Abstract
Senescence, the deterioration of morphological, physiological, and reproductive functions with age that ends with the death of the organism, was widely studied in plants. Genes were identified that are linked to the deterioration of cells, organs and the whole plant. It is, however, unclear whether those genes are the source of age dependent deterioration or get activated to regulate such deterioration. Furthermore, it is also unclear whether such genes are active as a direct consequence of age or because they are specifically involved in some developmental stages. At the individual level, it is the relationship between quantitative genetic variation, and age that can be used to detect the genetic signature of senescence. Surprisingly, the latter approach was only scarcely applied to plants. This may be the consequence of the demanding requirements for such approaches and/or the fact that most research interest was directed toward plants that avoid senescence. Here, I review those aspects in turn and call for an integrative genetic theory of senescence in plants. Such conceptual development would have implications for the management of plant genetic resources and generate progress on fundamental questions raised by aging research.
Keywords: aging, ROS, SAG, SEN, PAG, additive genetic variation, mutation accumulation, antagonistic pleiotropy
Senescence is not a simple concept. Senescence is the decline of organismal functions with age. For physiologists, it can be defined as the degradation of cells or organs and their associated functionality. For demographers, senescence is defined by the positive relationship between the probability to die and age. For evolutionary biologists, senescence is also defined as a decline in function and an increase in the chance of dying with age. Their focus is, however, made on the decline in the strength of natural selection with age at the population level. In the present review, my aim is not to discuss the knowledge accumulated on plant senescence in every discipline. It is not to animate a semantic debate for the sake of providing a unique definition of senescence. Here, my focus is on genetic data and its involvement with plant senescence. I will explore that question at the level of cells, organs, and individuals within populations. I will then discuss how recent methodological developments in quantitative genetics could be used to develop an integrative framework and investigate the links observed between those scales.
GENES INVOLVED WITH CELL SENESCENCE
Senescence at the cellular level is the deterioration of the cell that is due to the age of the cell and not to the age of the whole organism. In plant cells, as in animal cells, life in aerobic conditions induces the production of reactive oxygen species (ROS) such as hydrogen peroxide (H2O2), and nitric oxide (NO). They are toxic and despite the action of antioxidant mechanisms, their toxicity leads to the death of cells (Van Breusegem and Dat, 2006). Their production is inevitable because they are the consequence of membrane linked electron transport in plant cells. ROS are generated during the production of energy through respiration and photosynthesis. In Arabidopsis cells, the expression level of the gene LSC54 was found to increase with the age of cells. The activity of LSC54 was also found to be linked with the level of oxidative stress in the cell. One might think that the activity of LSC54 is generating oxidative stress but in fact, the activity of LSC54 is induced by increased levels of ROS (Navabpour et al., 2003). Such genes probably become active in response to ROS signaling in order to regulate oxidative mechanisms of deterioration.
Many genes are involved with cell senescence. They can be found under the name of senescence associated genes (SAG) or senescence enhanced genes (SEN). Their activity is generally linked to protein and/or lipid degradation. For example SAG101 codes for acyl hydrolase and the activity of SEN3 is linked to the degradation of leaf tissues (Buchanan-Wollaston et al., 2003). Proteins such as RuBisCO, which is the key protein for carbon fixation during photosynthesis, are also degraded during the deterioration of leaf cells. ROS can be responsible for such degradation. However, the activity of protease genes involved with the degradation of RuBisCO was also found to increase during cell senescence in leaves. Several other genes that encode for enzymes are responsible for the degradation of membrane lipids during cell deterioration. Whether the activity of the many genes involved with cell senescence is directly causing cell deterioration or not remains unclear. This is because some of the genes that are active during cell deterioration are likely to be directly involved in such deterioration whereas others are likely to be involved in regulatory mechanisms counteracting deterioration (Buchanan-Wollaston, 1997). Whatever the scenario, detecting the activity of such genes implies that cell deterioration is underway because the cell is becoming older.
GENES INVOLVED WITH ORGAN SENESCENCE
Many genes that are involved in the senescence of plant cells are also found to play a role in organ senescence (Table 1). This is partly because many studies on organ senescence investigate how leaf tissues deteriorate with leaf age at the cellular level (e.g., ROS accumulation; Thomas and Stoddart, 1980; Smart et al., 1995; Gan and Amasino, 1997; Bleecker, 1998). Leaf senescence intervenes right after growth and involves the deterioration of the mesophyll from the tip to the base of the leaf (Jansson and Thomas, 2008). Regulatory gene activity was found to vary between the early growth of the organ and its death (Guo and Gan, 2011). MicroRNAs such as miR156 were identified in primordia of rice leaves where they target transcription factors of the SQUAMOSA PROMOTER BINDING-LIKE (SPBL) family and affect the duration of juvenile leaf character expression. The interplay of those MicroRNAs with the expression of miR164 was also found to affect age related cell death in Arabidopsis (Kim et al., 2009).
Table 1.
Genes that are known to play a role in organ senescence (compiled from Bleecker, 1998; Thomas et al., 2009; Thomas, 2013, and references therein).
| Gene | Related function | Reference |
|---|---|---|
| Arabidopsis RCCR (Red Chlorophyll Catabolite Reductase) | Chlorophyll degradation | Wüthrich et al. (2000), Pružinská et al. (2007) |
| Arabidopsis PaO | Chlorophyll degradation | Oshio and Hase (1969), Rodoni et al. (1997) |
| Arabidopsis Sgr1 (Stay green) | chlorophyll degradation | Guiamet and Giannibelli (1996), Armstead et al. (2006), Park et al. (2007), Sato et al. (2007) |
| Arabidopsis Wrky53 | Cell and tissue specialization | Miao et al. (2004), Uauy et al. (2006) |
| Arabidopsis AtNAP | Cell and tissue specialization | Guo and Gan (2006) |
| Auxenochlorella dee4 – amino acid permease | Cell and tissue specialization | Hörtensteiner et al. (2000) |
| Arabidopsis Fibrillin | Cell and tissue specialization | Yang et al. (2006), Brehelin et al. (2007) |
| Arabidopsis Carotenoid Cleavage Dioxygenase8 | Plastid trans-differentiation | Snowden et al. (2005) |
| Brassica dnaJ chaperone OrI | Plastid trans-differentiation | Lu et al. (2006) |
| Arabidopsis dnaJ chaperone OrII | Plastid trans-differentiation | Giuliano and Diretto (2007) |
| Zea Bronze1 | Vacuole function | Furtek et al. (1988) |
| Zea myb C1 anthocyanin TF | Vacuole function | Paz-Ares et al. (1990) |
| Arabidopsis AtMRP2 ATP transporter | Vacuole function | Hinder et al. (1996), Lu et al. (1998) |
| Zea See2 | Vacuole function | Smart et al. (1995), Carter et al. (2004) |
| Squamosa Promoter Binding-Like (SPBL) gene family | Leaf development | Poethig (2010), Bergonzi and Albani (2011), Wang et al. (2011), Xie et al. (2012) |
| TFL1 (Terminal Flower 1) | juvenile period in perennials | Bergonzi and Albani (2011) |
| SnRK1 (Snf1-Related Kinase1 | Sugar mediated regulation of aging | Thelander et al. (2004), Baena-Gonzalez et al. (2007) |
| Hexokinase1 (HXK1) | Sugar mediated regulation of aging | Jongebloed et al. (2004), Parrott et al. (2007), Smeekens et al. (2010) |
| Arabidopsis “old” mutant | Sugar mediated regulation of aging | Schippers et al. (2008) |
| Arabidopsis AtMYB2 | Bud outgrowth during monocarpic senescence | Guo and Gan (2011) |
| Arabidopsis TERT (Telomerase Reverse Transcriptase) | Telomere attrition | Watson and Riha (2010) |
| RTBP1 (Rice Telomere Binding Protein) 1 | Telomere attrition | McKnight and Shippen (2004), Hong et al. (2007) |
| ATGs (Autophagy genes) | Cell degradation | Thompson et al. (2005) |
| TOR (Target of Rapamycin) | Growth and development | Blagosklonny and Hall (2009) |
At the level of the leaf organ, the assimilation and storage of carbon and nitrogen during leaf development stops when leaf senescence starts. It is important to note that the timing of leaf senescence depends on leaf age and on the environmental resources available (Wingler et al., 2006). Leaves start their life by being a sink for the products of photosynthesis and accumulate Carbon and Nitrogen until their function changes when senescence starts. At that time, they stop being a sink and become a source of Carbon and Nitrogen for the rest of the plant. Leaves reach an advanced stage of deterioration when they transfer the products of photosynthesis to the whole plant. The shift toward leaf senescence is associated with the increased transcription of photosynthesis-associated (PAG) genes (Hensel et al., 1993). A whole network of regulatory functions associated with genes was found to be active during leaf senescence (e.g., CWinv, HXK1, TOR, and SnRK1, see Thomas, 2013). Other genes such as the HYS1/CPR5 have also been identified because they are activated during leaf senescence but they are in fact counteracting the deterioration (Yoshida et al., 2002). Delayed leaf senescence was also found in etr1-1 mutant plants. The specificity of those plants is to be that are insensitive to the plant hormone ethylene (Grbić and Bleecker, 1995). Furthermore, many other SAG and SEG genes were identified using mutant lines and Next Generation Sequencing methods (see Buchanan-Wollaston et al., 2003).
It remains unclear whether most genes involved with leaf senescence or organ senescence are directly part of a genetic program coding for senescence at these biological scales. One alternative scenario is that they exhibit an activity that is associated with the degradation of organs because they are participating to genetic mechanisms that are in fact counteracting degradation (Nam, 1997). However, whatever the reasons, the activity of these genes can therefore be used to detect organ senescence. Whether those genes are truly involved with organ senescence or are part of a developmental program of organ turnover throughout the plant life also remains unclear. It is interesting to note that leaf characteristics (defense, photosynthesis and the age at which they fall) have evolved under domestication (El-Sharkawy, 2004; Mondolot et al., 2008; Pujol et al., 2008b). These results suggest that leaf senescence may be integrated in the evolution of senescence at the whole plant level. It is nevertheless difficult to figure out how those genes can help infer how senescence affects the whole plant (Bleecker, 1998). Indeed, many of the genetic mechanisms presented above are shared by plants characterized by completely different life histories such as annual plants, short lived perennial monocarpic plants (i.e., semelparous: a single reproductive episode precedes death) and long lived perennial polycarpic (i.e., iteroparous: multiple flowering events throughout a lifetime) plants (Munné-Bosch, 2008).
GENES PLAYING A ROLE IN WHOLE PLANT SENESCENCE
Whether the activity of the genes and the epigenetic changes presented below are relevant to the study of senescence at the whole plant level is at the center of a debate between, on one hand, physiologists, and system biologists, and in the other hand, demographers, and evolutionary biologists. How to define senescence at the level of the whole plant? Is it chronological age and/or a suite of developmental stages that lead to mortality? This is not a simple question because those two chronological factors are often confounded. Physiologists and system biologists study senescence by analysing developmental modifications. Demographers and evolutionary biologists study senescence by assessing the age trajectory of fitness components. The genetic mechanisms presented below play a role in the chronological succession of developmental stages. A doubt therefore remains on the direct relationship between the activity of those genes and age. At the level of the whole plant, TERMINAL FLOWER 1 (TFL1) genes are involved with developmental changes. TFL1 genes are suspected to play a role in senescence because they regulate the length of the juvenile period in perennial plants. The life stage-dependent activity of those genes affects, albeit only indirectly, the expression of the LEAFY genes and the APETALA 1 genes that determine the development of flowers from meristematic tissue (Ratcliffe et al., 1998). In Arabidopsis, the AGAMOUS gene, which is known to play a crucial role in the development of flowers, was only found to be active during the flowering period (Yanofsky et al., 1990). Other genes are known to be responsible for an increased regulation of branching during successive developmental phases from germination to death in Arabidopsis (Guo and Gan, 2011). Epigenetic methylation may also play a role in the whole plant senescence. Changes in DNA methylation that drive the silencing of MuDR transposable elements were indeed observed during transitions between life stages in maize (Li et al., 2010). Furthermore, the global level of DNA methylation was also found to vary across life stages in the giant redwood tree (Monteuuis et al., 2008). It would be a great breakthrough to determine whether the chronological genetic and epigenetic changes documented above are directly playing a role in the genetic programming of senescence. To this aim, it would be necessary to disentangle age and stage specific organismal changes (Roach, 1993; Caswell and Salguero-Gómez, 2013).
HOW TO PREDICT SENESCENCE AT THE INDIVIDUAL LEVEL
Senescence at the level of the whole organism, be it a plant or an animal, affects complex physiological, and reproductive functions. Complex characters often depend on a large genetic basis formed by more than a few genes. Quantitative genetics offers an opportunity to analyze the genetic architecture of complex traits. Using quantitative genetics to identify the signature of senescence may therefore prove useful in plants. To understand the value of using quantitative genetics in order to identify senescence at the level of the whole plant, one must go back to the basics of the evolutionary theory of senescence. In plants, as in animals, the deterioration with age of fitness related characters can evolve if older individuals contribute less in the gene pool of the next generation than younger individuals. As first suggested by Fisher (1930) and Haldane (see Medawar, 1952, p. 3), selection against age related malfunctions is not efficient if the underlying genes have already been transmitted to the next generation. This idea was further developed by Medawar (1952) and Williams (1957) who discussed the underlying genetic mechanisms. At the level of the genetic architecture of fitness related characters, theory predicts that the deleterious mutations responsible for senescence and expressed in old age will accumulate in the genome (“Mutation Accumulation” theory) as selection fails to remove them (Medawar, 1952). Another non exclusive expectation regarding the genetic architecture of senescence is that genes coding for a fitness advantage in young age, but having detrimental effects in older age (“Antagonistic Pleiotropy” theory) will be favored by selection (Williams, 1957).
In Hamilton (1966) claimed that the evolution of senescence could be universal. However, in plants, a recent analysis of demographic data bases showed that most plant species were not affected by a decay in fertility and by an increase in the risk of mortality with age (Baudisch et al., 2013). The age dependent decrease of the individual probability of survival that is expected in the case senescence was nevertheless found to be common in perennial plant species (Silvertown et al., 2001). Furthermore, longitudinal demographic surveys show that demographic senescence can occur in wild plant populations (Picó and Retana, 2008; Roach et al., 2009; Roach, 2012). Whether the demographic assumptions underlying Hamilton’s claim apply to most plants is still debated. The evolution of senescence does not depend only on survival. Reproductive success is also a key parameter that must be taken into account because it molds the relative contribution of individuals to the gene pool of the next generation (Partridge and Barton, 1993). In a scenario where a higher reproductive success is favored in younger age, theory also predicts that senescence will evolve.
In the 1990s, Roach inventoried cases of indirect phenotypic evidence suggesting that senescence had evolved in Papaver, Solidago, Poa, and Geranium plant species (Roach, 1993). At the genetic level, two theories (Mutation Accumulation and Antagonistic Pleiotropy) predict specific modifications of the additive genetic variability of fitness traits with age (Flatt, 2012; Rose et al., 2012). These predicted changes are the consequence of the failure of natural selection to remove genes with deleterious effects in old age (Greer, 2012). This is because such genes were already transmitted to the next generation. Under the evolutionary theory of senescence, age is correlated with a decline in fitness. We also expect aging rates to be genetically based. Finally, both the mutation accumulation and antagonistic pleiotropy mechanisms are expected to drive an increase in additive genetic variance with age. Negative genetic correlations between early and late age classes are expected to bring evidence for antagonistic pleiotropy. In animals, especially in birds, and ungulates, quantitative genetics was used to confirm these expectations already several times (Charmantier et al., 2006a,b; Brommer et al., 2007; Kruuk et al., 2008; Monaghan et al., 2008; Nussey et al., 2008; Wilson et al., 2008). It is likely that age dependent specific patterns of quantitative genetic variation, in other words Gene by Age interactions (G × A), can also be used to detect senescence in plants.
THE EVOLUTIONARY QUANTITATIVE GENETICS OF SENESCENCE IN PLANTS
Pujol et al. (2014) detected the quantitative genetic signature of senescence in Silene latifolia, a short lived perennial plant species. They found that the reproductive performance of individuals was declining with age. They also found that the age dependent deterioration of reproduction had an additive genetic basis. Finally, they found that additive genetic variation for reproductive characters increased with age, which is predicted under both Mutation Accumulation and Antagonistic Pleiotropy hypotheses. However, they did not find negative genetic correlations between young and old age classes as expected under the Antagonistic Pleiotropy hypothesis. This investigation was rendered possible by the use of a pedigree based random regression animal model which is a method first designed to estimate the quantitative genetic variation of phenotypic plasticity (Wilson et al., 2005; Nussey et al., 2007b; Brommer et al., 2008; Pujol and Galaud, 2013). Such finding calls for the replication of such studies in other plant species across the plant kingdom.
Roach (1993) was already calling for investigating the quantitative genetic architecture of senescence in plants. Shefferson and Roach (2012) found evidence for genetic variation in population growth rates. There is now evidence that variation in individual aging rates has a genetic basis (Pujol et al., 2014). Why are such studies so rare? First, the longitudinal surveys that are necessary for such studies are very demanding. Those surveys are labor and time intensive and require large sample sizes. Another non exclusive explanation is that research projects on plant senescence are often investigating how plants can escape senescence rather than analyzing the senescence process in plants that are negatively affected by aging. Already Harper (1977) was mentioning that plants did not show apparent senescence as a consequence of their indeterminate growth. Ever since then, many efforts were logically directed toward identifying what could cause plants to avoid senescence (Watkinson, 1992; Orive, 1995; Pedersen, 1995; Gardner and Mangel, 1997; Vaupel et al., 2004; Munné-Bosch, 2008; Garcia et al., 2011; Martínez, 2012; Morales et al., 2013; Salguero-Gómez et al., 2013). As a consequence, it is not surprising that the quantitative genetic signature of senescence may have been overlooked in plants. Some insights could certainly be gained by comparing the effect of age on the quantitative genetic basis of fitness related traits in species whose demography and/or physiology suggest that senescence is absent.
Knowledge on additive genetic variation at fitness related traits can be used to predict the evolutionary potential of a population (Etterson, 2004; Pujol and Pannell, 2008; Pujol et al., 2008a; Dibattista et al., 2009; Pujol et al., 2010). This is because additive genetic effects are expected to underlie most phenotypic evolutionary changes under selection (Hill et al., 2008). Today, methods are available to describe the age dependent additive genetic variation of fitness related traits in animals and plants (Charmantier et al., 2006a,b; Brommer et al., 2007, 2010; Nussey et al., 2007a, 2008; Wilson et al., 2007; Monaghan et al., 2008; Wilson, 2008; Morrissey et al., 2010, 2012; Pujol et al., 2014). Knowing such age dependent patterns has implications for the study of the adaptive potential of populations because populations that are characterized by different age structures have different evolutionary properties (Ronce and Promislow, 2010).
It is important to note that genes with additive effects on the phenotype are probably not the only genes forming the genetic architecture of senescence. Ignoring other components of the genetic architecture of fitness related traits such as dominance and epistasis may result in missing an important part of an evolutionary process (Barton and Turelli, 1989; Fenster et al., 1997; Barton and Keightley, 2002; Wilson and Reale, 2006; Reid et al., 2011; Hansen, 2013; Mackay, 2013). Quantitative genetics models including dominance effects have already been proposed (e.g., Wolak, 2012; Vitezica et al., 2013; Sun et al., 2014). An alternative way to approach dominance is to study inbreeding depression because it is the expression of recessive deleterious mutations. Under the Mutation Accumulation hypothesis (Medawar, 1952), inbreeding depression should depend on age (Charlesworth and Hughes, 1996). In a landmark meta-analysis of the magnitude and timing of inbreeding expression in plants, Husband and Schemske (1996) conjectured that the accumulation of late-acting inbreeding depression usually found across plant taxa was likely the result of the expression of mildly deleterious recessive mutations that could not be purged by natural selection (because the mutations had small effects). However, the patterns they observed are also consistent with a weaker action of selection with age as expected under the evolutionary theory of senescence. The magnitude of inbreeding depression was often compared among populations and environments (Husband and Schemske, 1996; Pujol and McKey, 2006; Pujol et al., 2009; Facon et al., 2011). It would be insightful for the understanding of the evolution of senescence in plants to accumulate case studies that compare inbreeding depression between age classes in the litterature.
Environmental variability can interact with genetic variability to shape the phenotype (Wilson et al., 2005; Kruuk and Hadfield, 2007; Nussey et al., 2007b; Pujol and Galaud, 2013). However, only few studies quantified how the environment interacts with the genetic architecture of senescence (Nussey et al., 2007a). In their study, Nussey et al. (2007a) found that age did interact with both quantitative genetic variation and environmental variation. Their findings imply that environmental constraints have the potential to amplify the magnitude of senescence. It would be worth testing whether such results can be found in other taxa and correspond to a general property of senescence. Also, age dependent epigenetic changes were found to be associated with senescence (Li et al., 2010). It is now widely acknowledged that epigenetic variation can be transmitted to the next generation and that such inheritance system can influence phenotypic evolution (Danchin et al., 2011; Mesoudi et al., 2013; Singh et al., 2014). Since quantitative genetic statistical models can also accommodate such non genetic source of transgenerational variation (Day and Bonduriansky, 2011; Danchin et al., 2013; Townley and Ezard, 2013), it would be worth verifying whether senescence related expectations on genetic variation also apply to transgenerational epigenetic variation.
TOWARD AN INCLUSIVE GENETIC THEORY OF SENESCENCE
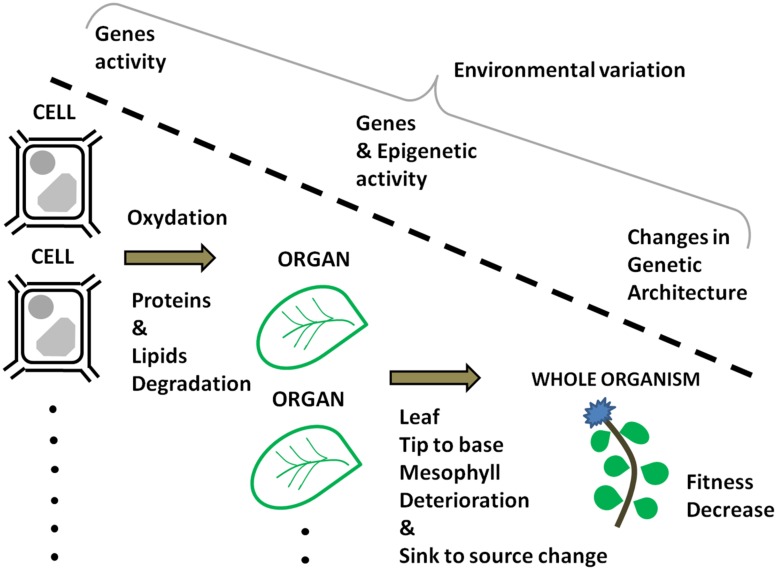
At present, prediction of plant senescence based on geneti and environmental data is hampered by the lack of integration between the scales at which senescence is studied, i.e., cells, organs, and individuals (Figure 1). One if not the most important challenge for the future is to understand the genetic interactions between those scales. Are the genes involved with senescence at the cell and the organ level similar to those that are affected by the increase with age of deleterious mutations balanced by selection? Quantitative genetics offers an opportunity to advance in that direction. Quantitative genetics provides us with the framework to predict the phenotype of individuals on the basis of our knowledge of their genetic architecture (Falconer and Mackay, 1996; Roff, 1997; Lynch and Walsh, 1998; Mackay, 2001). The usual approach in quantitative genetics is to use statistical models in order to relate phenotypic resemblance to genetic similarities. Those genetic similarities can be established on the basis of pedigree information and/or molecular genomic data (Abney, 2008; Carré et al., 2013; Gay et al., 2013; Bérénos et al., 2014). This framework allows us to accommodate multi-trait analyses, longitudinal analyses, and nested genetic effects (Lande and Arnold, 1983; Roff, 1996; Delph et al., 2004, 2005; Etterson, 2004; Clements et al., 2011; Teplitsky et al., 2014). As a result, age-dependent variation in cell, organ, and individual characteristics can be adequately studied using models accommodating longitudinal data such as a random regression animal models (Fischer et al., 2004; Schaeffer, 2004; Wilson et al., 2005; Nussey et al., 2007b; Brommer et al., 2008; Pujol and Galaud, 2013). Such models can evaluate genetic effects at different biological scales by including multi-trait and nested variables. In this way, it is possible to study the dynamics of senescence and the genetic correlations among the genetic components of senescence at multiple biological scales. Only then will we be able to test the hypothesis that cellular, organ, and individual senescence are interdependent (Bleecker, 1998) and to bring an answer to the question: are genes underlying senescence at multiple biological scales belonging to the same genetic program that was shaped by the failure of natural selection in old age? Access to genome wide decrypted information may help answering that question. The combination of next generation sequencing techniques to novel methodological developments in quantitative genetics (Beraldi et al., 2007; Brown et al., 2012; Gay et al., 2013; Bérénos et al., 2014) has a lot of unexploited potential for the genetic study of senescence. Using those methods, DNA sequence variation data can be used to quantify how molecular polymorphism is expressed in different compartments (cells, organs, etc.) in function of the age of the plant. Collectively, the elements presented above illustrate how the genetic knowledge on senescence is scattered across separated research domains. The time is ripe to integrate those disciplines in one approach that will include multiple biological scales. Only then will we be able to identify the relationship between the genes involved with senescence at the cell, organ, and individual levels (Figure 1).
FIGURE 1.
Age-dependent information at different levels of organization that can be integrated within a quantitative genetics framework as to identify their relationship.
CONCLUSION
The genetic prediction of senescence in plants can be seen as an equation. The genetic architecture of a plant would be on one side of that equation. On the other side of this multivariate equation would be the age dependent deterioration of cells, organs, and functions that define the fitness performance of plants. The reviewed elements presented above imply that several links between the two sides of the equation do exist. However, this equation remains incomplete. As I underlined above in various sections of the text, many genetic, and environmental effects remain to be investigated. It is also crucial to identify the genetic links between the biological scales at which organismal functions decline. Quantitative genetics offers a unique opportunity to build such an integrative framework. Understanding the genetic architecture of senescence in plant populations is equivalent to evaluating the adaptive potential of fitness changes with age. Such studies should therefore be an asset for the management of plant genetic resources and the conservation of endangered plant species.
In fact, attempts to reconcile the separated components of the genetic architecture of senescence in plants are scarce. The potential implications of such studies conducted across the plant kingdom in the light of the immense variation it conceals in terms of genomic complexity, life spans, reproductive systems, and life histories are immense. In plants, it is easier to find closely related species that display large phenotypic resemblances but have very different life histories. Genetic studies on senescence in animals and humans are a top research priority that has a direct interest for the people (Zimniak, 2012; Yashin et al., 2013). It is understandable that the interest for these studies outweighs the interest for genetic research on plant senescence. One should nevertheless not neglect that adopting an inclusive approach of senescence in plants that integrates multiple biological scales would bring alternative insights on issues raised by aging research in animals and humans.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Thanks to Anthony Convey M.A. in classics and ancient history, teacher at Kent house primary academy in London, the five anonymous reviewers and the topic editor for their invaluable help in editing this paper. BP’s work was supported by the French Laboratory of Excellence projects “LABEX TULIP” and “LABEX CEBA” and the Toulouse Initiative of Excellence “IDEX UNITI” (ANR-10-LABX-41, ANR-10-LABX-25-01, and ANR11-IDEX-0002-02) and by the CAPA ANR grant to BP (ANR-13-JSV7-0002).
REFERENCES
- Abney M. (2008). Identity-by-descent estimation and mapping of qualitative traits in large, complex pedigrees. Genetics 179 1577–1590 10.1534/genetics.108.089912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstead I., Donnison I., Aubry S., Harper J., Hörtensteiner S., James C., et al. (2006). From crop to model to crop: identifying the genetic basis of the staygreen mutation in the Lolium/Festuca forage and amenity grasses. New Phytol. 172 592–597 10.1111/j.1469-8137.2006.01922.x [DOI] [PubMed] [Google Scholar]
- Baena-Gonzalez E., Rolland F., Thevelein J. M., Sheen J. (2007). A central integrator of transcription networks in plant stress and energy signalling. Nature 448 938–942 10.1038/nature06069 [DOI] [PubMed] [Google Scholar]
- Barton N., Keightley P. (2002). Understanding quantitative genetic variation. Nat. Rev. Genet. 3 11–21 10.1038/nrg700 [DOI] [PubMed] [Google Scholar]
- Barton N. H., Turelli M. (1989). Evolutionary quantitative genetics: how little do we know? Annu. Rev. Genet. 23 337–370 10.1146/annurev.ge.23.120189.002005 [DOI] [PubMed] [Google Scholar]
- Baudisch A., Salguero-Gómez R., Jones O. R., Wrycza T., Mbeau-Ache C., Franco M., et al. (2013). The pace and shape of senescence in angiosperms. J. Ecol. 101 596–606 10.1111/1365-2745.12084 [DOI] [Google Scholar]
- Beraldi D., Mcrae A. F., Gratten J., Slate J., Visscher P. M., Pemberton J. M. (2007). Mapping quantitative trait loci underlying fitness-related traits in a free living sheep population. Evolution 61 1403–1416 10.1111/j.1558-5646.2007.00106.x [DOI] [PubMed] [Google Scholar]
- Bérénos C., Ellis P. A., Pilkington J. G., Pemberton J. M. (2014). Estimating quantitative genetic parameters in wild populations: a comparison of pedigree and genomic approaches. Mol. Ecol. 23 3434–3451 10.1111/mec.12827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergonzi S., Albani M. C. (2011). Reproductive competence from an annual and a perennial perspective. J. Exp. Bot. 62 4415–4422 10.1093/jxb/err192 [DOI] [PubMed] [Google Scholar]
- Blagosklonny M. V., Hall M. N. (2009). Growth and aging: a common molecular mechanism. Aging 1 357–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleecker A. (1998). The evolutionary basis of leaf senescence: method to the madness? Curr. Opin. Plant Biol. 1 73–78 10.1016/S1369-5266(98)80131-3 [DOI] [PubMed] [Google Scholar]
- Brehelin C., Kessler F., van Wijk K. J. (2007). Plastoglobules: versatile lipoprotein particles in plastids. Trends Plant Sci. 12 260–266 10.1016/j.tplants.2007.04.003 [DOI] [PubMed] [Google Scholar]
- Brommer J. E., Rattiste K., Wilson A. J. (2008). Exploring plasticity in the wild: laying date–temperature reaction norms in the common gull Larus canus. Proc. Biol. Sci. 275 687–693 10.1098/rspb.2007.0951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brommer J. E., Rattiste K., Wilson A. (2010). The rate of ageing in a long-lived bird is not heritable. Heredity 104 363–370 10.1038/hdy.2009.125 [DOI] [PubMed] [Google Scholar]
- Brommer J. E., Wilson A. J., Gustafsson L. (2007). Exploring the genetics of aging in a wild passerine bird. Am. Nat. 170 643–650 10.1086/521241 [DOI] [PubMed] [Google Scholar]
- Brown E. A., Pilkington J. G., Nussey D. H., Watt K. A., Hayward A. D., Tucker R., et al. (2012). Detecting genes for variation in parasite burden and immunological traits in a wild population: testing the candidate gene approach. Mol. Ecol. 22 757–773 10.1111/j.1365-294X.2012.05757.x [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V. (1997). The molecular biology of leaf senescence. J. Exp. Biol. 307 181–199. [Google Scholar]
- Buchanan-Wollaston V., Earl S., Harrison E., Mathas E., Navabpour S., Page T., et al. (2003). The molecular analysis of leaf senescence – a genomics approach. Plant Biotechnol. J. 1 3–22 10.1046/j.1467-7652.2003.00004.x [DOI] [PubMed] [Google Scholar]
- Carré C., Gamboa F., Pujol B., Manfredi E. (2013). Genetic links among individuals: from genealogies to molecular markers. Acta Bot. Gall. 160 221–226 10.1080/12538078.2013.809316 [DOI] [Google Scholar]
- Carter C., Pan S., Zouhar J., Avila E. L., Girke T., Raikhel N. V. (2004). The vegetative vacuole proteome of Arabidopsis thaliana reveals predicted and unexpected proteins. Plant Cell 16 3285–3303 10.1105/tpc.104.027078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caswell H., Salguero-Gómez R. (2013). Age, stage and senescence in plants. J. Ecol. 101 585–595 10.1111/1365-2745.12088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., Hughes K. A. (1996). Age-specific inbreeding depression and components of genetic variance in relation to the evolution of senescence. Proc. Natl. Acad. Sci. U.S.A. 93 6140–6145 10.1073/pnas.93.12.6140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charmantier A., Perrins C., Mccleery R. H., Sheldon B. C. (2006a). Age-dependent genetic variance in a life-history trait in the mute swan. Proc. Biol. Sci. 273 225–232 10.1098/rspb.2005.3294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charmantier A., Perrins C., Mccleery R. H., Sheldon B. C. (2006b). Quantitative genetics of age at reproduction in wild swans: support for antagonistic pleiotropy models of senescence. Proc. Natl. Acad. Sci. U.S.A. 103 6587–6592 10.1073/pnas.0511123103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements M. N., Clutton-Brock T. H., Guinness F. E., Pemberton J. M., Kruuk L. E. B. (2011). Variances and covariances of phenological traits in a wild mammal population. Evolution 65 788–801 10.1111/j.1558-5646.2010.01161.x [DOI] [PubMed] [Google Scholar]
- Danchin E., Charmantier A., Champagne F. A., Mesoudi A., Pujol B., Blanchet S. (2011). Beyond DNA: integrating inclusive inheritance into an extended theory of evolution. Nat. Rev. Genet. 12 475–486 10.1038/nrg3028 [DOI] [PubMed] [Google Scholar]
- Danchin E., Pujol B., Wagner R. H. (2013). The double pedigree: a method for studying culturally and genetically inherited behavior in tandem. PLoS ONE 8:e61254 10.1371/journal.pone.0061254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day T., Bonduriansky R. (2011). A unified approach to the evolutionary consequences of genetic and nongenetic inheritance. Am. Nat. 178 E18–E36 10.1086/660911 [DOI] [PubMed] [Google Scholar]
- Delph L. F., Gehring J. L., Arntz A. M., Levri M., Frey F. M. (2005). Genetic correlations with floral display lead to sexual dimorphism in the cost of reproduction. Am. Nat. 166 S31–S41 10.1086/444597 [DOI] [PubMed] [Google Scholar]
- Delph L. F., Gehring J. L., Frey F. M., Arntz A. M., Levri M. (2004). Genetic constraints on floral evolution in a sexually dimorphic plant revealed by artificial selection. Evolution 58 1936–1946 10.1111/j.0014-3820.2004.tb00481.x [DOI] [PubMed] [Google Scholar]
- Dibattista J. D., Feldheim K. A., Garant D., Gruber S. H., Hendry A. P. (2009). Evolutionary potential of a large marine vertebrate: quantitative genetic parameters in a wild population. Evolution 63 1051–1067 10.1111/j.1558-5646.2008.00605.x [DOI] [PubMed] [Google Scholar]
- El-Sharkawy M. A. (2004). Cassava biology and physiology. Plant Mol. Biol. 56 481–501 10.1007/s11103-005-2270-7 [DOI] [PubMed] [Google Scholar]
- Etterson J. R. (2004). Evolutionary potential of Chamaecrista fasciculata in relation to climate change. II. Genetic architecture of three populations reciprocally planted along an environmental gradient in the great plains. Evolution 58 1459–1471 10.1111/j.0014-3820.2004.tb01727.x [DOI] [PubMed] [Google Scholar]
- Facon B., Hufbauer R. A., Tayeh A., Loiseau A., Lombaert E., Vitalis R., et al. (2011). Inbreeding depression is purged in the invasive insect Harmonia axyridis. Curr. Biol. 21 424–427 10.1016/j.cub.2011.01.068 [DOI] [PubMed] [Google Scholar]
- Falconer D. S., Mackay T. F. C. (1996). Introduction to Quantitative Genetics, 4th Edn. Harlow: Longmans Green. [Google Scholar]
- Fenster C., Galloway L., Chao L. (1997). Epistasis and its consequences for the evolution of natural populations. Trends Ecol. Evol. 12 282–286 10.1016/S0169-5347(97)81027-0 [DOI] [PubMed] [Google Scholar]
- Fischer T. M., Van der Werf J. H. J., Banks R. G., Ball A. J. (2004). Description of lamb growth using random regression on field data. Livest. Prod. Sci. 89 175–185 10.1016/j.livprodsci.2004.02.004 [DOI] [Google Scholar]
- Fisher R. A. (1930). The Genetical Theory of Natural Selection. Oxford: Oxford University Press. [Google Scholar]
- Flatt T. (2012). A new definition of aging? Front. Genet. 3:148 10.3389/fgene.2012.00148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furtek D., Schiefelbein J. W., Johnston F., Nelson O. E. (1988). Sequence comparisons of 3 wild-type bronze-1 alleles from Zea mays. Plant Mol. Biol. 11 473–481 10.1007/BF00039028 [DOI] [PubMed] [Google Scholar]
- Gan S., Amasino R. M. (1997). Making sense of senescence. Plant Physiol. 113 313–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M. B., Dahlgren J. P., Ehrlén J. (2011). No evidence of senescence in a 300-year-old mountain herb. J. Ecol. 99 1424–1430 10.1111/j.1365-2745.2011.01871.x [DOI] [Google Scholar]
- Gardner S. N., Mangel M. (1997). When can a clonal organism escape senescence? Am. Nat. 150 462–490 10.1086/286076 [DOI] [PubMed] [Google Scholar]
- Gay L., Siol M., Ronfort J. (2013). Pedigree-free estimates of heritability in the wild: promising prospects for selfing populations. PLoS ONE 8:e66983 10.1371/journal.pone.0066983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano G., Diretto G. (2007). Of chromoplasts and chaperones. Trends Plant Sci. 12 529–531 10.1016/j.tplants.2007.09.008 [DOI] [PubMed] [Google Scholar]
- Grbić V., Bleecker A. B. (1995). Ethylene regulates the timing of leaf senescence in Arabidopsis. Plant J. 8 595–602 10.1046/j.1365-313X.1995.8040595.x [DOI] [Google Scholar]
- Greer L. F. (2012). Aging: the fading signal of natural selection. Front. Genet. 3:155 10.3389/fgene.2012.00155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiamet J. J., Giannibelli M. V. (1996). Nuclear and cytoplasmic ‘stay-green’ mutations of soybean alter the loss of leaf soluble proteins during senescence. Physiol. Plant. 96 655–661 10.1111/j.1399-3054.1996.tb00239.x [DOI] [Google Scholar]
- Guo Y., Gan S. (2006). AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J. 46 601–612 10.1111/j.1365-313X.2006.02723.x [DOI] [PubMed] [Google Scholar]
- Guo Y., Gan S. (2011). AtMYB2 regulates whole plant senescence by inhibiting cytokinin-mediated branching at late stages of development in Arabidopsis. Plant Physiol. 156 1612–1619 10.1104/pp.111.177022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton W. D. (1966). The moulding of senescence by natural selection. J. Theor. Biol. 12 12–45 10.1016/0022-5193(66)90184-6 [DOI] [PubMed] [Google Scholar]
- Hansen T. F. (2013). Why epistasis is important for selection and adaptation. Evolution 67 3501–3511 10.1111/evo.12214 [DOI] [PubMed] [Google Scholar]
- Harper J. (1977). The Population Biology of Plants. London: The Academic Press. [Google Scholar]
- Hensel L. L., Grbić V., Baumgarten D. A., Bleecker A. B. (1993). Developmental and age-related processes that influence the longevity and senescence of photosynthetic tissues in Arabidopsis. Plant Cell 5 553–564 10.1105/tpc.5.5.553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W. G., Goddard M. E., Visscher P. M. (2008). Data and theory point to mainly additive genetic variance for complex traits. PLoS Genet. 4:e1000008 10.1371/journal.pgen.1000008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinder B., Schellenberg M., Rodoni S., Ginsburg S., Vogt E., Martinoia E., et al. (1996). How plants dispose of chlorophyll catabolites. Directly energized uptake of tetrapyrrolic breakdown products into isolated vacuoles. J. Biol. Chem. 271 27233–27236 10.1074/jbc.271.44.27233 [DOI] [PubMed] [Google Scholar]
- Hong J.-P., Byun M. Y., Koo D.-H., An K., Bang J.-W., Chung I. K., et al. (2007). Suppression of RICE TELOMERE BINDING PROTEIN1 results in severe and gradual developmental defects accompanied by genome instability in rice. Plant Cell 19 1770–1781 10.1105/tpc.107.051953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörtensteiner S., Chinner J., Matile P., Thomas H., Donnison I. S. (2000). Chlorophyll breakdown in Chlorella protothecoides: characterization of degreening and cloning of degreening-related genes. Plant Mol. Biol. 42 439–450 10.1023/A:1006380125438 [DOI] [PubMed] [Google Scholar]
- Husband B. C., Schemske D. W. (1996). Evolution of the magnitude and timing of inbreeding depression in plants. Evolution 50 54–70 10.2307/2410780 [DOI] [PubMed] [Google Scholar]
- Jansson S., Thomas H. (2008). Senescence: developmental program or timetable? New Phytol. 179 575–579 10.1111/j.1469-8137.2008.02471.x [DOI] [PubMed] [Google Scholar]
- Jongebloed U., Szederkenyi J., Hartig K., Schobert C., Komor E. (2004). Sequence of morphological and physiological events during natural ageing and senescence of a castor bean leaf: sieve tube occlusion and carbohydrate back-up precede chlorophyll degradation. Physiol. Plant. 120 338–346 10.1111/j.0031-9317.2004.0245.x [DOI] [PubMed] [Google Scholar]
- Kim J. H., Woo H. R., Kim J., Lim P. O., Lee I. C., Choi S. H., et al. (2009). Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 323 1053–1057 10.1126/science.1166386 [DOI] [PubMed] [Google Scholar]
- Kruuk L. E. B., Hadfield J. D. (2007). How to separate genetic and environmental causes of similarity between relatives. J. Evol. Biol. 20 1890–1903 10.1111/j.1420-9101.2007.01377.x [DOI] [PubMed] [Google Scholar]
- Kruuk L. E. B., Slate J., Wilson A. J. (2008). New answers for old questions: the evolutionary quantitative genetics of wild animal populations. Annu. Rev. Ecol. Evol. Syst. 39 525–548 10.1146/annurev.ecolsys.39.110707.173542 [DOI] [Google Scholar]
- Lande R., Arnold S. J. (1983). The measurement of selection on correlated characters. Evolution 37 1210–1226 10.2307/2408842 [DOI] [PubMed] [Google Scholar]
- Li H., Freeling M., Lisch D. (2010). Epigenetic reprogramming during vegetative phase change in maize. Proc. Natl. Acad. Sci. U.S.A. 107 22184–22189 10.1073/pnas.1016884108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S., Van Eck J., Zhou X., Lopez A. B., O’Halloran D. M., Cosman K. M., et al. (2006). The cauliflower or gene encodes a DNAJ cysteine-rich domain-containing protein that mediates high levels of beta-carotene accumulation. Plant Cell 18 3594–3605 10.1105/tpc.106.046417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y.-P., Li Z.-S., Drozdowicz Y.-M., Hörtensteiner S., Martinoia E., Rea P. A. (1998). AtMRP2, an Arabidopsis ATP binding cassette transporter able to transport glutathione S-conjugates and chlorophyll catabolites: functional comparisons with AtMRP1. Plant Cell 10 267–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Walsh B. (1998). Genetics and Analysis of Quantitative Traits. Sunderland, MA: Sinauer. [Google Scholar]
- Mackay T. (2001). The genetic architecture of quantitative traits. Annu. Rev. Genet. 35 303–339 10.1146/annurev.genet.35.102401.090633 [DOI] [PubMed] [Google Scholar]
- Mackay T. F. C. (2013). Epistasis and quantitative traits: using model organisms to study gene-gene interactions. Nat. Rev. Genet. 15 22–33 10.1038/nrg3627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez D. E. (2012). Negligible senescence. Front. Genet. 3:151 10.3389/fgene.2012.00151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight T. D., Shippen D. E. (2004). Plant telomere biology. Plant Cell 16 794–803 10.1105/tpc.160470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medawar P. B. (1952). An Unsolved Problem of Biology. London: H.K. Lewis. [Google Scholar]
- Mesoudi A., Blanchet S., Charmantier A., Danchin E., Fogarty L., Jablonka E., et al. (2013). Is non-genetic inheritance just a proximate mechanism? A corroboration of the extended evolutionary synthesis. Biol. Theory 7 189–195 10.1007/s13752-013-0091-5 [DOI] [Google Scholar]
- Miao Y., Laun T., Zimmermann P., Zentgraf U. (2004). Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol. Biol. 55 853–867 10.1007/s11103-005-2142-1 [DOI] [PubMed] [Google Scholar]
- Monaghan P., Charmantier A., Nussey D. H., Ricklefs R. E. (2008). The evolutionary ecology of senescence. Funct. Ecol. 22 371–378 10.1111/j.1365-2435.2008.01418.x [DOI] [Google Scholar]
- Mondolot L., Marlas A., Barbeau D., Gargadennec A., Pujol B., McKey D. (2008). Domestication and defence: foliar tannins and C/N ratios in cassava and a close wild relative. Acta Oecol. 34 147–154 10.1016/j.actao.2008.05.009 [DOI] [Google Scholar]
- Monteuuis O., Doulbeau S., Verdeil J.-L. (2008). DNA methylation in different origin clonal offspring from a mature Sequoiadendron giganteum genotype. Trees 22 779–784 10.1007/s00468-008-0238-3 [DOI] [Google Scholar]
- Morales M., Oñate M., García M. B., Munné-Bosch S. (2013). Photo-oxidative stress markers reveal absence of physiological deterioration with ageing in Borderea pyrenaica, an extraordinarily long-lived herb. J. Ecol. 101 555–565 10.1111/1365-2745.12080 [DOI] [Google Scholar]
- Morrissey M. B., Kruuk L. E. B., Wilson A. J. (2010). The danger of applying the breeder’s equation in observational studies of natural populations. J. Evol. Biol. 23 2277–2288 10.1111/j.1420-9101.2010.02084.x [DOI] [PubMed] [Google Scholar]
- Morrissey M. B., Parker D. J., Korsten P., Pemberton J. M., Kruuk L. E. B., Wilson A. J. (2012). The prediction of adaptive evolution: empirical application of the secondary theorem of selection and comparison to the breeder’s equation. Evolution 66 2399–2410 10.1111/j.1558-5646.2012.01632.x [DOI] [PubMed] [Google Scholar]
- Munné-Bosch S. (2008). Do perennials really senesce? Trends Plant Sci. 13 216–220 10.1016/j.tplants.2008.02.002 [DOI] [PubMed] [Google Scholar]
- Nam H. G. (1997). The molecular genetic analysis of leaf senescence. Curr. Opin. Biotechnol. 8 200–207 10.1016/S0958-1669(97)80103-6 [DOI] [PubMed] [Google Scholar]
- Navabpour S., Morris K., Allen R., Harrison E., A-H-Mackerness S., Buchanan-Wollaston V. (2003). Expression of senescence-enhanced genes in response to oxidative stress. J. Exp. Bot. 54 2285–2292 10.1093/jxb/erg267 [DOI] [PubMed] [Google Scholar]
- Nussey D. H., Kruuk L. E. B., Morris A., Clutton-Brock T. H. (2007a). Environmental conditions in early life influence ageing rates in a wild population of red deer. Curr. Biol. 17 R1000–R1001 10.1016/j.cub.2007.10.005 [DOI] [PubMed] [Google Scholar]
- Nussey D. H., Wilson A. J., Brommer J. E. (2007b). The evolutionary ecology of individual phenotypic plasticity in wild populations. J. Evol. Biol. 20 831–844 10.1111/j.1420-9101.2007.01300.x [DOI] [PubMed] [Google Scholar]
- Nussey D. H., Wilson A. J., Morris A., Pemberton J., Clutton-Brock T., Kruuk L. E. B. (2008). Testing for genetic trade-offs between early- and late-life reproduction in a wild red deer population. Proc. Biol. Sci. 275 745–750 10.1098/rspb.2007.0986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orive M. E. (1995). Senescence in organisms with clonal reproduction and complex life histories. Am. Nat. 145 90–108 10.1086/285729 [DOI] [Google Scholar]
- Oshio Y., Hase E. (1969). Studies on red pigments excreted by cells of Chlorella protothecoides during the process of bleaching induced by glucose or acetate. II. Mode of formation of the red pigments. Plant Cell Physiol. 10 51–59. [Google Scholar]
- Park S.-Y., Yu J.-W., Park J.-S., Li J., Yoo S.-C., Lee N.-Y., et al. (2007). The senescenceinduced staygreen protein regulates chlorophyll degradation. Plant Cell 19 1649–1664 10.1105/tpc.106.044891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrott D. L., McInnerney K., Feller U., Fischer A. M. (2007). Steam-girdling of barley (Hordeum vulgare) leaves leads to carbohydrate accumulation and accelerated leaf senescence, facilitating transcriptomic analysis of senescence-associated genes. New Phytol. 176 56–69 10.1111/j.1469-8137.2007.02158.x [DOI] [PubMed] [Google Scholar]
- Partridge L., Barton N. H. (1993). Optimally, mutation and the evolution of ageing. Nature 362 305–311 10.1038/362305a0 [DOI] [PubMed] [Google Scholar]
- Paz-Ares J., Ghosal D., Saedler H. (1990). Molecular analysis of the C1-I allele from Zea mays: a dominant mutant of the regulatory C1 locus. EMBO J. 9 315–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen B. (1995). An evolutionary theory of clonal senescence. Theor. Popul. Biol. 47 292–320 10.1006/tpbi.1995.1013 [DOI] [Google Scholar]
- Picó F. X., Retana J. (2008). Age-specific, density-dependent and environment-based mortality of a short-lived perennial herb. Plant Biol. 10 374–381 10.1111/j.1438-8677.2008.00044.x [DOI] [PubMed] [Google Scholar]
- Poethig R. S. (2010). The past, present, and future of vegetative phase change. Plant Physiol. 154 541–544 10.1104/pp.110.161620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pružinská A., Anders I., Aubry S., Schenk N., Tapernoux-Lüthi E., Müller T., et al. (2007). In vivo participation of red chlorophyll catabolite reductase in chlorophyll breakdown. Plant Cell 19 369–387 10.1105/tpc.106.044404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol B., Galaud J.-P. (2013). A practical guide to quantifying the effect of genes underlying adaptation in a mixed genomics and evolutionary ecology approach. Acta Bot. Gall. 160 197–204 10.1080/12538078.2013.799045 [DOI] [Google Scholar]
- Pujol B., Marrot P., Pannell J. R. (2014). A quantitative genetic signature of senescence in a short-lived perennial plant. Curr. Biol. 24 744–747 10.1016/j.cub.2014.02.012 [DOI] [PubMed] [Google Scholar]
- Pujol B., McKey D. (2006). Size asymmetry in intraspecific competition and the density-dependence of inbreeding depression in a natural plant population: a case study in cassava (Manihot esculenta Crantz, Euphorbiaceae). J. Evol. Biol. 19 85–96 10.1111/j.1420-9101.2005.00990.x [DOI] [PubMed] [Google Scholar]
- Pujol B., Obbard D. J., Pannell J. R. (2010). Symptoms of population range expansion: lessons from phenotypic and genetic differentiation in hexaploid Mercurialis annua. Plant Ecol. Divers. 3 103–108 10.1080/17550874.2010.516027 [DOI] [Google Scholar]
- Pujol B., Pannell J. R. (2008). Reduced responses to selection after species range expansion. Science 321 96 10.1126/science.1157570 [DOI] [PubMed] [Google Scholar]
- Pujol B., Wilson A. J., Ross R. I. C., Pannell J. R. (2008a). Are QST-FST comparisons for natural populations meaningful? Mol. Ecol. 17 4782–4785 10.1111/j.1365-294X.2008.03958.x [DOI] [PubMed] [Google Scholar]
- Pujol B., Salager J. L., Beltran M., Bousquet S., McKey D. (2008b). Photosynthesis and leaf structure in domesticated cassava (Euphorbiaceae) and a close wild relative : have leaf photosynthetic parameters evolved under domestication ? Biotropica 40 305–312 10.1111/j.1744-7429.2007.00373.x [DOI] [Google Scholar]
- Pujol B., Zhou S.-R., Sanchez-Vilas J., Pannell J. R. (2009). Reduced inbreeding depression after species range expansion. Proc. Natl. Acad. Sci. U.S.A. 106 15379–15383 10.1073/pnas.0902257106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe O. J., Amaya I., Vincent C. A., Rothstein S., Carpenter R., Coen E. S., et al. (1998). A common mechanism controls the life cycle and architecture of plants. Development 125 1609–1615. [DOI] [PubMed] [Google Scholar]
- Reid J. M., Arcese P., Sardell R. J., Keller L. F. (2011). Additive genetic variance, heritability, and inbreeding depression in male extra-pair reproductive success. Am. Nat. 177 177–187 10.1086/657977 [DOI] [PubMed] [Google Scholar]
- Roach D. A. (1993). Evolutionary senescence in plants. Genetica 91 53–64 10.1007/bf01435987 [DOI] [Google Scholar]
- Roach D. A. (2012). Age, growth and size interact with stress to determine life span and mortality. Exp. Gerontol. 47 782–786 10.1016/j.exger.2012.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach D. A., Ridley C. E., Dudycha J. L. (2009). Longitudinal analysis of Plantago: age-by-environment interactions reveal aging. Ecology 90 1427–1433 10.1890/08-0981.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodoni S., Mühlecker W., Anderl M., Kräutler B., Moser D., Thomas H., et al. (1997). Chlorophyll breakdown in senescent chloroplasts. Cleavage of pheophorbide a in two enzymic steps. Plant Physiol. 115 669–676 10.1104/pp.115.2.669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roff D. A. (1996). The evolution of genetic correlations: an analysis of patterns. Evolution 50 1392–1403 10.2307/2410877 [DOI] [PubMed] [Google Scholar]
- Roff D. A. (1997). Evolutionary Quantitative Genetics. New York: Chapman and Hall; 10.1007/978-1-4615-4080-9 [DOI] [Google Scholar]
- Ronce O., Promislow D. (2010). Kin competition, natal dispersal and the moulding of senescence by natural selection. Proc. Biol. Sci. 277 3659–3667 10.1098/rspb.2010.1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M. R., Flatt T., Graves J. L., Greer L. F., Martinez D. E., Matos M., et al. (2012). What is aging? Front. Genet. 3:134 10.3389/fgene.2012.00134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salguero-Gómez R., Shefferson R. P., Hutchings M. J. (2013). Plants do not count… or do they? New perspectives on the universality of senescence. J. Ecol. 101 545–554 10.1111/1365-2745.12089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y., Morita R., Nishimura M., Yamaguchi H., Kusaba M. (2007). Mendel’s green cotyledon gene encodes a positive regulator of the chlorophyll-degrading pathway. Proc. Biol. Sci. 104 14169–14174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer L. R. (2004). Application of random regression models in animal breeding. Livest. Prod. Sci. 86 35–45 10.1016/S0301-6226(03)00151-9 [DOI] [Google Scholar]
- Schippers J. H. M., Nunes-Nesi A., Apetrei R., Hille J., Fernie A. R., Dijkwel P. P. (2008). The Arabidopsis onset of leaf death 5 mutation of quinolinate synthase affects nicotinamide adenine dinucleotide biosynthesis and causes early ageing. Plant Cell 20 2909–2925 10.1105/tpc.107.056341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shefferson R. P., Roach D. A. (2012). The triple helix of Plantago lanceolata: genetics and the environment interact to determine population dynamics. Ecology 93 793–802 10.1890/11-0742.1 [DOI] [PubMed] [Google Scholar]
- Silvertown J., Franco M., Perez-Ishiwara R. (2001). Evolution of senescence in iteroparous perennial plants. Evol. Ecol. Res. 3 393–412. [Google Scholar]
- Singh D. P., Saudemont B., Guglielmi G., Arnaiz O., Gout J.-F., Prajer M., et al. (2014). Genome-defence small RNAs exapted for epigenetic mating-type inheritance. Nature 509 447–452 10.1038/nature13318 [DOI] [PubMed] [Google Scholar]
- Smart C., Hosken S., Thomas H., Greaves J., Blair B., Schuch W. (1995). The timing of maize leaf senescence and characterization of senescence-related cDNAs. Physiol. Plant. 93 673–682 10.1111/j.1399-3054.1995.tb05116.x [DOI] [Google Scholar]
- Smeekens S., Ma J., Hanson J., Rolland F. (2010). Sugar signals and molecular networks controlling plant growth. Curr. Opin. Plant Biol. 13 274–279 10.1016/j.pbi.2009.12.002 [DOI] [PubMed] [Google Scholar]
- Snowden K. C., Simkin A. J., Janssen B. J., Templeton K. R., Loucas H. M., Simons J. L., et al. (2005). The decreased apical dominance1/Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE8 gene affects branch production and plays a role in leaf senescence, root growth, and flower development. Plant Cell 17 746–759 10.1105/tpc.104.027714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C., VanRaden P. M., Cole J. B., O’Connell J. R. (2014). Improvement of prediction ability for genomic selection of dairy cattle by including dominance effects. PLoS ONE 9:e103934 10.1371/journal.pone.0103934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplitsky C., Tarka M., Møller A. P., Nakagawa S., Balbontín J., Burke T. A., et al. (2014). Assessing multivariate constraints to evolution across ten long-term avian studies. PLoS ONE 9:e90444 10.1371/journal.pone.0090444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelander M., Olsson T., Ronne H. (2004). Snf1-relatedproteinkinase1isneededfor growth in a normal day–night light cycle. EMBO J. 23 1900–1910 10.1038/sj.emboj.7600182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas H. (2013). Senescence, ageing and death of the whole plant. New Phytol. 197 696–711 10.1111/nph.12047 [DOI] [PubMed] [Google Scholar]
- Thomas H., Huang L., Young M., Ougham H. (2009). Evolution of plant senescence BMC Evol. Biol. 9:163 10.1186/1471-2148-9-163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas H., Stoddart J. L. (1980). Leaf senescence. Annu. Rev. Plant Physiol. 31 83–111 10.1146/annurev.pp.31.060180.000503 [DOI] [Google Scholar]
- Thompson A. R., Doelling J. H., Suttangkakul A., Vierstra R. D. (2005). Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 138 2097–2110 10.1104/pp.105.060673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townley S., Ezard T. H. G. (2013). A G matrix analogue to capture the cumulative effects of nongenetic inheritance. J. Evol. Biol. 26 1234–1243 10.1111/jeb.12089 [DOI] [PubMed] [Google Scholar]
- Uauy C., Distelfeld A., Fahima T., Blechl A., Dubcovsky J. (2006). A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 314 1298–1301 10.1126/science.1133649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Breusegem F., Dat J. F. (2006). Reactive oxygen species in plant cell death. Plant Physiol. 141 384–390 10.1104/pp.106.078295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaupel J. W., Baudisch A., Dölling M., Roach D. A., Gampe J. (2004). The case for negative senescence. Theor. Popul. Biol. 65 339–351 10.1016/j.tpb.2003.12.003 [DOI] [PubMed] [Google Scholar]
- Vitezica Z. G., Varona L., Legarra A. (2013). On the additive and dominant variance and covariance of individuals within the genomic selection scope. Genetics 195 1223–1230 10.1534/genetics.113.155176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.-W., Park M. Y., Wang L.-J., Koo Y., Chen X.-Y., Weigel D., et al. (2011). miRNA control of vegetative phase change in trees. PLoS Genet. 7:e1002012 10.1371/journal.pgen.1002012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkinson A. (1992). Plant senescence. Trends Ecol. Evol. 7 417–420 10.1016/0169-5347(92)90024-6 [DOI] [PubMed] [Google Scholar]
- Watson J. M., Riha K. (2010). Comparative biology of telomeres: where plants stand. FEBS Lett. 584 3752–3759 10.1016/j.febslet.2010.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams G. C. (1957). Pleiotropy, natural selection, and the evolution of senescence. Evolution 11 398–411 10.2307/2406060 [DOI] [Google Scholar]
- Wilson A. J. (2008). Why h2 does not always equal V A/V P? J. Evol. Biol. 21 647–650 10.1111/j.1420-9101.2008.01500.x [DOI] [PubMed] [Google Scholar]
- Wilson A. J., Charmantier A., Hadfield J. D. (2008). Evolutionary genetics of ageing in the wild: empirical patterns and future perspectives. Funct. Ecol. 22 431–442 10.1111/j.1365-2435.2008.01412.x [DOI] [Google Scholar]
- Wilson A. J., Kruuk L. E. B., Coltman D. W. (2005). Ontogenetic patterns in heritable variation for body size: using random regression models in a wild ungulate population. Am. Nat. 166 E177–E192 10.1086/497441 [DOI] [PubMed] [Google Scholar]
- Wilson A. J., Nussey D. H., Pemberton J. M., Pilkington J. G., Morris A., Pelletier F., et al. (2007). Evidence for a genetic basis of aging in two wild vertebrate populations. Curr. Biol. 17 2136–2142 10.1016/j.cub.2007.11.043 [DOI] [PubMed] [Google Scholar]
- Wilson A. J., Reale D. (2006). Ontogeny of additive and maternal genetic effects: lessons from domestic mammals. Am. Nat. 167 E23–E38 10.1086/498138 [DOI] [PubMed] [Google Scholar]
- Wingler A., Purdy S., Maclean J. A., Pourtau N. (2006). The role of sugars in integrating environmental signals during the regulation of leaf senescence. J. Exp. Bot. 57 391–399 10.1093/jxb/eri279 [DOI] [PubMed] [Google Scholar]
- Wolak M. E. (2012). nadiv: an R package to create relatedness matrices for estimating non-additive genetic variances in animal models. Methods Ecol. Evol. 3 792–796 10.1111/j.2041-210X.2012.00213.x [DOI] [Google Scholar]
- Wüthrich K. L., Bovet L., Hunziker P. E., Donnison I. S., Hörtensteiner S. (2000). Molecular cloning, functional expression and characterisation of RCC reductase involved in chlorophyll catabolism. Plant J. 21 189–198 10.1046/j.1365-313x.2000.00667.x [DOI] [PubMed] [Google Scholar]
- Xie K., Shen J., Hou X., Yao J., Li X., Xiao J., et al. (2012). Gradual increase of miR156 regulates temporal expression changes of numerous genes during leaf development in rice. Plant Physiol. 158 1382–1394 10.1104/pp.111.190488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Sulpice R., Himmelbach A., Meinhard M., Christmann A., Grill E. (2006). Fibrillin expression is regulated by abscisic acid response regulators and is involved in abscisic acid-mediated photoprotection. Proc. Natl. Acad. Sci. U.S.A. 103 6061–6066 10.1073/pnas.0501720103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky M. F., Ma H., Bowman J. L., Drews G. N., Feldmann K. A., Meyerowitz E. M. (1990). The protein encoded by the Arabidopsis homeotic gene Agamous resembles transcription factors. Nature 346 35–39 10.1038/346035a0 [DOI] [PubMed] [Google Scholar]
- Yashin A. I., Arbeev K. G., Wu D., Arbeeva L. S., Kulminski A. M., Akushevich I., et al. (2013). How the quality of GWAS of human lifespan and health span can be improved. Front. Genet. 4:125 10.3389/fgene.2013.00125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S., Ito M., Nishida I., Watanabe A. (2002). Identification of a novel gene HYS1/CPR5 that has a repressive role in the induction of leaf senescence and pathogen-defence responses in Arabidopsis thaliana. Plant J. 29 427–437 10.1046/j.0960-7412.2001.01228.x [DOI] [PubMed] [Google Scholar]
- Zimniak P. (2012). What is the proximal cause of aging? Front. Genet. 3:189 10.3389/fgene.2012.00189 [DOI] [PMC free article] [PubMed] [Google Scholar]