Abstract
CASMI (Critical Assessment of Small Molecule Identification) is a contest in which participants identify the molecular formula and chemical structure of challenging molecules using blind mass spectra as the challenge data. Seven research teams participated in CASMI2013. The winner of CASMI2013 was the team of Andrew Newsome and Dejan Nikolic, the University of Illinois at Chicago, IL, USA. The team identified 15 among 16 challenge molecules by manually interpreting the challenge data and by searching in-house and public mass spectral databases, and chemical substance and literature databases. MAGMa was selected as the best automated tool of CASMI2013. In some challenges, most of the automated tools successfully identified the challenge molecules, independent of the compound class and magnitude of the molecular mass. In these challenge data, all of the isotope peaks and the product ions essential for the identification were observed within the expected mass accuracy. In the other challenges, most of the automated tools failed, or identified solution candidates together with many false-positive candidates. We then analyzed these challenge data based on the quality of the mass spectra, the dissociation mechanisms, and the compound class and elemental composition of the challenge molecules.
Keywords: metabolomics, automated identification, data quality, mass spectrometry, challenge molecules
INTRODUCTION
Mass spectrometry (MS) is a key analytical method that is extensively used in proteomics and metabolomics. The target molecules in proteomics are proteins. Proteins consist of only 20 amino acids and their amino acid sequences can be predicted from the DNA sequences of their genes. Hence, the amino acid sequences of proteins and their peptide fragments, detected by MS, can be uniquely identified by referring to genomic DNA sequence databases. On the other hand, in the case of metabolomics, the targets are small molecules that have a molecular mass less than ca. 1,500 Da in biological samples. Tissues and blood of the human body, for example, typically contain not only endogenously synthesized human metabolites but also exogenous natural and artificial small molecules derived from foods and drugs. No complete list of small molecules in human cells, tissues, and body fluids is available and they are not predictable from human genomic DNA sequences.
To date, the identification of small molecules by MS has solely depended on searching databases that collect mass spectral data of various small molecules. When the electrospray ionization-tandem mass spectrometry (ESI-MS/MS) data of a small molecule in a biological sample match with that of a small molecule in databases, the small molecule could well be identified. MassBank,1) METLIN,2) NIST12 MS/MS Database,3) and HMDB4) are spectral databases that are available for identifying small molecules. However, the number of small molecules that have the mass spectral data in these databases is only a small fraction of that of those detectable by MS. Consequently, most of the molecules detected have been left unidentified, as unknown molecules. Thus, the identification of small molecules has been the bottleneck of metabolomics.
MS using a time-of-flight (TOF) mass spectrometry and Fourier transform (FT) mass spectrometry can accurately measure the m/z of ions.5) Accurate mass data have greatly improved the accuracy of identification. In recent years, the volume of accurate ESI-MS/MS data has rapidly increased in MassBank and other databases. However, the number of small molecules that have the accurate spectral data has not improved as rapidly. One reason for this is that the commercial availability of standard reagents of metabolites has limited the number of such small molecules in mass spectral databases.
To improve the number of small molecules with mass spectral data, MassBank, a public repository, helps researchers in depositing their accurate mass spectral data by providing Record Editor as a tool for preparing MassBank-format records from raw mass spectral data. Researchers at Eawag, Switzerland, have developed a fully automated R package tool, RMassBank,6) to promote the preparation and submission of data to MassBank. The tool prepares MassBank-format records and provides chemical annotation to product ions. The Eawag group has deposited over 6,000 accurate ESI-MS/MS data entries with chemical annotations on MassBank. Elsewhere, in Japan, the Shimadzu Corporation and Eisai Co., Ltd. have collaborated to develop Mass++ as a tool for assisting in the generation of MassBank formatted records.7) Furthermore, the mass spectral database also becomes the chemical resource for the in silico modeling of collision-induced dissociations (CID) and for analyzing the empirical relationships between product ions or neutral loss molecules and chemical substructures.
For the manual and automated identification of small molecules, chemical substance databases are as essential as mass spectral databases. Once the molecular formula of unknown molecules can be calculated from mass spectral data, comprehensive chemical substance databases such as CAS,8) PubChem,9) and ChemSpider10) are the sources of candidate molecules. For metabolomics, metabolite databases such as KEGG11) and KNApSAcK12) are better sources, although they do not collect all of the known metabolites that plants, microorganisms, and other living organisms endogenously produce in species-specific metabolic pathway networks.13)
Such technical progress in MS, coupled with the increase in accurate mass spectral data and in the number of chemical substances in databases, makes automated tools for use in identification a reality. Researchers in metabolomics now hold the opinion that, in the near future, automated tools for the identification of small molecules will replace the low throughput manual identification methodology.
The first CASMI (Critical Assessment of Small Molecule Identification) was initiated by Emma Schymanski and Steffen Neumann in 2012.14) The contest participants, manually or by using automated tools, interpreted the challenges that were blind data by analyzing challenge molecules by single stage (MS1) and second stage (MS2) mass spectrometry and then identified the molecular formula (Category 1) and the chemical structure (Category 2) of the challenge molecules. When the output of the manual or automated methods indicated two or more solution candidates for a challenge, the participants submitted a list of solution candidates sorted by the level of confidence for the identification. CASMI provided an opportunity to evaluate the performance of automated tools and to compare them with that of the manual method.
The second contest, CASMI2013, was organized by 10 Japanese researchers, who are the authors of this article. The organizers carefully prepared MS1 and MS2 data as challenges based on the following three points of view. (1) All of the ions that are necessary for the identification were observed in each set of challenge data. A lack of any one of such ions might significantly affect the accuracy of identification. (2) Compound class, elemental composition and the molecular mass of the challenge molecules are as diverse as possible. (3) Challenge data might include experimental inadequacy of MS due to instrumental instability and instrument type that researchers encounter in their laboratory.
CASMI2013 provided no challenge category for GC-MS data, although the first CASMI did.
METHODS
Materials and analytical methods of the CASMI2013 challenges
As a general rule, CASMI provides one or more sets of MS1 and MS2 data and metadata for each challenge.
MS1 and MS2 data of the challenge molecules are given as “MSpos” and “MSMSpos” files or “MSneg” and “MSMSneg” files for the positive or negative ion mode, respectively, on the “Challenge data” page of the CASMI2013 web page (http://www.casmi-contest.org/2013/challenges.shtml). Metadata, including the analytical method of MS, mass accuracy, LC retention time, and the biological origin of the challenge molecule, if available, are specified in the “AnalyticalMethods” file for each challenge molecule. All mass spectra and metadata were deposited on MassBank with the record IDs from MSJ00001 to MSJ00034.
Evaluation of the performance of the methods
The CASMI2013 organizers adopted the contest rules and the evaluation measures that were established for the first CASMI.14) For each challenge, participants attempted to identify the molecular formula (Category 1) and/or chemical structure (Category 2) of the challenge molecules by interpreting the challenge MS1 and MS2 data using manual processing methods or automated tools. When participants submitted two or more solution candidates in an entry to the Categoriy 1 or 2 challenges, they were required to sort the solution candidates in the order of the confidence level based on their own scoring methods. Because two or more candidates could have the same score, incorrect candidates might be scored equal to or better than the correct candidate. In the present study, such incorrect candidates are false-positive candidates and unfavorable because they might significantly affect real-life identification when automatic methods are used.
For an entry to a challenge, first, the number of the candidates with scores better than the correct candidate (BC) and that of those with a score equal to the correct candidate (EC) are calculated. Second, the rank of the correct candidate is calculated by Eq. (1):
| (1) |
where the rank is equivalent to the absolute rank, rankWorstCase, defined in the first contest.14) As the correct candidate at rank=1 is the most reliable candidate in the entry, the rank is an index of the accuracy of the identification method.
The rank of the correct candidate is calculated for each entry to a challenge. The entry that is evaluated as the best rank among the entries wins the challenge. When two or more entries result in the same best rank or rank=1 for the challenge, all win the challenge, equally. The team that wins the most challenges in the two categories is the winner of CASMI2013.
The rank and statistics calculations of all the entries were performed by the organizers of the first CASMI.
CHALLENGE MOLECULES AND MASS SPECTRA
Sixteen organic molecules with their corresponding MS1 and MS2 data were selected as the challenge molecules, and the Categories 1 and 2 challenge data, respectively, to assess individual methods in CASMI2013. Challenge molecules included metabolites, agrochemicals, environmental chemicals, and synthetically prepared small molecules. The mass range of the challenge molecules was between 210 Da (Challenge 4) and 1,442 Da (Challenge 7). The molecular formulae of most of the challenge molecules consisted of various combinations of C and H, or neither, or one or more N, O, P, S, Cl, and F atoms. The formula of Challenge 15 contained 17 fluorine atoms. Most of the challenge data were analyzed with a mass accuracy within 10 ppm. In Challenges 15 and 16, MS2 data were nominal mass data. MS1 data for Challenges 7, 8, 13, and 14 were unavailable. The organizers did not invite the entry for these four challenges to Category 1; instead, they provided their molecular formulae. As a result, the entry called for 12 challenges in Category 1 and 16 challenges in Category 2. Thus, the challenge molecules had sufficient chemical and physical diversity to enable us to adequately evaluate the performance of both the manual and automated methods.
Aim of the challenges, and ions leading to the solutions
This section is not a textbook “answers to questions” section, where college students learn how to interpret mass spectra. In this section, each challenge consists of “Aim of the challenge” and “Ions leading to the solution.” The latter assures the participants that the MS1 and MS2 data of the challenge provide all the isotope peaks and product ions that are necessary for identifying the molecular formulae and chemical structures of compounds.
Challenge 1. Aim of the challenge: Due to the physicochemical properties of the challenge molecule, only a few product ions were observed in MS2 data. In such a case, the solution is refined to remove false-positive candidates by considering the biological origin of the challenge molecule.
Ions leading to the solution: MS1 data suggest that the neutral molecule formed [M+H]+ and [M+Na]+ ions. It contains an odd number of N atoms. The relative intensities of the isotope peaks of [M+H]+, which are 1, 0.22, and 0.023, suggest that the precursor ion consists of 19±2 carbon and 1 or 3 nitrogen atoms. The molecular formulae C10H9O3+, C9H5O2+, and C8H5O+ are assigned to product ions m/z 177.0548, 145.0284, and 117.0337, with a mass accuracy <1.8 ppm. The [M+H]+ ion has at least 3 oxygen atoms. The molecular formula of the [M+H]+ ion is C18H20NO4+, with an error of 7.3 ppm.
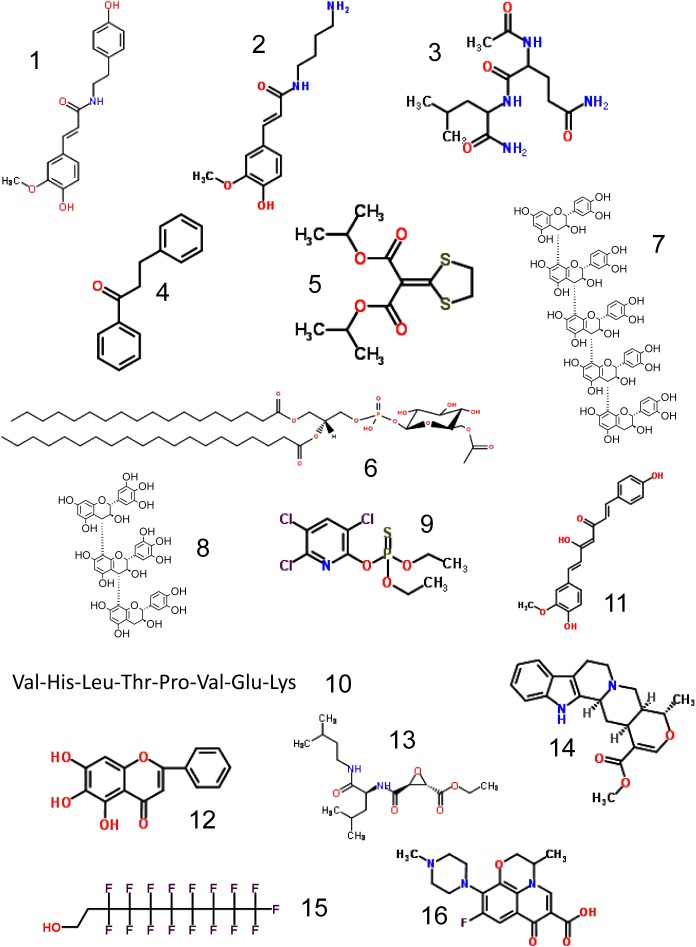
A KNApSAcK search with C18H19NO4 as keyword returned a hit list. Only N-trans-feruloyltyramine (1 in Fig. 1), isolated from three Solanaceae plants, appeared in the list. The feruloyl group is the source of the above three product ions. The cis isomer and isoferuloyltyramine are known in other plant species. The molecular formula and mass of the challenge molecule are C18H19NO4 and 313.1314 Da.
Fig. 1. The chemical structures of CASMI2013.
Challenge 2. Aim of the challenge: Considering that the major product ions are common to those observed in Challenge 1, the challenge molecule has a substructure similar to the previous challenge.
Ions leading to the solution: Three isotope peaks of [M+H]+, with relative intensities of 1, 0.17, and 0.01, suggest that the molecular ion consists of 15±1 carbon atoms. The [M+H]+ ion, m/z 265.1524, which generated the product ion, m/z 248.1260, by the loss of NH3, has a primary amino group. The [M+H]+ ion consists of none or an even number of nitrogen atoms because the mass of the ion is an odd number, 265.
The product ions, m/z 177.0546 and 145.0276, are suggestive of a feruloyl group, although Challenge 2 lacks the third product ion, m/z 117.0337, observed in Challenge 1. From the observed mass, a possible molecular formula of the [M+H]+ ion is C14H21N2O3+, with an error of 8.5 ppm.
The challenge molecule was isolated from Solanaceae plants and contains aromatic structures. With the molecular formula C14H20N2O3 as a query, KNApSAcK gives N-trans-feruloylputrescine (2 in Fig. 1). The molecular formula and mass of the challenge molecule are C14H20N2O3 and 264.1474 Da.
Challenge 3. Aim of the challenge: Four similar substructures are included in the chemical structure. This is the reason why only three product ions are observed in the challenge data.
Ions leading to the solution: MS1 data show three peaks at m/z 301.1843 [M+H]+, m/z 323.1669 [M+Na]+, and m/z 339.1369 [M+K]+. The following four conditions lead to C8H24N6O6, C9H20N10O2, and C13H24N4O4 as the candidate molecular formulae: (1) m/z 301.1843 [M+H]+ within a mass tolerance of ≤10 ppm, (2) the presence of an amide group in the chemical structure, indicating the presence of N and O atoms, (3) the nitrogen rule, and (4) a degree of unsaturation >1. A PubChem search with C8H24N6O6 and C9H20N10O2 resulted in no small molecules. A search with C13H24N4O4 resulted in more than 400 small molecules. Thus, only C13H24N4O4 is a possible molecular formula.
MS2 data are less informative: only three product ions, m/z 284.1600, 171.0774, and 131.1189, were observed. The ion m/z 284.1600, which is ca. 17 Da less than the precursor ion, suggests that the challenge molecule contains one or more terminal amide groups and/or primary amino groups. No ion that is ca. 18 Da less than the precursor ion was observed. This suggests that the compound contains no OH groups.
The product ions m/z 131.1189 and 171.0774 are complementary fragments generated by the cleavage of the same covalent bond. The challenge molecule is neither a urea derivative, RN–C(=O)–NR′, nor a diacylamine derivative, RC(=O)–NR′–C(=O)R″, because these chemical structures would give many more product ions due to the cleavage of the covalent bond between C(=O) and N atoms. Instead, it is a peptide as no other product ion was observed. The solution is N–Acetyl–Gln–Leu–amide (3 in Fig. 1). The two peptide fragments: m/z 131.1189 [H3N–C5H10–CONH2]+ and 171.0774 [Ac–HN–C4H7NO–CO]+ are y- and b-type ions, respectively, which are preferentially generated by the low-energy CID of peptides. N–Acetyl–Gln–Ile-amide might give spectra that are identical to or similar to those of Challenge 3. The molecular formula and mass of the challenge molecule are C13H24N4O4 and 300.17974 Da.
Challenge 4. Aim of the challenge: The challenge molecule, which has a low molecular mass, has a simple chemical structure: it consists of only C, H, and O atoms. Consistent with such chemical simplicity, the simple fragmentation pattern provides less information. Hence, high-resolution data help in the assignment of product ions. In addition, the intent of this challenge is to evaluate how automated methods deal with the migration of two hydrogen atoms from an aliphatic carbon atom to a carbonyl oxygen atom. The loss of the carbonyl oxygen atom as H2O is suggestive of such a migration.
Ions leading to the solution: The isotope pattern shows that the challenge molecule contains neither halogen nor sulfur atoms. The product ion, m/z 91.05422, suggests a benzyl or tropylium cation, C7H7+. The differences between the ions, m/z 91.05422, 105.07002, and 133.06489, indicate a methylene group, CH2, and a carbonyl group, CO, respectively. Note that ion m/z 105.07002 does not indicate a benzoyl cation because a benzoyl cation should be m/z 105.03349. The m/z 193.10131 ion indicates a 2H migration followed by the loss of H2O from the precursor ion. Dihydrochalcone (1,3-diphenyl-1-1propanone) (4 in Fig. 1) is the solution. 2-(2-Methylphenyl)-1-phenylethanone and the isomer, 2-(4-methylphenyl)-1-phenylethanone, might give spectra that are the same as those of Challenge 4 by low-energy CID. The molecular formula and mass of the challenge molecule are C15H14O and 210.1044 Da.
Challenge 5. Aim of the challenge: The important objectives of this challenge are to determine the correct molecular formula of the molecule, which contains two sulfur atoms and has a symmetrical chemical structure.
Ions leading to the solution: The isotope pattern suggests that the molecule contains two sulfur atoms. The high-resolution mass of [M+H]+ can indicate the possible composition with ease. The major fragmentations are the loss of an isopropyl group (m/z 249.02495) and the cleavage of an ester bond (m/z 231.01417). The other peaks, at m/z 172.97256, 188.96743, and 206.97805, are assigned as different combinations of these two fragmentations. The solution is isoprothiolane (5 in Fig. 1). Dipropyl 1,3-dithiolan-2-ylidene propanedioate might give spectra that are the same as those of Challenge 5 by low-energy CID. The molecular formula and mass of the challenge molecule are C12H18O4S2 and 290.0647 Da.
Challenge 6. Aim of the challenge: MS1 and MS2 data obviously do not provide sufficient ions to deduce a single molecular formula and/or a chemical structure. This is a situation that researchers often encounter in the laboratory. If the participants are experienced in MS, they may manually work out a likely molecular formula and possible chemical structures from such poor data by extracting other available information. However, the CASMI2013 organizers are also interested in how automated methods can be used to reach the correct molecular formula starting from several candidate formulae, including the correct one. As this molecule came from animal tissue, limiting the elements that constitute the molecule to C, H, N, O, P, and S is a reasonable assumption. Limiting the elements to C, H, N, O, P, and S and narrowing the mass tolerance to 3 ppm are not sufficient to refine the possible molecular formulae; many candidates remain. However, if one takes advantage of the accurate mass of the MS2 data, it is possible to reduce the number of possible molecular formulae to just a few. Here, some common knowledge of organic chemistry and MS is required.
Ions leading to the solution: Lower mass product ions, m/z 152.99581, 283.26412, and 311.2954, suggest their chemical formulae uniquely within the elemental composition C, H, N, O, P, and S and a mass tolerance ≤3 ppm. As this is a negative ion spectrum, m/z 283.26412 (C18H35O2−) and 311.2954 (C20H39O2−), the ions are carboxylate anions. These two carboxylate anions should be independent of each other, i.e., the smaller anion is never derived from the larger anion, as the loss of C2H4 from a saturated hydrocarbon chain is not likely to occur under the low-energy CID MS2 conditions. The ion m/z 152.99581 (C3H6O5P−) is likely to be a phosphate anion, –OP(=O)O2−, and the three carbons in this ion are not likely to be a part of the carboxylate anions.
Based on the elemental compositions of the three lower mass product ions, the precursor ion should contain: C41 (18+20+3); H78 (35+39+6−2), where the minus 2 allows for the possibility of double H-transfer; O7 (2+2+5−2), where the minus 2 allows for the possibility of forming two ester bonds; and with a minimum of P1.
With the minimum atoms constraint and the assumption that the precursor ion is an even-electron anion with a minimum of two double bonds (two carbonyl groups), the possible formulae for the precursor ion can be limited to 11, including C49H92O14P− (Table 1). A few of them may be excluded when the composition of the counterpart to [C41H78O7P] is considered. For example, the first candidate is unlikely, since the counterpart represents a phosphine-type moiety, which is highly susceptible to oxidation. On the other hand, the second candidate (C49H92O14P−) is a good candidate formula, since the counterpart contains a reasonable number of C, H, and O atoms. As the list became a manageable size at this point, a further in-depth inspection of the candidates, including their chemical and biological relevance, becomes possible. The solution is phosphatidyl-6-acetyl-glucose (18 : 0/20 : 0) (6 in Fig. 1). The molecular formula and mass of the challenge molecule are C49H93O14P and 936.6303 Da.
Table 1. Candidate chemical formulae of the precursor anion of Challenge 6.
| No. | Candidate formulae | Calculated mass | Error1) (mDa) | Error1) (ppm) | RDB2) | Counterpart to [C41H78O7P] |
|---|---|---|---|---|---|---|
| 1 | C53 H94 O7 P3− | 935.62180 | 0.595 | 0.636 | 9 | C12 H16 P2 |
| 2 | C49 H92 O14 P− | 935.62302 | −0.631 | −0.674 | 5 | C8 H14 O7 |
| 3 | C46 H84 N10 O8 P− | 935.62167 | 0.718 | 0.767 | 11 | C5 H6 N10 O |
| 4 | C44 H89 N8 O9 P2− | 935.62333 | −0.938 | −1.002 | 6 | C3 H11 N8 O2 P |
| 5 | C46 H92 N6 O7 P S2− | 935.62121 | 1.184 | 1.2657 | 5 | C5 H14 N6 S2 |
| 6 | C50 H96 O9 P S2− | 935.62389 | −1.501 | −1.6045 | 4 | C9 H18 O2 S2 |
| 7 | C53 H92 O9 P S− | 935.62052 | 1.871 | 2.000 | 9 | C12 H14 O2 S |
| 8 | C50 H88 N4 O10 P− | 935.62436 | −1.968 | −2.103 | 10 | C9 H10 N4 O3 |
| 9 | C45 H88 N6 O12 P− | 935.62034 | 2.055 | 2.196 | 6 | C4 H10 N6 O5 |
| 10 | C43 H88 N10 O8 P S− | 935.62504 | −2.655 | −2.837 | 6 | C2 H10 N10 O S |
| 11 | C50 H98 O7 P3 S− | 935.62517 | −2.777 | −2.968 | 4 | C9 H20 P2 S |
1) Error is the difference between the observed mass, m/z 935.62239, and the calculated mass. 2) RDB is Rings plus Double Bonds. The number is calculated for neutral molecule corresponding to each candidate anion, i.e., a proton is added to each candidate formula to calculate RDB.
Challenge 7. Aim of the challenge: The challenge molecule is a pentamer of catechin with a large molecular mass, 1,442 Da.
Ions leading to the solution: The molecular formula of the [M+H]+ ion is C75H63O+30. Five product ions, m/z 1443.345, 1155.2795, 867.2144, 579.1493, and 291.0875, show the neutral loss of the same molecule, mass 288.0644±0.001727. This suggests that the solution is a pentamer of C15H12O6, calculated mass 288.06339; e.g., H–(C15H12O6)5–H. The solution is a catechin pentamer, the C4–C8 flavan bond of which is cleaved by CID. Other isomers such as mixed polymers of catechin and/or epicatechin that form C4–C8 and/or C4–C6 bonds would be expected to give similar product ions in MS2 data. The solution is cinnamtannin A3 (7 in Fig. 1). The molecular formula and mass of the challenge molecule are C75H62O30 and 1,442.3326 Da.
Challenge 8. Aim of the challenge: The challenge molecule is a trimer consisting of two gallocatechin groups and one catechin group, in a specific sequence.
Ions leading to the solution: The molecular formula of the [M+H]+ ion is given as C45H39O+20; calculated m/z 899.20293. The most likely molecular formula of the product ions, m/z 747.1633, 731.1682, 609.1281, 595.1481, 443.0999, 441.0839, and 291.0895, are C37H31O+17, C37H31O+16, C30H25O+14, C30H27O+13, C22H19O+10, C22H17O+10, and C15H15O6+, with a mass accuracy of +8.5 ppm on average. The solution is prodelphinidin C2 (8 in Fig. 1), which consists of two gallocatechin groups and one catechin group. The product ions, m/z 747.1633, 731.1682, and 443.0999, suggest that the precursor ion, [M+H]+, is dissociated by cleavage of the C4–C8 flavan bond by a retro Diels–Alder reaction.15) The sequence order of gallocatechin and the catechin groups in the solution is suggested by m/z 611.134, a dimer of gallocatechin, and 595.1481, gallocatechin–catechin or catechin–gallocatechin. Two other product ions, m/z 747.1561 and 731.1682, are suggestive of gallocatechin–gallocatechin–catechin. The molecular formula and mass of the challenge molecule are C45H38O20 and 898.1957 Da.
Challenge 9. Aim of the challenge: Researchers usually take stepwise procedures to identify a molecule by a manual method. It will be interesting to determine whether stepwise procedures are also taken when automated methods are used.
Ions leading to the solution: Three isotope peaks at m/z 349.93369 ([M+H]+, monoisotopic mass of the precursor ion), 351.93064, and 353.92753, observed at ca. 2 mass unit difference, with relative intensities of 100, 96, and 30, strongly suggest the presence of three chlorine atoms; these give the calculated relative intensities of 100, 98, and 32. Two isotope peaks at m/z 349.93369 and 350.93724 are observed at relative intensities of 100 and 8.2, which suggests 8±1 as the number of carbon atoms. The number of carbon atoms is much lower than the number that would generally be expected from the molecular mass of the precursor ion.
The monoisotopic mass of the precursor ion, m/z 349.93369, is slightly less than the nominal mass of 350. Hence, it appears that the molecule contains more heteroatoms, in addition to three chlorine atoms. The molecule also contains heteroatoms such as O, P, and S. Furthermore, the molecule contains an odd number of nitrogen atoms because the nominal mass of the precursor ion, 350, is an even number. Finally, the molecular formula is estimated as C9H12Cl3NO3PS+, which gives the calculated m/z 349.9336 with an accuracy of 0.4 ppm.
Two higher mass product ions, m/z 321.90224 and 293.87101, suggest two sequential losses of a neutral molecule, C2H4, with the calculated mass of 28.0313, from [M+H]+. Similarly, the difference between the product ion, m/z 303.89174, and [M+H]+, and that of two product ions, m/z 321.90224 and 275.86044, corresponds to the loss of a neutral molecule, C2H6O, with the calculated mass of 46.0419. The molecule should therefore contain two ethoxy groups.
The chemical formula H4O3PS+, with the calculated m/z 114.96133, is assigned to a lower mass product ion, m/z 114.96142, which is [S=P(OH)3+H]+. Three ions, m/z 171.02398, 142.99266, and 114.96142, are produced by the successive loss of a neutral molecule, C2H4. Thus, a diethyl thiophosphate, [S=P(OH)(OCH2CH3)2+H]+, is assigned to the product ion, m/z 171.02398. The counterpart of the molecule is found as the ion m/z 197.92748; it is a trichlorohydroxypyridine group, [C5H2Cl3NO+H]+, with a calculated mass of 197.92747 Da. Finally, the solution is chlorpyrifos (9 in Fig. 1). As the position of the three chlorine atoms and a hydroxyl group on the pyridine ring cannot be determined by low-energy CID, at least 12 possible isomers (including the solution) are possible that might give spectra that are the same as those of Challenge 9 by low-energy CID. The molecular formula and mass of the challenge molecule are C9H11Cl3NO3PS and 348.9263 Da.
Challenge 10. Aim of the challenge: This challenge tests whether automated methods are available for the determination of the amino acid sequence of oligopeptides.
Ions leading to the solution: The nominal mass of the precursor ion, m/z 922.53505, is an even number. The challenge molecule contains an odd number of nitrogen atoms. On considering the decimal fraction of the precursor ion, 0.53505, it is likely that the molecule consists of as many hydrogen atoms as typical organic molecules having a molecular mass of ca. 922. The relative intensities of the isotope peaks, m/z 922.53505 to m/z 925.54322, which are 1, 0.483, 0.139, and 0.027, suggest that the precursor ion consists of ca. 45 carbon atoms, as well as hydrogen, nitrogen, and oxygen atoms. The molecular formula of the precursor ion is predicted as C42H72N11O+12. It has the calculated mass of 922.53564 Da, which deviates by 0.6 ppm from the observed peak.
The mass difference between any pair of major product ions suggests that they are peptide fragments. Generally, the assignment of amino acid residues to the MS2 data is made easier by removing the product ions that were generated from other product ions by the loss of a H2O molecule. For example, ignore the peaks m/z 904.52459, 805.45627, 758.41915, and 668.39760 that were generated from the peaks m/z 922.53499, 823.46710, 776.42976, and 686.40800, by dehydration. As the challenge data were analyzed by low-energy CID, mainly y-, a-, and b-type cleavages generated the peptide fragments. For example, the above three cleavages at the site between ‘VHLTPV’ and ‘EK’ give the ions m/z 276.15538, 647.38730, and 619.3926, which are actually observed in the challenge data. If the amino acid sequence is ‘VHLTPEVK,’ the corresponding three ions should be m/z 246.1812, 677.3617, and 649.3668. Only the ion m/z 677.36166 is observed as a small peak. Thus, the amino acid sequence of the solution is uniquely determined as ‘VHLTPVEK’ (10 in Fig. 1). ‘VHITPVEK’ might give spectra that are the same as those in Challenge 10 by low-energy CID. The molecular formula and mass of the challenge molecule are C42H71N11O12 and 921.5283 Da.
Challenge 11. Aim of the challenge: Curcuminoids are popular molecules. This challenge is of interest to chemists concerned with natural products derived from plants, whose knowledge and experience should enable them to deal with the challenge successfully. It will be interesting to see how automated tools can be used to identify the chemical structure of a curcuminoid.
Ions leading to the solution: Four pairs of major product ions, m/z 217.0506 and 187.0414, m/z 175.0426 and 145.0308, m/z 173.0625 and 143.0534, and m/z 149.0630 and 119.0538, have a mass difference of 30.0 Da; i.e., that of OCH3–H. These product ion pairs are typically observed in MS2 data of the molecules containing both feruloyl and coumaroyl moieties. The challenge solution is demethoxycurcumin (11 in Fig. 1), which is one of the major constituents of turmeric.
Jiang et al.16) proposed a fragmentation scheme of curcuminoids in ESI-MS/MS. According to the scheme, the diketo isomer gives all of the observed product ions through its two diketo–enol tautomers. In their proposed scheme, only a mixture of the two diketo–enol tautomers could generate all the observed product ions, because the two tautomers give different sets of product ions. The molecular formula and mass of the challenge molecule are C20H18O5 and 338.1154 Da.
Challenge 12. Aim of the challenge: The challenge molecule is a flavone. It is also a popular secondary metabolite produced by plants.
Ions leading to the solution: The product ion at m/z 251.0347 in the MS2 data is assigned as [M−H−H2O]−, which is observed only when the flavone has a vicinal hydroxyl group. Successive losses of CO and/or CO2 give major product ions such as m/z 241.0529 [M−H−CO]−, 225.052 [M−H−CO2]−, 223.0396 [M−H−H2O−CO]−, 197.0581 [M−H−CO−CO2]−, 195.0397 [M−H−H2O−2CO]−, and 169.0675 [M−H−2CO−CO2]−.17,18) In the literature,17) a product ion m/z 171 has been reported to be a characteristic ion of flavones with a trihydroxyl group on the A ring, although it is not observed in Challenge 12.
The solution is baicalein (C15H10O5) (12 in Fig. 1), which is one of the major constituents of Scutellariae Radix (the root of Scutellaria baicalensis). The molecular formula and mass of the challenge molecule are C15H10O5 and 270.0528 Da.
Challenges 13 and 14. Aim of the challenges: The aims of these two challenges are quite different from the other challenges. The organizers did not show which of the observed ions in the MS2 data are essential for solving the problem. We have no proof of the quality of the challenge data.
The molecules in challenges 13 and 14 are secondary metabolites and their derivatives are found in plants or microorganisms. In Challenge 13, as the mass error of the acquired data were more than 5 ppm, the m/z values of the observed fragment peaks were artificially generated by applying mass errors of less than 5 ppm to the theoretical m/z values. In Challenge 14, the observed mass error of the m/z values was within 5 ppm. The CASMI2013 organizers are also interested in how the participants will find the solutions to these two challenges by using manual or automated methods. The solutions to Challenges 13 and 14 are aloxistatin (13 in Fig. 1) and tetrahydroalstonine (14 in Fig. 1), respectively. The molecular formula and mass of the challenge molecule 13 are C17H30N2O5 and 342.2155 Da, and those of 14 are C21H24N2O3 and 352.1787 Da.
Challenge 15. Aim of the challenge: Fluorochemicals, which were introduced about 60 years ago, have truly unique chemical and physical properties. The number of newly developed fluorochemicals is constantly increasing. Coupled with this increase, knowledge of their ionization and fragmentations in MS has increased. The total number of fluorine atoms in the challenge molecule is 17, which might exceed the maximum number of halogens that automated methods would be able to deal with. In addition, the loss of HF and CF2 was observed.
Ions leading to the solution: Product ions that are related by the neutral losses of 20.006 Da and 49.997 Da are observed in the MS1 scan analysis and HCD analyses, respectively. These losses correspond to the molecules of HF and CF2. The elimination of HF is familiar to the fragmentation of polyfluoroalkyl groups. Since four successive HF eliminations are observed in “MSneg” of the challenge, m/z 462.99742, 442.99065, 422.98506, 402.9787, and 382.97277, it is inferred that the polyfluoroalkyl chain of the [M−H]− ion contains four hydrogen atoms. The neutral loss between two major product ions, m/z 354.97782 and 382.97277, [M−H−4HF]−, is 27.9949 Da, which corresponds to the loss of CO. If the elements constituting the challenge molecule are limited to C, H, O, and F atoms, then C10H4F17O− is deduced as the chemical formula of [M−H]−. The precursor ion shows a negative mass defect although it has a relatively high molecular mass. The possibility that it includes many atoms with negative mass defects, such as F, S, and P, should not be excluded.
All of the product ions observed in “MSnegCE40” data, where CE40 is 40 V of collision energy, are mutually related by a loss of CF2, 49.9968 Da; e.g., m/z 392.97464 and 342.97800. In addition, the product ion m/z 68.99463 is a CF3− ion. These product ions suggest that the compound contains a linear perfluorinated alkyl chain rather than a branched one because the latter would not give successive ions that are related by a loss of CF2. Therefore, the most likely neutral molecule is n-C8F17C2H4OH. The ion m/z 506.98634 is an adduct of [M−H]− with CO2. MS1 and MS2 data similar to the challenge data have been reported in the literature.19) The solution is 2-(perfluorooctyl)ethanol (15 in Fig. 1). The molecular formula and mass of the challenge molecule are C10H5F17O and 464.0069 Da.
Challenge 16. Aim of the challenge: Ions that are observed with a very weak peak intensity on high spectral resolution instruments are often important in terms of the identification of small molecules. This challenge evaluates how manual and automated methods chemically interpret ions with a weak peak intensity.
Ions leading to the solution: A characteristic neutral loss observed in product ions is 43.99 Da, which suggests the loss of CO2. The other loss is 57.0581 Da. When MassBank was searched by a query of the peak difference 57.0581 Da, with a tolerance of 0.005 Da, it retrieved MS2 data for about 60 candidate molecules. These molecules are heterocyclic compounds containing nitrogen atom(s), such as piperidine.
When the product ions that are commonly observed in both “MSposCE40” (analyzed on Orbitrap) and “MSMSposCE40” (analyzed using QqQ) data were extracted, the chemical formulae are assignable under the assumption that the ions consist of C, H, N, and O atoms and were observed at a mass error <0.001 as follows; m/z 362.1526, [M+H]+; 318.1626, [M+H−CO2]+; 261.10451, [M+H−C4H7NO2]+; 247.08881, [M+H−C5H9NO2]+; 245.07316, [M+H−C5H11NO2]+; 233.07312, [M+H−C6H11NO2]+; 221.07309, [M+H−C7H11NO2]+; 219.05744, [M+H−C7H13NO2]+; 205.04175, [M+H−C8H15NO2]+; 194.02574, [M+H−C9H16N2O]+; 193.04172, [M+H−C9H15NO2]+; 179.03857, [M+H−C9H15N2O2]+; 178.03072, [M+H−C9H16N2O2]+; 122.04078, [M+H−C11H16N2O4]+; 70.06578 [C4H8N]+; and 58.06577, [C3H8N]+. Only 199.05144 cannot be determined under this assumption. The difference between 219.05744 and 199.05144 is 20.006. This difference is specific to the fragmentation of the molecules that have fluorine atom(s), similar to Challenge 15. If this molecule includes one fluorine atom, then 122.04078 can be determined to be C7H5NF+. Therefore, the chemical formula of the precursor ion, C18H21N3O4F+, can be deduced.
A search of ChemSpider returned more than 200 candidate molecules with the chemical formula C18H20FN3O4. Although ofloxacin (16 in Fig. 1) is most likely the correct candidate, the possibility of other isomers with heterocyclic substructures cannot be ruled out. The molecular formula and mass of the challenge molecule are C18H20FN3O4 and 361.1438 Da.
Commended candidate structures for Category 2
Because most of the CASMI2013 challenge MS2 data were analyzed under low-energy CID conditions, all of the alkyl chain isomers, such as propyl and isopropyl isomers, are likely to give the same or similar MS2 data. Challenges 3, 5, and 10 represent such cases. Commended structures might also be possible in other challenges, e.g., branched-chain fatty acids in Challenge 6. However, these alternatives were not added to the list of commended candidates because branched-chain fatty acids are quite rare in animals.
Challenges 1 and 2 are another case. The feruloyl group is a common substructure in the two solutions. The common substructure has isomers at the double bond and at the ring substituent positions. The cis isomer and isoferuloyl isomer are known metabolites in plants. Furthermore, all of the positional isomers of substituted aromatic molecules such as in Challenges 4, 7, 8, and 9 could give the same MS2 data.
PARTICIPATING TEAMS AND THEIR METHODS
In this section, the seven teams that participated in CASMI2013 are introduced, and their methods are summarized. The participating teams briefly described their methods of identifying the challenge molecules in the metadata that were submitted with their solution candidates. The metadata are collected in section S1 in Supplementary Information. Detailed descriptions of the automated and manual methods used by the participants have been published as research articles in the CASMI2013 special issue in Mass Spectrometry. The participants, as the authors, prepared the research articles after the solutions were announced; hence, they were able to report the reasons for why they won some challenges and failed others. The participating teams in CASMI2013 were as follows.
(1) The team of Andrew Newsome and Dejan Nikolic, the University of Illinois at Chicago, IL, USA, team ‘Newsome,’ participated in CASMI2013 with a manual method.20) (2) The team of Lars Ridder and Justin J. J. van der Hooft, Wageningen University, Wageningen, The Netherlands, and the University of Glasgow, UK, team ‘Ridder,’ participated in the contest with an automated tool, MAGMa.21–24) (3) The team of Kai Dührkop and Sebastian Böcker, Friedrich-Schiller-University, Jena, Germany, team ‘Dührkop,’ participated in only Category 1 with an automated tool, SIRIUS.25–27) This team also participated in the first CASMI.28) (4) The team of Emma Schymanski and Steffen Neumann, Eawag, Dübendorf, Switzerland, and the Leibniz Institute of Plant Biochemistry, Halle (Saale), Germany, team ‘Schymanski,’ participated with the solution candidates prepared by three automated tools: MOLGEN-MS/MS,29) MetFrag,30) and MetFusion.31,32) This team also participated in the first CASMI.14) (5) Daniel L. Sweeney, MathSpec, Inc., IL, USA, team ‘Sweeney,’ participated in only the Category 2 challenges with a commercially available automated tool, Rational Numbers FragSearch,33) and a manual method. He has constructed his own in-house accurate mass fragmentation database.34–36) (6) The team of Felicity Allen and Russ Greiner, the University of Alberta, Alberta, Canada, team ‘Allen,’ participated in Categories 1 and 2 challenges with an automated method, CFM.37,38) The team missed an opportunity to submit their paper to the CASMI2013 special issue. (7) Tsubasa Miyazaki and Hisayuki Horai, Ibaraki National College of Technology, Ibaraki, Japan, participated in Category 2 challenges with manually prepared solution candidates. They submitted no correct candidate.
The winner of CASMI2013 and the best automated method of CASMI2013
In Category 1, the participants identified the molecular formula of the challenge molecules by interpreting the challenge MS1 and MS2 data. The CASMI2013 organizers provided 12 MS1 data for the Category 1 challenges. For the other challenges, Challenges 7, 8, 13, and 14, the organizers provided molecular formulae instead. Five teams submitted their solution candidates to 12 challenges. The results of Category 1 by the participants are summarized in Table 2. Statistics of the results of Category 1 by participants are summarized in Table S1 in Supplementary Information.
Table 2. Rank and win of the correct candidate in Category 1. The number in each cell shows the rank of the correct candidate submitted by each team. For each challenge, the team that submitted the correct candidate in the best rank won the challenge. The rank that won the challenge is highlighted in bold. “—” or blank shows that the team has a submission with no correct candidate or no submission to the challenge, respectively. Newsome, Schymanski, Allen, Dührkop, and Ridder are team names (see Text).
| Challenges | Newsome | Schymanski | Allen | Dührkop | Ridder |
|---|---|---|---|---|---|
| 1 | 1 | 2 | 1 | 1 | 1 |
| 2 | 1 | 1 | — | 1 | 1 |
| 3 | 1 | 1 | — | 1 | 1 |
| 4 | 1 | 1 | 1 | 1 | 1 |
| 5 | 1 | 1 | 3 | 1 | 1 |
| 6 | 1 | 8 | 1 | 1 | |
| 9 | 1 | 1 | 1 | 1 | 1 |
| 10 | 1 | 45 | 1 | 1 | 1 |
| 11 | 1 | 4 | 1 | ||
| 12 | 1 | 9 | — | ||
| 15 | 1 | — | — | ||
| 16 | 1 | — | 2 | 1 | |
| Win/Submit1) | 12/12 | 5/12 | 4/8 | 10/12 | 8/8 |
| Methods2) | Manual | MOLGEN-MS/MS | CFM | SIRIUS | MAGMa |
1) “Win/Submit”: “Win” and “Submit” are the number of wins and that of the submitted challenges, respectively. 2) When all or a part of the candidates were prepared by automated methods, the method names are shown.
Team Newsome manually interpreted the challenges and submitted the solution candidate at rank=1 to all the Category 1 challenges. The team won all the submissions (12/12), where m and n in “(m/n)” are the number of the challenges the team won and the number of those the team submitted, respectively. Team Newsome obtained the most wins with the best accuracy (rank=1), thereby being the winner of Category 1.
Four teams, using automated tools, MOLGEN-MS/MS (team Schymanski), CFM (team Allen), SIRIUS (team Dührkop), and MAGMa (team Ridder), participated in Category 1 challenges. Team Dührkop submitted the solution candidates for all of the challenges and won 10 challenges (10/12) with the correct candidate at rank=1. This team has significantly improved the accuracy of SIRIUS after the first CASMI, in which the team obtained a result of 5/14 with an older version of the same tool.27) Team Ridder participated with an automated tool, MAGMa, and won all eight challenges at rank=1, i.e., with no mistakes, 8/8. Team Schymanski and team Allen had fewer wins. The CASMI2013 organizers selected SIRIUS (team Dührkop) as the CASMI2013 best automated tool of Category 1 because the most wins were achieved with SIRIUS with a competitive accuracy to MAGMa.
In Category 2, participants identified the chemical structure of the challenge molecules by interpreting the challenge MS2 data. The CASMI2013 organizers provided 16 sets of MS2 data as the Category 2 challenges. Four teams submitted their solution candidates by interpreting the challenge data with their automated tools and the other two teams by the manual method. The results of Category 2 by participants are summarized in Table 3. Statistics of the results for Category 2 by participants are summarized in Table S2 in Supplementary Information.
Table 3. Rank and win of the correct candidate in Category 2. The number in each cell shows the rank of the correct candidate submitted by each team. The rank that won the challenge is highlighted in bold. “—” or blank shows that the team has a submission with no correct candidate or no submission to the challenge, respectively. Newsome, Sweeney, Schymanski, Allen, Ridder, and Miyazaki are team names (see Text).
| Challenges | Newsome | Sweeney | Schymanski | Allen | Ridder | Miyazaki |
|---|---|---|---|---|---|---|
| 1 | 1 | 1 | 9 | 12 | 1 | — |
| 2 | 1 | 1 | 44 | — | 3 | — |
| 3 | — | 21 | — | 17 | — | |
| 3 (ile) | 1 | 21 | — | 2 | — | |
| 4 | 1 | — | 238 | 18 | 78 | — |
| 4 (4mp) | — | — | 299 | 4 | 75 | — |
| 4 (2mp) | — | — | 293 | 4 | 76 | — |
| 5 | 1 | 1 | 4 | 9 | 2 | — |
| 5 (propyl) | 2 | — | 1 | 42 | 1 | — |
| 6 | 1 | 1 | 1 | 1 | — | |
| 7 | 1 | *1 | 17 | 23 | 1 | — |
| 8 | 1 | *2 | 1 | 1 | 1 | — |
| 9 | 1 | 1 | 1 | 2 | 1 | — |
| 10 | 1 | *1 | 1 | 1 | 1 | — |
| 11 | 2 | 6 | 21 | |||
| 11 (tautomer1) | 3 | 5 | 1 | |||
| 11 (tautomer2) | 1 | — | 22 | |||
| 12 | 1 | 3 | 35 | |||
| 13 | 12 | 24 | 42 | |||
| 14 | 1 | 2 | 1 | 761 | 5 | |
| 15 | 1 | 1 | — | |||
| 16 | 1 | 1 | — | 100 | ||
| Win/Submit1) | 15/15 | 9/14 | 8/16 | 2/12 | 7/12 | 0/10 |
| Methods2) | Manual | Rational Numbers FragSearch (*manual) | MetFrag/MetFusion | CFM | MAGMa | Manual |
1) “Win/Submit”: “Win” and “Submit” are the number of wins and that of the submitted challenges, respectively. 2) When all or most of the candidates were prepared by automated methods, the name of the method is specified.
Team Newsome submitted the chemical structures as the solution candidates prepared by manually interpreting 15 challenges. The team obtained 15 wins at rank=1. Therefore, team Newsome, which obtained 12/12 in Category 1 and 15/15 in Category 2, had the most wins at rank=1 among all of the teams that participated in CASMI2013. The winner of CASMI2013 is Andrew Newsome and Dejan Nikolic; they are the winner of both Categories 1 and 2, by the manual method. The CASMI2013 organizers commend the team of Andrew Newsome and Dejan Nikolic for their outstanding record in CASMI2013.
Four other teams participated in Category 2 with the candidate structures by automatically interpreting the Category 2 challenge data. MAGMa (team Ridder) resulted in 8/8 in Category 1 and 7/12 in Category 2. CFM (team Allen) resulted in 4/8 in Category 1 and 2/12 in Category 2. MAGMa (team Ridder) and CFM (team Allen) were the automated tools used in both Categories 1 and 2. MAGMa resulted in a total of 15 wins and was better in terms of the accuracy of calculating the molecular formulae and identifying the chemical structures than that of CFM. The CASMI2013 organizers therefore selected MAGMa (team Ridder) as the best automated tool of CASMI2013 because it achieved better results in both Categories 1 and 2.
Four automated tools were included in Category 2. Team Sweeney participated only in Category 2 with Rational Numbers FragSearch and achieved 7/11. However, the CASMI2013 organizers did not nominate the Rational Numbers FragSearch for the best automated tool of Category 2 because it is a patented and not an open-source freeware that other researchers would be permitted to freely modify. The use of CFM (team Allen) resulted in only two wins among the automated tools. Those of MetFrag/MetFusion (team Schymanski) and MAGMa (team Ridder) resulted in 8/16 and 7/12, respectively. These two tools are competitive with each other. MAGMa resulted in fewer wins. Four fewer entries to the challenge were made with MAGMa, but there were more correct candidates, at better ranks, than with MetFrag/MetFusion. The CASMI2013 organizers selected MAGMa as the CASMI2013 best automated tool of Category 2 by vote. Eight of the nine CASMI2013 organizers voted for MAGMa. The organizers considered fewer false-positive candidates rather than the number of wins as an important feature for the automated tools. This will be discussed further at the latter part of the present study.
The CASMI2013 organizers will encourage entries from more teams developing commercial tools in the next contest because CASMI provides an opportunity to assess the performance of commercial tools as well as open-source, free tools.
CHALLENGE DATA AND PERFORMANCE OF AUTOMATED TOOLS
Of particular interest are the results tabulated in Tables 2 and 3. The most automated tools won, or submitted, the correct candidates to some challenges, while they failed to win, or made no entries, to other challenges. It is most likely that such a dependency of the contest results on the challenges is derived from some aspects of the challenge data and molecules rather than from differences in the algorithm of the automated tools.
We analyzed and discussed the challenges from a technical point of view of the challenge MS2 data and from a chemical point of view of the challenge molecules because we consider that these two aspects of the challenge data and molecules can have a significant influence on the performance of automated tools. We analyzed the results contained in Tables 2 and 3 as the primary source of the performance. In addition, the preprints of the participants’ research articles in the CASMI2013 special issue, which the authors of the preprints kindly provided us with to help us in our analysis, are another important source for understanding the performance of automated tools.
Quality of the challenge data, compound class, and dissociation mechanism of challenge molecules
Most automated tools won Challenges 1–6, 9, and 10 in Category 1 (Table 2), and Challenges 1 and 5–10 in Category 2 (Table 3). The molecules of these challenges are an amide of ferulic acid (Challenge 1), agrochemicals consisting of Cl, N, O, P, and S atoms (Challenges 5 and 9), a phospholipid of C18 and C20 alkyl chain carboxylic acids (Challenge 6), catechin pentamers and trimers (Challenges 7 and 8), and an oligopeptide (Challenge 10). The molecular masses range from 290 Da (Challenge 5) to 1,442 Da (Challenge 7). Ions in these challenge data were observed within a mass tolerance of ≤10 ppm.
These results proved that most automated tools are able to successfully interpret the challenge data and correctly identify molecular formulae and chemical structures of major classes of metabolites having molecular masses approaching >1,000 Da when the isotope peaks and all the product ions essential to lead to the solution are observed in the challenge data within the expected or better mass accuracy.
On the other hand, although most automated tools won Challenges 2–4 in Category 1, they failed to win the challenges in Category 2 because their submissions contained the correct candidates but with many false-positive candidates also included (Table 3). The solutions to these challenges are small molecules having molecular masses ranging from 210 Da to 300 Da; an amide of ferulic acid (Challenge 2), a derivative of a dipeptide (Challenge 3), and dihydrochalcone (Challenge 4).
The molecule of Challenge 2 (2 in Fig. 1) shares a common feruloyl substructure with that of Challenge 1 (1 in Fig. 1). However, one of the product ions relevant to the common substructure was not observed in the Challenge 2 data. This might have affected the automated methods, causing them to give low scoring positions to the correct candidate in the list of candidates. We suggest that a knowledge of the biological source of the molecule, Solanaceae plants, could point to the correct candidate. The solution to Challenge 3 is an amide derivative of a dipeptide. The chemical derivatization generated four identical substructures, –NH–C(=O)–C, in the molecule. As a result of the recurring substructure, the challenge molecule gave only three product ions on MS2 analysis. To Challenge 4, MetFrag/MetFusion, MAGMa, and CFM gave the solution, but with 236, 77, and 16 false-positive candidates, which were erroneous candidates at better ranks than the correct solution. The solution to Challenge 4 is the smallest molecule, 210 Da; it consists of two phenyl rings and one carbonyl group. Although different combinations of two phenyl rings and a carbonyl group within a chemical structure generate many possible chemical structures other than the actual solution, team Newsome manually interpreted the Challenge 4 data, which analyzed the challenge molecule within a mass tolerance of ≤3 ppm, and finally refined the three chemical structures, which are the solution structure and two highly commended structures (see “Aim of the challenges, and ions leading to the solutions” in the present article and the report by team Newsome20)). Automated tools could reduce the number of false-positive candidates by integrating the knowledge of relationships between product ions and chemical substructures, and by integrating the two hydrogen migrations during CID.
There is another reason for why automated tools were unsuccessful for Categories 1 and 2 of Challenges 11 and 12. The use of “AnalyticalMethods” for the two challenges gave no mass accuracy about their MS1 and MS2 data. Team Ridder failed to identify two challenges. The team failed to interpret the challenges by expecting a mass accuracy of 10 ppm or 0.002 Da. After the announcement of the solutions, the team retried and then successfully identified the challenge molecule with MAGMa by relaxing the mass accuracy to 0.005 Da.24) In Challenge 12, the exact mass of the product ions in the MS1 and MS2 data occasionally deviated beyond an expected accuracy range. The deviation fatally affected MAGMa and other automated tools.
There might be two ways by which automated tools can overcome the problem of larger mass deviations from the expected mass accuracy. One way is to relax the mass tolerance, because automated tools tend to postulate too narrowly in mass tolerance.24) The other way is to integrate experts’ knowledge of relationships between compound classes and product ions. Researchers often observe a set of product ions that are attributable to a common chemical substructure shared by a class of chemical compounds. Such a set of product ions could be detectable even in the case of larger mass deviations. Team Newsome accumulated such product ions of various classes of metabolites on its own in-house mass spectral database.20) The database supported the team’s winning the contest. MS2 data for Challenges 11 and 12 showed product ions characteristic to curcuminoids and flavones, respectively. Although a literature17) reported that flavones with a trihydroxyl group on the A ring can be characterized based on a product ion at m/z 171, the ion was not observed in Challenge 12. Although CASMI challenges should be free from mass deviation and missing product ions, researchers often encounter such experimental inadequacy of MS data, due to instrumental instability and instrument type. It would be desirable that automated tools be sufficiently robust to overcome such experimentally unsatisfactory MS data.
In addition to mass accuracy, these two challenges presented a complication to the automated tools. As pointed out by team Schymanski,32) the solution to Challenge 11 is either one of three tautomers of demethoxycurcumin that are the same chemical entity but that have different but interchangeable structures under the experimental conditions in which the challenge data were obtained. The present version of MetFrag/MetFusion treated the solution as a mixture of three different candidates and gave them the same or different ranks.32) Challenge 12 presented a similar problem to automated tools. A series of product ions in Challenge 12 was generated by the successive loss of CO or H2O+CO, which is associated with the chemical rearrangement of the flavone substructures. Generally, one chemical rearrangement modifies two substructures of the solution chemical structure and affects the solution candidates. Such chemical rearrangements and modification of substructures are not integrated in MAGMa.24)
Challenge 13, Category 2 is the challenge that team Newsome refrained from entering and hence missed full wins in CASMI2013. Only three automated methods, MetFrag/MetFusion, CFM, and MAGMa, submitted candidates. The results in Category 2 of Challenge 13 were unsatisfactory. All of the product ions in the MALDI-QIT MS2 data were observed within the 5 ppm range of the calculated mass and are reproducible on a different analytical platform, ESI-Q-TOF MS/MS. No ions other than [M+H]+ of aloxistatin were observed in the MS1 data. However, as described in the section “Aim of the challenge” of Challenges 13 and 14, it is not certain whether all the product ions necessarily leading to the solution were observed in the MS2 data in this challenge. It is likely that the MS2 data lacked some product ions necessary to provide a solution.
On the day after the solutions to the CASMI2013 challenges were announced, team Newsome contacted us by email regarding the solution to Challenge 13. According to the team, they arrived at aloxistatin as the lead candidate before the announcement but did not submit the structure because the product ion at m/z 246.1331, which is a base peak in the challenge, is uninterpretable from the chemical structure of aloxistatin. We also noticed that the molecular formula of the ion, C11H20NO5+, could not have been produced directly from the precursor ion of aloxistatin, [M+H]+, even if rearrangement reactions were taken into consideration. A few days later, team Newsome provided a hypothesis, whereby, in a hydration reaction at the epoxide group of aloxistatin, a glycol derivative of aloxistatin is formed inside the mass spectrometer, and the glycol is the source of the product ion.20) The CASMI2013 organizers regret this error and express our apologies to all of the other participants that Challenge 13 was, in fact, MS2 data of a mixture of two different small molecules. The lesson we learned is that the quality of the challenge data should be confirmed by manual interpretation of the data as well as by analysis on different analytical platforms.
There are other reasons for the poor results for Challenges 15 and 16 in Categories 1 and 2. The solution to Challenge 15 contains 17 fluorine atoms in the molecular formula. Generally, endogenous small molecules contain no fluorine atoms.39) Furthermore, in automated tools, the number of fluorine atoms might exceed the default number of fluorine atoms. After the solution was announced, MAGMa successfully solved the challenge, after adding the fluorine atom to their default settings.24)
The solution to Challenge 16, ofloxacin, contains a ring system in which three hexagonal rings are fused in its chemical structure. The covalent bonds consisting of the fused ring should be cleaved at least five times before the precursor ion releases product ions from this moiety. When the default cleavage time was retrospectively increased from 3 to 5 times, MAGMa improved the rank of the correct candidate up to 3.24)
MetFusion integrating MassBank for the identification process retrospectively found that four MS2 data sets for ofloxacin were unknowingly deposited in MassBank during the submission period of CASMI20132): MassBank IDs UF407501–UF407504 were contributed by a research group from the Helmholtz Centre for Environmental Research (UFZ), Leipzig, Germany.
Searching chemical substance databases such as PubChem with a molecular formula or a molecular mass as a query results in the retrieval of thousands of candidate molecules. Manually evaluating such a long list of candidates is nearly an impossible task. Team Newsome reported that an empirical rule was helpful for refining a long list of candidates in Challenges 11, 13, and 16.20) According to the rule,40) the most useful candidates for further evaluation are at the top of the list when the candidates are refined by sorting the number of references associated with each molecule. Team Ridder reported that MAGMa integrated the empirical rule as a filter for refining the correct candidate and worked effectively for some challenges, but not for others, in which case the correct candidate was discarded from the list.24) It is clear that the rule was not always useful and in some cases, was even detrimental in refining the correct candidate.
What makes the manual method perform so well?
Team Newsome won all 15 of the Category 2 challenges at rank=1 (Table 3). The team chemically annotated as many product ions as possible in its refining and confirmed the solution candidate.20) When the challenge molecules contain isomers, the team attempted to identify product ions that specify either of the isomers. For example, there are four possible isomers of the Challenge 1 molecule, trans and cis, and feruloyl and isoferuloyl. The team accumulated QTOF-MS/MS data for the feruloyl and isoferuloyl compounds in its own in-house database to date, and found peak patterns corresponding to each of the isomers. However, the pattern in the ITTOF-MS/MS data of Challenge 1 was not sufficiently obvious to permit a specific isomer to be identified. The team thought that their knowledge of peak patterns and compound classes on one instrument might be inapplicable to different types of instruments. The team analyzed each isomer on the same type of instrument, from the same manufacturer, as was used for the challenge data, and found a slight difference in the identity of the solution. The team concluded that a database search or computational approaches alone would most likely result in both isomers being identified as matches with the data. In another case, after the solution to Challenge 13 was announced, the team purchased a commercially available sample of the challenge molecule for MS/MS analysis with a QTOF instrument. They managed to observe a key product ion, m/z 88.1130, that was anticipated but not found in the MS2 data of Challenge 13,20) presumably due to the low m/z cutoff of the ion-trap instrument used for the acquisition of the challenge data. Furthermore, during the chemical annotation of the product ions observed in the data for Challenge 13, the team found that some product ions should not be produced from the precursor ion of the challenge molecule. The team proposed a hypothesis in which the challenge molecule is not a single molecule but, rather, a mixture of the solution and derivative molecules, the latter of which were generated inside the instrument during the measurement by a CASMI2013 organizer.
A lesson to be learned from team Newsome is that most of the product ions observed in the mass spectra of small molecules are expected and can be explained based on an empirical knowledge of the fragmentation mechanisms of the class of molecules. When they are not explained, the solution candidate is most likely incorrect, and should then be discarded. When a new mechanism is found, this is added to the knowledge databases as new knowledge.
As shown in Table 3, the correct candidate for a CASMI challenge was almost always in the list of solution candidates generated by automated tools because the list was retrieved from chemical substance databases based on the molecular formula or the mass of the molecular ion of the challenge molecule as a search key. We are of the opinion that, for such automated tools, the solution to this problem is to reduce the number of false-positive candidates. To accomplish this, we encourage the participants to evaluate the performance of their automated tools by assessing the total number of the predicted ions that match the ions observed in the challenge MS2 data. It is likely that team Schymanski attempted to do this by overlaying the predicted peaks on the observed peaks as described in their article.32)
Mass spectra for better performance of automated tools
When researchers in metabolomics wish to successfully use the current versions of automated tools for identifying a metabolite, the following three analytical requirements in terms of mass spectral data acquisition need to be considered. First, MS2 data observes different as many product ions as possible, in proportion to the complexity of the chemical structure of the target molecule. Second, the observed m/z values should be within an expected mass accuracy, whether a low or high mass accuracy. Third, only ions that are generated from a single molecular ion are observed in one MS2 data set. In other words, good quality mass spectral data are required for success. Researchers could satisfy these analytical requirements by carefully maintaining good instrumental stability and by analyzing a single sample on different types of instruments and under different CID conditions, as far as possible.
Furthermore, target molecules must satisfy the following chemical requirements: they undergo no rearrangement reactions during the dissociation reactions, and no tautomer, nor multiple-fused ring structures are involved. As the researchers had no chemical information on the target molecules prior to their identification, automated tools have to integrate or refer to the databases of such empirical knowledge. The CASMI2013 organizers therefore encourage all researchers in biological and environmental studies to deposit their mass spectral data, analyzed on different instrument types and under different analytical conditions, to MassBank or other public databases, so as to accumulate a body of empirical knowledge that covers much wider varieties of chemical structures.
Acknowledgments
The authors express their thanks to all of the CASMI2013 participants, with their manual and automated methods, for enhancing the value of the contest, and to the organizing team of the first CASMI, for preparing the CASMI2013 web page, and for the automated evaluation and the statistical calculations. CASMI2013 was sponsored by the Spectral Data Division of the Mass Spectrometry Society of Japan and partly supported from a grant to Professor Shigehiko Kanaya from the National Bioscience Database Center, Japan. This work was partly supported by JSPS KAKENHI Grant Number 25513011.
T. Nishioka, et al., Winners of CASMI2013: Automated Tools and Challenge Data, Mass Spectrom (Tokyo) 2014; 3(3): S0039; DOI: 10.5702/massspectrometry.S0039
Supplementary Information
References
- 1) H. Horai, M. Arita, S. Kanaya, Y. Nihei, T. Ikeda, K. Suwa, Y. Ojima, K. Tanaka, S. Tanaka, K. Aoshima, Y. Oda, Y. Kakazu, M. Kusano, T. Tohge, F. Matsuda, Y. Sawada, M. Y. Hirai, H. Nakanishi, K. Ikeda, N. Akimoto, T. Maoka, H. Takahashi, T. Ara, N. Sakurai, H. Suzuki, D. Shibata, S. Neumann, T. Iida, K. Tanaka, K. Funatsu, F. Matsuura, T. Soga, R. Taguchi, K. Saito, T. Nishioka. MassBank: A public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 45: 703–714, 2010. [DOI] [PubMed] [Google Scholar]
- 2) R. Tautenhahn, K. Cho, W. Uritboonthai, Z. Zhu, G. J. Patti, G. Siuzdak. An accelerated workflow for untargeted metabolomics using the METLIN database. Nat. Biotechnol. 30: 826–828, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3) http://chemdata.nist.gov/mass-spc/msms-search/
- 4) D. S. Wishart, T. Jewison, A. C. Guo, M. Wilson, C. Knox, Y. Liu, Y. Djoumbou, R. Mandal, F. Aziat, E. Dong, S. Bouatra, I. Sinelnikov, D. Arndt, J. Xia, P. Liu, F. Yallou, T. Bjorndahl, R. Perez-Pineiro, R. Eisner, F. Allen, V. Neveu, R. Greiner, A. Scalbert. HMDB 3.0—The Human Metabolome Database in 2013. Nucleic Acids Res. 41(D1): D801–D807, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5) J. H. Gross. Mass Spectrometry: A Textbook. Springer-Verlag, New York, 2004, pp. 518.
- 6) M. A. Stravs, E. L. Schymanski, H. P. Singer, J. Hollender. Automatic recalibration and processing of tandem mass spectra using formula annotation. J. Mass Spectrom. 48: 89–99, 2013. [DOI] [PubMed] [Google Scholar]
- 7) S. Tanaka, Y. Fujita, H. E. Parry, A. C. Yoshizawa, K. Morimoto, M. Murase, Y. Yamada, J. Yao, S. Utsunomiya, S. Kajihara, M. Fukuda, M. Ikawa, T. Tabata, K. Takahashi, K. Aoshima, Y. Nihei, T. Nishioka, Y. Oda, K. Tanaka. Mass++: A visualization and analysis tool for mass spectrometry. J. Proteome Res. 13: 3846–3853, 2014. [DOI] [PubMed] [Google Scholar]
- 8) http://www.cas.org/
- 9) http://pubchem.ncbi.nlm.nih.gov/
- 10) http://www.chemspider.com/
- 11) M. Kanehisa, S. Goto, Y. Sato, M. Kawashima, M. Furumichi, M. Tanabe. Data, information, knowledge and principle: Back to metabolism in KEGG. Nucleic Acids Res. 42(D1): D199–D205, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12) F. M. Afendi, T. Okada, M. Yamazaki, A. Hirai-Morita, Y. Nakamura, K. Nakamura, S. Ikeda, H. Takahashi, M. Altaf-Ul-Amin, L. K. Darusman, K. Saito, S. Kanaya. KNApSAcK family databases: Integrated metabolite-plant species databases for multifaceted plant research. Plant Cell Physiol. 53: e1, 2012. [DOI] [PubMed] [Google Scholar]
- 13) F. Matsuda. Rethinking mass spectrometry-based small molecule identification strategies in metabolomics. Mass Spectrom. (Tokyo) 3: S0038, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14) E. Schymanski, S. Neumann. CASMI: And the Winner is .... Metabolites 3: 412–439, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15) K. Krohn, A. Ishtiaq, J. Markus, L. C. Matthias, K. Dietmar. Stereoselective synthesis of benzylated prodelphinidins and their diastereomers with use of the Mitsunobu Reaction in the preparation of their gallocatechin precursors. Eur. J. Org. Chem. 2010: 2544–2554, 2010. [Google Scholar]
- 16) H. Jiang, A. R. Somogyi, N. E. Jacobsen, B. N. Timmermann, D. R. Gang. Analysis of curcuminoids by positive and negative electrospray ionization and tandem mass spectrometry. Rapid Commun. Mass Spectrom. 20: 1001–1012, 2006. [DOI] [PubMed] [Google Scholar]
- 17) W. Wu, Z. Liu, F. Song, S. Liu. Structural analysis of selected characteristic flavones by electrospray tandem mass spectrometry. Anal. Sci. 20: 1103–1105, 2004. [DOI] [PubMed] [Google Scholar]
- 18) Y. Wang, L. Yang, Y. Q. He, C. H. Wang, E. W. Welbeck, S. W. A. Bligh, Z. T. Wang. Characterization of fifty-one flavonoids in a Chinese herbal prescription Longdan Xiegan Decoction by high-performance liquid chromatography coupled to electrospray ionization tandem mass spectrometry and photodiode array detection. Rapid Commun. Mass Spectrom. 22: 1767–1778, 2008. [DOI] [PubMed] [Google Scholar]
- 19) U. Berger, I. Langlois, M. Oehme, R. Kallenborn. Comparison of three types of mass spectrometers for HPLC/MS analysis of perfluoroalkylated substances and fluorotelomer alcohols. Eur. J. Mass Spectrom. (Chichester, Eng.) 10: 579–588, 2004. [DOI] [PubMed] [Google Scholar]
- 20) A. G. Newsome, D. Nikolic. CASMI 2013: Identification of small molecules by tandem mass spectrometry combined with database and literature mining. Mass Spectrom. (Tokyo) 3: S0034, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21) http://www.emetabolomics.org/magma
- 22) L. Ridder, J. J. J. van der Hooft, S. Verhoeven, R. C. H. de Vos, R. van Schaik, J. Vervoort. Substructure-based annotation of high-resolution multistage MSn spectral trees. Rapid Commun. Mass Spectrom. 26: 2461–2471, 2012. [DOI] [PubMed] [Google Scholar]
- 23) L. Ridder, J. J. J. van der Hooft, S. Verhoeven, R. C. H. de Vos, R. J. Bino, J. Vervoort. Automatic chemical structure annotation of an LC-MSn based metabolic profile from green tea. Anal. Chem. 85: 6033–6040, 2013. [DOI] [PubMed] [Google Scholar]
- 24) L. Ridder, J. J. J. van der Hooft, S. Verhoeven. Automatic compound annotation from mass spectrometry data using MAGMa. Mass Spectrom. (Tokyo) 3: S0033, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25) S. Böcker, F. Rasche. Towards de novo identification of metabolites by analyzing tandem mass spectra. Bioinformatics 24: 49–55, 2008. [DOI] [PubMed] [Google Scholar]
- 26) S. Bocker, M. C. Letzel, Z. Lipták, A. Pervukhin. SIRIUS: Decomposing isotope patterns for metabolite identification. Bioinformatics 25: 218–224, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27) K. Dührkop, F. Hufsky, S. Böcker. Molecular formula identification using isotope pattern analysis and calculation of fragmentation trees. Mass Spectrom. (Tokyo) 3: S0037, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28) K. Dührkop, K. Scheubert, S. Böcker. Molecular formula identification with SIRIUS. Metabolites 3: 506–516, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29) M. Meringer, E. Schymanski. Small molecule identification with MOLGEN and mass spectrometry. Metabolites 3: 440–462, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30) S. Wolf, S. Schmidt, M. Muller-Hannemann, S. Neumann. In silico fragmentation for computer assisted identification of metabolite mass spectra. BMC Bioinformatics 11: 148, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31) M. Gerlich, S. Neumann. MetFusion: Integration of compound identification strategies. J. Mass Spectrom. 48: 291–298, 2013. [DOI] [PubMed] [Google Scholar]
- 32) E. L. Schymanski, M. Gerlich, C. Ruttkies, S. Neumann. Solving CASMI 2013 with MetFrag, MetFusion and MOLGEN-MS/MS. Mass Spectrom. (Tokyo) 3: S0036, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33) D. L. Sweeney. A systematic computational approach for identifying small molecules from accurate-mass fragmentation data. American Laboratory News 39: 12–14, 2007. [Google Scholar]
- 34) D. L. Sweeney. Small molecules as mathematical partitions. Anal. Chem. 75: 5362–5373, 2003. [DOI] [PubMed] [Google Scholar]
- 35) D. L. Sweeney. A representation of molecules as sets of masses of complementary subgroups and contiguous complementary subgroups. United States Patent Application 2011017161, published July 14, 2011.
- 36) D. L. Sweeney. A data structure for rapid mass spectral searching. Mass Spectrom. (Tokyo) 3: S0035, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37) http://sourceforge.net/projects/cfm-id/
- 38) F. Allen, R. Greiner, D. Wishart. Competitive Fragmentation Modeling of ESI-MS/MS spectra for metabolite identification. arXiv.org>cs>arXiv:1312.0264v2[cs.CE], 2014.
- 39) T. Kind, O. Fiehn. Seven Golden Rules for heuristic filtering of molecular formulas obtained by accurate mass spectrometry. BMC Bioinformatics 8: 105, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40) J. Little, A. Williams, A. Pshenichnov, V. Tkachenko. Identification of known unknowns utilizing accurate mass data and ChemSpider. J. Am. Soc. Mass Spectrom. 23: 179–185, 2012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.