Abstract
Long-term potentiation (LTP) is a form of synaptic plasticity that is an excellent model for the molecular mechanisms that underlie memory. LTP, like memory, is persistent, and both are widely believed to be maintained by a coordinated genomic response. Recently, a novel class of non-coding RNA, microRNA, has been implicated in the regulation of LTP. MicroRNA negatively regulate protein synthesis by binding to specific messenger RNA response elements. The aim of this review is to summarize experimental evidence for the proposal that microRNA play a major role in the regulation of LTP. We discuss a growing body of research which indicates that specific microRNA regulate synaptic proteins relevant to LTP maintenance, as well as studies that have reported differential expression of microRNA in response to LTP induction. We conclude that microRNA are ideally suited to contribute to the regulation of LTP-related gene expression; microRNA are pleiotropic, synaptically located, tightly regulated, and function in response to synaptic activity. The potential impact of microRNA on LTP maintenance as regulators of gene expression is enormous.
Keywords: long-term potentiation, microRNA, maintenance, synaptic plasticity, memory
INTRODUCTION
Long-term potentiation (LTP) is a form of synaptic plasticity whereby high frequency stimulation (HFS) induces a long-lasting enhancement of synaptic transmission. LTP is widely accepted as an excellent model for the molecular mechanisms that mediate long-term information storage in the brain. Many of its key properties are analogous to those of long-term memory, including input specificity, rapid induction, and co-operativity (Abraham and Williams, 2008). Most importantly, LTP is persistent: indeed an in vivo study suggests that LTP can last for at least a year after induction (Abraham et al., 2002). Although the potential for persistent LTP was noted in the earliest studies (Bliss and Gardner-Medwin, 1973), we still do not understand how the mechanisms underlying the stabilization of synaptic change allow LTP to persist for periods of days or weeks.
MicroRNA (miRNA) are endogenous non-coding RNA that act as post-transcriptional inhibitors of protein synthesis. They function by base-pairing with miRNA response elements (MREs) located in target messenger RNA (mRNA). This occurs within the ribonuclear protein complex known as the RNA-induced silencing complex (RISC; Kawamata and Tomari, 2010). To date, 2588 unique mature human miRNA have been annotated (miRBase 21). MiRNA are predicted to regulate the activity of more than 60% of human protein-coding genes (Friedman et al., 2009), although others predict far fewer in humans (30%; Lewis et al., 2005) and in Caenorhabditis elegans (10%; Lall et al., 2006). Even considering the most conservative estimates, the potential impact of miRNA activity on protein expression is profound. MiRNA are present in all body tissues (Lee et al., 2008), and their stability in circulating fluids suggests that they play an important role in cell–cell communication (Chen et al., 2012) and have utility as biomarkers of disease (Etheridge et al., 2011). Further, specific miRNA are crucial for development and function of both neurons and glia (Sayed and Abdellatif, 2011) and miRNA dysfunction is associated with neurodegenerative diseases, including Alzheimer’s disease (AD), which is characterized by memory impairment (Delay et al., 2012; Kim et al., 2014).
While recent reviews have detailed miRNA regulation at synapses (Siegel et al., 2011; Sim et al., 2014), their potential role as biomarkers of neurological disease (Sheinerman and Umansky, 2013) and highlighted the involvement of miRNA in a wide range of synaptic processes, from neurotransmission (Higa et al., 2014) to morphology (McNeill and Van Vactor, 2012), miRNA function in LTP has not been addressed specifically. To address this gap in the literature, this review will summarize experimental evidence for the proposal that miRNA play a major role in the regulation of LTP.
LONG-TERM POTENTIATION
LTP is not a unitary phenomenon: multiple forms exist distinguished by stimulation paradigm, experimental preparation and brain region. While LTP can be induced in the visual cortex and a variety of cortical and subcortical structures (Abraham et al., 2002; Dityatev and Bolshakov, 2005; Zhao et al., 2005; Cooke and Bear, 2014), the majority of LTP research focuses on the hippocampus, a memory-related structure with a robust circuitry that lends itself well to experimentation. Here, LTP can be triggered not only by electrical stimulation (e.g., theta-burst, delta-burst), activation of metabotropic glutamate receptors (mGluRs) and brain-derived neurotrophic factor/tropomyosin receptor kinase B (BDNF/TrkB) signaling (Bortolotto et al., 1994; Balschun et al., 1999; Huang et al., 2013; Schildt et al., 2013) but by learning itself (Whitlock et al., 2006). This, alongside the remarkable persistence of LTP at hippocampal perforant path-dentate gyrus synapses, provides compelling evidence for the involvement of LTP-type plasticity in memory function.
Long-term potentiation induced at perforant path synapses in awake freely moving animals consists of three temporally and mechanistically distinct canonical phases: LTP1, LTP2, and LTP3 (Abraham and Otani, 1991). LTP1 refers to the initial, rapid strengthening of synapses, independent of protein synthesis and lasting for a few hours at most (Racine et al., 1983; Abraham and Otani, 1991). The putative cellular mechanisms that underlie LTP1, including enhanced release of neurotransmitter (Bayazitov et al., 2007), protein kinase activation (Sacktor et al., 1993; Lisman et al., 2012), and trafficking of both α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) and N-methyl-D-aspartate (NMDA) subtypes of glutamate receptors to synapses (Williams et al., 1998; Hayashi et al., 2000; Shi et al., 2001), are likely to only explain short term synaptic strengthening. In contrast, the latter two phases require new protein synthesis. LTP2, the intermediate phase, requires synaptically localized protein synthesis but not new gene transcription, and has an average decay time constant of a few days (Otani et al., 1989; Kang and Schuman, 1996). However, as dendrites cannot maintain LTP for long periods of time when physically separated from their cell bodies (Kang and Schuman, 1996), translation of extant mRNA alone cannot explain LTP stabilization. The most long-lasting phase, LTP3, is dependent on new gene transcription as well as translation and can last for periods of weeks when induced at perforant path synapses (Nguyen and Kandel, 1996; Abraham et al., 2002). Here, the gene expression changes that accompany LTP maintenance have been intensively studied (Abraham and Williams, 2003).
Indeed, recent microarray studies have identified many LTP-induced, co-regulated genes, supporting the concept that LTP persistence involves regulation of coordinated gene networks (Lee et al., 2005; Park et al., 2006; Wibrand et al., 2006; Havik et al., 2007). Our recent studies have shown that the LTP-related transcriptional response is not limited to periods of minutes following LTP but extends to hours and days (Ryan et al., 2011, 2012). Bioinformatic analysis of this data set using Ingenuity Pathway Analysis (IPA) algorithms predicts this ongoing transcriptional response contributes both to dynamic alteration of synapses, through regulation of calcium dynamics, protein kinases, and synaptogenesis, and higher level regulation of gene expression. Intriguingly, this analysis predicted that miRNA functioned as regulatory hubs in these networks. This result suggests that complex processes such as LTP, involving the coordinated regulation of gene networks, may require fine-tuning of protein synthesis by miRNA, acting alongside other translational regulators (Bramham and Wells, 2007; Sossin and Lacaille, 2010; Jung et al., 2014).
MicroRNA BIOGENESIS AND FUNCTION: KEY CHARACTERISTICS RELEVANT TO LTP
We propose that miRNA are ideally suited to control the rapid, coordinated and region-specific translation that underlies LTP maintenance. The characteristics of miRNA biogenesis and function that are of particular interest in this regard are tight spatial and temporal control of miRNA expression, their ability to function in a combinatorial manner with other miRNA, and the ability of a single miRNA to coordinate the expression of many mRNA related by their MRE sequences.
microRNA function
Mature miRNA range from 15 to 34 nucleotides in length and their average length is 22 nucleotides (miRBase 21). The functional impact of miRNA binding depends primarily on the thermo-stability of base-pairing between the target mRNA MRE and the miRNA 5′-seed sequence (nucleotides 2–8; Bartel, 2009); the 3′ region of the miRNA affects function to a lesser degree (Doench and Sharp, 2004). MiRNA usually interact with MREs in the 3′-untranslated region (UTR) of target mRNA (Bartel, 2009); yet the 5′-UTR and protein-coding regions can also contain functional MREs (Lytle et al., 2007; Forman et al., 2008), though the effect may be marginal (Hafner et al., 2010). In animals, complementarity between miRNA and their mRNA targets is typically imperfect (Bartel, 2004); this creates an internal bulge structure that distorts the helix, thereby preventing mRNA cleavage but allowing translational repression (Zeng et al., 2003; Rana, 2007). The exact mechanisms of RISC-induced translational repression are still under debate (reviewed by Morozova et al., 2012), however, translational repression is generally coupled ultimately with mRNA degradation via deadenylation (Eulalio et al., 2009) and decapping (Behm-Ansmant et al., 2006). Infrequently, imperfect miRNA:mRNA base-pairing can cause translational repression without mRNA degradation, which may be reversible (Bhattacharyya et al., 2006; Schratt et al., 2006). In contrast, perfect or near-perfect base-pairing can result in mRNA degradation via endonucleolytic cleavage, but this is very rare in animals (Yekta et al., 2004). Intriguingly, miRNA have been shown to up-regulate translation under some circumstances in mammals (Vasudevan et al., 2007; Orom et al., 2008).
microRNA biogenesis
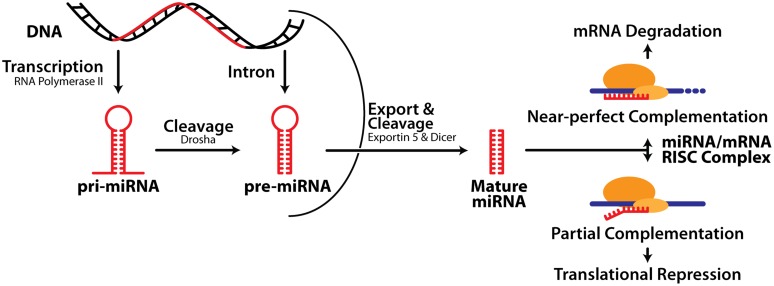
MiRNA biogenesis consists of three major processes: transcription of a much longer primary transcript (pri-miRNA), followed by two consecutive cleavage events initiated by the RNase III enzymes Drosha and Dicer, which generate a precursor miRNA (pre-miRNA) and ultimately liberate the mature miRNA (Figure 1). Note that both the 5′ or 3′ strands of the miRNA hairpin can release a functional miRNA and these are denoted as, e.g., miR-132-5p or miR-132-3p respectively. Precise control of miRNA expression can be modulated at multiple points throughout this pathway (Thomson et al., 2006). In addition, pri- and pre-miRNA can be modified via RNA editing, affecting their activity (Peng et al., 2012). There is also evidence that turnover of some neuronal miRNA is activity-dependent (Krol et al., 2010a), and tightly regulated (reviewed in Krol et al., 2010b), although this is less well understood than miRNA biogenesis.
FIGURE 1.
MicroRNA biogenesis and mechanism of action. See text for details.
Tight spatial and temporal control
As miRNA are functional immediately after transcription and processing, they can respond more rapidly to external stimuli than transcription factors, which require translation into protein, transportation to the nucleus, and often post-translational modification to regulate gene expression. Tight spatial and temporal control of miRNA activity could allow miRNA to regulate translation selectively at individual synapses. This hypothesis is supported by the observation that both pri- and pre-miRNA, as well as their cleavage partners Drosha, DGCR8 and Dicer, are located at mouse hippocampal post-synaptic densities (PSDs; Lugli et al., 2005, 2012). Under normal conditions, Dicer was shown to be inactive in the PSD; however, following synaptic activation, the release of calcium caused calpain, a proteolytic enzyme, to activate Dicer, thereby triggering the conversion of pre-miRNA into active, mature miRNA (Lugli et al., 2005). Furthermore, pri-miRNA were found to be highly enriched with RNA transport granules, suggesting they can be transported alongside mRNA and processed at the PSD (Lugli et al., 2012). These important studies suggest that the expression of mature miRNA can be locally regulated by activity, providing a possible mechanism for translational regulation specific to activated synapses, thus contributing to the input specificity of LTP.
microRNA are highly pleiotropic
Individual miRNA can coordinately regulate the translation of many mRNA that share the same or similar MRE sequences, as complete complementation is not required for miRNA activity (Krek et al., 2005). Indeed, Selbach et al. (2008) demonstrated that one miRNA can not only promote degradation of hundreds of mRNA transcripts, but also repress the production of hundreds of additional proteins at the level of translation. Furthermore, the short sequences involved in miRNA:mRNA interactions make them well-suited for combinatorial effects with other miRNA or RNA-binding proteins that associate with separate sites on the same target mRNA (Krek et al., 2005). There is experimental evidence that multiple miRNA regulate the expression of the activity related cytoskeletal protein (Arc); in some cases in an additive manner (Wibrand et al., 2012). Additionally, there is evidence that miRNA act co-operatively with fragile X mental retardation protein (FMRP), an RNA-binding protein, known to regulate the translation of plasticity-associated mRNA (Xu et al., 2008). FMRP inhibits translation of Arc and CaMKIIα (calcium/calmodulin-dependent protein kinase IIα; Bassell and Warren, 2008) and Fmr1 knockout mice exhibit impaired LTP at CA1 synapses (Lauterborn et al., 2007). FMRP is also associated with a number of miRNA in the mouse brain, including miR-132-3p, miR-125b-5p, miR-138-5p, and miR-124-3p (Edbauer et al., 2010), the last mediated by the FMRP Drosophila homolog dFMR1 to inhibit dendritic arbor (Xu et al., 2008). Furthermore, mouse models of fragile X syndrome showed regulation of miR-9a-5p and miR-124-3p by the FMRP family protein, FXR1P, which forms a complex with Dicer and affects miRNA processing (Xu et al., 2011). Interestingly, FXR1P has been found to be subjected to miRNA inhibition, creating a feedback loop that is becoming more evident in gene networks (Cheever et al., 2010). These feedback loops imply a homeostatic role for miRNA. It is noteworthy that many miRNA targets are regulatory genes such as transcription factors (Tsang et al., 2007) thus extending the range of genes regulated by one miRNA beyond those that it interacts with directly.
MicroRNA ARE REGULATED IN RESPONSE TO LONG-TERM POTENTIATION
If miRNA regulate LTP-related protein synthesis, we would expect to see a change in the level of active, mature miRNA in response to LTP induction. LTP induced in vitro has been shown to regulate miRNA levels (Park and Tang, 2009; Lee et al., 2012a), however, the Bramham laboratory was the first to investigate miRNA expression after LTP induction in the dentate gyrus in vivo (Wibrand et al., 2010, 2012). Using anesthetized rats, they confirmed differential expression of three miRNA in dentate gyrus tissue 2 h after induction: miR-132-3p and miR-212-3p were up-regulated, and miR-219a-5p was down-regulated. Surprisingly, when HFS was delivered in the presence of an NMDA receptor antagonist expression of all three regulated miRNA was enhanced, despite ablation of LTP, suggesting that NMDA receptor activity represses miRNA levels. In contrast, regulation of the miRNA was blocked by a group 1 mGluR antagonist, which also prevented activity-dependent depotentiation, leading to the conclusion that differential expression of miRNA was functionally correlated with reversal of LTP.
By contrast, our recent work using an in vivo awake rat model (Abraham et al., 2002; Bowden et al., 2012) and Affymetrix miRNA arrays showed that at 20 min post-LTP the majority of the 65 differentially expressed mature miRNA transcripts were down-regulated, including miR-132-3p and miR-34a-5p (Joilin et al., 2014). These data contrast with the work of Wibrand et al. (2010, 2012), who found no apparent rapid regulation of miR-34a-5p or miR-132-3p, and Pai et al. (2014), who reported rapid up-regulation of miR-34a-5p, with no change in miR-132-3p levels. These discrepancies may potentially be accounted for by differences in stimulation paradigms and normalization procedures. Our data, interpreted alongside the generalized up-regulation of mRNA transcripts 20 min post-LTP (Ryan et al., 2011, 2012), suggest that down-regulation of miRNA may contribute to the long-term changes that lead to LTP persistence by rapidly releasing inhibition of target mRNA transcripts. We have also quantified miRNA expression at later time points using our in vivo awake rat model. While miR-132-3p had returned to baseline by 5 h, miR-34a-5p remained down-regulated (Ryan et al., 2012; Joilin et al., 2014) alongside miR-24-3p (Ryan et al., 2012); a finding consistent with the prediction that miR-34a-5p and miR-24-3p target mRNA that are up-regulated 5 h post-LTP (Ryan et al., 2012). As expression of these miRNA returned to baseline by 24 h, these results support the hypothesis that translational suppression of mRNA is released during the late phase of LTP to allow consolidation of LTP. Interestingly, using reverse transcription-quantitative polymerase chain reaction (RT-qPCR), we showed that the observed down-regulation of miR-34a-5p and miR-132-3p was mediated by NMDA receptors. Indeed, NMDA receptor antagonism in combination with tetanic stimulation led to a highly variable increase in miR-34a-5p, revealing an accord with Wibrand et al. (2010) and suggesting that in awake animals, the NMDA receptor-mediated reduction of miRNA levels out-competes an independent process working to increase them.
Very recently, Pai et al. (2014) have questioned the assumption that the quantity of total miRNA accurately represents miRNA activity. Instead, they have quantified miRNA bound to Ago2, one of the four Argonaute family proteins that can anchor mature miRNA within the RISC and allow miRNA activity. Using Ago2 immunoprecipitation, locked nucleic acid (LNA)-based microarrays and RT-qPCR validation, they demonstrated differential expression of Ago2-bound miRNA after LTP induction in the dentate gyrus of anesthetized rats (see Table 1 for details). This differential expression was quantitatively and qualitatively different to that measured in whole dentate gyrus lysate. Ago2 immunoprecipitate/whole lysate ratios indicated that eight miRNA associated with Ago2 in response to LTP induction, while three miRNA dissociated from Ago2 in response to LTP induction.
Table 1.
Differential expression of miRNA after LTP induction.
| Study | Organism | Preparation | LTP induction | Time-points post-LTP induction | Identification of candidate miRNA | Control | Criteria for differential expression | Differentially expressed miRNA determined by RT-qPCR validation |
|---|---|---|---|---|---|---|---|---|
| Joilin et al. (2014) | 4–6 months old Sprague Dawley rat | Dentate gyrus; LTP in vivo, un-anesthetized | HFS to PP-GC synapses | 20 min, 5 and 24 h | Affymetrix microarray: 65/6,703 miRNA had FC ≥ 1.15; n = 4 | Unstimulated hemisphere (within animal) | FC ≥ 1.15; p < 0.05: paired t-test | miR-34a-5p down-regulated at 20 min (FC = 0.44 ± 0.07; n = 8) and 5 h (FC = 0.53 ± 0.16; n = 5); back to baseline at 24 h miR-132-3p down-regulated at 20 min (FC = 0.37 ± 0.11; n = 8); back to baseline at 5 h and 24 h |
| Lee et al. (2012a) | 4–5 weeks old Sprague Dawley rat | Hippocampal slice | Chemical LTP | 30, 60, 120 min | Agilent microarray: 5/287 miRNA had FC ≥ 1.5 at at least one time-point; n = 1 | Slices that did not undergo LTP induction | FC ≥ 1.5; p < 0.05: one-way ANOVA and LSD post hoc | miR-188-5p up-regulated at 60 min (FC = 1.75 ± 0.3; n = 3); no regulation at 30 min or 120 min. Four other miRNA (unidentified) were not regulated at any time-points |
| Pai et al. (2014) | Adult Sprague Dawley rat | Dentate gyrus; LTP in vivo, anesthetized. Total lysate and Ago2 IP examined | HFS to PP-GC synapses | 30, 120 min | miRCURY microarray: 44/376 miRNA differentially expressed (p < 0.05: Student’s t-test with Dunn–Bonferroni correction); n = 3 | Unstimulated hemisphere (within animal) | p < 0.05; one-way ANOVA | Total lysate: miR-384-3p, miR-29b-3p, miR-219a-5p, miR-592-5p, miR-20a-5p, let-7f-5p, miR-338-3p, miR-212-3p, miR-34a-5p, miR-19a-3p, miR-326-3p up-regulated at 30 min; miR-223-3p down-regulated at 30 min. Ago2 IP: miR-384-3p, miR-29b-3p, miR-219a-5p, miR-592-5p, miR-20a-5p, let-7f-5p, miR-338-3p, miR-330-5p, miR-223-3p, miR-34a-5p, miR-19a-3p up-regulated at 30 min; miR-212-3p down-regulated at 30 min |
| Park and Tang (2009) | 7–8 weeks old C57/BL6 mice | Hippocampal slice | Chemical LTP | 0, 15, 30, 60, 120 min | GenoExplorer microarray: 55/237 miRNA had mean FC over all time-points ≥ 2; n = 1 | Slices that did not undergo LTP induction | FC ≥ 2 | miR-181b-5p not regulated at any time-point (n = 10); miR-128a-3p not regulated at any time-point (n = 10) |
| Ryan et al. (2012) | 4–6 months old Sprague Dawley rat | Dentate gyrus; LTP in vivo, un-anesthetized | HFS to PP-GC synapses | 5 h | IPA network analysis: miRNA identified as hubs in LTP-related gene networks | Unstimulated hemisphere (within animal) | FC ≥ 1.15; p < 0.05: paired t-test | miR-34a-5p down-regulated (FC = 0.53 ± 0.16; n = 5); miR-24-3p down-regulated (FC = 0.59 ± 0.12; n = 9) |
| Wibrand et al. (2010) | Adult Sprague Dawley rat | Dentate gyrus; LTP in vivo, anesthetized | HFS to PP-GC synapses | 10, 120 min | LC Sciences microarray: 21/237 miRNA had FC ≥ 1.2 at 120 min; n = 2 | Unstimulated hemisphere (within animal) | FC ≥ 1.20; p < 0.05: t-test | miR-132-3p up-regulated at 120 min (FC = 1.38; n = 8); miR-212-3p up-regulated at 120 min (FC = 1.26; n = 8); miR-219a-5p down-regulated at 120 min (FC = 0.68; n = 8). None of the three miRNA were regulated at 10 min (n = 5) |
| Wibrand et al. (2012) | Adult Sprague Dawley rat | Dentate gyrus; LTP in vivo, anesthetized | HFS to PP-GC synapses | 30, 120 min | Arc-targeting miRNA | Unstimulated hemisphere (within animal) | FC ≥ 1.20; p < 0.05: t-test | miR-132-3p up-regulated at 120 min (FC = ∼1.6; n = 5); not regulated at 30 min. No regulation of miR-19a-3p, miR-34a-5p, miR-326-3p, miR-193a-3p at 30 min or 120 min |
Ago2 IP, Argonaute 2 immunoprecipitates; FC, fold change; GC, granule cell; h, hour; HFS, high frequency stimulation; IPA, Ingenuity Pathway Analysis; LSD, least significant difference; LTP, long-term potentiation; min, minute; PP, perforant path; RT-qPCR, real time-quantitative polymerase chain reaction. Data are presented as mean ± SEM.
Collectively, these data (summarized in Table 1) demonstrate that individual miRNA are regulated between 20 min and 5 h after LTP induction, supporting the theory that miRNA mediate LTP-related protein synthesis. There is little overlap in the miRNA found to be differentially expressed in these seven studies, this may however be due to variation in the models used to induce LTP or differences in statistical analyses. MiR-34a-5p and miR-132-3p are the only miRNA that have been reported as differentially expressed in response to LTP by multiple laboratories. One study characterized the plasticity properties of a miR-132-3p/miR-212-3p double knockout mouse, and reported enhanced theta burst LTP in hippocampal slices, but no effect on tetanic LTP (Remenyi et al., 2013). Expression of both mRNA and protein products of potential miR-132-3p targets was unchanged, which may explain why tetanic LTP was not affected.
MicroRNA REGULATE LTP-RELATED GENES
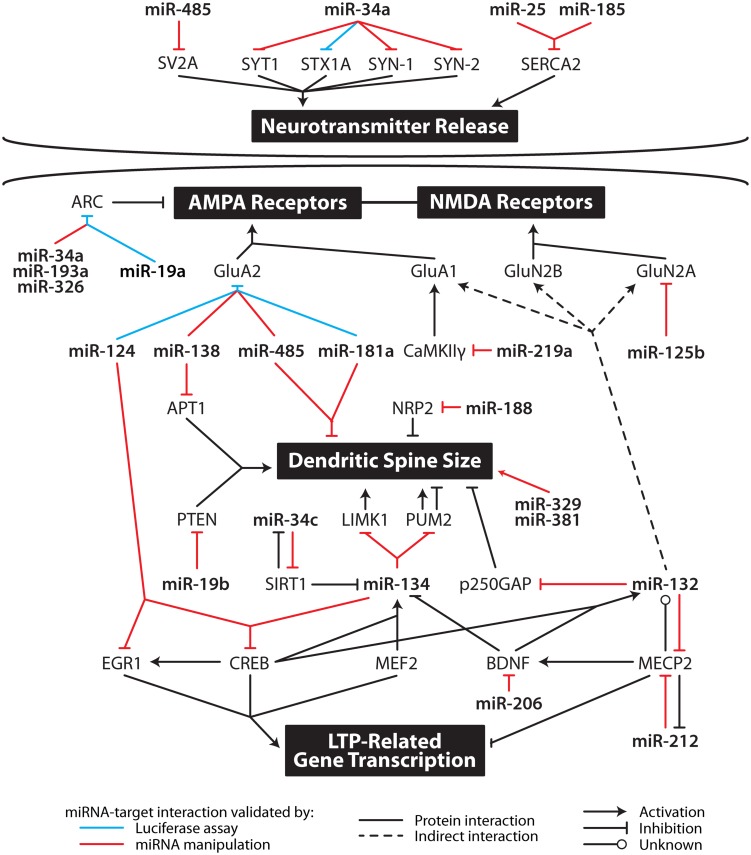
The putative molecular mechanisms underpinning LTP maintenance include enhanced release of neurotransmitter, alongside enhanced post-synaptic responsiveness driven initially by activation of protein kinases and glutamate receptor trafficking, which is ultimately underpinned by altered gene expression and structural reorganization of synaptic connections (Abraham and Williams, 2003; Lynch, 2004). The following sections discuss the mounting evidence suggesting that individual miRNA interact with transcripts coding for proteins important to all aspects of the maintenance of LTP. These studies are summarized in Figure 2 and Table 2.
FIGURE 2.
Multi-level contribution of microRNA to synaptic plasticity. MicroRNA likely influence the translation of multiple mRNA important in specific aspects of synaptic plasticity including neurotransmitter release, AMPA and NMDA receptor subunit levels, dendritic spine size, and gene transcription. APT1, acyl-protein thioesterase 1; ARC, activity-related cytoskeleton-associated protein; BDNF, brain-derived neurotrophic factor; CaMKIIγ, calcium/calmodulin-dependent protein kinase II gamma; CREB, cAMP response element-binding protein; EGR1, early growth response 1; GluA1, glutamate receptor, ionotropic, AMPA 1; GluA2, glutamate receptor, ionotropic, AMPA 2; GluN2A, glutamate receptor, ionotropic, NMDA 2A; GluN2B, glutamate receptor, ionotropic, NMDA 2B; LIMK1, LIM domain kinase 1; MECP2, methyl CpG binding protein 2; MEF2, myocyte enhancer factor-2; NRP2, neurophilin 2; PTEN, phosphatase and tensin homolog; PUM2, pumilio homolog 2; SERCA, sarco/endoplasmic reticulum Ca2+ ATPase; SIRT1, sirtuin 1; STX1A, syntaxin 1a; SV2A, synaptic vesicle glycoprotein 2A; SYN1, synapsin I; SYN2, synapsin II; SYT1, synaptotagmin I.
Table 2.
Evidence for miRNA regulation of LTP-related genes.
| Function | miRNA | mRNA | miRNA:mRNA interaction | Further information | Reference |
|---|---|---|---|---|---|
| Pre-synaptic vesicle release | miR-485-5p | SV2A | Direct inhibition in vitro | MiR-485-5p decreased the number of vesicles released after depolarization | Cohen et al. (2011) |
| miR-34a-5p | SYN1, SYN2 | Inhibition in vitro | MiR-34a-5p down-regulated 20 min (Joilin et al., 2014) and 5 h after LTP induction (Ryan et al., 2012); not regulated 30 min or 2 h after LTP induction (Wibrand et al., 2012) | Agostini et al. (2011) | |
| miR-34a-5p | SYT1, STX-1A | Direct inhibition in vitro | Increased miR-34a-5p correlated with decreased SYT1 and STX-1A in human Alzheimer’s disease (AD) brain. MiR-34a-5p down-regulated 20 min (Joilin et al., 2014) and 5 h after LTP induction (Ryan et al., 2012); not regulated 30 min or 2 h after LTP induction (Wibrand et al., 2012) | Agostini et al. (2011) | |
| miR-25-3p miR-185-5p | SERCA2 | Decreased miR-25-3p and miR-185-5p correlated with increased SERCA2 in vivo | Decreased miR-25-3p and miR-185-5p linked to increased LTP in Schizophrenia mouse model | Earls et al. (2012) | |
| Glutamate receptor regulation | miR-34a-5p miR-326-3p miR-19a-3p miR-193a-3p | Arc | Direct inhibition in vitro | MiR-34a-5p, miR-326-3p, miR-19a-3p, miR-193a-3p not regulated 30 min or 2 h after LTP induction. MiR-34a-5p down-regulated 20 min (Joilin et al., 2014) and 5 h after LTP induction in vivo (Ryan et al., 2012); not regulated 30 min or 2 h after LTP induction (Wibrand et al., 2012) | Wibrand et al. (2012) |
| miR-181a-5p | GluA2 | Direct inhibition in vitro (minimal) | MiR-181a-5p reduced spine volume and density in hippocampal neurons | Edbauer et al. (2010), Saba et al. (2012) | |
| miR-138-5p | GluA2 | Inhibition in vitro | MiR-138-5p reduced spine volume in hippocampal neurons (APT-1 dependent) | Siegel et al. (2009) | |
| miR-485-5p | GluA2 | Inhibition in vitro | MiR-485-5p reduced spine density and increased immature spines in hippocampal neurons | Cohen et al. (2011) | |
| miR-124-3p | GluA2 | Direct inhibition in vitro (minimal) | – | Edbauer et al. (2010) | |
| miR-132-3p | GluA1, GluN2A, GluN2B | Indirect up-regulation in vitro | MiR-132-3p up-regulated 2 h after LTP induction (Wibrand et al., 2010, 2012); miR-132-3p down-regulated 20 min after LTP induction (Joilin et al., 2014) | Kawashima et al. (2010) | |
| miR-219a-5p | GluA1 | Direct inhibition of CaMKIIy in vivo, which regulates GluA1 | MiR-219a-5p down-regulated 2 h after LTP induction (Wibrand et al., 2010) | Kocerha et al. (2009) | |
| miR-125b-5p | GluN2A | Direct inhibition in vitro | MiR-125b-5p reduced spine width and increased length in hippocampal neurons (FMRP- dependent) | Edbauer et al. (2010) | |
| miR-212-3p | BDNF | Indirect inhibition via MeCP2 in vivo | – | Im et al. (2010) | |
| miR-132-3p | BDNF | miR-132-3p indirectly inhibits BDNF via MeCP2; BDNF indirectly up-regulates miR-132-3p via TrkB and ERK1/2 | MiR-132-3p up-regulated 2 h after LTP induction (Wibrand et al., 2010, 2012); miR-132-3p down-regulated 20 min after LTP induction (Joilin et al., 2014) | Klein et al. (2007), Kawashima et al. (2010) | |
| miR-206-3p | BDNF | Direct inhibition in vivo | MiR-206-3p up-regulated in Tg2576 AD mice and human AD brain samples. Intraventricular injection of miR-206-3p antagomir increased BDNF and improved memory | Lee et al. (2012b) | |
| Transcription | miR-132-3p | MeCP2 | Reciprocal inhibition: MeCP2 inhibits miR-132-3p in vitro; miR-132-3p inhibits MeCP2 in vivo; MeCP2 KO decreased miR-132-3p in vivo | MiR-132-3p up-regulated 2 h after LTP induction (Wibrand et al., 2010, 2012); miR-132-3p down-regulated 20 min after LTP induction (Joilin et al., 2014) | Klein et al. (2007), Hansen et al. (2010), Im et al. (2010), Tognini et al. (2011) |
| miR-212-3p | MeCP2 | Reciprocal inhibition: MeCP2 inhibits miR-132 in vitro; miR-212-3p inhibits MeCP2 in vivo | MiR-212-3p up-regulated 2 h after LTP induction (Wibrand et al., 2010) | Im et al. (2010) | |
| miR-34c-5p | SIRT1 | Direct inhibition in vivo | Increased miR-34c-5p correlated with decreased SIRT1 in memory impairment mouse models (APPS1-21 and aged) | Zovoilis et al. (2011) | |
| miR-124-3p | CREB, EGR1 | Direct inhibition in vitro | – | Rajasethupathy et al. (2009), Yang et al. (2012) | |
| miR-134-5p | CREB | Direct inhibition in vitro | – | Gao et al. (2010) | |
| Activity-dependent dendritogenesis | miR-134-5p | Pum2 | Direct inhibition in vitro | Interaction only occurs after neuronal activity, not under basal conditions; miR-134-5p buffers Pum2 in a narrow range that is critical for activity-dependent dendritogenesis. MiR-134-5p decreased spine volume in hippocampal neurons | Schratt et al. (2006), Fiore et al. (2009) |
| miR-134-5p | Limk1 | Direct inhibition in vitro | Inhibition of Limk1 in dendrites relieved by BDNF | Schratt et al. (2006) | |
| miR-132-3p | p250GAP | Direct inhibition in vitro and in vivo | p250GAP regulates existing spine growth via Rac1 and kalirin-7; miR-132-3p affects spine size in vivo. MiR-132-3p up-regulated 2 h after LTP induction (Wibrand et al., 2010, 2012); miR-132-3p down-regulated 20 min after LTP induction (Joilin et al., 2014) | Impey et al. (2010), Mellios et al. (2011) | |
| miR-138-5p | APT1 | Direct inhibition in vitro | MiR-138-5p and APT1 co-localized at the synapse; miR-138-5p reduced spine volume in hippocampal neurons (APT-1 dependent) | Banerjee et al. (2009), Siegel et al. (2009) | |
| miR-19b-3p | PTEN | Inhibition in vitro | MiR-19b-3p up-regulated after fear conditioning; miR-19b-3p increased total neurite length in hippocampal neurons | Kye et al. (2011) | |
| miR-188-5p | Nrp2 | Direct inhibition in vitro | MiR-188-5p has no effect on dendritogenesis in hippocampal neurons MiR-188-5p is up-regulated 1 h after LTP induction (Lee et al., 2012a) | Lee et al. (2012a) |
AD, Alzheimer’s disease; APT1, acyl-protein thioesterase 1; ARC, activity-related cytoskeleton-associated protein; BDNF, brain-derived neurotrophic factor; CaMKIIγ, calcium/calmodulin-dependent protein kinase II gamma; CREB, cAMP response element-binding protein; EGR1, early growth response 1; GluA1, glutamate receptor, ionotropic, AMPA 1; GluA2, glutamate receptor, ionotropic, AMPA 2; GluN2A, glutamate receptor, ionotropic, NMDA 2A; GluN2B, glutamate receptor, ionotropic, NMDA 2B; LIMK1, LIM domain kinase 1; LTP, long-term potentiation; MECP2, methyl CpG binding protein 2; NRP2, neurophilin 2; PTEN, phosphatase and tensin homolog; PUM2, pumilio homolog 2; SERCA2, sarco/endoplasmic reticulum Ca2+ ATPase; SIRT1, sirtuin 1; STX1A, syntaxin 1a; SV2A, synaptic vesicle glycoprotein 2A; SYN1, synapsin I; SYN2, synapsin II; SYT1, synaptotagmin I.
MicroRNA REGULATE PRESYNAPTIC VESICLE RELEASE
MiRNA have been found within biochemical preparations that are enriched for presynaptic terminals (Xu et al., 2013). Intriguingly, three miRNA (miR-29a-3p, miR-99a-5p, miR-125a-5p) were shown to be released from synaptosomes in a calcium-dependent manner, and miR-125a-5p was shown to be endocytosed. Collectively, these findings led the authors to propose that miRNA release and uptake from nerve terminals may contribute to intercell communication. Other miRNA have also been implicated in the regulation of neurotransmitter release. MiR-25-3p and miR-185-5p target sarco/endoplasmic reticulum Ca2+ ATPase (SERCA2), which is involved in the maintenance of Ca2+ in the endoplasmic reticulum (Earls et al., 2012). Interestingly, SERCA2 is over-expressed in a mouse model of schizophrenia, concomitant with decreased miR-25-3p and miR-185-5p expression. As a result, calcium in the presynaptic terminal was increased, leading to increased neurotransmitter release and increased LTP. Restoration of these two miRNA attenuated the enhanced LTP.
Furthermore, two miRNA, miR-485-5p and miR-34a-5p, have been shown to interact with a number of transcripts that code for synaptic vesicle proteins (Table 2). Of these, synapsin 1a (SYN1), synaptotagmin I (SYT1), and syntaxin 1a (STX1A) have been implicated in LTP; SYN1 and SYT1 are regulated in response to LTP (reviewed in Abraham and Williams, 2003) and knockout of STX1A, a t-SNARE protein, impairs LTP in hippocampal slices (Fujiwara et al., 2006). While differential expression of miR-485-5p after LTP induction has not been reported, as outlined above, miR-34a-5p is down-regulated rapidly following LTP, the functional result of which may be to release inhibition of the synthesis of presynaptic vesicle proteins thereby contributing to enhanced release of neurotransmitter. Consistent with this hypothesis, miR-34a-5p is up-regulated in human AD, concomitant with SYT1 and STX1A down-regulation (Agostini et al., 2011).
MicroRNA REGULATE GLUTAMATE RECEPTOR SUBUNITS
AMPA and NMDA glutamate receptors both play critical roles in LTP. Early work from our laboratory has shown that the levels of both AMPA and NMDA receptor subunits are dynamically regulated in the dentate gyrus in response to perforant path LTP (Williams et al., 1998, 2003, 2007; Kennard et al., 2009, 2014). Interestingly, the rapid up-regulation of AMPA receptors is not due to an increase in their synthesis, but to movement of receptor subunits into the membrane from an extrasynaptic pool (Shi et al., 1999; Williams et al., 2007; Granger et al., 2013). However, protein synthesis still plays an important role as it can affect NMDA receptor expression (Williams et al., 2007) putatively through the regulation of associated chaperone molecules. To date glutamate receptor subunits have been shown to be regulated by seven miRNA (miR-132-3p, miR-124-3p, miR-125b-5p, miR-138-5p, miR-181a-5p, miR-219a-5p, and miR-485-5p); including indirect up-regulation mediated through miR-132-3p (Kocerha et al., 2009; Siegel et al., 2009; Edbauer et al., 2010; Kawashima et al., 2010; Cohen et al., 2011; Saba et al., 2012). Recently, Wibrand et al. (2012) showed that a group of miRNA, including miR-34a-5p, regulates Arc, which is involved with trafficking AMPA receptors out of the synapse (Wibrand et al., 2012). These miRNA were not found to be regulated 30 min or 2 h after LTP induction; however, in situ hybridization and analysis of synaptoneurosome preparations showed that these Arc-targeting miRNA were synaptically expressed, suggesting basal inhibition of Arc.
MiRNA may also influence glutamate receptor subunit expression via regulation of BDNF, which is thought to contribute to the sustained structural and functional changes underlying LTP (Panja and Bramham, 2014). MiR-206-3p directly inhibits BDNF, and is up-regulated in AD (Lee et al., 2012b). Intra-ventricular injection of miR-206-3p antagomir increased BDNF expression in a mouse model of AD and improved memory function (Lee et al., 2012b). Additionally, miR-212-3p decreases BDNF protein expression in the dorsal striatum in vivo via inhibition of methyl CpG binding protein 2 (MeCP2; Im et al., 2010). In vitro, MeCP2 induces BDNF III mRNA expression and miR-132-3p decreases BDNF III mRNA expression in an MeCP2-dependent manner (Klein et al., 2007). This inhibition is significant as miR-132-3p inhibition can partially reduce BDNF-induced up-regulation of glutamate receptor subunits (Kawashima et al., 2010). Interestingly, BDNF has been shown to up-regulate miR-132-3p via the kinases TrkB and ERK1/2, suggesting a form of self-regulation through miRNA (Kawashima et al., 2010). These interactions between MeCP2, BDNF, and miR-132-3p/miR-212-3p are complex and need to be investigated further in the context of LTP. Together these data suggest that miRNA act co-operatively to regulate the expression of glutamate receptor subunits post-LTP.
MicroRNA REGULATE LTP-RELATED TRANSCRIPTION FACTORS
While synthesis of proteins from extant mRNA is sufficient for LTP2, the concurrent and additional activation and regulation of transcription leads to the long-term stability of memory and LTP3 in vivo. Five miRNA (miR-34c-5p, miR-124-3p, miR-132-3p, miR-134-5p, and miR-212-3p) target proteins that regulate transcription in response to synaptic activity.
The BDNF-related gene MeCP2 is a putative dendritic growth regulator that mediates transcriptional repression. Li et al. (2011) have shown that a lack of activity-induced phosphorylation of MeCP2 causes enhanced LTP. As described above, both miR-132-3p and miR-212-3p interact with MeCP2 (Klein et al., 2007; Im et al., 2010). Excluding the role of BDNF in the circuit, there is evidence of reciprocal inhibition between MeCP2 and miR-132-3p/212-3p (Klein et al., 2007; Hansen et al., 2010; Im et al., 2010; Tognini et al., 2011). Furthermore, MeCP2 knockdown in HEK293 cells increased miR-212-3p and miR-132-3p expression (Im et al., 2010). This conflicts with other evidence showing MeCP2 knockout in vivo decreased miR-132-3p levels (Klein et al., 2007), but may be explained by the different preparations used. Further studies are needed to investigate these complex interactions involving MeCP2 in the context of LTP.
Another transcriptional repressor linked to LTP is Sirtuin 1 (SIRT1; Michan et al., 2010) which can be directly inhibited by miR-34c-5p (Zovoilis et al., 2011). In a study comparing young and aged mice, endogenous miR-34c-5p decreased SIRT1 protein in the hippocampus of aged, but not young, mice in vivo (Zovoilis et al., 2011). Over-expression of miR-34c-5p in the young mice caused a decrease in SIRT1 protein. Furthermore, in two memory impairment mouse models (aged and APPS1-21), an increase in miR-34c-5p was correlated with decreased SIRT1 protein (Zovoilis et al., 2011). Intrahippocampal injection of miR-34 inhibitors restored the level of SIRT1 protein in the APPS1-21 mice. The finding that miR-34c-5p is differentially expressed in a mouse model of AD supports the hypothesis that miR-34c-5p down-regulation is necessary for memory. In keeping with this hypothesis, increased miR-34c-5p expression was correlated with impaired contextual fear conditioning in mouse models of aging and AD and was rescued with miR-34c seed inhibitors (Zovoilis et al., 2011). However, while SIRT1 was the only target of miR-34c-5p that was investigated in this study, the authors note that other targets of miR-34c-5p are likely to be involved.
Interestingly, while miR-34c-5p regulates SIRT1, SIRT1 itself regulates miR-134-5p. In mouse neural cells, knockdown of SIRT1 increased miR-134-5p expression, which in turn inhibited cAMP response element binding protein (CREB) protein expression (Gao et al., 2010). In vivo miR-134-5p over-expression in the mouse hippocampus mimicked this effect (Gao et al., 2010). Furthermore, miR-134-5p is also regulated by an activity-dependant transcription factor, myocyte enhancer factor-2 (Mef2), which negatively regulates the number of excitatory synapses in mature hippocampal neurons (Flavell et al., 2006). Endogenous Mef2 is required for the depolarization-induced transcription of the miR-379-410 cluster, which contains miR-134 (Fiore et al., 2009). Inhibition of Mef2 in rat hippocampal neurons followed by depolarization caused a decrease in the precursors of miR-134-5p, which may lead to decreased mature miR-134-5p, affecting its ability to regulate its targets such as CREB. However, these interactions of miRNA and memory-related transcription regulators are yet to be studied in an LTP paradigm.
As a key regulator of activity-dependent dendritic morphogenesis, CREB mediates LTP-induced transcription, in part though the activation of an array of other transcription factors, which bind to specific response elements in immediate early genes (IEGs) such as early growth response 1 (EGR1). Both CREB and EGR1 have been linked to miRNA. MiR-124-3p directly decreased EGR1 expression in vivo (Yang et al., 2012) and CREB expression in vitro (Rajasethupathy et al., 2009). MiR-134-3p directly decreased CREB expression in vitro (Gao et al., 2010). These miRNA:mRNA interactions are intriguing, but are yet to be validated in a mammalian LTP model.
MicroRNA REGULATE DENDRITOGENESIS
The majority of excitatory synapses in the mammalian brain are formed on specialized protrusions from dendrites, known as spines. Dendritic spines exhibit actin-dependent morphological plasticity and their size has been correlated with synaptic strength (Matsuzaki et al., 2004). LTP3 consolidation, and long-term storage of memories, may be achieved by an increase in the size, and therefore strength, of potentiated synapses. Dendritic morphogenesis may allow synapses to maintain their enhanced strength for long periods, in spite of continual turnover of their constituent proteins. Specific miRNA regulate transcripts that code for proteins involved in the regulation of dendritic spine morphology, which may underlie LTP maintenance.
MiR-134-5p interacts with the translational regulatory protein, pumilio homolog 2 (Pum2), which mediates activity-dependent dendritogenesis. Pum2 is an RNA-binding protein that regulates translation and mRNA stability by binding the 3′UTR of mRNA targets. MiR-134-5p directly targets Pum2 in membrane-depolarized rat cortical neurons, but not under basal conditions, suggesting only newly processed miR-134-5p interacts with Pum2 (Fiore et al., 2009). Furthermore, miR-134-5p buffers Pum2 protein levels within a narrow range necessary for activity-dependent dendritogenesis (Fiore et al., 2009), suggesting that miR-134-5p-mediated regulation of Pum2 may be critical for the structural changes that underlie LTP maintenance. This proposal is supported by in vivo evidence (Christensen et al., 2010).
In contrast, miR-134-5p has been found to decrease spine volume in hippocampal neurons under normal conditions (Schratt et al., 2006). MiR-134-5p has been shown to interact with LIM domain kinase 1 (Limk1), a serine/threonine kinase that regulates actin polymerization by inactivating cofilin neurons (Schratt et al., 2006). Limk1 knockout mice exhibit dendritic spine structural abnormalities and enhanced LTP (Meng et al., 2002). Over-expression of miR-134-5p in normal mice impaired LTP persistence and performance in learning paradigms including fear conditioning, Morris water maze, and novel object recognition (Gao et al., 2010). Performance in these tasks and LTP persistence were rescued when miR-134-5p was knocked down. However, whether this rescue was mediated by Pum2 and/or Limk1, or other miR-134-5p targets is unknown.
MiR-132-3p regulates existing spine growth in neurons in vitro by directly inhibiting translation of p250GAP in a Rac1- and kalirin-7-dependent manner (Edbauer et al., 2010; Impey et al., 2010; Mellios et al., 2011). In vitro and in vivo studies suggest that the effect of miR-132-3p on spine morphology differs according to the developmental stage of the neuron: miR-132-3p triggers spine formation during the spine development phase, and increases the volume of existing spines once the majority of spines have developed (Edbauer et al., 2010; Hansen et al., 2010; Impey et al., 2010; Magill et al., 2010; Mellios et al., 2011; Tognini et al., 2011). Whether these effects of miR-132-3p on existing spines contribute past development to underlie LTP-related structural changes remains to be seen. Furthermore, over-expression of miR-132-3p leads to the formation of spines with low plasticity properties: the mushroom spines of stable mature spines, and immature filopodia (Mellios et al., 2011; Tognini et al., 2011; Tognini and Pizzorusso, 2012). This suggests that like miR-134-5p, an optimal range of miR-132-3p expression is required for plasticity-induced spine changes to occur.
MiR-138-5p has been found to decrease spine size in rat hippocampal neurons by directly targeting an enzyme co-localized at the synapse, called acyl-protein thioesterase 1 (APT1; Banerjee et al., 2009; Siegel et al., 2009). APT1 catalyzes the depalmitoylation of signaling proteins, a lipid modification that can affect not just the function of the protein but its localization; the latter is particularly important considering the extent of neuronal processes. By inhibiting APT1, miR-138-5p was found to increase the membrane localization of the G-protein Gα13, thus activating the downstream Rho signaling pathway, which has been implicated in spine morphology regulation (Tada and Sheng, 2006). MiRNA also regulate other pathways involved in spine morphogenesis, in addition to G-protein signaling pathways. MiR-19b-3p inhibits PTEN (phosphatase and tensin homolog), a modulator of the AKT-mTOR signaling pathway, which controls dendritic development and synapse formation (Kye et al., 2011). MiR-19b-3p was up-regulated in mouse CA1 tissue 3 h after fear conditioning and in cultured hippocampal neurons 1 h after NMDA stimulation (but not bicuculline stimulation). Based on this research, Kye et al. (2011) proposed that learning induces miR-19b-3p, which inhibits PTEN and prevents it dephosphorylating phosphoinositide-3-kinase (PI3K). This in turn releases inhibition of the mTOR pathway, leading to increased protein synthesis. In keeping with this theory, miR-19b-3p has been shown to increase total neurite length in hippocampal neurons (Kye et al., 2011). A number of other miRNA that are up-regulated in response to fear conditioning and neural stimulation are known to increase mTOR-dependent protein synthesis via PTEN or Fox01 regulation (miR-21-5p, miR-22-3p, miR-27a-3p, miR-106b-5p, miR-139-5p), but these miRNA were not investigated further in Kye et al.’s (2011).
As mentioned above, miR-188-5p is down-regulated 2 h after LTP induction in rat hippocampal slices (Lee et al., 2012a). In this study, it was found that miR-188-5p directly inhibited neuropilin 2 (Nrp2), a transmembrane receptor protein for class 3 semaphorins, which have been shown to be chemorepellents against axonal growth cones, and therefore may act in a similar way against growth cones for dendritic spines. An increase in Nrp2 induced a decrease in spine density in hippocampal neurons, but miR-188-5p application rescued this effect. Over-expression of miR-188-5p does not have the expected effect of increasing spine density; the authors suggested this may have been due to a saturation effect (Lee et al., 2012a).
Five other miRNA have been shown to regulate dendritic spine morphology in DIV 10–15 rat hippocampal neurons: miR-125b-5p (Edbauer et al., 2010), miR-181a-5p (Saba et al., 2012), miR-485-5p (Cohen et al., 2011), miR-329-3p, and miR-381-3p (Fiore et al., 2009). The latter study is particularly interesting, as it reported that miR-329-3p and miR-381-3p were necessary for activity-dependent increases in dendritic complexity, but application of these miRNA had no effect under basal conditions. This result suggests that these miRNA may play a role in activity-dependent dendritogenesis.
Collectively, these results indicate that a sub-set of miRNA regulates translation of several proteins that are involved in dendritic morphogenesis. While, in some cases, these interactions have been investigated in the context of LTP, research into the role of miRNA in dendritic morphogenesis has focused on the genesis of new spines in developing neurons. These same miRNA:mRNA interactions may also play a role in the growth of existing spines in adult neurons during LTP maintenance, but further research is required to test this hypothesis.
DISCUSSION
This review has summarized current knowledge regarding the possible involvement of miRNA in LTP maintenance. We presented evidence that a subset of miRNA are differentially expressed between 20 min and 5 h after induction of LTP in vivo and converging lines of evidence suggesting that miRNA likely regulate the expression of proteins relevant to all aspects of LTP maintenance.
Key properties of miRNA – pleiotropism, tight regulation, synaptic localization, and responsiveness to activity – make them ideal candidates as regulators of the co-ordinated gene expression that underpins LTP maintenance. Some of these properties are shared with other non-coding RNA, which have also been implicated in the maintenance of memory (Mercer et al., 2008). The hypothesis that miRNA regulate LTP maintenance is underscored by the studies presented in this paper and clearly demonstrated in Figure 2, which shows the influence of individual miRNA on multiple LTP-related genes. For example, miR-132-3p has been linked to glutamate receptor expression, dendritogenesis and transcription factor regulation. In keeping with these results, miR-132-3p has been implicated in learning behavior in vivo: over-expression of miR-132-3p in mouse forebrain neurons was associated with deficits in novel object recognition (Hansen et al., 2010) and contextual fear conditioning increased pri-miR-132 expression (Nudelman et al., 2010).
There is strong evidence that miRNA fine-tune translation locally at activated synapses (Schratt et al., 2006; Bredy et al., 2011): miR-134-5p inhibits translation of Limk1 at the synapse until synaptic activation, at which point miR-134-5p is inactivated and Limk1 protein is expressed, leading to dendritic spine growth. Conversely, synaptic activation could trigger miRNA to suppress translation of synaptic proteins that prevent LTP maintenance. Either or both of these mechanisms would allow miRNA to couple synaptic activity to the selective protein synthesis that contributes to LTP2 and/or LTP3. This is consistent with the reported changes in miRNA expression post-LTP and the localisation of miRNA and proteins involved in miRNA biogenesis and function at synapses (Lugli et al., 2005, 2008; Schratt et al., 2006; Kye et al., 2007; Aschrafi et al., 2008; Siegel et al., 2009; Natera-Naranjo et al., 2010). There is, however, no direct evidence that miRNA affect local protein synthesis at the synapse and thereby contribute to the input specificity of LTP.
MiRNA may also affect the translational ability of mRNA transcripts as they are being transported from the soma to the axonal terminals or dendritic spines (Schratt et al., 2006). There is evidence that miRNA may be bound to mRNA in structures similar to processing-bodies (P-bodies) for transportation, allowing the miRNA to suppress translation of the mRNA until after synaptic activation (Schratt et al., 2006). The transcripts are likely to encode proteins that stabilize the changes at synapses that allow LTP to persist. For example, over-expression of miR-132-3p can result in increased levels of the NMDA receptor subunits GluN2A and GluN2B (Kawashima et al., 2010) and our own research has shown that post-LTP there is a rapid protein synthesis-dependent, transcription-independent increase in their expression (Williams et al., 2007).
That miRNA play a role in LTP-maintenance outside of the synapse is supported by evidence that miRNA influence the expression of LTP-related transcription regulators such as CREB, EGR1, MeCP2, SIRT1 and Mef2, contributing to the coordinate genomics response which underpins LTP3 by allowing tight regulation of a subset of genes post-LTP induction (Ryan et al., 2012; Joilin et al., 2014).
It is important to note that regulation of miRNA in response to LTP induction does not prove that they contribute to LTP maintenance. There are currently no reports of a direct effect of altering miRNA activity on LTP maintenance. Indeed, Dicer knockdown and concomitant down-regulation of miRNA had no effect on LTP (Konopka et al., 2010), however, this could be attributed to the short recording time of 50 min after HFS. By contrast, over-expression of individual miRNA in vivo prior to LTP induction was associated with a decrease in LTP (Gao et al., 2010; Scott et al., 2012), although these studies could not determine whether this affected LTP maintenance or LTP induction as the miRNA levels were altered prior to the induction of LTP. We also cannot rule out the possibility that miRNA regulation is occurring outside of neurons (e.g., in glia), or at synapses that are not undergoing LTP. Indeed, the observed changes in miRNA expression could be due to related forms of activity-dependent synaptic plasticity, such as LTD or depotentiation. For example, NMDA receptor-dependent LTD requires miR-191a-5p and miR-135a-3p for its maintenance (Hu et al., 2014), and electroconvulsive shock (ECS), which shares many properties with LTP, can induce differential expression of a large number of miRNA (Eacker et al., 2011). These forms of synaptic plasticity are likely to be activated concurrently to scale synapses in neighboring spines, dendrites, or neurons. Further research accounting for cell type and/or sub-cellular compartments is needed to resolve these issues.
An important next step for this field is to confirm that the miRNA:mRNA interactions presented here are indeed physiologically relevant to LTP maintenance, either in vitro or in vivo. Likewise, conditional regulation of miRNA levels or activity is required to discriminate between effects on the early and late phases of LTP. Furthermore, investigation of the mechanisms underpinning altered miRNA turnover and/or synthesis, particularly with regard to rapid down-regulation of miRNA is required. To establish the full cohort of LTP-related miRNA, high-throughput broad screens of differentially expressed miRNA at multiple time points after LTP induction are needed. Alongside this the development of improved target prediction algorithms would facilitate identification of relevant targets of these miRNA, and thereby increase our understanding of the functional significance of miRNA regulation in response to LTP induction.
SUMMARY
We conclude that to date, there is insufficient evidence to confirm that miRNA contribute to the maintenance of LTP; seven studies from four groups have reported differential expression of miRNA in response to LTP induction and converging lines of evidence suggest that they are ideally suited to this purpose. The functional significance of miRNA in the maintenance of LTP remains to be determined; this is the next step in this challenging and exciting area of research.
AUTHOR CONTRIBUTIONS
All authors were involved in the conception and writing of the review.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. W. C. Abraham for valuable discussions regarding this review. The review was constructed during the tenure of a Post-graduate Scholarship of the New Zealand Neurological Foundation awarded to BR and a Helen Rosa Thacker Scholarship for Neurological Research to GJ.
REFERENCES
- Abraham W. C., Logan B., Greenwood J. M., Dragunow M. (2002). Induction and experience-dependent consolidation of stable long-term potentiation lasting months in the hippocampus. J. Neurosci. 22 9626–9634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham W. C., Otani S. (1991). “Macromolecules and the maintenance of long-term potentiation,” in Kindling and Synaptic Plasticity: The Legacy of Graham Goddard ed. Morrell F. (Boston: Birkhauser) 92–109. [Google Scholar]
- Abraham W. C., Williams J. M. (2003). Properties and mechanisms of LTP maintenance. Neuroscientist 9 463–474 10.1177/1073858403259119 [DOI] [PubMed] [Google Scholar]
- Abraham W. C., Williams J. M. (2008). LTP maintenance and its protein synthesis-dependence. Neurobiol. Learn. Mem. 89 260–268 10.1016/j.nlm.2007.10.001 [DOI] [PubMed] [Google Scholar]
- Agostini M., Tucci P., Killick R., Candi E., Sayan B. S., Rivetti di Val Cervo P., et al. (2011). Neuronal differentiation by TAp73 is mediated by microRNA-34a regulation of synaptic protein targets. Proc. Natl. Acad. Sci. U.S.A. 108 21093–21098 10.1073/pnas.1112061109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschrafi A., Schwechter A. D., Mameza M. G., Natera-Naranjo O., Gioio A. E., Kaplan B. B. (2008). MicroRNA-338 regulates local cytochrome c oxidase IV mRNA levels and oxidative phosphorylation in the axons of sympathetic neurons. J. Neurosci. 28 12581–12590 10.1523/JNEUROSCI.3338-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balschun D., Manahan-Vaughan D., Wagner T., Behnisch T., Reymann K. G., Wetzel W. (1999). A specific role for group I mGluRs in hippocampal LTP and hippocampus-dependent spatial learning. Learn. Mem. 6 138–152. [PMC free article] [PubMed] [Google Scholar]
- Banerjee S., Neveu P., Kosik K. S. (2009). A coordinated local translational control point at the synapse involving relief from silencing and MOV10 degradation. Neuron 64 871–884 10.1016/j.neuron.2009.11.023 [DOI] [PubMed] [Google Scholar]
- Bartel D. (2004). MicroRNAs: genomics, biogenesis, mechanisms, and function. Cell Press 116 281–297. [DOI] [PubMed] [Google Scholar]
- Bartel D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136 215–233 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassell G. J., Warren S. T. (2008). Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron 60 201–214 10.1016/j.neuron.2008.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayazitov I. T., Richardson R. J., Fricke R. G., Zakharenko S. S. (2007). Slow presynaptic and fast postsynaptic components of compound long-term potentiation. J. Neurosci. 27 11510–11521 10.1523/JNEUROSCI.3077-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I., Rehwinkel J., Doerks T., Stark A., Bork P., Izaurralde E. (2006). mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 20 1885–1898 10.1101/gad.1424106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya S. N., Habermacher R., Martine U., Closs E. I., Filipowicz W. (2006). Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 125 1111–1124 10.1016/j.cell.2006.04.031 [DOI] [PubMed] [Google Scholar]
- Bliss T. V., Gardner-Medwin A. R. (1973). Long-lasting potentiation of synaptic transmission in the dentate area of the unanaesthetized rabbit following stimulation of the perforant path. J. Physiol. 232 357–374 10.1113/jphysiol.1973.sp010274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolotto Z. A., Bashir Z. I., Davies C. H., Collingridge G. L. (1994). A molecular switch activated by metabotropic glutamate receptors regulates induction of long-term potentiation. Nature 368 740–743 10.1038/368740a0 [DOI] [PubMed] [Google Scholar]
- Bowden J. B., Abraham W. C., Harris K. M. (2012). Differential effects of strain, circadian cycle, and stimulation pattern on LTP and concurrent LTD in the dentate gyrus of freely moving rats. Hippocampus 22 1363–1370 10.1002/hipo.20972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramham C. R., Wells D. G. (2007). Dendritic mRNA: transport, translation, and function. Nat. Rev. Neurosci. 8 776–789 10.1038/nrn2150 [DOI] [PubMed] [Google Scholar]
- Bredy T. W., Lin Q., Wei W., Baker-Andresen D., Mattick J. S. (2011). MicroRNA regulation of neural plasticity and memory. Neurobiol. Learn. Mem. 96 89–94 10.1016/j.nlm.2011.04.004 [DOI] [PubMed] [Google Scholar]
- Cheever A., Blackwell E., Ceman S. (2010). Fragile X protein family member FXR1P is regulated by microRNAs. RNA 16 1530–1539 10.1261/rna.2022210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Liang H., Zhang J., Zen K., Zhang C. Y. (2012). Secreted microRNAs: a new form of intercellular communication. Trends Cell Biol. 22 125–132 10.1016/j.tcb.2011.12.001 [DOI] [PubMed] [Google Scholar]
- Christensen M., Larsen L. A., Kauppinen S., Schratt G. (2010). Recombinant adeno-associated virus-mediated microRNA delivery into the postnatal mouse brain reveals a role for miR-134 in dendritogenesis in vivo. Front. Neural Circuits 3:16 10.3389/neuro.04.016.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. E., Lee P. R., Chen S., Li W., Fields R. D. (2011). MicroRNA regulation of homeostatic synaptic plasticity. Proc. Natl. Acad. Sci. U.S.A. 108 11650–11655 10.1073/pnas.1017576108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke S. F., Bear M. F. (2014). How the mechanisms of long-term synaptic potentiation and depression serve experience-dependent plasticity in primary visual cortex. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369 20130284 10.1098/rstb.2013.0284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delay C., Mandemakers W., Hebert S. S. (2012). MicroRNAs in Alzheimer’s disease. Neurobiol. Dis. 46 285–290 10.1016/j.nbd.2012.01.003 [DOI] [PubMed] [Google Scholar]
- Dityatev A. E., Bolshakov V. Y. (2005). Amygdala, long-term potentiation, and fear conditioning. Neuroscientist 11 75–88 10.1177/1073858404270857 [DOI] [PubMed] [Google Scholar]
- Doench J. G., Sharp P. A. (2004). Specificity of microRNA target selection in translational repression. Genes Dev. 18 504–511 10.1101/gad.1184404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eacker S. M., Keuss M. J., Berezikov E., Dawson V. L., Dawson T. M. (2011). Neuronal activity regulates hippocampal miRNA expression. PLoS ONE 6:e25068 10.1371/journal.pone.0025068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earls L. R., Fricke R. G., Yu J., Berry R. B., Baldwin L. T., Zakharenko S. S. (2012). Age-dependent microRNA control of synaptic plasticity in 22q11 deletion syndrome and schizophrenia. J. Neurosci. 32 14132–14144 10.1523/JNEUROSCI.1312-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edbauer D., Neilson J. R., Foster K. A., Wang C.-F., Seeburg D. P., Batterton M. N., et al. (2010). Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron 65 373–384 10.1016/j.neuron.2010.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etheridge A., Lee I., Hood L., Galas D., Wang K. (2011). Extracellular microRNA: a new source of biomarkers. Mutat. Res. 717 85–90 10.1016/j.mrfmmm.2011.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A., Huntzinger E., Nishihara T., Rehwinkel J., Fauser M., Izaurralde E. (2009). Deadenylation is a widespread effect of miRNA regulation. RNA 15 21–32 10.1261/rna.1399509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore R., Khudayberdiev S., Christensen M., Siegel G., Flavell S. W., Kim T. K., et al. (2009). Mef2-mediated transcription of the miR379-410 cluster regulates activity-dependent dendritogenesis by fine-tuning Pumilio2 protein levels. EMBO J. 28 697–710 10.1038/emboj.2009.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell S. W., Cowan C. W., Kim T. K., Greer P. L., Lin Y., Paradis S., et al. (2006). Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science 311 1008–1012 10.1126/science.1122511 [DOI] [PubMed] [Google Scholar]
- Forman J. J., Legesse-Miller A., Coller H. A. (2008). A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc. Natl. Acad. Sci. U.S.A. 105 14879–14884 10.1073/pnas.0803230105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman R. C., Farh K. K., Burge C. B., Bartel D. P. (2009). Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19 92–105 10.1101/gr.082701.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T., Mishima T., Kofuji T., Chiba T., Tanaka K., Yamamoto A., et al. (2006). Analysis of knock-out mice to determine the role of HPC-1/syntaxin 1A in expressing synaptic plasticity. J. Neurosci. 26 5767–5776 10.1523/JNEUROSCI.0289-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Wang W.-Y., Mao Y.-W., Gräff J., Guan J.-S., Pan L., et al. (2010). A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature 466 1105–1109 10.1038/nature09271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granger A. J., Shi Y., Lu W., Cerpas M., Nicoll R. A. (2013). LTP requires a reserve pool of glutamate receptors independent of subunit type. Nature 493 495–500 10.1038/nature11775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M., Landthaler M., Burger L., Khorshid M., Hausser J., Berninger P., et al. (2010). Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141 129–141 10.1016/j.cell.2010.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen K. F., Sakamoto K., Wayman G. A., Impey S., Obrietan K. (2010). Transgenic miR132 alters neuronal spine density and impairs novel object recognition memory. PLoS ONE 5:e15497 10.1371/journal.pone.0015497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havik B., Rokke H., Dagyte G., Stavrum A. K., Bramham C. R., Steen V. M. (2007). Synaptic activity-induced global gene expression patterns in the dentate gyrus of adult behaving rats: induction of immunity-linked genes. Neuroscience 148 925–936 10.1016/j.neuroscience.2007.07.024 [DOI] [PubMed] [Google Scholar]
- Hayashi Y., Shi S. H., Esteban J. A., Piccini A., Poncer J. C., Malinow R. (2000). Driving AMPA receptors into synapses by LTP and CaMKII: requirement for GluR1 and PDZ domain interaction. Science 287 2262–2267 10.1126/science.287.5461.2262 [DOI] [PubMed] [Google Scholar]
- Higa G. S., de Sousa E., Walter L. T., Kinjo E. R., Resende R. R., Kihara A. H. (2014). MicroRNAs in neuronal communication. Mol. Neurobiol. 49 1309–1326 10.1007/s12035-013-8603-7 [DOI] [PubMed] [Google Scholar]
- Hu Z., Yu D., Gu Q. H., Yang Y., Tu K., Zhu J., et al. (2014). miR-191 and miR-135 are required for long-lasting spine remodelling associated with synaptic long-term depression. Nat. Commun. 5 3263 10.1038/ncomms4263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S. H., Wang J., Sui W. H., Chen B., Zhang X. Y., Yan J., et al. (2013). BDNF-dependent recycling facilitates TrkB translocation to postsynaptic density during LTP via a Rab11-dependent pathway. J. Neurosci. 33 9214–9230 10.1523/JNEUROSCI.3256-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im H. I., Hollander J. A., Bali P., Kenny P. J. (2010). MeCP2 controls BDNF expression and cocaine intake through homeostatic interactions with microRNA-212. Nat. Neurosci. 13 1120–1127 10.1038/nn.2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impey S., Davare M., Lesiak A., Fortin D., Ando H., Varlamova O., et al. (2010). An activity-induced microRNA controls dendritic spine formation by regulating Rac1-PAK signaling. Mol. Cell Neurosci. 43 146–156 10.1016/j.mcn.2009.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joilin G., Guevremont D., Ryan B., Claudianos C., Cristino A. S., Abraham W. C., et al. (2014). Rapid regulation of microRNA following induction of long-term potentiation in vivo. Front. Mol. Neurosci. 7:98 10.3389/fnmol.2014.00098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H., Gkogkas C. G., Sonenberg N., Holt C. E. (2014). Remote control of gene function by local translation. Cell 157 26–40 10.1016/j.cell.2014.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H., Schuman E. M. (1996). A requirement for local protein synthesis in neurotrophin-induced hippocampal synaptic plasticity. Science 273 1402–1406 10.1126/science.273.5280.1402 [DOI] [PubMed] [Google Scholar]
- Kawamata T., Tomari Y. (2010). Making RISC. Trends Biochem. Sci. 35 368–376 10.1016/j.tibs.2010.03.009 [DOI] [PubMed] [Google Scholar]
- Kawashima H., Numakawa T., Kumamaru E., Adachi N., Mizuno H., Ninomiya M., et al. (2010). Glucocorticoid attenuates brain-derived neurotrophic factor-dependent upregulation of glutamate receptors via the suppression of microRNA-132 expression. Neuroscience 165 1301–1311 10.1016/j.neuroscience.2009.11.057 [DOI] [PubMed] [Google Scholar]
- Kennard J. T., Guevremont D., Mason-Parker S. E., Abraham W. C., Williams J. M. (2009). Increased expression, but not postsynaptic localisation, of ionotropic glutamate receptors during the late-phase of long-term potentiation in the dentate gyrus in vivo. Neuropharmacology 56 66–72 10.1016/j.neuropharm.2008.07.044 [DOI] [PubMed] [Google Scholar]
- Kennard J. T., Guevremont D., Mason-Parker S. E., Abraham W. C., Williams J. M. (2014). Redistribution of ionotropic glutamate receptors detected by laser microdissection of the rat dentate gyrus 48 h following LTP induction in vivo. PLoS ONE 9:e92972 10.1371/journal.pone.0092972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. H., Yeo S. H., Park J. M., Choi J. Y., Lee T. H., Park S. Y., et al. (2014). Genetic markers for diagnosis and pathogenesis of Alzheimer’s disease. Gene 545 185–193 10.1016/j.gene.2014.05.031 [DOI] [PubMed] [Google Scholar]
- Klein M. E., Lioy D. T., Ma L., Impey S., Mandel G., Goodman R. H. (2007). Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nat. Neurosci. 10 1513–1514 10.1038/nn2010 [DOI] [PubMed] [Google Scholar]
- Kocerha J., Faghihi M. A., Lopez-Toledano M. A., Huang J., Ramsey A. J., Caron M. G., et al. (2009). MicroRNA-219 modulates NMDA receptor-mediated neurobehavioral dysfunction. Proc. Natl. Acad. Sci. U.S.A. 106 3507–3512 10.1073/pnas.0805854106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka W., Kiryk A., Novak M., Herwerth M., Parkitna J. R., Wawrzyniak M., et al. (2010). MicroRNA loss enhances learning and memory in mice. J. Neurosci. 30 14835–14842 10.1523/JNEUROSCI.3030-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A., Grun D., Poy M. N., Wolf R., Rosenberg L., Epstein E. J., et al. (2005). Combinatorial microRNA target predictions. Nat. Genet. 37 495–500 10.1038/ng1536 [DOI] [PubMed] [Google Scholar]
- Krol J., Busskamp V., Markiewicz I., Stadler M. B., Ribi S., Richter J., et al. (2010a). Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell 141 618–631 10.1016/j.cell.2010.03.039 [DOI] [PubMed] [Google Scholar]
- Krol J., Loedige I., Filipowicz W. (2010b). The widespread regulation of microRNA biogenesis, function, and decay. Nat. Rev. Genet. 11 597–610 10.1038/nrg2843 [DOI] [PubMed] [Google Scholar]
- Kye M. J., Liu T., Levy S. F., Xu N. L., Groves B. B., Bonneau R., et al. (2007). Somatodendritic microRNAs identified by laser capture and multiplex RT-PCR. RNA 13 1224–1234 10.1261/rna.480407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kye M. J., Neveu P., Lee Y. S., Zhou M., Steen J. A., Sahin M., et al. (2011). NMDA mediated contextual conditioning changes miRNA expression. PLoS ONE 6:e24682 10.1371/journal.pone.0024682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lall S., Grun D., Krek A., Chen K., Wang Y. L., Dewey C. N., et al. (2006). A genome-wide map of conserved microRNA targets in C. elegans. Curr. Biol. 16 460–471 10.1016/j.cub.2006.01.050 [DOI] [PubMed] [Google Scholar]
- Lauterborn J. C., Rex C. S., Kramar E., Chen L. Y., Pandyarajan V., Lynch G., et al. (2007). Brain-derived neurotrophic factor rescues synaptic plasticity in a mouse model of fragile X syndrome. J. Neurosci. 27 10685–10694 10.1523/JNEUROSCI.2624-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E. J., Baek M., Gusev Y., Brackett D. J., Nuovo G. J., Schmittgen T. D. (2008). Systematic evaluation of microRNA processing patterns in tissues, cell lines, and tumors. RNA 14 35–42 10.1261/rna.804508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., Kim J. H., Kwon O. B., An K., Ryu J., Cho K., et al. (2012a). An activity-regulated microRNA, miR-188, controls dendritic plasticity, and synaptic transmission by downregulating neuropilin-2. J. Neurosci. 32 5678–5687 10.1523/JNEUROSCI.6471-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. T., Chu K., Jung K. H., Kim J. H., Huh J. Y., Yoon H., et al. (2012b). miR-206 regulates brain-derived neurotrophic factor in Alzheimer disease model. Ann. Neurol. 72 269–277 10.1002/ana.23588 [DOI] [PubMed] [Google Scholar]
- Lee P. R., Cohen J. E., Becker K. G., Fields R. D. (2005). Gene expression in the conversion of early-phase to late-phase long-term potentiation. Ann. N. Y. Acad. Sci. 1048 259–271 10.1196/annals.1342.023 [DOI] [PubMed] [Google Scholar]
- Lewis B. P., Burge C. B., Bartel D. P. (2005). Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120 15–20 10.1016/j.cell.2004.12.035 [DOI] [PubMed] [Google Scholar]
- Li H., Zhong X., Chau K. F., Williams E. C., Chang Q. (2011). Loss of activity-induced phosphorylation of MeCP2 enhances synaptogenesis, LTP, and spatial memory. Nat. Neurosci. 14 1001–1008 10.1038/nn.2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisman J., Yasuda R., Raghavachari S. (2012). Mechanisms of CaMKII action in long-term potentiation. Nat. Rev. Neurosci. 13 169–182 10.1038/nrn3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugli G., Larson J., Demars M. P., Smalheiser N. R. (2012). Primary microRNA precursor transcripts are localized at post-synaptic densities in adult mouse forebrain. J. Neurochem. 123 459–466 10.1111/j.1471-4159.2012.07921.x [DOI] [PubMed] [Google Scholar]
- Lugli G., Larson J., Martone M. E., Jones Y., Smalheiser N. R. (2005). Dicer and eIF2c are enriched at postsynaptic densities in adult mouse brain and are modified by neuronal activity in a calpain-dependent manner. J. Neurochem. 94 896–905 10.1111/j.1471-4159.2005.03224.x [DOI] [PubMed] [Google Scholar]
- Lugli G., Torvik V. I., Larson J., Smalheiser N. R. (2008). Expression of microRNAs and their precursors in synaptic fractions of adult mouse forebrain. J. Neurochem. 106 650–661 10.1111/j.1471-4159.2008.05413.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. (2004). Long-term potentiation and memory. Physiol. Rev. 84 87–136 10.1152/physrev.00014.2003 [DOI] [PubMed] [Google Scholar]
- Lytle J. R., Yario T. A., Steitz J. A. (2007). Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5’ UTR as in the 3’ UTR. Proc. Natl. Acad. Sci. U.S.A. 104 9667–9672 10.1073/pnas.0703820104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magill S. T., Cambronne X. A., Luikart B. W., Lioy D. T., Leighton B. H., Westbrook G. L., et al. (2010). microRNA-132 regulates dendritic growth and arborization of newborn neurons in the adult hippocampus. Proc. Natl. Acad. Sci. U.S.A. 107 20382–20387 10.1073/pnas.1015691107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki M., Honkura N., Ellis-Davies G. C. R., Kasai H. (2004). Structural basis of long-term potentiation in single dendritic spines. Nature 429 761–766 10.1038/nature02617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill E., Van Vactor D. (2012). MicroRNAs shape the neuronal landscape. Neuron 75 363–379 10.1016/j.neuron.2012.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellios N., Sugihara H., Castro J., Banerjee A., Le C., Kumar A., et al. (2011). miR-132, an experience-dependent microRNA, is essential for visual cortex plasticity. Nat. Neurosci. 14 1240–1242 10.1038/nn.2909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y., Zhang Y., Tregoubov V., Janus C., Cruz L., Jackson M., et al. (2002). Abnormal spine morphology and enhanced LTP in LIMK-1 knockout mice. Neuron 35 121–133 10.1016/S0896-6273(02)00758-4 [DOI] [PubMed] [Google Scholar]
- Mercer T. R., Dinger M. E., Mariani J., Kosik K. S., Mehler M. F., Mattick J. S. (2008). Noncoding RNAs in long-term memory formation. Neuroscientist 14 434–445 10.1177/1073858408319187 [DOI] [PubMed] [Google Scholar]
- Michan S., Li Y., Chou M. M., Parrella E., Ge H., Long J. M., et al. (2010). SIRT1 is essential for normal cognitive function and synaptic plasticity. J. Neurosci. 30 9695–9707 10.1523/JNEUROSCI.0027-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozova N., Zinovyev A., Nonne N., Pritchard L. L., Gorban A. N., Harel-Bellan A. (2012). Kinetic signatures of microRNA modes of action. RNA 18 1635–1655 10.1261/rna.032284.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natera-Naranjo O., Aschrafi A., Gioio A. E., Kaplan B. B. (2010). Identification and quantitative analyses of microRNAs located in the distal axons of sympathetic neurons. RNA 16 1516–1529 10.1261/rna.1833310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen P. V., Kandel E. R. (1996). A macromolecular synthesis-dependent late phase of long-term potentiation requiring cAMP in the medial perforant pathway of rat hippocampal slices. J. Neurosci. 16 3189–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudelman A. S., DiRocco D. P., Lambert T. J., Garelick M. G., Le J., Nathanson N. M., et al. (2010). Neuronal activity rapidly induces transcription of the CREB-regulated microRNA-132, in vivo. Hippocampus 20 492–498 10.1002/hipo.20646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom U. A., Nielsen F. C., Lund A. H. (2008). MicroRNA-10a binds the 5’UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell 30 460–471 10.1016/j.molcel.2008.05.001 [DOI] [PubMed] [Google Scholar]
- Otani S., Marshall C. J., Tate W. P., Goddard G. V., Abraham W. C. (1989). Maintenance of long-term potentiation in rat dentate gyrus requires protein synthesis but not messenger RNA synthesis immediately post-tetanization. Neuroscience 28 519–526 10.1016/0306-4522(89)90001-8 [DOI] [PubMed] [Google Scholar]
- Pai B., Siripornmongcolchai T., Berentsen B., Pakzad A., Vieuille C., Pallesen S., et al. (2014). NMDA receptor-dependent regulation of miRNA expression and association with Argonaute during LTP in vivo. Front. Cell Neurosci. 7:285 10.3389/fncel.2013.00285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panja D., Bramham C. R. (2014). BDNF mechanisms in late LTP formation: a synthesis and breakdown. Neuropharmacology 76(Pt.C) 664–676 10.1016/j.neuropharm.2013.06.024 [DOI] [PubMed] [Google Scholar]
- Park C. S., Gong R., Stuart J., Tang S.-J. (2006). Molecular network and chromosomal clustering of genes involved in synaptic plasticity in the hippocampus. J. Biol. Chem. 281 30195–30211 10.1074/jbc.M605876200 [DOI] [PubMed] [Google Scholar]
- Park C. S., Tang S.-J. (2009). Regulation of microRNA expression by induction of bidirectional synaptic plasticity. J. Mol. Neurosci. 38 50–56 10.1007/s12031-008-9158-3 [DOI] [PubMed] [Google Scholar]
- Peng Z., Cheng Y., Tan B. C., Kang L., Tian Z., Zhu Y., et al. (2012). Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat. Biotechnol. 30 253–260 10.1038/nbt.2122 [DOI] [PubMed] [Google Scholar]
- Racine R. J., Milgram N. W., Hafner S. (1983). Long-term potentiation phenomena in the rat limbic forebrain. Brain Res. 260 217–231 10.1016/0006-8993(83)90676-5 [DOI] [PubMed] [Google Scholar]
- Rajasethupathy P., Fiumara F., Sheridan R., Betel D., Puthanveettil S. V., Russo J. J., et al. (2009). Characterization of small RNAs in Aplysia reveals a role for miR-124 in constraining synaptic plasticity through CREB. Neuron 63 803–817 10.1016/j.neuron.2009.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana T. M. (2007). Illuminating the silence: understanding the structure and function of small RNAs. Nat. Rev. Mol. Cell Biol. 8 23–36 10.1038/nrm2085 [DOI] [PubMed] [Google Scholar]
- Remenyi J., van den Bosch M. W., Palygin O., Mistry R. B., McKenzie C., Macdonald A., et al. (2013). miR-132/212 knockout mice reveal roles for these miRNAs in regulating cortical synaptic transmission and plasticity. PLoS ONE 8:e62509 10.1371/journal.pone.0062509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan M. M., Mason-Parker S. E., Tate W. P., Abraham W. C., Williams J. M. (2011). Rapidly induced gene networks following induction of long-term potentiation at perforant path synapses in vivo. Hippocampus 21 541–553 10.1002/hipo.20770 [DOI] [PubMed] [Google Scholar]
- Ryan M. M., Ryan B., Kyrke-Smith M., Logan B., Tate W. P., Abraham W. C., et al. (2012). Temporal profiling of gene networks associated with the late phase of long-term potentiation in vivo. PLoS ONE 7:e40538 10.1371/journal.pone.0040538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saba R., Storchel P. H., Aksoy-Aksel A., Kepura F., Lippi G., Plant T. D., et al. (2012). Dopamine-regulated microRNA MiR-181a controls GluA2 surface expression in hippocampal neurons. Mol. Cell Biol. 32 619–632 10.1128/MCB.05896-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacktor T. C., Osten P., Valsamis H., Jiang X., Naik M. U., Sublette E. (1993). Persistent activation of the zeta isoform of protein kinase C in the maintenance of long-term potentiation. Proc. Natl. Acad. Sci. U.S.A. 90 8342–8346 10.1073/pnas.90.18.8342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayed D., Abdellatif M. (2011). MicroRNAs in development and disease. Physiol. Rev. 91 827–887 10.1152/physrev.00006.2010 [DOI] [PubMed] [Google Scholar]
- Schildt S., Endres T., Lessmann V., Edelmann E. (2013). Acute and chronic interference with BDNF/TrkB-signaling impair LTP selectively at mossy fiber synapses in the CA3 region of mouse hippocampus. Neuropharmacology 71 247–254 10.1016/j.neuropharm.2013.03.041 [DOI] [PubMed] [Google Scholar]
- Schratt G. M., Tuebing F., Nigh E. A., Kane C. G., Sabatini M. E., Kiebler M., et al. (2006). A brain-specific microRNA regulates dendritic spine development. Nature 439 283–289 10.1038/nature04367 [DOI] [PubMed] [Google Scholar]
- Scott H. L., Tamagnini F., Narduzzo K. E., Howarth J. L., Lee Y. B., Wong L. F., et al. (2012). MicroRNA-132 regulates recognition memory and synaptic plasticity in the perirhinal cortex. Eur. J. Neurosci. 36 2941–2948 10.1111/j.1460-9568.2012.08220.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M., Schwanhausser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. (2008). Widespread changes in protein synthesis induced by microRNAs. Nature 455 58–63 10.1038/nature07228 [DOI] [PubMed] [Google Scholar]
- Sheinerman K. S., Umansky S. R. (2013). Circulating cell-free microRNA as biomarkers for screening, diagnosis, and monitoring of neurodegenerative diseases and other neurologic pathologies. Front. Cell Neurosci. 7:150 10.3389/fncel.2013.00150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S., Hayashi Y., Esteban J. A., Malinow R. (2001). Subunit-specific rules governing AMPA receptor trafficking to synapses in hippocampal pyramidal neurons. Cell 105 331–343 10.1016/S0092-8674(01)00321-X [DOI] [PubMed] [Google Scholar]
- Shi S. H., Hayashi Y., Petralia R. S., Zaman S. H., Wenthold R. J., Svoboda K., et al. (1999). Rapid spine delivery and redistribution of AMPA receptors after synaptic NMDA receptor activation. Science 284 1811–1816 10.1126/science.284.5421.1811 [DOI] [PubMed] [Google Scholar]
- Siegel G., Obernosterer G., Fiore R., Oehmen M., Bicker S., Christensen M., et al. (2009). A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat. Cell Biol. 11 705–716 10.1038/ncb1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel G., Saba R., Schratt G. (2011). microRNAs in neurons: manifold regulatory roles at the synapse. Curr. Opin. Genet. Dev. 21 491–497 10.1016/j.gde.2011.04.008 [DOI] [PubMed] [Google Scholar]
- Sim S. E., Bakes J., Kaang B. K. (2014). Neuronal activity-dependent regulation of microRNAs. Mol. Cells 37 511–517 10.14348/molcells.2014.0132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sossin W. S., Lacaille J. C. (2010). Mechanisms of translational regulation in synaptic plasticity. Curr. Opin. Neurobiol. 20 450–456 10.1016/j.conb.2010.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada T., Sheng M. (2006). Molecular mechanisms of dendritic spine morphogenesis. Curr. Opin. Neurobiol. 16 95–101 10.1016/j.conb.2005.12.001 [DOI] [PubMed] [Google Scholar]
- Thomson J. M., Newman M., Parker J. S., Morin-Kensicki E. M., Wright T., Hammond S. M. (2006). Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 20 2202–2207 10.1101/gad.1444406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognini P., Pizzorusso T. (2012). MicroRNA212/132 family: molecular transducer of neuronal function and plasticity. Int. J. Biochem. Cell Biol. 44 6–10 10.1016/j.biocel.2011.10.015 [DOI] [PubMed] [Google Scholar]
- Tognini P., Putignano E., Coatti A., Pizzorusso T. (2011). Experience-dependent expression of miR-132 regulates ocular dominance plasticity. Nat. Neurosci. 14 1237–1239 10.1038/nn.2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang J., Zhu J., van Oudenaarden A. (2007). MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol. Cell 26 753–767 10.1016/j.molcel.2007.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S., Tong Y., Steitz J. A. (2007). Switching from repression to activation: microRNAs can up-regulate translation. Science 318 1931–1934 10.1126/science.1149460 [DOI] [PubMed] [Google Scholar]
- Whitlock J. R., Heynen A. J., Shuler M. G., Bear M. F. (2006). Learning induces long-term potentiation in the hippocampus. Science 313 1093–1097 10.1126/science.1128134 [DOI] [PubMed] [Google Scholar]
- Wibrand K., Messaoudi E., Havik B., Steenslid V., Lovlie R., Steen V. M., et al. (2006). Identification of genes co-upregulated with Arc during BDNF-induced long-term potentiation in adult rat dentate gyrus in vivo. Eur. J. Neurosci. 23 1501–1511 10.1111/j.1460-9568.2006.04687.x [DOI] [PubMed] [Google Scholar]
- Wibrand K., Pai B., Siripornmongcolchai T., Bittins M., Berentsen B., Ofte M. L., et al. (2012). MicroRNA regulation of the synaptic plasticity-related gene arc. PLoS ONE 7:e41688 10.1371/journal.pone.0041688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibrand K., Panja D., Tiron A., Ofte M. L., Skaftnesmo K.-O., Lee C. S., et al. (2010). Differential regulation of mature and precursor microRNA expression by NMDA and metabotropic glutamate receptor activation during LTP in the adult dentate gyrus in vivo. Eur. J. Neurosci. 31 636–645 10.1111/j.1460-9568.2010.07112.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J. M., Guevremont D., Kennard J. T., Mason-Parker S. E., Tate W. P., Abraham W. C. (2003). Long-term regulation of N-methyl-D-aspartate receptor subunits and associated synaptic proteins following hippocampal synaptic plasticity. Neuroscience 118 1003–1013 10.1016/S0306-4522(03)00028-9 [DOI] [PubMed] [Google Scholar]
- Williams J. M., Guevremont D., Mason-Parker S. E., Luxmanan C., Tate W. P., Abraham W. C. (2007). Differential trafficking of AMPA and NMDA receptors during long-term potentiation in awake adult animals. J. Neurosci. 27 14171–14178 10.1523/JNEUROSCI.2348-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J. M., Mason-Parker S. E., Abraham W. C., Tate W. P. (1998). Biphasic changes in the levels of N-methyl-D-aspartate receptor-2 subunits correlate with the induction and persistence of long-term potentiation. Brain Res. Mol. Brain Res. 60 21–27 10.1016/S0169-328X(98)00154-5 [DOI] [PubMed] [Google Scholar]
- Xu J., Chen Q., Zen K., Zhang C., Zhang Q. (2013). Synaptosomes secrete and uptake functionally active microRNAs via exocytosis and endocytosis pathways. J. Neurochem. 124 15–25 10.1111/jnc.12057 [DOI] [PubMed] [Google Scholar]
- Xu X. L., Li Y., Wang F., Gao F. B. (2008). The steady-state level of the nervous-system-specific microRNA-124a is regulated by dFMR1 in Drosophila. J. Neurosci. 28 11883–11889 10.1523/JNEUROSCI.4114-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X. L., Zong R., Li Z., Biswas M. H., Fang Z., Nelson D. L., et al. (2011). FXR1P but not FMRP regulates the levels of mammalian brain-specific microRNA-9 and microRNA-124. J. Neurosci. 31 13705–13709 10.1523/JNEUROSCI.2827-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Shu X., Liu D., Shang Y., Wu Y., Pei L., et al. (2012). EPAC null mutation impairs learning and social interactions via aberrant regulation of miR-124 and Zif268 translation. Neuron 73 774–788 10.1016/j.neuron.2012.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekta S., Shih I., Bartel D. P. (2004). MicroRNA-directed cleavage of HOXB8 mRNA. Science 304 594–596 10.1126/science.1097434 [DOI] [PubMed] [Google Scholar]
- Zeng Y., Yi R., Cullen B. R. (2003). MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc. Natl. Acad. Sci. U.S.A. 100 9779–9784 10.1073/pnas.1630797100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M. G., Toyoda H., Lee Y. S., Wu L. J., Ko S. W., Zhang X. H., et al. (2005). Roles of NMDA NR2B subtype receptor in prefrontal long-term potentiation and contextual fear memory. Neuron 47 859–872 10.1016/j.neuron.2005.08.014 [DOI] [PubMed] [Google Scholar]
- Zovoilis A., Agbemenyah H. Y., Agis-Balboa R. C., Stilling R. M., Edbauer D., Rao P., et al. (2011). microRNA-34c is a novel target to treat dementias. EMBO J. 30 4299–4308 10.1038/emboj.2011.327 [DOI] [PMC free article] [PubMed] [Google Scholar]