.7 Å2) than in those of human CYP2C9 (11.8 ± 6.1 and 14.0 ± 8.6 Å2). In both the soluble and membrane-bound simulations of CYP51, the binding cavity residues have low mobility (average B-factor less than 10 Å2). The mobility of SRS1 of CYP51 is lower than that of CYP2C9 in both soluble and membrane-bound simulations and one CYP2E1 simulation but similar to that in the other CYP2E1 simulations. The C helix region of the binding cavity has lower mobility in CYP51 than in CYP2C9 and CYP2E1 in the soluble simulations. In the membrane-bound simulations, the C helix of T. brucei CYP51 is less mobile than that of model 2 of CYP2C9. The C helix of model 1 of CYP2C9 has lower mobility in this region because this region is partly embedded in the membrane. The SRS4 and SRS5 regions of all the CYPs studied have low mobility (average B-factor mostly less than 10 Å2) in all the simulations.
The average computed B-factor of the 2f tunnel entrance residues of T. brucei CYP51 (25.4 ± 15.6 Å2) and of human CYP51 (23.1 ± 15.1 Å2) is higher than that of human CYP2E1 (12.8 ± 7.3 and 9.9 ± 5.3 Å2) and of human CYP2C9 (16.9 ± 9.3 and 13.7 ± 11.2 Å2; Figure 2A). That of CYP51s is also higher than that of the 2a tunnel entrance residues of human CYP2E1 (18.9 ± 14.2 and 20.1 ± 16.1 Å2) and of human CYP2C9 (18.7 ± 8.1 and 19.8 ± 12.2 Å2). In the membrane-bound simulations, the 2f tunnel entrance residues (αA′, SRS2, and SRS6) also have higher mobility in T. brucei CYP51 (29.3 ± 14.6 Å2) than the 2a tunnel entrance residues (β1, B–B′ loop, and G′ helix) in human CYP2C9 (17.8 ± 8.0 and 17.3 ± 14.1 Å2). The A′ helix and SRS6 of T. brucei CYP51 are more mobile than those of CYP2C9 and of CYP2E1 (Table 3).
The differences between the computed B-factors of the CYPs studied are small, and the overall mobility of these CYPs does not differ a lot. However, the binding cavity residues of CYP51s are, in general, less mobile than those of CYP2C9 and CYP2E1 and the tunnel entrance residues of CYP51s more mobile than those of CYP2C9 and CYP2E1.
Ligand tunnels in CYP51
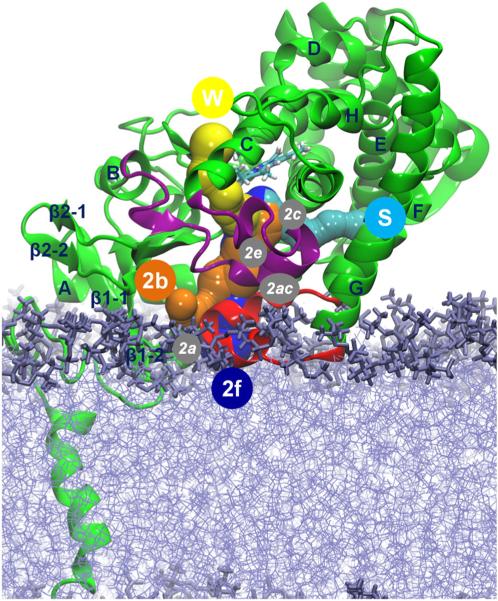
Figure 4 shows the tunnels between the active site and the protein surface (Cojocaru et al., 2007). Tunnel 2f (between helices A′, F″, and the tip of the β4 hairpin), leading to the membrane, and tunnels 2b (between the B–B′ loop and the β1–2 and β1–4 sheets), S (the solvent tunnel, between F, I helices, and the β4 hairpin), and W (the water tunnel, helix C and the β-bulge segment), leading to the solvent, were observed to open in simulations of membrane-bound and soluble forms of the T. brucei CYP51. Tunnel 2a (between the F–G loop, B–B′ loop, and β1–1 sheet), pointing to the bilayer, and tunnels 2c (between the G and I helices, B′–C loop), 2ac (between the B′ helix and G helix), and 2e (through the B–C loop) were closed throughout the simulations of the membrane-bound form of T. brucei CYP51. Some opening of tunnels 2e and 2ac was seen in the simulations of the soluble form of T. brucei CYP51 (Figure 5).
Figure 4.
Cartoon representation of T. brucei CYP51 showing the ligand tunnels of the membrane-bound CYP51 analyzed in CAVER3.0 (Chovancova et al., 2012). The important secondary structure elements are labeled. The B–C loop is shown in purple and the F–G loop in red. The bilayer is shown in violet. Tunnels that opened at least in one snapshot during the 60-ns simulation of the membrane-bound ligand-free CYP51 are shown by sphere representations and labeled by circles with white letters. Tunnel 2f (blue) leads to the membrane, and tunnels 2b (orange), S (cyan), and W (yellow) lead to the solvent. Tunnels 2a, 2c, 2e, and 2ac that were not open in the 60-ns simulation are labeled by white italic letters in gray circles.
Figure 5.
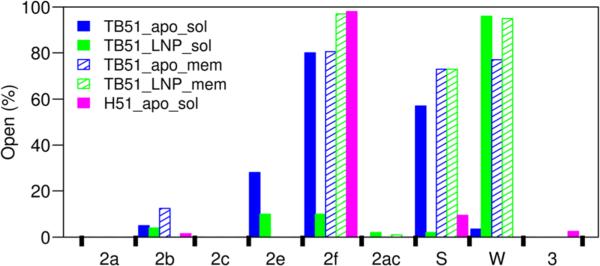
Percentage of the snapshots in which the ligand tunnels were open in the simulations is shown for the ligand-free soluble T. brucei CYP51 (solid blue), MCP-bound soluble T. brucei CYP51 (solid green), ligand-free membrane-bound T. brucei CYP51 (hatched blue), MCP-bound membrane-bound T. brucei CYP51 (hatched green), and ligand-free soluble human CYP51 (solid magenta). In CYP51s, the tunnels are located as follows: 2a—between the F–G loop, B–B′ loop, and β1–1 sheet; b—between the B–B′ loop, and the β1–2 and β1–4 sheets; 2c—between the G and I helices, B′–C loop; 2ac—between the B′ helix and G helix; 2e— through the B–C loop; 2f—between helices A′, F″, and the tip of the β4 hairpin; 3—between the F and G helices; S—between F, I helices, and the β4 hairpin; and W—helix C and the β-bulge segment.
Protein flexibility influences the opening and closing of ligand tunnels
In the simulations of T. brucei CYP51, dynamic opening and closing of the ligand tunnels were observed. The 2f tunnel is open in the crystal structures and was mostly open in the simulations (for 10–97% of the snapshots analyzed). The S tunnel is closed in the crystal structure (PDB id: 3G1Q) but was found to be open in the simulations (for 2–73% of the snapshots analyzed). These two tunnels were very often open in the simulations of the ligand-free soluble form and the ligand-free membrane-bound form. However, although these two tunnels were open in the simulation of the MCP-bound membrane-bound form of T. brucei CYP51, they were mostly closed in that of the MCP-bound soluble form. This indicates that the membrane stabilized the opening of tunnels 2f and S when the ligand was bound in the active site. The common entrance residues of tunnels 2f and S (SRS2 and SRS6) are mobile in the MD simulations (discussed previously), and thus, these two tunnels were able to open and close in the simulations. The B–C loop and C helix modulate the opening and closing of most family 2 tunnels, including 2a, 2b, 2c, 2e, and 2ac. These tunnels are closed in the crystal structures, and because of the low mobility of the B–C loop and C helix region (discussed previously), these tunnels were mostly closed in the simulations (Figure 5). However, tunnel W, which is open in the crystal structures (Table 4), stayed open throughout most of the simulations.
Table 4.
Accessibility of the water tunnel to solvent and hydrogen bonding of the two heme propionate groups in the crystal structures of CYP51
| Heme A-propionate |
Heme B-propionate |
||||||
|---|---|---|---|---|---|---|---|
| Organism | PDB ID | Liganda | Accessibilityb | Conformationc | H-bondingd | Conformationc | H-bondingd |
| T. brucei | 3TIK, 3P99, 2WV2, 4G3J | + | + | Distal | Y103, R361 | Proximal | Y116, R124, H420 |
| 3G1Q | - | + | Distal | Y103, R361 | Proximal | Y116, R124, H420 | |
| 2X2N | + | + | Distal | Y103, R361 | Parallel | Y116, H420 | |
| 3GW9, 4G7G | + | + | Distal | R361 | Proximal | Y116, R124, H420 | |
| T. cruzi | 4H60, 3KSW | + | + | Distal | Y103, R361 | Parallel | H420 |
| 2WX2 | + | + | Distal | Y103, R361 | Parallel | Y116, H420 | |
| 3KHM | + | + | Distal | R361 | Proximal | R124, H420 | |
| 3K10 | + | + | Distal | Y103, R361 | Distal | Y116, H420 | |
| 2WUZ | + | - | Distal | Y103, R361 | Proximal | Y116, R124, H420 | |
| L infantum | 3L4D | + | + | Distal | Y103, R361 | Proximal | Y116, R124, H420 |
| Human | 3JUS, 3LD6 | + | - | Distal | Y131, R382 | Distal | Y145, K156, H447 |
| 3JUV | - | + | Proximal | R382 | Distal | Y145 | |
| M. tuberculosis H37Rv | 2W09e | + | + | Distal | R326 | Proximal | ---- |
| 2W0Ae | + | + | Distal | Q72, R326 | Distal | H392 | |
| 2W0B | + | + | Distal | Y76, R326 | Proximal | K97, H392 | |
| 2VKU | + | + | Distal | Y76, R326 | Distal (R)f | Ligands | |
| 2CI0e | + | + | Distal | R326 | Distal&Proximalg | H392 | |
| 2CIBe | + | + | Distal | R326 | Distal | H392 | |
| M. tuberculosis | 2BZ9, 1 H5Ze | - | + | Distal | R326 | Proximal | ---- |
| 1U13 | - | + | Distal | Y76, R326 | Proximal | K97, H392 | |
| 1X8Ve | + | + | Distal | Y76, R326 | Proximal | K97, H392 | |
| 1EA1, 1E9X | + | + | Distal | Y76, R362 | Proximal | R95, H392 | |
If there is more than one chain, chain A is used.
With (+) or without (-) ligand in the active site.
The water tunnel (tunnel W) is accessible (+) or inaccessible (-) to the solvent (with a probe radius of 1.4 Å in PyMOL).
The propionate is on the distal side (distal) or the proximal side (proximal) of the heme plane, or roughly parallel (parallel) to it.
The hydrogen-bonding partners of the heme propionate groups. A hydrogen bond is considered to be formed when the distance between the two non-hydrogen atoms is less than 3.3 Å. The residue number is the residue number in the corresponding PDB structures. “----” indicates that the propionate has no hydrogen bonding with surrounding residues. “Ligands” indicate that the propionate makes hydrogen bonds to the ligands.
There are residues missing in the B-C loop and C helix region of the crystal structures. 2W09, 2W0A, 2CI0, and 1X8V have the B′-C loop missing. 2CIB has the B′-C loop and the beginning of the C helix missing, and 1H5Z has the C helix missing.
The B-propionate has rotated 120° toward the 1 helix because of the interaction with the two ligands (one in the binding site and the other in the ligand tunnel 1).
There are two conformations of the B-propionate group in the crystal structure. One conformation is on the distal and one on the proximal side of the heme plane.
Tunnels 2f and S can serve as major ligand tunnels in CYP51
The open tunnels of CYP51 are different from those of the human drug-metabolizing CYPs. In the simulations of CYP2C9, all the common tunnels (2a, 2b, 2c, 2ac, 2e, and S) were mostly open in at least one of the eight simulations of the two models of the soluble and membrane-bound CYP2C9 models. The 2f tunnel was not open in most of the CYP2C9 simulations (Cojocaru et al., 2011). The ligand tunnel analysis of the simulations of the membrane-bound models of six human drug-metabolizing CYPs (CYP1A2, CYP2A6, CYP2C9, CYP2E1, and CYP3A4) shows that tunnel S was open in all six simulated CYPs, tunnels 2b and 2c were open in five of the simulated CYPs, tunnel W in four, and tunnels 2e and 2f in three (Berka et al., 2013). However, tunnels 2f, S, and W were found to be wide open in all our simulations of T. brucei CYP51, and the other tunnels were mostly closed. In the simulation of human CYP51, only tunnels 2f and S were wide open.
The 2f tunnel has been proposed to serve as the substrate access channel for CYP51 (Lepesheva et al., 2010a; Lepesheva and Waterman, 2011). Our simulations support the idea that tunnel 2f can serve as the ligand entrance tunnel for hydrophobic ligands, such as the CYP51 substrates, to enter the buried active site from the membrane, where the sterol biosynthetic pathway takes place. The ligands can exit by either tunnel 2f to the membrane or tunnel S to the solvent, depending on the properties of the ligands.
Open water tunnel and multiple conformations of heme propionate groups
B–C loop rigidity keeps the W tunnel open
The W tunnel is directed toward the proximal surface of CYP51, and the residues lining this tunnel are mostly in the proximal region of the B–C loop and the B and C helices. This tunnel cannot serve as a substrate/product tunnel because it is on the proximal (opposite from the substrate binding cavity) side of CYP51, which is the expected binding face of CYP51 for the CYP reductase. The W tunnel was found open only in the simulations of the membrane-bound models of six human drug-metabolizing CYPs but was closed in the simulations of the soluble models (Berka et al., 2013). The open tunnel W in the membrane-bound form of CYP51 suggests that it is possible that tunnel W has to be open to facilitate the interaction with the reductase. This is different from the crystal structures of most human CYPs that have this tunnel closed. In the human drug-metabolizing CYPs, the opening and closing of this tunnel can be observed because the proximal side is flexible. However, in CYP51, because of the rigidity of the C helix and β-bulge segment (Figure 2), the residues lining the W tunnel are not able to facilitate the opening and closing motions of this tunnel. As a result, this tunnel stays open in both the crystal structures (Table 4) and simulations (Figure 5).
The two heme propionate groups adopt different conformations
Conformational changes of the A-propionate group have been observed in experimental and computational studies of mutants of the prokaryotic cytochrome P450 BM3 (Chen et al., 2004; Stjernschantz et al., 2008). In these studies, the heme A-propionate group was shown to adopt different conformations upon ligand binding. However, conformational changes of the two propionate groups have not been reported in MD simulations of wild-type CYPs. Here, we discuss the different conformations of the two propionate groups of CYP51 in crystal structures and in our simulations of the wild-type T. brucei CYP51.
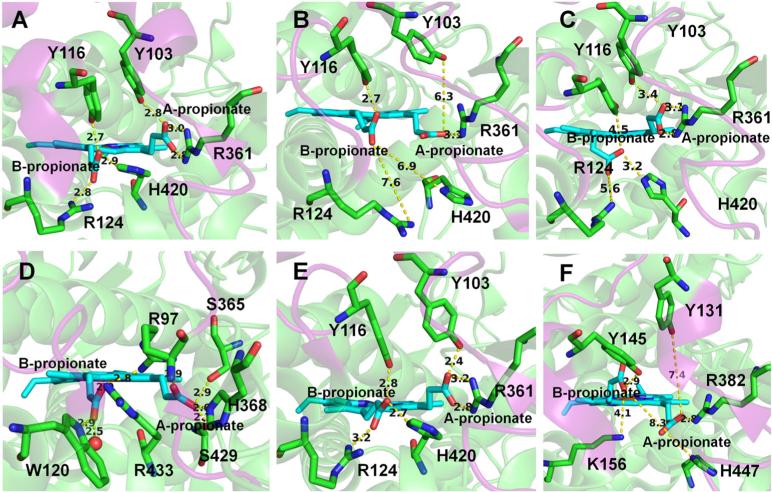
The B-propionate group of the heme connects the W tunnel and the active site. The two heme propionate groups show different conformations in the CYP51 crystal structures from different organisms (Table 4 and Figure 6). Three crystallographic conformations of the two heme propionate groups are shown in Figures 6A, E, and F and two conformations from the simulations in Figures 6B and C. Although small conformational changes of the A-propionate group have been seen in the mutants of cyto-chrome P450 BM3 (Chen et al., 2004; Stjernschantz et al., 2008), such high flexibility of the A-propionate group, and the flexibility of the B-propionate group, has not been observed in simulations of other wild-type CYPs. In our simulations, we found that the heme propionate groups of T. brucei CYP51 are able to oscillate between different conformations (Figure S8), but they prefer the conformation shown in Figure 6B, which was shown to be occupied in all crystal structures of the membrane-bound CYPs except those of CYP51 and CYP46A1 (Denisov et al., 2012). The conformation in Figure 6C was not found in the crystal structures but was observed in our simulations. The propionate groups might have different conformations when binding to different ligands in the active site.
Figure 6.
Different conformations and hydrogen-bonding interactions of the two heme propionate groups in the crystal structures and simulations. The distance between the two heme propionate groups and important hydrogen-bonding partners is shown. (A) The most predominant conformation of the propionate groups observed in the crystal structures of CYP51 enzymes. The coordinates are from T. brucei CYP51 (PDB ID: 3G1Q). (B, C) Two alternative conformations of the propionate groups seen in the snapshots at 60 and 15 ns in the simulations of the membrane-bound ligand-free and MCP-bound T. brucei CYP51, respectively. (D) The conformation in the crystal structure and simulations of CYP2C9 (PDB ID: 1R9O). (E, F) Two different conformations in the crystal structures of T. cruzi (PDB ID: 2WX2) and human CYP51 (PDB ID: 3JUV), respectively.
The conformational changes of the A-propionate group of cytochrome P450 BM3 have been shown to be an effect of mutations but not substrate binding and suggested to influence heme reduction, catalytic efficiency, and electron transfer (Stjernschantz et al., 2008). Our simulations suggest that the heme propionate groups of CYP51 have higher flexibility than those of most of the other CYPs. The high flexibility of the two propionate groups may compensate the rigidity of the binding cavity residues of CYP51. This flexibility may be important for ligand binding, electron transfer, and heme reduction in CYP51.
Opening of the W tunnel is affected by the conformation of the heme B-propionate group and is facilitated by ligand binding in the active site
Tunnel W stayed open to a 1.4-Å radius probe in the simulations of T. brucei CYP51. We checked the width of tunnel W using CAVER with a bigger probe radius (2.0 Å) for the simulations of T. brucei CYP51. The influence of the conformation of the heme propionate groups on the width of tunnel W was analyzed. In the ligand-free simulations, tunnel W stayed closed to the 2.0-Å radius probe in the soluble simulations and was open in 3.5% of snapshots from the membrane-bound simulation. The heme propionate groups mostly adopted a conformation similar to that in Figure 6B. However, in the ligand-bound simulations, they adopted a conformation similar to Figure 6C, and tunnel W was open to the 2.0-Å radius probe in 28% of the soluble simulation and 65% of the membrane-bound simulation.
This analysis suggests that the conformation of the B-propionate group can influence the opening width of tunnel W. In the presence of ligands in the active site, tunnel W tends to be more open. Hence, the presence of a substrate in the active site might facilitate the water transfer between the active site and the solvent necessary for the catalytic cycle.
A hydrogen bond network determines the conformation of the heme propionate groups
When the two heme propionate groups are in different conformations, they have different hydrogen-bonding partners (Figure 6 and Table 4). The two propionate groups in CYP51 have fewer hydrogen-bonding partners than the propionate groups in the other CYPs studied. In the crystal structures of CYP51, the A-propionate has a maximum of three hydrogen bonds (two with R361), whereas the B-propionate has a maximum of three hydrogen bonds (Table 4 and Figure 6A, E, and F). However, in CYP2C9 (PDB id: 1R9O), the A-propionate forms hydrogen bonds with R97, S365, H368, and S429, whereas the B-propionate forms hydrogen bonds with R97, W120, R433, and one water molecule (represented by a red sphere) (Figure 6D). Fewer hydrogen-bonding partners might be one reason for the flexibility of the two heme propionate groups in CYP51.
Conformational changes of the two propionate groups, which are driven by the changes of the interactions with neighboring residues, were seen in our simulations of T. brucei CYP51. The orientation of the A-propionate group is mainly driven by the hydrogen-bonding interaction with R361. The A-propionate group changed its conformation from that in Figure 6A to that in Figure 6B when the two hydrogen bonds with R361 were disrupted by water molecules in the active site in the ligand-free simulation. However, when the hydrophobic ligand MCP is bound in the active site, the water molecules were expelled from the active site. To regain the two hydrogen bonds with R361, the A-propionate group changed its conformation from that in Figure 6B to that in Figure 6C. Whereas the conformation of the A-propionate group is largely determined by the interaction with one residue (R361), the conformation of the B-propionate group is determined by its interaction with several neighboring residues, including R124 and H420. In the simulation of the membrane-bound MCP-bound T. brucei CYP51, the transition of the B-propionate group from the conformation in Figure 6B to that in Figure 6C was facilitated by reforming the hydrogen bond with H420.
The hydrogen-bonding network between the two heme propionates and the neighboring binding cavity residues plays a crucial role in maintaining the catalytic activity of CYPs (Hasemann et al., 1995; Guallar et al., 2003). Changes of the hydrogen bond network may affect the binding of ligands, the electron transfer process, and the release of water after the catalytic reaction.
Functional implications for drug design
The rigidity of the binding cavity residues, from the comparison of CYP51 (T. brucei and human), CYP2C9 (human), and CYP2E1 (human), is consistent with the high substrate selectivity of CYP51. The rigidity restricts the reorientation of the binding site residues upon ligand binding. Thus, only ligands that can fit rather perfectly into the binding cavity can bind to the active site of CYP51. This indicates that, for antiparasitic drug design, the inhibitors have to fit the binding pocket of CYP51 in a similar way to the substrates to have inhibitory effects.
The higher mobility of the tunnel entrance residues of CYP51 than that of other CYPs can facilitate antiparasitic drug design. The reason for the high mobility could be that as the substrates of CYP51 are large compared with those of CYP2C9 and CYP2E1, the tunnel entrance residues have to open wider to allow the large substrates to enter the active site of CYP51. This indicates that the design of large molecules that are able to enter the active site of CYP51 but not of human drug-metabolizing CYPs such as CYP2C9 and CYP2E1 is possible. However, CYP3A4 is able to bind to large inhibitors, and this would have to be taken into consideration.
Comparison of the dynamics of T. brucei CYP51 and human CYP51 indicates that there are limited differences in the SRS regions of the two proteins. Therefore, inhibitors that can only inhibit parasitic CYP51s, and not the human ortholog, may result from the differences in the substrate-contacting residues, such as F105 (L in human), M106 (T in human), and F290 (L in human), rather than differences in the flexibility of the binding cavity residues.
CONCLUSION
A model of the membrane-bound T. brucei CYP51 was built and simulated with an improved version of the protocol that has been applied to CYP2C9. The model is in good agreement with available experimental data on CYPs. The F–G loop region is more rigid in the simulation of the membrane-bound T. brucei CYP51 than in that of the soluble T. brucei CYP51 because this region is embedded in the membrane.
We found that, unlike other membrane-bound CYPs (represented by CYP2C9 and CYP2E1) that have high flexibility in the active site and a large range of ligands, the CYP51 active site is relatively rigid in simulations of both soluble and membrane-bound forms. Longer and repeated simulations would be desirable to gain a complete understanding of the dynamics of different CYPs. However, despite their limited length, our simulations show distinct dynamics in the different CYP forms.
Opening and closing of ligand tunnels were observed in CYP51. The simulations suggest that the 2f tunnel is used as a ligand access tunnel and that both 2f and S can potentially be used as ligand egress tunnels. The simulations of the membrane-bound form of T. brucei CYP51 indicate that ligands need to enter tunnel 2f from the membrane and can exit via 2f back to the membrane or via S to the solvent. High flexibility is seen in the residues at the surface entrance to tunnel 2f in T. brucei CYP51. The rigid B–C loop keeps tunnel W open in both crystal structures and simulations of T. brucei CYP51. The conformation of the two heme propionate groups affects the opening width of tunnel W. Tunnel W opened wider in the presence of ligands, which indicates a role for this tunnel in water delivery between the active site and the bulk solvent. The hydrogen-bonding network determines the conformation of the two heme propionate groups and can further affect the binding and unbinding of ligands. The conformational changes of the heme's two propionate groups may be important for ligand binding, electron transfer, and heme reduction of CYP51.
Access of the ligands to the binding site of CYP51 is facilitated by the flexibility of the 2f tunnel, but the relative rigidity of the binding site means that high complementarity in shape and chemical properties is required for binding in the active site. The tunnels provide another potential target binding site for drugs apart from the active site. Both tunnel 2f and tunnel W are more open in CYP51 than in the other human CYPs studied and might serve as sites for CYP51-specific ligands.
From the comparison of the dynamics of T. brucei CYP51 and human CYP51, the reason for the specific inhibition of parasitic CYP51 does not appear to be the difference in the flexibility of the active site but may arise from the differences in the interaction properties of the binding site residues.
Supplementary Material
Acknowledgements
We gratefully acknowledge the support of the Klaus Tschira Foundation. XY acknowledges the kind help of Dr. Jonathan C. Fuller, for his help with the lipid parameter calculations, and the support of the Hartmut Hoffmann-Berling International Graduate School of Molecular and Cellular Biology, Heidelberg University. VC acknowledges the support of the Max Planck Society. GM acknowledges the support of German Academic Exchange Service (Deutscher Akademischer Austauschdienst). OMHS-A acknowledges the Sigrid Juselius Foundation and the Tor, Joe and Pentti Borg Foundation for contributing to the infrastructure and Prof. Mark Johnson of Åbo Akademi University for providing excellent computing facilities. GIL acknowledges the support of the US National Institutes of Health, grant GM067871.
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found in the online version of this article at the publisher's web site.
REFERENCES
- Allen WJ, Lemkul JA, Bevan DR. GridMAT-MD: a grid-based membrane analysis tool for use with molecular dynamics. J. Comput. Chem. 2009;30:1952–8. doi: 10.1002/jcc.21172. [DOI] [PubMed] [Google Scholar]
- Andriani G, Amata E, Beatty J, Clements Z, Coffey BJ, Courtemanche G, Devine W, Erath J, Juda CE, Wawrzak Z, Wood JT, Lepesheva GI, Rodriguez A, Pollastri MP. Antitrypanosomal lead discovery: identification of a ligand-efficient inhibitor of Trypanosoma cruzi CYP51 and parasite growth. J. Med. Chem. 2013;56:2556–67. doi: 10.1021/jm400012e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama Y, Noshiro M, Gotoh O, Imaoka S, Funae Y. Sterol 14-Demethylase P450 ( P45014DM *) Is One of the Most Ancient and Conserved P450 Species. J. Biochem. 1996;933:926–933. doi: 10.1093/oxfordjournals.jbchem.a021331. [DOI] [PubMed] [Google Scholar]
- Bayburt TH, Sligar SG. Single-molecule height measurements on microsomal cytochrome P450 in nanometer-scale phospholipid bilayer disks. Proc. Natl. Acad. Sci. U. S. A. 2002;99:6725–30. doi: 10.1073/pnas.062565599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylon JL, Lenov IL, Sligar SG, Tajkhorshid E. Characterizing the membrane-bound state of cytochrome P450 3A4: structure, depth of insertion, and orientation. J. Am. Chem. Soc. 2013;135:8542–51. doi: 10.1021/ja4003525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayly CI, Cieplak P, Cornell W, Kollman PA. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J. Phys. Chem. 1993;97:10269–10280. [Google Scholar]
- Berendsen HJC, van der Spoel D, van Drunen R. GROMACS: A message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995;91:43–56. [Google Scholar]
- Berka K, Hendrychová T, Anzenbacher P, Otyepka M. Membrane position of ibuprofen agrees with suggested access path entrance to cytochrome P450 2C9 active site. J. Phys. Chem. A. 2011;115:11248–55. doi: 10.1021/jp204488j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berka K, Paloncýová M, Anzenbacher P, Otyepka M. Behavior of Human Cytochromes P450 on Lipid Membranes. J. Phys. Chem. B. 2013;117:11556–11564. doi: 10.1021/jp4059559. [DOI] [PubMed] [Google Scholar]
- Black SD, Martin ST, Smith CA. Membrane Topology of Liver Micro-somal Cytochrome P450 2B4 Determined via Monoclonal Antibodies Directed to the Halt-Transfer Signal. Biochemistry. 1994;33:6945–6951. doi: 10.1021/bi00188a025. [DOI] [PubMed] [Google Scholar]
- Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Walker RC, Zhang W, Merz KM, Reberts B, Hayik S, Roitberg A, Seabra G, Swails J, Goetz AW, Kolossvary I, Wong KF, Paesani F, Vanicek J, Wolf RM, Liu J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Cai Q, Ye X, Wang J, Hsieh M-J, Cui G, Roe DR, Mathews DH, Seetin MG, Salomon-Ferrer R, Sagui C, Babin V, Luchko T, Gusarov S, Kovalenko A, Kollman PA. Amber 12. University of Califonia; San Francisco: 2012. [Google Scholar]
- Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Walker RC, Zhang W, Merz KM, Reberts B, Wang B, Hayik S, Roitberg A, Seabra G, Kolossvary I, Wong KM, Paesani F, Vanicek J, Liu J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Cai Q, Ye X, Wang J, Hsieh M-J, Cui G, Roe DR, Mathews DH, Seetin MG, Sagui C, Babin V, Luchko T, Gusarov S, Kovalenko A, Kollman PA. Amber 11. University of Califonia; San Francisco: 2010. [Google Scholar]
- Chen Z, Ost TWB, Schelvis JPM. Phe393 mutants of cytochrome P450 BM3 with modified heme redox potentials have altered heme vinyl and propionate conformations. Biochemistry. 2004;43:1798–808. doi: 10.1021/bi034920g. [DOI] [PubMed] [Google Scholar]
- Chovancova E, Pavelka A, Benes P, Strnad O, Brezovsky J, Kozlikova B, Gora A, Sustr V, Klvana M, Medek P, Biedermannova L, Sochor J, Damborsky J. CAVER 3.0: a tool for the analysis of transport pathways in dynamic protein structures. PLoS Comput. Biol. 2012;8:e1002708. doi: 10.1371/journal.pcbi.1002708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cojocaru V, Balali-Mood K, Sansom MSP, Wade RC. Structure and dynamics of the membrane-bound cytochrome P450 2C9. PLoS Comput. Biol. 2011;7:e1002152. doi: 10.1371/journal.pcbi.1002152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cojocaru V, Winn PJ, Wade RC. The ins and outs of cytochrome P450s. Biochim. Biophys. Acta. 2007;1770:390–401. doi: 10.1016/j.bbagen.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Denisov IG, Shih AY, Sligar SG. Structural differences between soluble and membrane bound cytochrome P450s. J. Inorg. Biochem. 2012;108:150–8. doi: 10.1016/j.jinorgbio.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32:W665–7. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupradeau F-Y, Pigache A, Zaffran T, Savineau C, Lelong R, Grivel N, Lelong D, Rosanski W, Cieplak P. The R.E.D. tools: advances in RESP and ESP charge derivation and force field library building. Phys. Chem. Chem. Phys. 2010;12:7821–39. doi: 10.1039/c0cp00111b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey B, David D. Determination of the Membrane Topology of the Phenobarbital-inducible Rat Liver Cytochrome P-450 Isoenzyme PB-4 Using Site-specific Antibodies. J. Cell Biol. 1987;104:209–219. doi: 10.1083/jcb.104.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh O. Substrate Recognition Sites in Cytochrome P450 Family 2 (CYP2) Proteins Inferred from Comparative Analyses of Amino Acid and Coding Nucleotide Sequences. J. Biol. Chem. 1992;267:83–90. [PubMed] [Google Scholar]
- Guallar V, Baik M, Lippard SJ, Friesner RA. Peripheral heme substituents control the hydrogen-atom abstraction chemistry in cytochromes P450. Proc. Natl. Acad. Sci. U. S. A. 2003;100:6998–7002. doi: 10.1073/pnas.0732000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haider S, Islam B, D'Atri V, Sgobba M, Poojari C, Sun L, Yuen T, Zaidi M, New MI. Structure-phenotype correlations of human CYP21A2 mutations in congenital adrenal hyperplasia. Proc. Natl. Acad. Sci. U. S. A. 2013;110:2605–10. doi: 10.1073/pnas.1221133110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargrove TY, Kim K, de Nazaré Correia Soeiro M, da Silva CF, da Gama Jaen Batista D, Batista MM, Yazlovitskaya EM, Waterman MR, Sulikowski GA, Lepesheva GI. CYP51 structures and structure-based development of novel, pathogen-specific inhibitory scaffolds. Int. J. Parasitol. Drugs Drug Resist. 2012a;2:178–186. doi: 10.1016/j.ijpddr.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargrove TY, Wawrzak Z, Liu J, Nes WD, Waterman MR, Lepesheva GI. Substrate preferences and catalytic parameters determined by structural characteristics of sterol 14alpha-demethylase (CYP51) from Leishmania infantum. J. Biol. Chem. 2011;286:26838–48. doi: 10.1074/jbc.M111.237099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargrove TY, Wawrzak Z, Liu J, Waterman MR, Nes WD, Lepesheva GI. Structural complex of sterol 14α-demethylase (CYP51) with 14α-methylenecyclopropyl-Delta7-24, 25-dihydrolanosterol. J. Lipid Res. 2012b;53:311–20. doi: 10.1194/jlr.M021865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris DL, Park J-Y, Gruenke L, Waskell L. Theoretical study of the ligand-CYP2B4 complexes: effect of structure on binding free energies and heme spin state. Proteins. 2004;55:895–914. doi: 10.1002/prot.20062. [DOI] [PubMed] [Google Scholar]
- Hasemann CA, Kurumbail RG, Boddupalli SS, Peterson JA, Deisenhofer J. Structure and function of cytochromes P450:a comparative analysis of three crystal structures. Structure. 1995;3:41–62. doi: 10.1016/s0969-2126(01)00134-4. [DOI] [PubMed] [Google Scholar]
- Headlam M. The F–G loop region of cytochrome P450scc (CYP11A1) interacts with the phospholipid membrane. Biochim. Biophys. Acta, Biomemb. 2003;1617:96–108. doi: 10.1016/j.bbamem.2003.09.007. [DOI] [PubMed] [Google Scholar]
- Hendrychova T, Berka K, Navratilova V, Anzenbacher P, Otyepka M. Dynamics and hydration of the active sites of mammalian cytochromes P450 probed by molecular dynamics simulations. Curr. Drug Metab. 2012;13:177–89. doi: 10.2174/138920012798918408. [DOI] [PubMed] [Google Scholar]
- Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–25. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Jójárt B, Martinek TA. Performance of the general amber force field in modeling aqueous POPC membrane bilayers. J. Comput. Chem. 2007;28:2051–8. doi: 10.1002/jcc.20748. [DOI] [PubMed] [Google Scholar]
- Jorgensen WL, Madura JD. Quantum and statistical mechanical studies of liquids. 25. Solvation and conformation of methanol in water. J. Am. Chem. Soc. 1983;105:1407–1413. [Google Scholar]
- Lepesheva GI, Hargrove TY, Anderson S, Kleshchenko Y, Furtak V, Wawrzak Z, Villalta F, Waterman MR. Structural insights into inhibition of sterol 14alpha-demethylase in the human pathogen Trypanosoma cruzi. J. Biol. Chem. 2010a;285:25582–90. doi: 10.1074/jbc.M110.133215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepesheva GI, Hargrove TY, Kleshchenko Y, Nes WD, Villalta F, Waterman MR. CYP51: A major drug target in the cytochrome P450 superfamily. Lipids. 2008;43:1117–25. doi: 10.1007/s11745-008-3225-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepesheva GI, Park H-W, Hargrove TY, Vanhollebeke B, Wawrzak Z, Harp JM, Sundaramoorthy M, Nes WD, Pays E, Chaudhuri M, Villalta F, Waterman MR. Crystal structures of Trypanosoma brucei sterol 14alpha-demethylase and implications for selective treatment of human infections. J. Biol. Chem. 2010b;285:1773–80. doi: 10.1074/jbc.M109.067470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepesheva GI, Waterman MR. CYP51--the omnipotent P450. Mol. Cell. Endocrinol. 2004;215:165–70. doi: 10.1016/j.mce.2003.11.016. [DOI] [PubMed] [Google Scholar]
- Lepesheva GI, Waterman MR. Sterol 14alpha-Demethylase (CYP51) as a Therapeutic Target for Human Trypanosomiasis and Leishmaniasis. Curr. Top. Med. Chem. 2011;11:2060–2071. doi: 10.2174/156802611796575902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl E, Hess B, van der Spoel D. GROMACS 3.0: a package for molecular simulation and trajectory analysis. J. Mol. Model. 2001;7:306–317. [Google Scholar]
- Lomize MA, Lomize AL, Pogozheva ID, Mosberg HI. OPM: orientations of proteins in membranes database. Bioinformatics. 2006;22:623–5. doi: 10.1093/bioinformatics/btk023. [DOI] [PubMed] [Google Scholar]
- Marrink S-J, de Vries AH, Mark AE. Coarse Grained Model for Semiquantitative Lipid Simulations. J. Phys. Chem. B. 2004;108:750–760. [Google Scholar]
- Marrink S-J, Risselada HJ, Yefimov S, Tieleman DP, de Vries AH. The MARTINI force field: coarse grained model for biomolecular simulations. J. Phys. Chem. B. 2007;111:7812–24. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- Miao Y, Baudry J. Active-site hydration and water diffusion in cytochrome P450cam: a highly dynamic process. Biophys. J. 2011;101:1493–503. doi: 10.1016/j.bpj.2011.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Yi Z, Cantrell C, Glass DC, Baudry J, Jain N, Smith JC. Coupled flexibility change in cytochrome P450cam substrate binding determined by neutron scattering, NMR, and molecular dynamics simulation. Biophys. J. 2012;103:2167–76. doi: 10.1016/j.bpj.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk BC, Tomasiak TM, Keniya MV, Huschmann FU, Tyndall JD, O'Connell JD, Cannon RD, McDonald JG, Rodriguez A, Finer-Moore JS, Stroud RM. Architecture of a single membrane spanning cytochrome P450 suggests constraints that orient the catalytic domain relative to a bilayer. Proc. Natl. Acad. Sci. 2014;111:3865–3870. doi: 10.1073/pnas.1324245111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monticelli L, Kandasamy SK, Periole X, Larson RG, Tieleman DP, Marrink S J. The MARTINI Coarse-Grained Force Field: Extension to Proteins. J. Chem. Theory Comput. 2008;4:819–834. doi: 10.1021/ct700324x. [DOI] [PubMed] [Google Scholar]
- Nelson DR. Cytochrome P450 and the individuality of species. Arch. Biochem. Biophys. 1999;369:1–10. doi: 10.1006/abbi.1999.1352. [DOI] [PubMed] [Google Scholar]
- Ohta Y, Kawato S, Tagashira H, Takemori S, Kominami S. Dynamic structures of adrenocortical cytochrome P-450 in proteoliposomes and microsomes: protein rotation study. Biochemistry. 1992;31:12680–12687. doi: 10.1021/bi00165a019. [DOI] [PubMed] [Google Scholar]
- Ozalp C, Szczesna-Skorupa E, Kemper B. Identification of membrane-contacting loops of the catalytic domain of cytochrome P450 2C2 by tryptophan fluorescence scanning. Biochemistry. 2006;45:4629–37. doi: 10.1021/bi051372t. [DOI] [PubMed] [Google Scholar]
- Pei J, Kim B-H, Grishin NV. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008;36:2295–300. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Periole X, Cavalli M, Marrink S-J, Ceruso MA. Combining an Elastic Network With a Coarse-Grained Molecular Force Field: Structure, Dynamics, and Intermolecular Recognition. J. Chem. Theory Comput. 2009;5:2531–2543. doi: 10.1021/ct9002114. [DOI] [PubMed] [Google Scholar]
- Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikuleva IA. Putative F-G loop is involved in association with the membrane in P450scc (P450 11A1). Mol. Cell. Endocrinol. 2004;215:161–4. doi: 10.1016/j.mce.2003.11.005. [DOI] [PubMed] [Google Scholar]
- Pronk S, Páll S, Schulz R, Larsson P, Bjelkmar P, Apostolov R, Shirts MR, Smith JC, Kasson PM, van der Spoel D, Hess B, Lindahl E. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–54. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzepiela AJ, Schäfer LV, Goga N, Risselada HJ, De Vries AH, Marrink SJ. Reconstruction of atomistic details from coarse-grained structures. J. Comput. Chem. 2010;31:1333–43. doi: 10.1002/jcc.21415. [DOI] [PubMed] [Google Scholar]
- Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Schrodinger LLC. The PyMOL Molecular Graphics System. 2010.
- Sgrignani J, Magistrato A. Influence of the membrane lipophilic environment on the structure and on the substrate access/egress routes of the human aromatase enzyme. A computational study. J. Chem. Inf. Model. 2012;52:1595–606. doi: 10.1021/ci300151h. [DOI] [PubMed] [Google Scholar]
- Stjernschantz E, van Vugt-Lussenburg BMA, Bonifacio A, de Beer SBA, van der Zwan G, Gooijer C, Commandeur JNM, Vermeulen NPE, Oostenbrink C. Structural rationalization of novel drug metabolizing mutants of cytochrome P450 BM3. Proteins. 2008;71:336–52. doi: 10.1002/prot.21697. [DOI] [PubMed] [Google Scholar]
- Van der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26:1701–18. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- Vanquelef E, Simon S, Marquant G, Garcia E, Klimerak G, Delepine JC, Cieplak P, Dupradeau F-Y. R.E.D. Server: a web service for deriving RESP and ESP charges and building force field libraries for new molecules and molecular fragments. Nucleic Acids Res. 2011;39:W511–7. doi: 10.1093/nar/gkr288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalta F, Dobish MC, Nde PN, Kleshchenko YY, Hargrove TY, Johnson CA, Waterman MR, Johnston JN, Lepesheva GI. VNI cures acute and chronic experimental Chagas disease. J. Infect. Dis. 2013;208:504–11. doi: 10.1093/infdis/jit042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang W, Kollman PA, Case DA. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 2006;25:247–60. doi: 10.1016/j.jmgm.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J. Comput. Chem. 2004;25:1157–74. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Gildenberg M, Ahuja S, Im S-C, Pearcy P, Waskell L, Ramamoorthy A. Probing the transmembrane structure and topology of microsomal cytochrome-p450 by solid-state NMR on temperature-resistant bicelles. Sci. Rep. 2013;3:2556. doi: 10.1038/srep02556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y. Cytochrome P450 of fungi: primary target for azole antifungal agents. Curr. Top. Med. Mycol. 1988;2:388–418. doi: 10.1007/978-1-4612-3730-3_11. [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Aoyama Y, Noshiro M, Gotoh O. Sterol 14-demethylase P450 (CYP51) provides a breakthrough for the discussion on the evolution of cytochrome P450 gene superfamily. Biochem. Biophys. Res. Commun. 2000;273:799–804. doi: 10.1006/bbrc.2000.3030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.