Abstract
In concentration-QTc modeling, oscillatory functions have been used to characterize biological rhythms in QTc profiles. Fitting such functions is not always feasible because it requires sufficient electrocardiograph sampling. In this study, drug concentration and QTc data were simulated using a published biological QTc model (oscillatory functions). Then, linear mixed-effect models and the biological model were fitted and evaluated in terms of biases, precisions, and qualities of inferences. The simpler linear mixed-effect model with day and time as a factor variables provided similar accuracy of the concentration-QTc slope estimates to the complex biological model and was able to accurately predict the drug-induced QTc prolongation with less than 1 ms bias, despite its empirical nature to account for biological rhythm. The current study may guide a concentration-QTc modeling strategy that can be easily prespecified, does not suffer from poor convergence, and achieves little bias in drug-induced QTc estimates.
Assessing proarrhythmic risk has been a key issue during drug development for the past decade1 due to reports of QT interval prolongation for nonantiarrhythmic drugs. In 2005, the US Food and Drug Administration (FDA) released the ICH E14 guidance, which proposed a thorough QT study (TQT study) be conducted to assess the risk of QT prolongation of new drugs.2 It did not take long for the TQT study to be criticized. Authors have reported that its primary statistical method, i.e., the intersection union test, can suffer from low power, the maximum mean difference from placebo provides a positively biased estimate of prolongation,3–5 and the result cannot be used directly to predict the outcomes in subjects at risk for increased exposures. Another criticism of the TQT trial is cost-effectiveness. The use of concentration-QTc (CQTc) analyses has increased significantly to address these issues. CQTc analyses provide increased efficiency in that the use of exposure facilitates integration of information across the sampling times and treatment groups, in contrast to the time point-based E14 analysis.6 Additionally, CQTc analyses can be performed with data from early development (phase I). In fact, a good number of studies have suggested the use of CQTc analysis of early phase studies as a substitute for a TQT study.7–9 The Consortium for Innovation and Quality in Pharmaceutical Development and the Cardiac Safety Research Consortium have collaborated recently to run a single ascending dose-like study with six marketed drugs, which were previously characterized by TQT studies, to investigate whether equivalent information about QT effects can be obtained from either a TQT intersection union test or a CQTc analysis.6
It is well known that QT intervals exhibit biological rhythms such as circadian patterns.10–12 Several authors have proposed oscillatory time functions to characterize biological rhythms in QTc profiles within a subject.10,13,14 However, fitting such functions is not always feasible, because the estimation of parameters governing these oscillations requires sufficient electrocardiograph (ECG) sampling. This can be a challenge even for a TQT study. Bayesian methods have even been proposed because of the dimensionality of the biological model.8,14,15 Other authors have proposed modeling baseline- and/or placebo-corrected QTc data (dQTc/ddQTc) to eliminate or reduce the correlation. While such strategies might work for a TQT study, generally these are not feasible solutions for Phase I study designs. Ultimately the goal of the analysis is to accurately estimate drug-induced QTc prolongation in the presence of such biological rhythms, and perform inference to establish what extent of prolongation can be precluded. Valid inference depends upon accurate confidence intervals (CIs). Prespecification of the model is essential for this reason. Precise prespecification of a biological model is difficult, because the number of oscillatory functions will likely be dictated by the study design. Given that the primary objective of the CQTc analysis is determining the extent of QTc prolongation as a function of concentration, and not estimating the magnitude of biological variability, complex models may not be necessary for decision making.
The objective of this simulation study was to evaluate specific linear mixed effect (LME) models for biases, precisions, and qualities of inferences under different pharmacokinetic (PK) profile scenarios and study designs in the presence of biologically varying QTc. Specifically, the performance of an LME model using sampling time as a factor variable was evaluated as an empirical way of addressing such variation in the data. To the best of our knowledge, such a study has not been conducted. Because biological rhythms are time-dependent processes, the performance of the LME model may be different depending on the Tmax of a drug. Therefore, three different Tmax magnitudes were investigated: short (Tmax ≈ 1 h), intermediate (Tmax ≈ 3 h), and long (Tmax ≈ 6 h). The amount of QTc prolongation (no QTc prolongation, 5 ms and 10 ms prolongation at geometric mean Cmax (peak plasma concentration) of supratherapeutic dose) and study design (TQT-like with rich ECG data and early phase studies with sparse ECG data) were also assessed. Ultimately, we pursue a modeling strategy that can be prespecified and achieve little bias in ddQTc estimates, and in the process, evaluate if a biological model is really necessary for decision making.
RESULTS
PK-Pharmacodynamic data simulation
Table 1 displays study designs (sampling times, sample sizes, doses administered, etc.) used in simulations. The PK-ECG matched sampling times in phase I were selected based on three different Tmax scenarios to capture the Cmax within each sampling scheme. In addition, inclusion of day 0 ECG sampling in phase I designs (phase I + day 0) was also investigated. For each simulated scenario, 1,000 QTc datasets were simulated using a published biological model.13
Table 1.
Description of studies used in the analysis
| Study | Description of study | N (number of subjects) | Treatment | ECG extraction and time-matched PK sample | |
|---|---|---|---|---|---|
| TQT | Three-way crossover, placebo-controlled study | 60 | Placebo, 25 and 250 mg | 0, 0.5, 1, 2, 2.5, 3, 4, 6, 8, 12, and 24 h | |
| Phase I | SAD | Parallel group, dose escalating study | 28 | Placebo, 25, 50, 100, and 200 mg | Short Tmax:0, 1, 2, 4, 8, and 24 h Intermediate Tmax: 0, 1.5, 3, 6, 12, and 24 h Long Tmax: 0, 3, 6, 9, 12, and 24 h |
| MAD | Parallel group, dose escalating study | 28 | Placebo, 20, 40, 60, and 80 mg | Short Tmax: 0, 1, and 2 h on day 1, 3, and 6. Intermediate Tmax: 0, 1.5, and 3 h on day 1, 3, and 6. Long Tmax: 0, 3, and 6 h on day 1, 3, and 6. | |
| Phase I + day 0 | SAD | Parallel group, baseline-controlled, dose escalating study | 28 | Placebo, 25, 50, 100, and 200 mg | Short Tmax:0, 1, 2, 4, 8, and 24 h on day 0 and 1 Intermediate Tmax: 0, 1.5, 3, 6, 12, and 24 h on day 0 and 1 Long Tmax: 0, 3, 6, 9, 12, and 24 h on day 0 and 1 |
| MAD | Parallel group, baseline-controlled, dose escalating study | 28 | Placebo, 20, 40, 60, and 80 mg | Short Tmax: 0, 1, and 2 h on day 0, 1, 3, and 6. Intermediate Tmax: 0, 1.5, and 3 h on day 0, 1, 3, and 6. Long Tmax: 0, 3, and 6 h on day 0, 1, 3, and 6. |
Assessment of the bias and estimation error
Figure 1 and Table 2 summarize the estimation error of drug effect parameters for the TQT study design with 5 ms true QTc prolongation. All LME models tended to have mildly negatively biased slope estimates, which was anticipated because of the incorporation of 20% measurement error in the concentration. The model with time as a factor variable (model 2) resulted in the least biased estimates across all three Tmax scenarios (−4.51% – −5.70% vs. −4.69% – −14.3%, Table 2). Incorporating an autoregressive residual error structure (model 3) did not reduce the slope estimate attenuation. The superiority of model 2 was confirmed by the assessment of coverage probability of the 90% CIs of the slope estimates (Figure 2a). Wald-type CIs maintained a nominal coverage rate of 90%, except for the intermediate Tmax case which just missed the nominal rate (89%, data not shown). Of interest, the bias in slope for the simple LME without time effects (model 1) was similar to model 2 for the short Tmax scenario, yet was worse for the intermediate and long Tmax scenarios. Between-subject variability (BSV) on slope was overestimated for all models including the biological model, except for the autoregressive model. Inter-occasion variability in the biological model used for simulation was not identified as day-to-day variability in LME models. Therefore, random effects on day were not incorporated. Residual variability was positively biased for all LME models (30.6%–36.2% bias, Figure 1) because these models did not capture structurally the subject-specific oscillation due to the biological rhythms. The full biological model consisting of two cosine functions suffered from overparametrization; only 40% of runs completed with a successful covariance step. A reduced biological model with one cosine function was fitted instead (>70% successful covariance step). The accuracy of the slope estimate for the reduced biological model was similar to model 2 (−3.87 – −7.15% bias). When the accuracy was evaluated for the full biological model including all converged runs (regardless of successful covariance step) the slope estimate was still negatively biased (−4.36% – −6.81% bias).
Figure 1.

Box and whisker plots of the estimation errors for drug effect model parameters in different Tmax scenarios of TQT study design. Models 1, 2, and 3 represent linear mixed effects models 1, 2, and 3, respectively, and the biological model represents a reduced model consisting of only one cosine function. Boxes denote the 25th and 75th percentiles and the filled circles inside the box denotes the median. Whiskers represent the 5th and 95th percentiles.
Table 2.
Biases (mean estimation error) of slope estimates (in %) and ddQTc estimates (in ms) by study design in 5ms QTc prolongation case
| Model | Tmax scenarios | ||||||
|---|---|---|---|---|---|---|---|
| Short Tmax | Intermediate Tmax | Long Tmax | Intermediate Tmax without measurement error | ||||
| Bias in slope (%) | Worst bias in ddQTc (ms) | Bias in slope (%) | Worst bias in ddQTc (ms) | Bias in slope (%) | Worst bias in ddQTc (ms) | Bias in slope (%) | |
| TQT study | |||||||
| 1 | −4.69 | −0.24 | −12.0 | −0.60 | −11.8 | −0.58 | −7.67 |
| 2 | −4.51 | −0.23 | −5.70 | −0.29 | −5.44 | −0.27 | −0.37 |
| 3 | −14.3 | −0.73 | −12.9 | −0.65 | −9.12 | −0.45 | −0.27 |
| Reduced biologicala | −3.87 | NE | −7.15 | NE | −5.94 | NE | −2.89 |
| Full biologicalb | −4.36 | NE | −6.81 | NE | −5.72 | NE | −2.10 |
| Phase I study | |||||||
| 1 | −24.9 | −1.00 | −49.7 | −1.98 | −40.0 | −1.58 | −45.8 |
| 2 | −11.8 | −0.48 | −14.0 | −0.55 | −18.7 | −0.74 | 0.103 |
| 3 | −12.6 | −0.51 | −14.9 | −0.59 | −20.8 | −0.82 | 0.652 |
| Phase I study + day 0 | |||||||
| 1 | −16.7 | −0.68 | −27.8 | −1.10 | −17.4 | −0.68 | NE |
| 2 | −10.0 | −0.41 | −8.80 | −0.35 | −10.8 | −0.42 | NE |
| 3 | −10.5 | −0.43 | −10.3 | −0.41 | −11.7 | −0.46 | NE |
NE, not evaluated.
Reduced biological model consist of one cosine function as a biological part.
All the converged runs were included regardless of successful covariance step.
Figure 2.

Coverage probability of slope estimates and false positive slopes. (a) Coverage probability of slope estimates containing the true slope calculated with 90% confidence intervals of slope estimates in 5 ms QTc prolongation scenario and (b) % false positive slopes of linear mixed effects models in the no QTc prolongation scenario (95% confidence interval excludes 0). Dashed line represents the 2.5% type 1 error.
For the pooled phase I study design, slope attenuation was more pronounced (<−10.0%) compared with the TQT study design, as indicated in Figure 3 and Table 2. The model 1 showed the most severe bias (−24.9% – −49.7%) and poor coverage of the true slope. Model 2 yielded the least biased slope estimates (−11.8% – −18.7% vs. −12.6% – −49.7%, Table 2) and reasonable coverage of true slope in all the Tmax scenarios (Figure 2a) despite its bias. Adding the autoregressive residual error component did not improve the prediction results in phase I designs as well. Because of the sparse sampling, BSV on slope could not be estimated and was fixed to zero for most runs, resulting in a skewed distribution as depicted in Figure 3. Residual variability increased for all LME models to a similar extent observed with the TQT study design (24.5%–35.5% bias, Figure 3). Neither the full nor reduced biological models were able to be applied to the phase I design (<8% successful covariance step), because of sparse sampling.
Figure 3.

Box and whisker plots of the estimation errors for drug effect model parameters in different Tmax scenarios of phase 1 study design. Models 1, 2, and 3 represent linear mixed effects models 1, 2, and 3, respectively. Boxes denote the 25th and 75th percentiles and closed circle inside the box denotes the median. Whiskers represent the 5th and 95th percentiles.
As shown, slope attenuation for the LME models is more pronounced for phase I studies compared with a TQT study, and model 2 resulted in the most accurate slope estimates for the short Tmax case among different Tmax scenarios. Compared with the TQT study, the phase I design in our analysis had (1) very limited samples per subject (n = 6 for single ascending dose and 9 for multiple ascending dose vs. 12 for TQT), (2) a smaller dose range (20–200 mg compared with 25 and 250 mg in TQT), (3) did not have time-matched baselines, and (4) was a parallel design (placebo was not given to all subjects). To evaluate these factors, an increased number of subjects (n = 104), number of observations per subject (n = 6 for single ascending dose and 15 for multiple ascending dose), and dose range (n = 20–300 mg) were tested. However, none of the conditions improved the accuracy of slope estimates (−13.6% – −16.8% bias for LME model 2, data not shown). Inclusion of a higher dose level only increased the precision of slope estimates (data not shown). However, when day 0 or baseline ECG sampling was included (phase 1 + day 0), the accuracy of slope estimate was significantly improved, especially for the long Tmax scenario (Table 2). Also, the coverage probability of the true slope was less sensitive to the selection of LME models when day 0 was included (Figure 2a). A two-way crossover, placebo-controlled phase 1 design was additionally investigated for testing sensitivity, and it further improved the accuracy of slope estimates with ≤−6.00% bias, a similar level of bias to the TQT study (data not shown). The simulation results were not affected by the magnitude of the slope (Supplement Table S1, which is available online). Thus, these results should apply to larger slopes that are of interest based on the Consortium for Innovation and Quality in Pharmaceutical Development and the Cardiac Safety Research Consortium proposals for phase I.6
For the no QTc prolongation case, absolute bias was reported (Table 3). There was no obvious bias observed across the LME models and this is because measurement error is not an issue when the true slope is 0. The false positive rate and 95% CIs of the slope estimates are displayed in Figure 2b. The results indicated that the false positive rate was not sensitive to the selection of LME models nor study design, having ≈ 2.5% error in most of the Tmax cases. The false positive rate was relatively high for the TQT short Tmax scenario (6%–8%), but the 95% CI lower bound was very small, approximately 10−6 – 10−4, for those false positive slopes. TQT study seems to underestimate the variability of slope, thereby resulting in tighter 95% CI and higher % false positive slopes.
Table 3.
Biases of slope estimates (in %) by study design in no QTc prolongation case
| Model | Tmax scenarios (bias in slopea, %) | ||
|---|---|---|---|
| Short Tmax | Intermediate Tmax | Long Tmax | |
| TQT study | |||
| 1 | −0.00493 | −0.0480 | −0.111 |
| 2 | −0.000685 | −0.00331 | −0.00163 |
| 3 | 0.000107 | −0.00186 | −0.000312 |
| Phase I study | |||
| 1 | −0.0579 | −0.268 | −0.428 |
| 2 | −0.000212 | −0.0190 | 0.0392 |
| 3 | 0.00271 | −0.0197 | 0.0394 |
| Phase I study + day 0 | |||
| 1 | −0.0323 | −0.134 | −0.183 |
| 2 | 0.00434 | 0.0149 | 0.0186 |
| 3 | 0.00449 | 0.0180 | 0.0206 |
Since true slope is zero, mean estimation error could not be calculated. Sum of (estimate - true) × 100 was reported instead.
When the 20% measurement error in PK was removed (evaluated using the intermediate Tmax scenario), accuracy of the slope estimate improved remarkably (< 1.00% bias) overall (Table 2). Only the simple LME model 1 without time factor still yielded negatively biased slope estimates. The negative bias in the slope estimates of LME models having time as a factor variable (model 2) resulted mainly due to measurement errors in the PK and not model misspecification.
Prediction of drug-induced QTc prolongation
Both the best LME model and biological model resulted in −3.87% – −7.15% bias of the slope estimates in the presence of 20% measurement error. Slope attenuation in LME models is a well-known issue when the independent variable includes measurement error. However, such bias may not be directly translated into the prediction of QTc prolongation, which is the main purpose of CQTc analysis. Therefore, the true ddQTc were compared with model-predicted ddQTc for the highest dose group of each study type (250 mg for TQT and 200 mg for phase 1). The comparisons are summarized by time (Figure 4). The worst bias among ddQTc estimates across time is reported in Table 2. As shown in Figure 4, 75% of the prediction errors between the true and model-predicted ddQTc were within 1 ms (i.e., in the interquartile range), for both the TQT and phase I + day 0 study designs. There was more variability in the phase I study results compared with those from the TQT study, but the accuracy and precision were significantly improved when day 0 was included. Overall, the worst biases were small, less than 1 ms across all the Tmax scenarios for phase I and less than 0.5 ms for the TQT and phase I + day 0 designs (Table 2).
Figure 4.

Box and whisker plots of the estimation errors for ddQTc in different Tmax scenarios of both TQT and phase 1 studies. Linear mixed effect model 2 was selected as the best model and difference between true ddQTc (true concentration × true slope) and model-predicted ddQTc (observed concentration x slope estimates) were calculated in ms. Boxes denote the 25th and 75th percentiles and closed circle inside the box denotes the median. Whiskers represent the 5th and 95th percentiles.
DISCUSSION
The present study evaluated the performance of LME models for assessing drug-induced QTc prolongation in the presence of physiologically induced QTc variability. The results indicated that the simpler LME model with time as a factor variable provided similar accuracy of the CQTc slope estimates to the complex biological or reduced biological models, despite its empirical nature (Table 2). Additionally, the LME model was able to predict accurately the drug-induced QTc prolongation (ddQTc), with less than 1 ms bias, even in the presence of a −4.51% – −18.7% bias in slope estimates.
CQTc analysis with true concentrations confirmed that bias in LME slope estimates were induced by measurement error, not by model misspecification. The slope in a linear model is known to be attenuated when an independent variable includes measurement error, because it is estimated with regression error.16 Bonate evaluated this, and reported that significant bias (<−10%) only occurred in the presence of assay measurement errors greater than 30%–40%.17 In contrast with that study, multiplicative measurement error, which is more likely in analytical assays, was used in our analysis. A magnitude of 20% was selected, because the FDA has set the limit of precision in analytical method to 15% coefficient of variation, allowing a 20% coefficient of variation for the lower limit of quantification.18 Our simulations showed slope attenuation was small (−4.51% – −5.70% in model 2) for all the Tmax scenarios of a TQT study design, which is in agreement with the work by Bonate et al. On the other hand, the extent of slope attenuation was more significant for phase I study designs and depended on the PK characteristics of the drug, observing greater bias in long Tmax scenario. Several methods have been proposed to correct for this bias.16,19–21 However, these require several unprovable assumptions, e.g., establishing a relationship between true independent variables and their observed errors. Also, methods requiring simulation-extrapolation as proposed by Cook et al.20,21 can be time consuming. Considering that the bias in ddQTc estimates from LME model is at most −0.74 ms for the long Tmax scenario, slope attenuation caused by 20% measurement error is not likely an issue for predicting QTc prolongation of a drug.
The reason that the ddQTc estimates were reasonable despite the bias in slope is that slope estimates are calibrated for the measurement error of observed concentrations.16 The predictive property of slope estimated in the presence of measurement errors is different when we use summary statistic of Cmax for ddQTc predictions, because mean Cmax does not include the same level of measurement errors as observed concentrations. Therefore, we did not consider inference on mean Cmax to test QTc prolongation of a drug here. One can instead look at the upper bound of the 1-sided 95% CI of predicted ddQTc across observed concentrations and find the concentration value at which certain levels of ddQTc increase are precluded. This is also helpful when unanticipated concentration increases exist, e.g., drug–drug interactions or genomic predisposition.
The stochastic approximation expectation maximization method with importance sampling as implemented in NONMEM was used to obtain the estimates of standard errors for the biological models in our analysis, because of numerical difficulties with the first-order conditional estimation method. The performance of stochastic approximation expectation maximization method was sensitive to the initial conditions as reported by Plan et al.,22 and only when we set initial values as the true values, could we get good convergence rates (>99%) for the TQT study design. However, the two-cosine model still suffered from over-parameterization, likely due to sparse sampling between 12 and 24 hours in our design. Our sampling scheme is reduced compared with the original study,13 yet is more consistent with standard TQT study designs. A reduced biological model having one cosine function was investigated as a substitute to the full model to improve such outcomes. Poor convergence rates and the need to modify the model based on a design are not conducive to rigorous inference, which is what is typically sought in QTc analyses. Prespecification of the model is ideal. However, such issues lead to difficulty in model prespecification (or confidence therein) or deviation from prespecified models resulting from poor convergence rates or overparameterization. As investigators make subjective changes to the model to avoid these issues, the robustness of the inference becomes increasingly suspect. The LME approach considered here is easily prespecified, does not suffer from poor convergence and can make similarly informed decisions about QTc prolongation as the biological model. The linear model also is easily implemented in phase I designs as discussed below.
Several phase I design modifications were investigated to improve performance — aiming to approach the TQT study results. The dose range, sample size, and number of samples per subject were relatively small in our analysis compared with typical First In Human (FIH) study designs to evaluate a worst case setting. Changing these did not significantly improve the results. However, when day 0 was included, the slope attenuation was significantly improved, having ≤ −10.8% bias in all the Tmax scenarios for the best LME model. It appears that including ECGs within a subject where the concentration was known to be 0 improved the accuracy. In this regard, the short Tmax case outcomes were as good as those for the phase I + day 0 study, because the PK profile contained more low concentration data (at later time points).
Model-based CQTc analysis of phase I data has been increasingly considered to detect QT prolongation of a drug as an alternative approach of TQT study. A successful outcome of the recent Consortium for Innovation and Quality in Pharmaceutical Development and the Cardiac Safety Research Consortium collaboration can add more evidence to obviate a need for TQT study for all NCEs, using CQTc assessment in early phase study. However, one criticism of the CQTc analysis is that the model selection criteria is not prospectively defined; rather the model is retrospectively determined by the observed data.6,9 In this regard, this current study may guide several points needed to be considered in establishing CQTc models that can be prespecified. Overall, the LME model with fixed effects for day and time to account for circadian rhythm resulted in the least biased estimates of the drug effect parameter and drug-induced QTc prolongation across all study designs with different PK characteristics. For a placebo-controlled crossover design, like a TQT study, biases of LME slope estimates were small (≤−5.70%), and accuracies of predicted QTc prolongations were acceptable in all the Tmax scenarios. For parallel group designs, the magnitude of slope estimate in CQTc linear model should be more carefully interpreted especially for long Tmax drugs, which do not include very low concentrations in their PK profile. When a placebo-controlled design cannot be implemented, including day 0 can improve the accuracy and precision of the slope parameter and QTc prolongation predictions.
METHODS
PK sata simulation
Concentrations for a hypothetical drug were simulated using a one compartment model with a typical clearance of 40 L/h, an apparent volume of distribution of 125 L and three different rate constants for absorption, 0.07 (long Tmax, i.e., 6 hours), 0.3 (intermediate Tmax, i.e., 3 hours), and 2 hours−1 (short Tmax, i.e., 1 hour). BSV of 33% was incorporated on all the population PK parameters (i.e., ). To account for measurement errors in drug concentrations, normally distributed residual errors with mean 0 and standard deviation of 20% were incorporated using a proportional error structure (i.e.,
). To account for measurement errors in drug concentrations, normally distributed residual errors with mean 0 and standard deviation of 20% were incorporated using a proportional error structure (i.e., ).
).
Table 1 describes the study designs used in the analysis. The phase I studies considered four dose cohorts consisting of five subjects on active treatment and two on placebo (28 subjects in total). In the multiple ascending dose study, treatments were given twice daily for 6 days. Steady state was achieved on the third day of dosing.
Pharmacodynamic data simulation
The CQTc model for simulation was based on a published biological QTc model13 with an additive concentration effect component. It was assumed that concentration and QTc exhibit a linear relationship because most of the observed concentrations are far less than the EC50. The drug effect was simulated in three different scenarios, no QTc prolongation (true slope of 0 ms/ng/mL), 5 ms prolongation (true slope of 0.00365, 0.0072, and 0.0172 ms/ng/mL for short, intermediate, and long Tmax respectively), and 10 ms prolongation at geometric mean Cmax of supratherapeutic dose (2 times the slopes above). The BSV on slope was assumed to be 33%. Mathematical expression of the pharmacodynamic model is the following:
 |
where QTc0 (378 ms for male and 395 ms for female) represents a typical baseline QTc interval (ms), An (0.0052 for A1 and 0.01 for A2) represents amplitudes (ms), ϕn (20.5 for ϕ1 and 14.2 for ϕ2) represents circadian rhythm phases (h) and t represents the clock time (h). Cp(t) was the true concentration, but observed concentration was used during estimation. η1, η2, η3, η4, and η7 are BSV which were normally distributed with mean 0 and 4.2, 69, 33, 38, and 33% coefficient of variation, respectively. BSV on ϕ2 (η5) was fixed to 0 and interoccasion variability of η6 was set to 44% coefficient of variation. The residual error, ε was normally distributed with mean 0 and standard deviation of 4 ms. An additive residual error model was used here, despite the proportional error used in the original publication, because the additive error assumption is more common in CQTc analyses. All the random effects were assumed to be uncorrelated.
CQTc model development
Three different LME models were considered. To address fluctuations in the mean due to circadian rhythms, fixed effects for day and sampling time were added as factor variables (model 2) to the simplest LME (model 1). Because the fixed effects might not capture the pattern completely, an autoregressive error structure for residual variance was incorporated additionally to mitigate the potential effects of correlated residuals (model 3). Random effects were incorporated on the intercept and slope using a diagonal covariance matrix as shown in the following equation. Random effects on day were not incorporated because the BSV estimates were nearly 0.
 |
where QTcijk represents a QTc response, i indexes the subject, j indexes day, and k indexes extraction time within day. The parameter βμ represents the baseline response, βF represents the baseline effect of females, βDj represents the day effect, and βTk represents the kth nominal time effect within a day. Bracketed expressions represent indicator functions which = 1 if the condition in brackets is true and = 0 otherwise. In model 3, σ2 is the residual variance, ρ represents the correlation parameter and s is the absolute difference between nominal times. Model parameters were estimated using maximum likelihood as implemented in the lme function in R version 3.0.1. (R Project for Statistical Computing).
The performance of the biological CQTc model was also evaluated. Because the data did not support the full biological model, a reduced biological model consisting of one cosine function was also fitted. The biological model parameters were estimated using stochastic approximation expectation maximization followed by importance sampling as implemented in NONMEM 7.3.0 (ICON Development Solutions).
Performance Comparison
The bias in drug effect parameters such as slope, BSV on slope, and residual variance were evaluated as a mean of percent estimation error:
 |
To evaluate the predictability of QTc prolongation, bias in model-predicted ddQTc was reported as a mean of estimation error in ms at sampling time t:
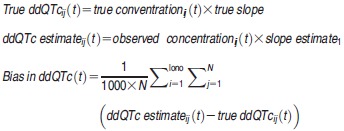 |
where i represents ith dataset and j represents jth individual. Coverage probability of the 90% CIs of the slope estimates for the true slope have also been calculated. In the case of no QTc prolongation, false positive slope rates (exclusion of 0) were calculated using 95% CIs of the slope estimates.
Author Contributions
Y.H. and M.M.H. wrote the manuscript. Y.H. and M.M.H. designed the research. Y.H. performed the research and analyzed the data.
Conflict of Interest
The authors declared no conflict of interest.
Study Highlights
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
✓ QT intervals exhibit biological rhythms and oscillatory functions have been proposed to characterize the patterns in Concentration-QTc (CQTc) modeling. However, fitting such functions is not always feasible because it requires sufficient ECG sampling, which is unlikely in early phase design.
WHAT QUESTION DOES THIS STUDY ADDRESS?
✓ This study assessed whether linear mixed-effect (LME) models accurately and precisely estimate CQTc slope and drug-induced QTc prolongation (ddQTc), and ultimately evaluated if a biological model (oscillatory functions) is necessary for decision making.
WHAT THIS STUDY ADDS TO OUR KNOWLEDGE
✓ The simpler LME model with time as a factor variable provided similar accuracy of the CQTc slope to the biological model and accurately predicted ddQTc with less than 1 ms bias.
HOW THIS MIGHT CHANGE CLINICAL PHARMACOLOGY AND THERAPEUTICS
✓ Even though CQTc analysis of phase 1 data has been increasingly considered, there was no consensus on the modeling strategy yet. This study proposed LME approach which is easily prespecified, does not have poor convergence issue, and achieves little bias in ddQTc estimates.
Supporting Information
Supplementary information accompanies this paper on the CPT: Pharmacometrics & Systems Pharmacology website (http://www.wileyonlinelibrary.com/psp4)
Supporting Information
Supporting Information
Supporting Information
References
- Fenichel RR, et al. Drug-induced torsades de pointes and implications for drug development. J. Cardiovasc. Electrophysiol. 2004;15:475–495. doi: 10.1046/j.1540-8167.2004.03534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Conference on Harmonisation. Guidance on E14 clinical evaluation of QT/QTc interval prolongation and proarrhythmic potential for non-antiarrhythmic drugs; availability. Notice. Fed. Regist. 2005;70:61134–1135. [PubMed] [Google Scholar]
- Florian JA, Tornoe CW, Brundage R, Parekh A. Garnett CE. Population pharmacokinetic and concentration—QTc models for moxifloxacin: pooled analysis of 20 thorough QT studies. J. Clin. Pharmacol. 2011;51:1152–1162. doi: 10.1177/0091270010381498. [DOI] [PubMed] [Google Scholar]
- Boos DD, Hoffman D, Kringle R. Zhang J. New confidence bounds for QT studies. Stat. Med. 2007;26:3801–3817. doi: 10.1002/sim.2826. [DOI] [PubMed] [Google Scholar]
- Hutmacher MM, Chapel S, Agin MA, Fleishaker JC. Lalonde RL. Performance characteristics for some typical QT study designs under the ICH E-14 guidance. J. Clin. Pharmacol. 2008;48:215–224. doi: 10.1177/0091270007311921. [DOI] [PubMed] [Google Scholar]
- Darpo B, et al. The IQ-CSRC Prospective Clinical Phase 1 Study: “Can early QT assessment using exposure response analysis replace the thorough QT study?”. Ann. Noninvasive Electrocardiol. 2014;19:70–81. doi: 10.1111/anec.12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah RR. Morganroth J. Early investigation of QTc liability: the role of multiple ascending dose (MAD) study. Drug Saf. 2012;35:695–709. doi: 10.1007/BF03261967. [DOI] [PubMed] [Google Scholar]
- Rohatagi S, Carrothers TJ, Kuwabara-Wagg J. Khariton T. Is a thorough QTc study necessary? The role of modeling and simulation in evaluating the QTc prolongation potential of drugs. J. Clin. Pharmacol. 2009;49:1284–1296. doi: 10.1177/0091270009341184. [DOI] [PubMed] [Google Scholar]
- Darpo B. Garnett C. Early QT assessment—how can our confidence in the data be improved? Br. J. Clin. Pharmacol. 2013;76:642–648. doi: 10.1111/bcp.12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrovsky V. Pharmacokinetic-pharmacodynamic modeling in the data analysis and interpretation of drug-induced QT/QTc prolongation. AAPS J. 2005;7:E609–E624. doi: 10.1208/aapsj070363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smetana P, Batchvarov V, Hnatkova K, Camm AJ. Malik M. Circadian rhythm of the corrected QT interval: impact of different heart rate correction models. Pacing Clin. Electrophysiol. 2003;26(Pt 2):383–386. doi: 10.1046/j.1460-9592.2003.00054.x. [DOI] [PubMed] [Google Scholar]
- Malik M, Hnatkova K, Schmidt A. Smetana P. Accurately measured and properly heart-rate corrected QTc intervals show little daytime variability. Heart Rhythm. 2008;5:1424–1431. doi: 10.1016/j.hrthm.2008.07.023. [DOI] [PubMed] [Google Scholar]
- Grosjean P. Urien S. Moxifloxacin versus placebo modeling of the QT interval. J. Pharmacokinet. Pharmacodyn. 2012;39:205–215. doi: 10.1007/s10928-012-9242-8. [DOI] [PubMed] [Google Scholar]
- Chain AS, Krudys KM, Danhof M. Della Pasqua O. Assessing the probability of drug-induced QTc-interval prolongation during clinical drug development. Clin. Pharmacol. Ther. 2011;90:867–875. doi: 10.1038/clpt.2011.202. [DOI] [PubMed] [Google Scholar]
- Florian J, Garnett CE, Nallani SC, Rappaport BA. Throckmorton DC. A modeling and simulation approach to characterize methadone QT prolongation using pooled data from five clinical trials in MMT patients. Clin. Pharmacol. Ther. 2012;91:666–672. doi: 10.1038/clpt.2011.273. [DOI] [PubMed] [Google Scholar]
- Carroll RJ, Ruppert D, Stefanski LA. Crainiceanu C. Measurement Error in Nonlinear Models: A Modern Perspective. 2nd. New York: Chapman & Hall; 2006. [Google Scholar]
- Bonate PL. Effect of assay measurement error on parameter estimation in concentration-QTc interval modeling. Pharm. Stat. 2013;12:156–164. doi: 10.1002/pst.1567. [DOI] [PubMed] [Google Scholar]
- U.S. Department of Health and Human Services FaDA. 2001. Center for Drug Evaluation and Research (CDER), Center for Veterinary Medicine (CVM). Guidance for Industry: Bioanalytical Method Validation ) [DOI] [PubMed]
- Stefanski LA. Measurement error models. J. Am. Stat. Assoc. 2000;95:1353–1358. [Google Scholar]
- Cook JR. Stefanski LA. Simulation-extrapolation estimation in parametric measurement error models. J. Am. Stat. Assoc. 1994;89:1314–1328. [Google Scholar]
- Stefanski LA. Cook JR. Simulation-extrapolation: the measurement error jackknife. J. Am. Stat. Assoc. 1995;90:1247–1256. [Google Scholar]
- Plan EL, Maloney A, Mentre F, Karlsson MO. Bertrand J. Performance comparison of various maximum likelihood nonlinear mixed-effects estimation methods for dose-response models. AAPS J. 2012;14:420–432. doi: 10.1208/s12248-012-9349-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Supporting Information
Supporting Information


